Abstract
The acid tolerance mechanism is important for Escherichia coli to resist acidic conditions encountered in mammalian host digestive tract environment. Here, we explored how the LuxR protein SdiA influenced E. coli acid tolerance ability in the context of the glutamate- and glutamine-dependent acid resistance system (AR2). First, using a growth and acid shock assay under different acid stresses, we demonstrated that the deletion of sdiA in SM10λpir or BW25113 led to impaired growth under the acidic environment of pH 3–6, which was restored by complementary expression of SdiA. Next, transcriptome sequencing and qPCR disclosed that the expression of glutamate decarboxylase W (GadW) and GadY, the key members of the AR2 system, were regulated by SdiA. Further, β-galactosidase reporter assays showed that the promoter activity of gadW and gadY was positively regulated by SdiA. Moreover, qPCR and β-galactosidase reporter assays confirmed that the regulation of SdiA on GadW, but not GadY, could be enhanced by quorum sensing (QS) signal molecules AHLs. Collectively, these data suggest that SdiA plays a crucial role in acid tolerance regulation of E. coli. Our findings provide new insights into the important contribution of quorum sensing system AHLs–SdiA to the networks that regulate acid tolerance.
Keywords: SdiA, glutamate decarboxylase W (GadW), glutamate decarboxylase Y (GadY), acid tolerance, Escherichia coli
Introduction
Enteric organisms, such as Escherichia coli, often colonize in the host’s gastrointestinal tract and cause disease. During this process, the acidic stress in the host’s stomach sets a barrier to enteric pathogens. E. coli has a remarkable ability to withstand low pH environment by activating several acid resistance (AR) systems, especially the glutamate-dependent AR system (AR2), which is the most effective AR system to cope with extreme acid stress (Lund et al., 2014; Zhao et al., 2018). The AR2 system has three structural components including two isoforms of glutamate decarboxylases (GadA and GadB), and the aminobutyrate (GABA) antiporter GadC. Among them, gadB and gadC genes are co-transcribed. The gadA gene is 2.1 Mb from gadBC and exists in the acid fitness island (AFI) (De Biase and Pennacchietti, 2012). This AFI comprises 14 genes related to AR. Among these genes, gadE, gadW, and gadX encode regulators of the AR2 system (Ma et al., 2003; Tramonti et al., 2008; Seo et al., 2015). In addition, a small RNA, GadY, is also involved in the regulation of AR2 system by stabilizing the gadX mRNA (Opdyke et al., 2004; Tramonti et al., 2008). The GadX–GadY–GadW circuit can strengthen the AR2 system by activating gadE transcription and directly binding to the promoter regions of gadA and gadBC (Tramonti et al., 2006). However, the regulation of this circuit remains unclear.
In adaptation to environmental change, quorum sensing (QS) system plays a significant role through chemical communication between bacterial cells. In Gram-negative bacteria, this communication mainly uses signal molecules, N-acyl homoserine lactones (AHLs), and cognate LuxR transcription factor (Schuster et al., 2013). The AHLs can recognize specific LuxRs by their variable acyl chains and regulate gene transcription (Fuqua and Greenberg, 2002). Some organisms, such as Escherichia and Salmonella, only have LuxR-type protein SdiA but do not produce AHLs. However, numerous studies have demonstrated that this orphan LuxR-type receptor is able to bind to DNA and regulates gene transcription with or without AHLs, and then involves in interspecies signaling (Yamamoto et al., 2001; Dyszel et al., 2010; Nguyen et al., 2015). Moreover, AHLs can improve the transcriptional regulation ability of SdiA through enhancing SdiA stability and DNA-binding affinity, thus regulating many gene transcription in an SdiA-dependent manner (Van Houdt et al., 2006; Lee et al., 2008; Nguyen et al., 2015). In addition, SdiA of enterohemorrhagic E. coli O157:H7 (EHEC) can be constitutively activated by the binding of molecule 1-octanoyl-rac-glycerol (OCL) in the absence of AHLs (Nguyen et al., 2015). As increasing investigations about SdiA have been done, it is generally accepted that SdiA plays a vital role in facing different environments including acid environment (Dyszel et al., 2010; Hughes et al., 2010). Nevertheless, the function of SdiA in AR and its underlying mechanisms remain largely unknown.
In this study, we focused on the role of SdiA in AR and explored how SdiA regulated the Gad system. We first confirmed the link between SdiA and acid tolerance by investigating the effect of the knockout and overexpression of sdiA on cell growth in media acidifed by citric acid. Subsequently, the transcriptome, qPCR and β-galactosidase reporter analysis were performed to shed some light on the mechanism. Our works offer a deeper understanding of the role of SdiA in the regulation of AR.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions
The bacterial strains and plasmids used in this study are listed in Supplementary Table S1. The construction of sdiA-deficient or -overexpression strains has been described in our previous study (Lu et al., 2017). Bacteria were grown in LB broth or on LB agar unless otherwise indicated. When necessary, antibiotics were used at the following concentrations: 100 μg/ml ampicillin (AMP) and 16 μg/ml chloramphenicol (CM).
Plasmid Construction
The plasmid pQF50 containing a promoter-less lacZ reporter was used for gadY and gadW promoter analysis. The DNA fragments of the gadY promoter spanning −245 to +13 and gadW promoter spanning −258 to −1 were PCR amplified and inserted into BamHI/Hind? sites upstream of the lacZ reporter in pQF50 to generate pQF50-PgadY and pQF50-PgadW, respectively. All constructs were confirmed by direct sequencing. Primers used in this study are listed in Supplementary Table S2.
RNA Sequencing and Bioinformatics Analysis
The wild-type strain, sdiA gene-deficient and -overexpression strains in SM10λpir were grown overnight (8∼10 h) at 37°C, and then 100 μl of the cultures were added into 3 ml of LB to grow to mid-exponential phase (0.5 McFarland standard). Then, total RNA was extracted using RNAprep Pure Cell/Bacteria Kit (Tiangen, Beijing, China), and the entire sequencing was conducted by the BGI Company. Experiments were performed in triplicate. The raw data had been submitted to SRA database of NCBI (accession number: PRJNA627821, SRP258248). The expression of Unigene was calculated by RPKM method (Reads Per kb per Million reads).
Real-Time PCR
Total RNA was extracted using RNAiso Plus reagent (Takara, Dalian, Liaoning, China). Reverse transcription (1 μg of total RNA) was performed with the PrimeScript RT Reagent Kit (code No. RR047A; Takara, Dalian, Liaoning, China). The cDNA was subjected to qPCR on a ViiATM 7 Dx system (Applied Biosystems, Foster, CA, United States) using SYBR Green qPCR Master Mix (Takara, Dalian, Liaoning, China). The expression levels of the target genes were normalized to the expression of an internal control gene (rpoD) using the 2–ΔΔCt method. The sequences of the primers are listed in Supplementary Table S2.
Growth Assay
Growth and acid shock assay were performed as described previously, with some modifications (Gao et al., 2018). E. coli strains were grown overnight (8∼10 h) in LBG medium (LB medium supplemented with 0.4% glucose) of pH 7.0 at 37°C. Then the bacteria was collected and re-cultured in LBG medium at pH 7.0 or LBG medium acidified by citric acid to pH 6, pH 5, pH 4, and pH 3 for 24 h at 37°C in a 96-well round bottom plate, and the OD600 values were determined at the indicated time points. All of the tests were carried out independently at least in triplicate.
Acid Shock Assay
Escherichia coli strains were grown overnight (8∼10 h) in LBG medium of pH 7.0 at 37°C. Then the bacteria was collected and re-cultured in LBG medium at pH 7.0 or LBG medium acidified by citric acid to pH 6, pH 5, pH 4, and pH 3 for 2 h at 37°C in a 96-well round bottom plate. After the acid shock, the cultures were serially diluted, plated on LB agar, incubated at 37°C overnight, and then photographed. All of the tests were carried out independently at least in triplicate.
β-Galactosidase Assays
β-Galactosidase assays were carried out by the Miller method when cells were grown to mid-log phase at 37°C at pH 7.0 (Giacomini et al., 1992). All of the tests were carried out independently at least in triplicate.
Treatment With AHLs
The indicated E. coli strains were grown overnight in LB medium at 37°C, and then 100 μl of the cultures was added into 3 ml of LB that contains DMSO (control) or C4-HSL and 3-oxo-C12-HSL (40 μM each) and grown for 6 h at 37°C, followed by RT-qPCR or b-galactosidase activity analysis. All of the tests were carried out independently at least in triplicate.
Statistical Analysis
Data of the results from multiple independent experiments were expressed as the mean ± standard deviation (SM). The differences between groups were analyzed using Student’s t-test when two groups were compared or one-way ANOVA when more than two groups were compared. All analyses were performed using GraphPad Prism, version 5 (GraphPad Software, Inc., San Diego, CA, United States). Differences with a value of 0.01 < P < 0.05 are represented by ∗, P < 0.01 are represented by ∗∗, and P < 0.001 are represented by ∗∗∗.
Results
SdiA Enhances Acid Tolerance Ability of E. coli
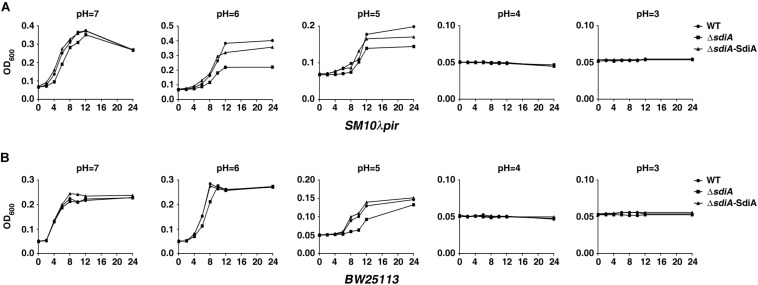
To assess the influence of SdiA on acid tolerance of E. coli, a series of growth assays under different acid stress (24 h of incubation in LBG medium acidified to pH 3–7 by citric acid) was performed in sdiA gene-deficient strain of SM10λpir and BW25113 (ΔsdiA), as well as sdiA-overexpression strain in ΔsdiA mutation (ΔsdiA/SdiA). For SM10λpir, when the pH was down to 6 or 5, deficiency of sdiA impaired the growth through the whole process, compared to the wild-type strain, which was restored by introduction of sdiA-overexpression plasmid (Figure 1A). For BW25113, compared to sdiA-deficient strain, wild-type and sdiA-overexpression strain also showed growth advantage in the early stage under pH 6 and all the stage under pH 5 (Figure 1B). For both SM10λpir and BW25113, when the pH was down to 4 or 3, the OD600 value of wild-type and modified strains showed no difference, as a result of no growth (Figures 1A,B).
FIGURE 1.
Growth of sdiA-deficient and overexpression strains under acid stress. (A,B) The indicated Escherichia coli SM10λpir and E. coli BW25113 strains in exponential phase were collected and re-cultured for 24 h in LBG medium with different initial pH values obtained by the addition of citric acid. The samples were collected at the indicated time points, and the OD600 values were determined. The curve of each strain growth was shown. WT, E. coli SM10λpir or E. coli BW25113 carrying pROp200; ΔsdiA, E. coli SM10λpirΔsdiA or E. coli BW25113ΔsdiA carrying pROp200; ΔsdiA-SidA, E. coli SM10λpirΔsdiA or E. coli BW25113ΔsdiA carrying pROp200-sdiA.
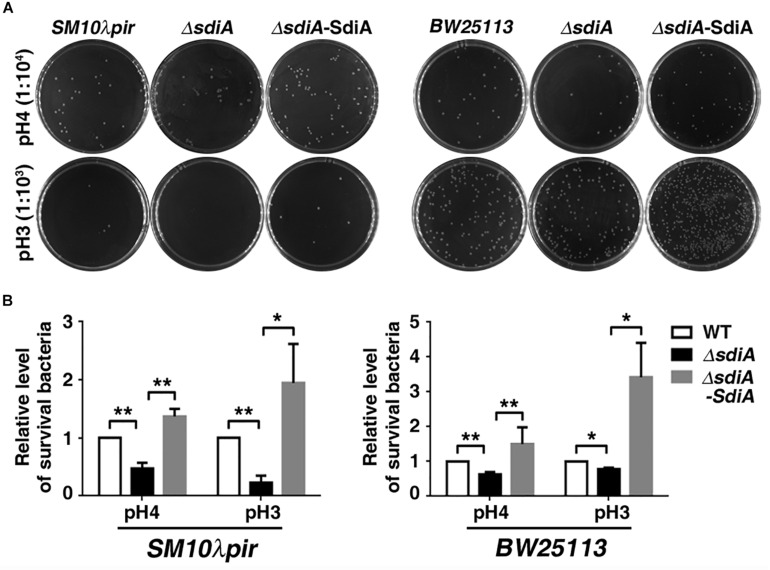
An acid shock assay (2 h of incubation in LBG medium acidified to pH 3–7 by citric acid) was further carried out to investigate the effect of SdiA on acid tolerance under extreme acid stress. The result showed that deficiency of sdiA resulted in decreased survival of both SM10λpir and BW25113 under the acid stress of pH 4 or 3, whereas overexpression of SdiA abrogated this effect (Figures 2A,B).
FIGURE 2.
Survival of sdiA-deficient and overexpression strains after acid shock. (A,B) E. coli SM10λpir and E. coli BW25113 wild-type strain, sdiA-deficiency strain (E. coli SM10λpirΔsdiA, E. coli BW25113ΔsdiA), sdiA-deficiency and overexpression strain (E. coli SM10λpirΔsdiA-SdiA, E. coli BW25113ΔsdiA-SdiA) (2.0 × 105) were incubated for 2 h in LBG medium acidified by citric acid. The images represent 1:104 (pH 4) or 1:103 (pH 3) of the serial dilutions of the cultures in 10-fold steps. *P < 0.05; **P < 0.01.
Collectively, these data suggest that SdiA plays a crucial role in acid tolerance ability of E. coli.
SdiA Promotes the Expression of GadW and GadY
Next, we explored the molecular mechanisms responsible for the function of SdiA that were observed above. The transcriptomes of SM10λpir, SM10λpirΔsdiA, and SM10λpirΔsdiA-SdiA were determined, and we mainly focused on AR2 system genes. The results showed that the transcription of gadA, gadB, gadC, gadE, and gadX was hardly changed in sdiA-knockouted strain, while overexpression of SdiA enhanced gadA, gadB, and gadC expression. This is consistent with the observation that many genes of E. coli that respond to plasmid-based expression of SdiA are largely more different than those that respond to chromosomal SdiA (Dyszel et al., 2010). Noteworthy, the expression of GadW and GadY was changed in both sdiA gene deficient and its compensatory strains, with the down-regulation in SM10λpirΔsdiA, and up-regulation upon SdiA overexpression (Table 1).
TABLE 1.
Transcript level of AR2 genes in SM10λpir sdiA deficient and complemented strains.
| WT | ΔsdiA | ΔsdiA-SdiA | |
| sdiA | 448 | 0 | 11416 |
| gadA | 327.5 | 262.23 | 462.62 |
| gadB | 58.5 | 56.77 | 138.38 |
| gadC | 93 | 61 | 159 |
| gadE | 449 | 360 | 470 |
| gadW | 387 | 147 | 372 |
| gadX | 751 | 563 | 791 |
| gadY | 14 | 8 | 17 |
The calculation of Unigene expression uses RPKM method (reads per kb per million reads).
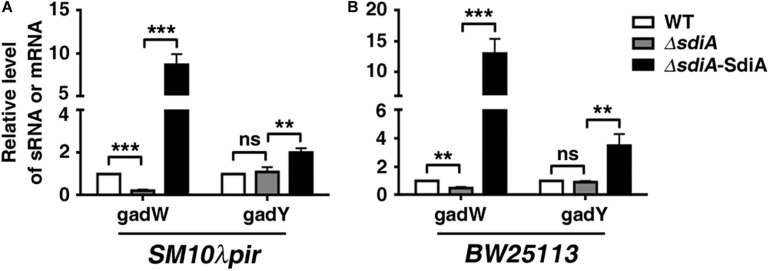
Based on the obvious change in transcriptome data, GadW and GadY were chosen for further analysis by quantitative RT-PCR. Conformably, deficiency of sdiA in both SM10λpir and BW25113 resulted in downregulation of GadW, which was reversed by restoration of sdiA. For GadY, the expression was barely changed in sdiA-deficient strain, but elevated in sdiA-overexpression strain (Figure 3). Taken together, these results implicate that SdiA may positively regulate GadW and GadY expression.
FIGURE 3.
SdiA promotes glutamate decarboxylase W (GadW) and GadY expression. (A) The influence of SdiA on GadW and GadY expression in E. coli SM10λpir. (B) The influence of SdiA on GadW and GadY expression in E. coli BW25113. E. coli SM10λpir or E. coli BW25113 (WT) and the sdiA-deficient strains (ΔsdiA) carrying pROp200, E. coli SM10λpirΔsdiA carrying pROp200-SdiA (ΔsdiA-SdiA) were cultured in LB for 6 h, followed by real-time PCR analysis; the rpoD gene was used as an internal control. **P < 0.01; ***P < 0.001.
SdiA Regulates Transcription Activity of GadW and GadY
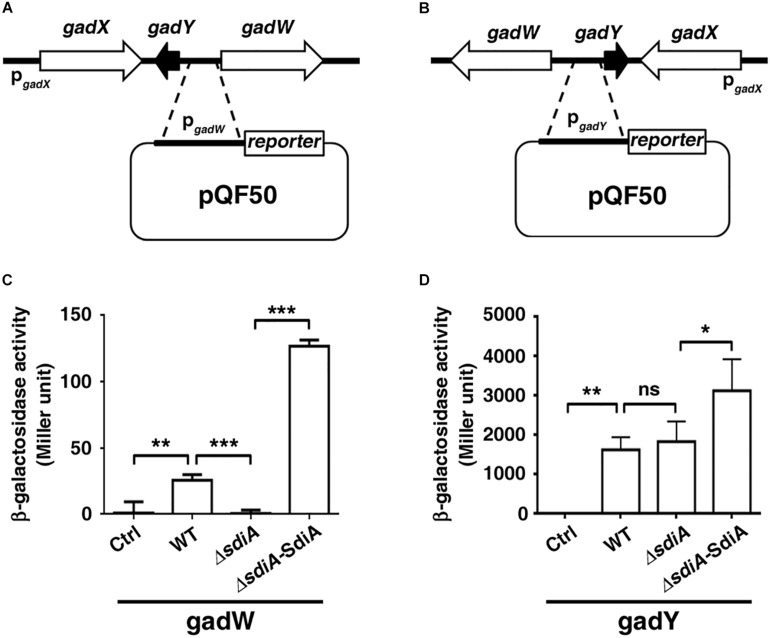
We subsequently evaluated the influence of SdiA on the transcription activity of the gadW and gadY promoter regions. The DNA fragment of gadW or gadY promoter was cloned upstream of the β-galactosidase gene in the pQF50-promoter reporter, respectively (Figures 4A,B). When transformed into BW25113 (without endogenous β-galactosidase), the β-galactosidase activity of pQF50-PgadW and pQF50-PgadY was greatly elevated, compared to that of the control. More importantly, deficiency of sdiA impaired the activity of pQF50-PgadW but hardly affected that of pQF50-PgadY, while overexpression of SdiA enhanced both promoter activity of gadW and gadY (Figures 4C,D). These observations were consistent with the above quantitative RT-PCR results.
FIGURE 4.
The promoter activity of gadW and gadY is regulated by SdiA. (A,B) Diagram of the gadW and gadY promoter and principles of the in vivo investigation of their activity. (C,D) The influence of SdiA on the promoter activity of gadW and gadY. The E. coli BW25113 strains carrying the reporter pQF50, pQF50-PgadW, or pQF50-PgadY combined with pROp200 or pROp200-SdiA were grown to mid-log phase, subjected to β-galactosidase activity assay. Values are mean ± SD of at least three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001.
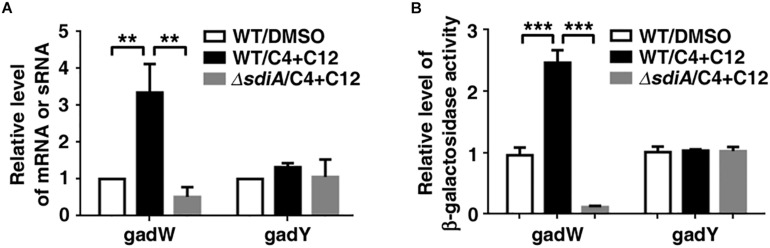
Moreover, it has been shown that transcriptional regulatory function of SdiA may be increased by AHLs. We further checked the influence of AHLs on the promoter activity and expression level of gadW and gadY. As shown in Figures 5A,B, treatment with 3-oxo-C12-HSL and C4-HSL not only enhanced the promoter activity of gadW but also promoted its expression, which were attenuated by deficiency of sdiA. However, this processing seemed to have no effect on the promoter activity and expression of GadY.
FIGURE 5.
The effect of N-acyl homoserine lactones (AHLs) on the expression of gadW and gadY. (A) AHLs promoted gadW but not gadY expression. E. coli SM10λpir was cultured in the presence of DMSO or 40 μM C4-HSL and 3-oxo-C12-HSL for 6 h, followed by real-time PCR analysis. (B) AHLs enhanced the promoter activity of gadW but not gadY. E. coli BW25113 was cultured in the presence of DMSO (Ctrl) or 40 μM C4-HSL and 3-oxo-C12-HSL for 6 h, followed by b-galactosidase activity analysis. Values are mean ± SD of at least three independent experiments. **P < 0.01; ***P < 0.001.
Taken together, our results implicate that SdiA may promote acid tolerance of E. coli at least partly through regulating GadW and GadY expression.
Discussion
In this study, we disclosed the role of SdiA in acid tolerance of E. coli. Our findings highlight the transcription control of SdiA on GadW and GadY, and provide new insights into the regulatory network of the acid resistance system AR2.
Previous studies have identified AFI as the key factor that improves the tolerance of E. coli strains to environmental acidic stress (Foster, 2004). One of the essential AFI regulators, GadE, was a LuxR-like protein. In the present study, we focused on another LuxR protein SdiA. Most recent publications about SdiA have reported its gene regulatory function (Dyszel et al., 2010; Nguyen et al., 2015; Sabag-Daigle et al., 2015). A few reports revealed the roles of SdiA in diverse biological processes (Smith et al., 2011), such as cell division (Sitnikov et al., 1996), antibiotic resistance (Yang et al., 2006; Tavío et al., 2010), virulence (Kanamaru et al., 2000), and biofilm formation (Culler et al., 2018). Here, gain- and loss-of-function studies showed that SdiA positively regulated mild and extreme acid (pH 3–6) tolerance ability of both E. coli strains SM10λpir and BW25113. Since acid stress can damage bacteria cells and impair their growth, and SdiA was activated during the transit of Salmonella through turtles (Smith et al., 2008), we conferred that SdiA might favor E. coli to resist acidic conditions encountered during transit through the mammalian host gastric environment and prolonged exposure to the mildly acidic environment of the host gut.
To date, many SdiA regulon members have been described (Dyszel et al., 2010; Abed et al., 2014; Shimada et al., 2014; Sabag-Daigle et al., 2015). Here, we report the identification of SdiA-regulated genes gadW and gadY through transcriptome sequencing and qPCR. Although the observation that the gad system can be activated by SdiA even in the absence of AHLs has been previously reported (Dyszel et al., 2010; Hughes et al., 2010), how SdiA directly activates transcription of the gad system is unclear. It seems that genes with specific DNA sequences (SdiA-box) 5′-AAAAG(N8)GAAAA-3′ in the promoter region may be the potential targets of SdiA (Yamamoto et al., 2001). Our bioinformatics analysis discovered a DNA motif 5′-AAAAT(N18)TAAAA-3′ and 5′-AAAAC(N18)CAAAA-3′ in the gadY–gadW intergenic region. Further, β-galactosidase activity test showed that SidA regulated the promoter activity containing the above-mentioned DNA motifs. However, the interaction between SdiA and these sites needed further validation. In addition, the β-galactosidase reporter system showed that the transcription activity of gadY arising from gadY–gadW intergenic region was much higher than that of gadW (Figure 4). Nonetheless, the influence of SdiA on the promoter activity of gadW was more remarkable than that of gadY, indicating that there might be other transcription factors that regulated gadY expression.
It has been shown that SdiA is already in a DNA-binding conformation in the absence of AHLs. AHLs enhance this protein’s affinity to DNA, allowing it to regulate transcription of genes that have lower-affinity sites to this protein, whose SdiA regulation occurs only in the presence of AHLs (Nguyen et al., 2015). This was consistent with our observation that SdiA alone can regulate GadW expression, while the presence of C4- and 3-oxo-C12 HSLs enhanced this effect. C4- and 3-oxo-C12 HSLs are the well-known AHLs from Pseudomonas aeruginosa. We have previously found that C4- and 3-oxo-C12 HSLs produced by P. aeruginosa can inhibit traI expression by activating E. coli SdiA. However, neither C4- nor 3-oxo-C12 is the most potent ligands for SdiA, thus the response of SdiA to these two AHLs in this system can possibly represent an artifact. For GadY, the expression was barely changed in chromosomal sdiA-deficient strain, but elevated in sdiA-overexpression strain, which might be explained by the use of high copy number plasmids in this study, and it has been well established that overexpression of SdiA can bypass their requirement for AHL (Michael et al., 2001; Dyszel et al., 2010). More importantly, the presence of AHLs has been suggested in bovine rumen and human gut, and EHEC senses rumen AHLs through SdiA to repress Lee expression and activate the gad AR system, which was necessary for efficient EHEC colonization of cattle fed a forage or grain diet (Sperandio, 2010; Sheng et al., 2013; Landman et al., 2018). Future studies are needed to illuminate the role of AHL-SdiA signaling in acid tolerance within the human gastrointestinal tract.
Data Availability Statement
All data sets generated for this study are included in the article/Supplementary Material.
Author Contributions
XM, SZ, and ZX performed the majority of the experiments. HL, QX, WZ, DZ, YFL, and FQ also conducted the experiments. XM, BH, CC, and YL participated in the experimental design and data analysis. CC and YL conceived the project, designed and under took the experiments, interpreted data, and wrote the manuscript. All authors read and approved the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are very grateful to Jiehu Chen for assistance with the bioinformatics and for helpful discussions.
Footnotes
Funding. This work was supported by the National Natural Science Foundation of China (Grant Nos. 81672081, 81871703, and 81702063); the Guangzhou Science Technology and Innovation Commission (Grant No. 201707010296); and the Guangdong Provincial Hospital of Traditional Chinese Medicine (Grant Nos. YN2018QJ01, YN2019QJ03, and YN2019MJ01).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01078/full#supplementary-material
References
- Abed N., Grépinet O., Canepa S., Hurtado-Escobar G. A., Guichard N., Wiedemann A., et al. (2014). Direct regulation of the pefI-srgC operon encoding the Rck invasin by the quorum-sensing regulator SdiA in Salmonella Typhimurium. Mol. Microbiol. 94 254–271. 10.1111/mmi.12738 [DOI] [PubMed] [Google Scholar]
- Culler H. F., Couto S. C. F., Higa J. S., Ruiz R. M., Yang M. J., Bueris V., et al. (2018). Role of SdiA on Biofilm Formation by Atypical Enteropathogenic Escherichia coli. Genes 9:E253. 10.3390/genes9050253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Biase D., Pennacchietti E. (2012). Glutamate decarboxylase-dependent acid resistance in orally acquired bacteria: function, distribution and biomedical implications of the gadBC operon. Mol. Microbiol. 86 770–786. 10.1111/mmi.12020 [DOI] [PubMed] [Google Scholar]
- Dyszel J. L., Soares J. A., Swearingen M. C., Lindsay A., Smith J. N., Ahmer B. M. M. (2010). E. coli K-12 and EHEC genes regulated by SdiA. PLoS One 5:e8946. 10.1371/journal.pone.0008946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster J. W. (2004). Escherichia coli acid resistance: Tales of an amateur acidophile. Nat. Rev. Microbiol. 2 898–907. 10.1038/nrmicro1021 [DOI] [PubMed] [Google Scholar]
- Fuqua C., Greenberg E. P. (2002). Listening in on bacteria: acyl-homoserine lactone signalling. Nat. Rev. Mol. Cell Biol. 3 685–695. 10.1038/nrm907 [DOI] [PubMed] [Google Scholar]
- Gao X., Yang X., Li J., Zhang Y., Chen P., Lin Z. (2018). Engineered global regulator H-NS improves the acid tolerance of E. coli. Microb. Cell Fact. 17:118. 10.1186/s12934-018-0966-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomini A., Corich V., Ollero F. J., Squartini A., Nuti M. P. (1992). Experimental conditions may affect reproducibility of the beta-galactosidase assay. FEMS Microbiol. Lett. 100 87–90. 10.1111/j.1574-6968.1992.tb14024.x [DOI] [PubMed] [Google Scholar]
- Hughes D. T., Terekhova D. A., Liou L., Hovde C. J., Sahl J. W., Patankar A. V., et al. (2010). Chemical sensing in mammalian host-bacterial commensal associations. Proc. Natl. Acad. Sci. U.S.A. 107 9831–9836. 10.1073/pnas.1002551107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamaru K., Kanamaru K., Tatsuno I., Tobe T., Sasakawa C. (2000). SdiA, an Escherichia coli homologue of quorum-sensing regulators, controls the expression of virulence factors in enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 38 805–816. 10.1046/j.1365-2958.2000.02171.x [DOI] [PubMed] [Google Scholar]
- Landman C., Grill J. P., Mallet J. M., Marteau P., Humbert L., Le Balc’h E., et al. (2018). Inter-kingdom effect on epithelial cells of the N-Acyl homoserine lactone 3-oxo-C12:2, a major quorum-sensing molecule from gut microbiota. PLoS One 13:e202587. 10.1371/journal.pone.0202587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Zhang X.-S., Hegde M., Bentley W. E., Jayaraman A., Wood T. K. (2008). Indole cell signaling occurs primarily at low temperatures in Escherichia coli. ISME J. 2 1007–1023. 10.1038/ismej.2008.54 [DOI] [PubMed] [Google Scholar]
- Lu Y., Zeng J., Wu B., Eaa S., Wang L., Cai R., et al. (2017). Quorum Sensing N-acyl Homoserine Lactones-SdiA Suppresses Escherichia coli-Pseudomonas aeruginosa conjugation through inhibiting traI expression. Front. Cell. Infect. Microbiol. 7:7. 10.3389/fcimb.2017.00007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund P., Tramonti A., De Biase D. (2014). Coping with low pH: molecular strategies in neutralophilic bacteria. FEMS Microbiol. Rev. 38 1091–1125. 10.1111/1574-6976.12076 [DOI] [PubMed] [Google Scholar]
- Ma Z., Gong S., Richard H., Tucker D. L., Conway T., Foster J. W. (2003). GadE (YhiE) activates glutamate decarboxylase-dependent acid resistance in Escherichia coli K-12. Mol. Microbiol. 49 1309–1320. 10.1046/j.1365-2958.2003.03633.x [DOI] [PubMed] [Google Scholar]
- Michael B., Smith J. N., Swift S., Heffron F., Ahmer B. M. (2001). SdiA of Salmonella enterica is a LuxR homolog that detects mixed microbial communities. J. Bacteriol. 183 5733–5742. 10.1128/JB.183.19.5733-5742.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen Y., Nguyen N. X., Rogers J. L., Liao J., MacMillan J. B., Jiang Y., et al. (2015). Structural and mechanistic roles of novel chemical ligands on the SdiA quorum-sensing transcription regulator. mBio 6:e02429-14. 10.1128/mBio.02429-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opdyke J. A., Kang J.-G., Storz G. (2004). GadY, a small-RNA regulator of acid response genes in Escherichia coli. J. Bacteriol. 186 6698–6705. 10.1128/JB.186.20.6698-6705.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabag-Daigle A., Dyszel J. L., Gonzalez J. F., Ali M. M., Ahmer B. M. M. (2015). Identification of sdiA-regulated genes in a mouse commensal strain of Enterobacter cloacae. Front. Cell. Infect. Microbiol. 5:47. 10.3389/fcimb.2015.00047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M., Sexton D. J., Diggle S. P., Greenberg E. P. (2013). Acyl-homoserine lactone quorum sensing: from evolution to application. Annu. Rev. Microbiol. 67 43–63. 10.1146/annurev-micro-092412-155635 [DOI] [PubMed] [Google Scholar]
- Seo S. W., Kim D., O’Brien E. J., Szubin R., Palsson B. O. (2015). Decoding genome-wide GadEWX-transcriptional regulatory networks reveals multifaceted cellular responses to acid stress in Escherichia coli. Nat. Commun. 6:7970. 10.1038/ncomms8970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng H., Nguyen Y. N., Hovde C. J., Sperandio V. (2013). SdiA aids enterohemorrhagic Escherichia coli carriage by cattle fed a forage or grain diet. Infect. Immun. 81 3472–3478. 10.1128/IAI.00702-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T., Shimada K., Matsui M., Kitai Y., Igarashi J., Suga H., et al. (2014). Roles of cell division control factor SdiA: Recognition of quorum sensing signals and modulation of transcription regulation targets. Genes Cells 19 405–418. 10.1111/gtc.12139 [DOI] [PubMed] [Google Scholar]
- Sitnikov D. M., Schineller J. B., Baldwin T. O. (1996). Control of cell division in Escherichia coli: regulation of transcription of ftsQA involves both rpoS and SdiA-mediated autoinduction. Proc. Natl. Acad. Sci. U.S.A. 93 336–341. 10.1073/pnas.93.1.336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. L., Fratamico P. M., Yan X. (2011). Eavesdropping by bacteria: the role of SdiA in Escherichia coli and Salmonella enterica serovar Typhimurium quorum sensing. Foodborne Pathog. Dis. 8 169–178. 10.1089/fpd.2010.0651 [DOI] [PubMed] [Google Scholar]
- Smith J. N., Dyszel J. L., Soares J. A., Ellermeier C. D., Altier C., Lawhon S. D., et al. (2008). SdiA, an N-acylhomoserine lactone receptor, becomes active during the transit of Salmonella enterica through the gastrointestinal tract of turtles. PLoS One 3:e2826. 10.1371/journal.pone.0002826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperandio V. (2010). SdiA sensing of acyl-homoserine lactones by enterohemorrhagic E. coli (EHEC) serotype o157:H7 in the bovine rumen. Gut Microbes 1 432–435. 10.4161/gmic.1.6.14177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavío M. M., Aquili V. D., Poveda J. B., Antunes N. T., Sánchez-Céspedes J., Vila J. (2010). Quorum-sensing regulator sdiA and marA overexpression is involved in in vitro-selected multidrug resistance of Escherichia coli. J. Antimicrob. Chemother. 65 1178–1186. 10.1093/jac/dkq112 [DOI] [PubMed] [Google Scholar]
- Tramonti A., De Canio M., De Biase D. (2008). GadX/GadW-dependent regulation of the Escherichia coli acid fitness island: transcriptional control at the gadY-gadW divergent promoters and identification of four novel 42 bp GadX/GadW-specific binding sites. Mol. Microbiol. 70 965–982. 10.1111/j.1365-2958.2008.06458.x [DOI] [PubMed] [Google Scholar]
- Tramonti A., De Canio M., Delany I., Scarlato V., De Biase D. (2006). Mechanisms of transcription activation exerted by GadX and GadW at the gadA and gadBC gene promoters of the glutamate-based acid resistance system in Escherichia coli. J. Bacteriol. 188 8118–8127. 10.1128/JB.01044-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Houdt R., Aertsen A., Moons P., Vanoirbeek K., Michiels C. W. (2006). N-acyl-L-homoserine lactone signal interception by Escherichia coli. FEMS Microbiol. Lett. 256 83–89. 10.1111/j.1574-6968.2006.00103.x [DOI] [PubMed] [Google Scholar]
- Yamamoto K., Yata K., Fujita N., Ishihama A. (2001). Novel mode of transcription regulation by SdiA, an Escherichia coli homologue of the quorum-sensing regulator. Mol. Microbiol. 41 1187–1198. 10.1046/j.1365-2958.2001.02585.x [DOI] [PubMed] [Google Scholar]
- Yang S., Lopez C. R., Zechiedrich E. L. (2006). Quorum sensing and multidrug transporters in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 103 2386–2391. 10.1073/pnas.0502890102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Zhou F., Xing Q., Cao Z., Liu J., Zhu G. (2018). The soluble transhydrogenase UdhA affecting the glutamate-dependent acid resistance system of Escherichia coli under acetate stress. Biol. Open 7:bio.031856. 10.1242/bio.031856 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data sets generated for this study are included in the article/Supplementary Material.