Abstract
Pre-B-cell leukemia transcription factor (PBX) proteins have important roles in the development of numerous organs. To date, four members of the PBX family have been identified to be involved in human cancer but little is known about their expression patterns and precise functions in breast cancer (BC) progression. The aim of the present study was to determine whether they have the potential to be prognostic biomarkers in patients with BC. The expression patterns of PBXs were evaluated using Oncomine, Cancer Cell Line Encyclopedia and Gene expression-based Outcome for Breast cancer Online algorithm analyses. The prognostic value of PBX1 was determined by Kaplan-Meier plotter analysis. It was observed that, among all PBX family members, only PBX1 was significantly upregulated in BC vs. normal tissues. Meta-analysis in the Oncomine database revealed that PBX1 was significantly upregulated in invasive breast carcinoma stroma, ductal breast carcinoma, invasive lobular breast carcinoma, invasive mixed breast carcinoma and male breast carcinoma compared with normal tissues. In addition, PBX1 was significantly correlated with forkhead box protein A1. Subtype analysis indicated that PBX1 overexpression was associated with luminal-like and hormone receptor-sensitive subtypes. In the survival analysis, a high expression level of PBX1 was associated with poor prognosis of patients with estrogen receptor (ER)-positive, luminal A and luminal B subtypes of BC. The results of the present study indicate that PBX1 may serve as a specific biomarker and essential prognostic factor for ER-positive, luminal A and luminal B subtypes of BC.
Keywords: Pre-B-cell leukemia transcription factor, breast cancer, biomarker, prognosis, database mining
Introduction
Breast cancer (BC) is one of the most common malignant diseases and has become the second leading cause of death in females (1-3). According to statistics from the World Health Organization and American Cancer Society, 2.09 million new cases of BC were diagnosed worldwide in 2018. BC has reached the highest age-standardized frequency (46.3 per 100,000) compared with other types of cancer. In addition, the prognosis of BC ranks third after thyroid and prostate cancer, with a 5-year survival rate of 80-85% (4). The incidence of BC, which seriously affects the quality of life of patients, is gradually increasing owing to an unhealthy lifestyle, enhanced awareness and new screening technologies (5,6). Although certain advances in therapeutic strategies against BC have been made, it remains a major public health problem. Clinically, it is difficult to observe the early symptoms of patients with cancer due to the pathophysiological characteristics of the tumor, which results in the majority of patients with BC being diagnosed at an advanced stage (7). Therefore, it is important to identify novel and valuable biomarkers for the diagnosis and prognosis of BC, and/or to provide novel therapeutic targets.
Pre-B-cell leukemia transcription factor (PBX) proteins are atypical homeodomain-containing transcription factors, which belong to the family of three amino-acid loop extension-class homeodomain proteins (8). They have an integral role in the development of multiple organs, including the appendicular skeleton, lung, heart, pancreas, spleen and kidney, by regulating the expression of crucial target genes (9). The mammalian system consists of four members, namely PBX1, PBX2, PBX3, and PBX4(10). PBX1 was initially identified due to its involvement in a chromosomal translocation in human pre-B cell acute lymphoid leukemia (11). PBX2 and PBX3 were subsequently identified based on their sequence homology with PBX1(12). PBX4 was first identified in the developing zebrafish brain (13). A large number of studies have demonstrated that the deregulation of PBX proteins is closely associated with the development and progression of numerous diseases, including cancer (14-18). PBX proteins have also been reported to be overexpressed in a variety of solid tumor types, including colorectal, gastric and ovarian cancers, as well as glioma (19-21). The estradiol acetate (E2A)-PBX1 fusion protein contributes to pre-B cell transformation in leukemogenesis by enhancing the transactivation activity of homeobox (HOX) family members (22). The HOXB7/PBX2 complex promotes the progression of cutaneous melanoma by transcriptionally activating microRNA (miR)-221 and -222(18). Furthermore, PBX4 was reported to be downregulated in leukemic cell lines and the samples from patients with acute lymphoblastic leukemia (ALL) (23). PBX3 has been indicated to be a key driver of mesenchymal transition and a potential therapeutic target in glioblastoma (24). In addition, it has been reported to be a prognostic biomarker in prostate cancer (25). Although PBX proteins have been demonstrated to act as crucial transcription factors in numerous malignancies (26-29), the roles of different PBX family members in the progression of BC have remained elusive.
The present study extensively analyzed the expression of distinct PBX family members in patients with BC based on publicly available clinical datasets in order to determine their expression patterns and prognostic value in BC. It was demonstrated that PBX1 is highly expressed in patients with BC and is associated with poor clinical outcomes in patients with estrogen receptor (ER)-positive, luminal A and luminal B subtypes of BC.
Materials and methods
Ethics statement
Informed consent for publication was not required, as all patient data used in the study were obtained from publicly available databases.
Oncomine database analysis
The Oncomine database (https://www.oncomine.org/resource/login.html; accession date, May 23, 2018) was used to determine the genes co-expressed with PBX1 and differences in the transcriptional levels of distinct PBXs in different types of cancer. Student's t-test was performed to determine significant differences between clinical tumor samples and control samples. The threshold of the P-value was set to 1x10-4 and the threshold of the fold change was set to 2.
Cancer cell line encyclopedia (CCLE) analysis
The expression of PBX1 in different types of cancer was analyzed using CCLE (https://portals.broadinstitute.org/ccle; accession date, May 26, 2018), which is an online database that provides public access to genomic data, analysis and visualization of 947 human cancer cell lines (30).
Gene expression-based outcome for breast cancer online algorithm (GOBO) analysis
GOBO (http://co.bmc.lu.se/gobo/gsa_cellines.pl; accession date, May 27 2018), a database that uses Affymetrix U133A microarray gene expression data from 1,881 patients with BC and a 51-sample BC cell line set (31), was used to determine the transcription level of PBX1 in different molecular subtypes of BC. Analysis of variance was performed to determine the significant difference among different molecular subtypes of BC, followed by Bonferroni's post-hoc test for multiple comparisons. The calculated P-values were adjusted for multiple testing by applying a false discovery rate adjustment of 0.2.
Survival analysis
Kaplan-Meier plotter (http://kmplot.com/analysis/) is an online database that contains updated gene expression data and survival information of patients with cancer. It may be used to evaluate the effect of genes on survival rate using clinical cancer specimens. The prognostic significance of PBX1 in BC specimens was assessed by displaying the relapse-free survival (RFS) in Kaplan-Meier plots (32).
Results
PBX1 is significantly overexpressed in patients with BC
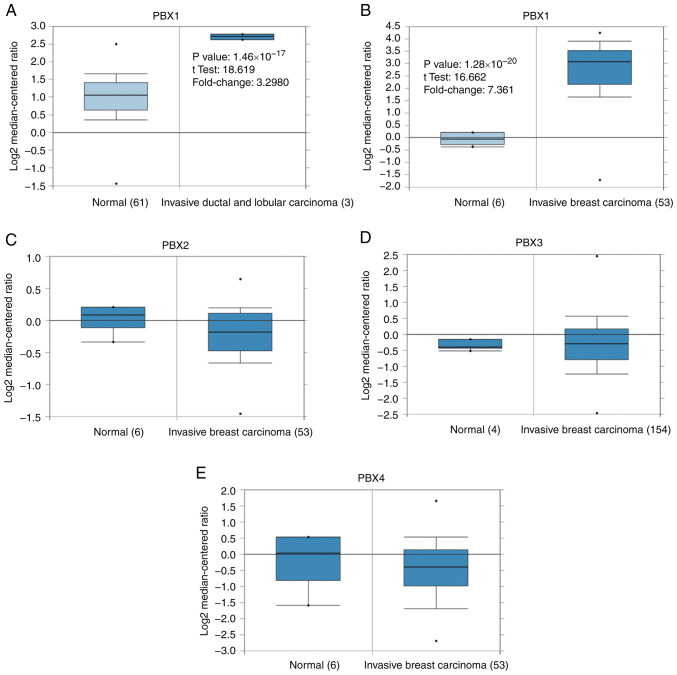
To date, four PBX members have been identified in mammalian cells. However, little is known about their roles in BC progression. To determine whether PBX family members are involved in BC progression, the expression of PBXs was determined in BC tissues and control tissues using the Oncomine database, which altogether includes 465, 449, 443 and 258 unique datasets for PBX1, PBX2, PBX3 and PBX4, respectively. The mRNA level of PBX1 in patients with BC was significantly upregulated in 8 datasets, whereas no significant difference in PBX2, PBX3 or PBX4 mRNA levels was observed between BC tissues and control tissues (Fig. 1). The PBX1 mRNA level was 3.298-fold elevated in BC tissues compared with control samples in an analysis of 593 specimens from The Cancer Genome Atlas (http://cancergenome.nih.gov) database (P=1.46x10-17; Fig. 2A). In Finak's dataset (33), PBX1 expression was 7.361-fold elevated in BC tissues compared with control samples (P=1.28x10-20), whereas no significant difference in PBX2, PBX3 or PBX4 mRNA levels was observed between BC samples and normal controls (Fig. 2B-E).
Figure 1.
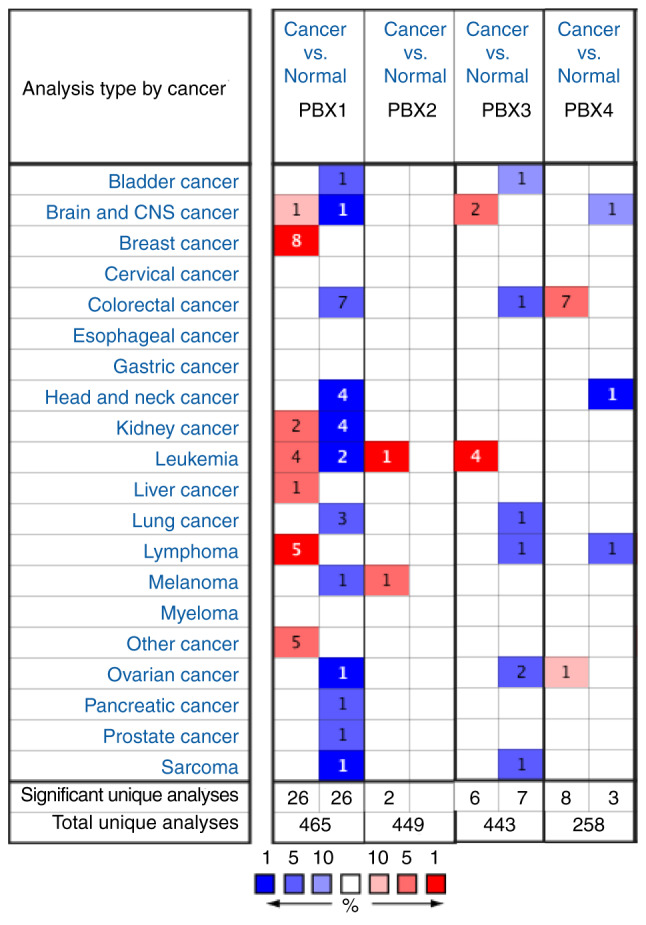
Analysis of PBX expression pattern among different types of cancer using the Oncomine database. The graph indicates the numbers of datasets with statistically significant overexpression (red) or downregulation (blue) of PBXs (cancer vs. normal). The threshold of the P-value was set to 1x10-4. The threshold of the fold change was set to 2. PBX, pre-B-cell leukemia transcription factor.
Figure 2.
Analysis of PBX family members in Oncomine datasets. Box plots derived from gene expression data in Oncomine comparing the mRNA expression of a specific PBX family member in normal and BC tissues. The threshold of the P-value was set to 1x10-4 and the fold change threshold was set to 2. Comparison of PBX1 mRNA expression by tissue type (cancer vs. normal) in the (A) TCGA database and (B) Finak's breast dataset. (C) Comparison of PBX2 mRNA expression by tissue type (cancer vs. normal) in Finak's breast dataset. (D) Comparison of PBX3 mRNA expression tissue type (cancer vs. normal) in the Gluck's breast dataset. (E) Comparison of PBX4 mRNA expression by tissue type (cancer vs. normal) in Finak's breast dataset. The black dots indicateoutliers. PBX, pre-B-cell leukemia transcription factor; BC, breast cancer; TCGA, The Cancer Genome Atlas.
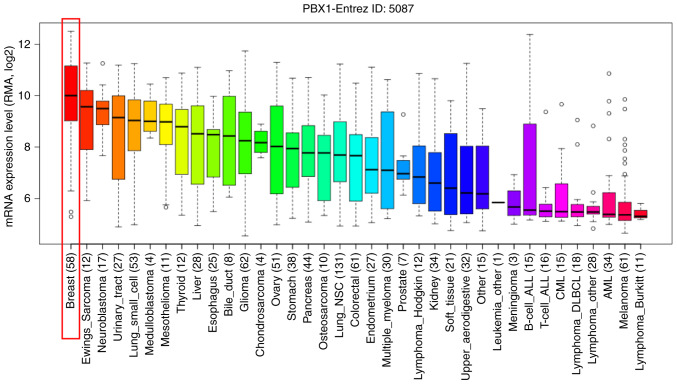
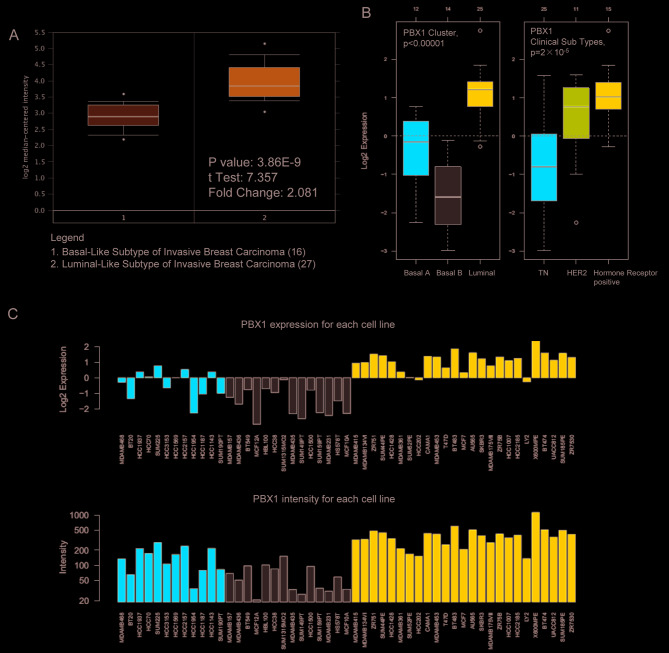
To further determine the exact expression pattern of PBX1, the mRNA expression of PBX1 in different cancer cell lines was examined using the CCLE database, which contains a large number of cancer cell lines and provides extensive information on the expression of PBX1 in numerous cancer subtypes with different tissue types of origin. As indicated in Fig. 3, the transcription level of PBX1 in BC cell lines was the highest among all of the cancer types. This result was consistent with the data from the Oncomine analysis, suggesting that PBX1 may have a crucial role in the carcinogenesis of BC.
Figure 3.
Analysis of PBX1 mRNA expression in the Cancer Cell Line Encyclopedia database. PBX1 was significantly overexpressed in BC cell lines and its expression level was the highest in BC cell lines among all of the different cancer cell lines. PBX, pre-B-cell leukemia transcription factor; BC, breast cancer; CML, chronic myeloid leukemia; AML, acute myeloid leukemia; DLBCL, diffuse large B-cell lymphoma; NSC, non-small cell.
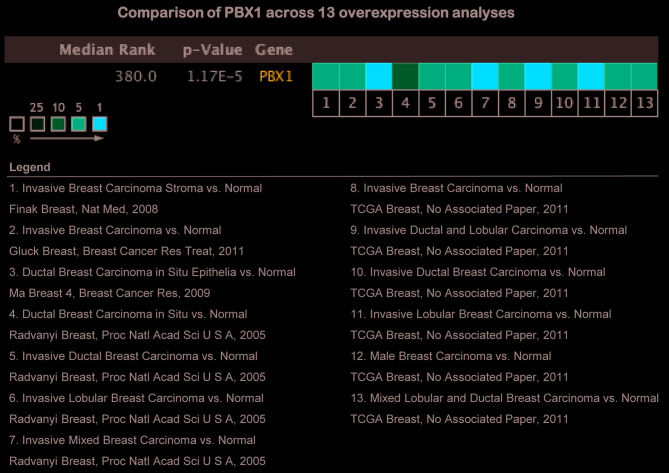
Since the above evidence indicated that PBX1 is highly expressed in BC tissues and cell lines, a pooled meta-analysis was performed to confirm the high expression of PBX1 in BC using the Oncomine database. A total of 13 analyses, including 939 samples, were included in the meta-analysis. Overall, the pooled analysis demonstrated that the mRNA level of PBX1 was significantly increased in the 13 clinical cohorts of patients with BC (Fig. 4). This result provided strong evidence that the expression of PBX1 is significantly increased in BC.
Figure 4.
Meta-analysis of gene expression profiling for the PBX1 gene in breast cancer using the Oncomine database. The P-value threshold was 1x10-4 and the threshold of the fold change was set to 2. The colored squares indicate the median rank for PBX1 across each analysis comparing breast cancer with normal tissue. The color intensity indicates the respective levels of PBX1. The red column indicates PBX1 mRNA upregulation. PBX, pre-B-cell leukemia transcription factor; TCGA, The Cancer Genome Atlas.
PBX1 expression is significantly correlated with forkhead box protein (FOX)A1 in BC
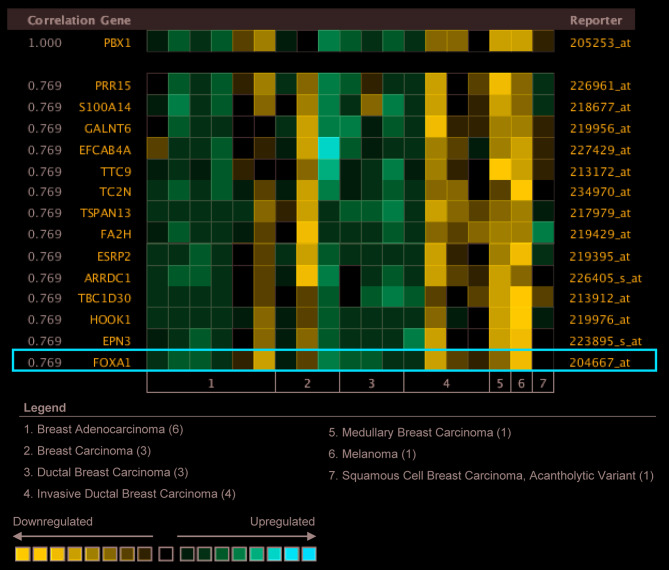
To gain further insight into the potential roles of PBX1 in BC, co-expression analysis of PBX1 mRNA was performed using the Oncomine database. The co-expression profile of PBX1 was examined using a cluster of 19,574 genes in 19 BC samples (34). As presented in Fig. 5, PBX1 expression was significantly correlated with FOXA1 (r=0.769). FOXA1 is a member of the FOX family of transcription factors, which has been reported to promote BC progression by enhancing the expression and activity of ERα (35,36). This result suggested that PBX1 might have a role in BC carcinogenesis by interacting with the FOXA1-ERα axis.
Figure 5.
Co-expression analysis of PBX1 among distinct subtypes of breast cancer. Oncomine analysis revealed that PBX1 expression was distinctively correlated with FOXA1 expression. The correlation between PBX1 and other proteins was presented as the ratio of the mRNA expression value of target protein to that of PBX1 mRNA. For comparison, the expression value of PBX1 mRNA was set to 1.000. PBX, pre-B-cell leukemia transcription factor; FOXA1, forkhead box protein 1.
Analysis of PBX1 expression pattern in distinct BC subtypes
The present study next examined the PBX1 expression pattern in distinct BC subtypes using the Oncomine database. The study by Farmer et al (37) revealed that PBX1 was 2.081-fold upregulated in luminal-like BC tissues compared with that in basal-like BC tissues (P=3.86x10-9; Fig. 6A). GOBO analysis was used to further examine the PBX1 expression pattern in distinct BC subtypes. As presented in Fig. 6B, PBX1 expression was significantly upregulated in the luminal-like subtype compared with that in the basal-A and basal-B subtypes of BC. Furthermore, the expression of PBX1 was significantly upregulated in the hormone receptor-sensitive subtype compared with those in the triple-negative (TN) and human epidermal growth factor receptor 2 (Her-2) subtypes. The PBX1 mRNA expression showed differences across 51 breast cancer cell lines (Fig. 6C).
Figure 6.
Analysis of PBX1 expression among distinct BC subtypes. (A) Box plot derived from Oncomine showed the comparison of PBX1 mRNA expression by different BC tissue subtypes (basal-like subtype vs. luminal-like subtype) in the Farmer's breast dataset. The black dots indicate outliers. (B) Gene expression-based Outcome for Breast cancer Online analysis revealed that the expression of PBX1 in luminal-like was significantly higher than in Basal A or Basal B subtypes of BC, and hormone receptor-positive subtype also express higher PBX1 compared with TN and HER2 subtypes. The white circles indicate outliers. (C) Gene expression-based Outcome for Breast cancer Online analysis showed the differential expression levels of PBX1 across 51 BC cell lines. Red bars represent Basal A BC cell lines. Grey bars represent Basal B BC cell lines. Blue bars represent hormone receptor-positive BC cell lines. PBX, pre-B-cell leukemia transcription factor; BC, breast cancer; TN, triple-negative; HER2, human epidermal growth factor receptor 2.
High PBX1 expression is associated with poor prognosis in patients with BC
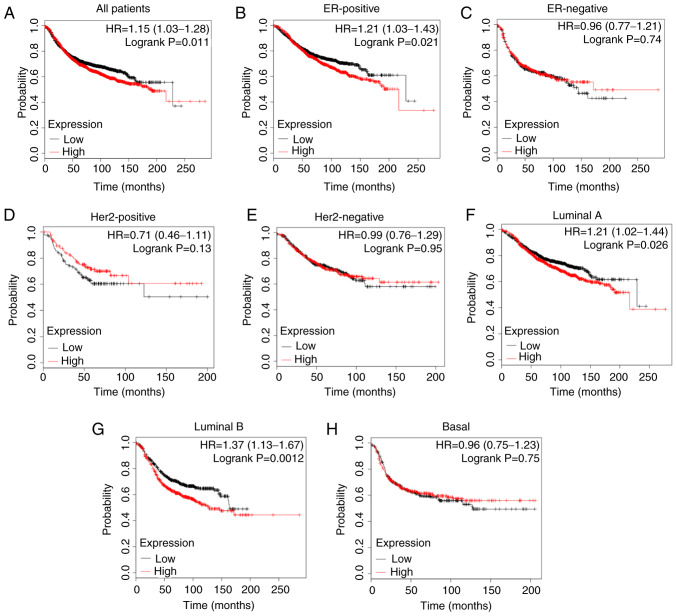
Since elevated PBX1 expression was closely associated with BC progression, the present study further determined the effect of PBX1 on the long-term survival of patients with BC using the Kaplan-Meier plotter, which is an online analysis tool that may be used to evaluate the effect of 54,000 genes on survival in 21 cancer types (32). As presented in Fig. 7A, a high expression level of PBX1 was closely associated with a shorter RFS in all patients with BC [hazard ratio (HR)=1.15, P=0.011]. Furthermore, the analysis of BC subtypes revealed that the overexpression of PBX1 was significantly associated with a shorter RFS in ER-positive (HR=1.21, P=0.021), luminal A (HR=1.21, P=0.026) and luminal B (HR=1.37, P=0.0012) subtypes, but not in ER-negative (HR=0.96, P=0.74), Her-2-positive (HR=0.71, P=0.13), Her2-negative (HR=0.99, P=0.95) or basal-like (HR=0.96, P=0.75) subtypes (Fig. 7B-H). These results indicated a crucial prognostic value of PBX1 in ER-positive, luminal A and luminal B subtypes of BC.
Figure 7.
Clinical significance of PBX1 expression in patients with BC. Low expression of PBX1 was significantly associated with longer RFS in (A) all patients with BC and (B) ER-positive subtype, but not in (C) the ER-negative subtype. Low expression of PBX1 was not associated with longer RFS in either (D) Her-2-positive or (E) Her-2-negative subtype. Low expression of PBX1 was significantly associated with longer RFS in (F) luminal A and (G) luminal B subtypes, but not in (H) basal-like subtype. PBX, pre-B-cell leukemia transcription factor; BC, breast cancer; RFS, relapse-free survival; ER, estrogen receptor; HR, hazard ratio; Her-2, human epidermal growth factor receptor 2.
Discussion
BC is one of the most common types of cancer among all malignant tumor types in females, with high incidence and mortality rates (38-40). Due to its pathological and anatomical features, it is difficult to observe the early onset of BC and tumors are frequently diagnosed in the late stages of the disease. The initiation and development of BC are immensely complex biological processes and the disease has a poor prognosis. Therefore, it is necessary to identify valuable predictive and prognostic biomarkers for early diagnosis and effective treatment of BC. In the present study, publicly available clinical datasets were evaluated to determine differentially expressed genes between cancer samples and control samples. The results suggested that PBX1 was significantly upregulated in BC samples compared with other PBX genes, indicating its specific role in patients with BC.
PBX proteins are homeodomain-containing transcription factors that act as HOX cofactors to regulate gene transcription during development (41-43). Growing evidence suggests that PBXs have oncogenic functions. For instance, PBX1 acts as a chimeric fusion partner in the oncoprotein E2A-PBX1 to promote the progression of pre-B cell ALL (44,45). It also confers prostate cancer cells resistance against common chemotherapeutic drugs, including doxorubicin and cisplatin, and its degradation promotes the apoptosis of prostate cancer cells (46). PBX2 has been reported to mediate the antiapoptotic function of miR-1915-3p in lung cancer (47). High expression of PBX2 has been identified as a poor prognostic indicator in gingival squamous cell carcinoma and non-small cell lung cancer (48,49). PBX3 has been suggested to be the target of multiple miRs, including miR-320a, miR-129-5p, and miR-526b, and mediates the antitumor effects of these miRs (50-52). Furthermore, PBX3 enhances the stability of Meis homeobox 1 (MEIS1) and dimerizes with MEIS1 to efficiently promote leukemogenesis in mouse models (53). It is also recognized as a prognostic factor in several types of cancer, including colorectal, cervical, gastric and prostate cancer (17,25,54,55). Low levels of PBX4 expression are observed in blood samples of patients with ALL (23). However, its function has not been reported in ALL progression. In the present study, data from the Oncomine analysis suggested that, among all PBX family members, only the expression level of PBX1 was higher in BC samples than in the corresponding control samples. Consistent with this, CCLE analysis indicated that the PBX1 expression levels in BC were the highest among all cancer types at the cellular level. The study further investigated the PBX1 expression pattern in distinct subtypes of BC. GOBO analysis indicated that PBX1 mRNA was significantly overexpressed in luminal-like subtypes of BC compared with the basal-A and basal-B subtypes. In addition, PBX1 mRNA was significantly overexpressed in the hormone receptor-sensitive subtype of BC compared with that in the TN and Her-2 clinical subtypes. Furthermore, meta-analysis also revealed that PBX1 overexpression was associated with BC. These results demonstrated that PBX1 is overexpressed in BC tissues and BC cell lines, and that PBX1 expression is closely associated with BC progression, indicating its unique role in BC. However, the detailed mechanisms of the role of PBX1 in BC remain unknown and require further elucidation.
It has been reported that PBX1 is correlated with ERα and promotes the expression of genes associated with the aggressive progression of ERα-positive BC by guiding estrogen-induced ERα recruitment to its target chromatin (56). Another study indicated that PBX1 also has an important role in ERα-mediated transcriptional response to estrogen-independent epidermal growth factor (EGF) signaling (57). Knockdown of PBX1 blocks the expression of a subset of EGF-ERα target genes in ERα-positive BC cells. Furthermore, upregulation of PBX1 was associated with an increased risk of metastatic progression of ERα-positive BC (57). In the present study, gene co-expression analysis revealed that PBX1 is closely associated with FOXA1 in BC patients. FOXA1 belongs to the FOX family of transcription factors and was first identified due to its role in the transcriptional regulation of genes expressing liver-specific transthyretin and α1-antitrypsin (58). FOXA1 is a prognostic marker and novel therapeutic target in BC (59). It has been reported that FOXA1 may act as an initiating factor to regulate the activity of ERα by enhancing the binding of ERα to its target genes (60). Furthermore, it directly binds to the promoter of ER and regulates the expression of ERα in BC cells (36). The present results suggest that PBX1 may have a crucial role in contributing to BC progression via the FOXA1 signaling pathway. In addition, PBX1 has been suggested to act as an initiating factor to guide ERα genomic activity to unique genomic regions, thus promoting the transcription of genes involved in BC progression (56,57).
The clinical significance of PBX1 expression in BC remains unclear. Therefore, the present study examined the association between the PBX1 expression pattern and the prognosis of different molecular subtypes of BC by Oncomine analysis and established the association of PBX1 expression with survival outcome. The pooled results indicated that high PBX1 expression was significantly associated with worse RFS in ER-positive, luminal A and luminal B subtypes of BC, but not in ER-negative, Her2-positive or basal-like BC subtypes, suggesting that PBX1 is a potential oncoprotein in BC. The results of the present study are consistent with those of Magnani et al (56,57). Overall, these data demonstrate that the expression level of PBX1 may be a useful biomarker of prognosis for ER-positive, luminal A and luminal B subtypes of BC.
In conclusion, PBX1 is significantly overexpressed in BC tissues compared with normal tissues and its high expression level is a predictor of poor prognosis of patients with ER-positive, luminal A and luminal B subtypes of BC. In addition, PBX1 is positively correlated with FOXA1. The present study suggests that PBX1 may be an important biomarker for BC. Large-scale and comprehensive studies are still required to further confirm the present results and to determine the clinical application of PBX1 in the prognostication of patients with BC.
Acknowledgements
Not applicable.
Funding
This work was supported by the National Natural Science Foundation of China (grant nos. 81702785 and 81802822), the Natural Science Foundation of Shandong Province (grant nos. ZR2017PH013 and ZR2018BH017) and the Qingdao Postdoctoral Application Research Funded Project (grant no. 2016067).
Availability of data and materials
All datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
XA, WD, HG, YZ, DD and YL analyzed the data. XA and YL designed the study and prepared the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Liu Y, Ao X, Jia Z, Bai XY, Xu Z, Hu G, Jiang X, Chen M, Wu H. FOXK2 transcription factor suppresses ERalpha-positive breast cancer cell growth through down-regulating the stability of ERα via mechanism involving BRCA1/BARD1. Sci Rep. 2015;5(8796) doi: 10.1038/srep08796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang M, Zhao F, Li S, Chang AK, Jia Z, Chen Y, Xu F, Pan H, Wu H. AIB1 cooperates with ERα to promote epithelial mesenchymal transition in breast cancer through SNAI1 activation. PLoS One. 2013;8(e65556) doi: 10.1371/journal.pone.0065556. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3.Ao X, Li S, Xu Z, Yang Y, Chen M, Jiang X, Wu H. Sumoylation of TCF21 downregulates the transcriptional activity of estrogen receptor-alpha. Oncotarget. 2016;7:26220–26234. doi: 10.18632/oncotarget.8354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mattiuzzi C, Lippi G. Current cancer epidemiology. J Epidemiol Glob Health. 2019;9:217–222. doi: 10.2991/jegh.k.191008.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang SY, Kim YS, Kim Z, Kim HY, Lee SK, Jung KW, Youn HJ. Korean Breast Cancer Society: Basic findings regarding breast cancer in Korea in 2015: Data from a breast cancer registry. J Breast Cancer. 2018;21:1–10. doi: 10.4048/jbc.2018.21.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drukteinis JS, Mooney BP, Flowers CI, Gatenby RA. Beyond mammography: New frontiers in breast cancer screening. Am J Med. 2013;126:472–479. doi: 10.1016/j.amjmed.2012.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y, Ao X, Ding W, Ponnusamy M, Wu W, Hao X, Yu W, Wang Y, Li P, Wang J. Critical role of FOXO3a in carcinogenesis. Mol Cancer. 2018;17(104) doi: 10.1186/s12943-018-0856-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Capellini TD, Zappavigna V, Selleri L. Pbx homeodomain proteins: TALEnted regulators of limb patterning and outgrowth. Dev Dyn. 2011;240:1063–1086. doi: 10.1002/dvdy.22605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Welsh IC, Hart J, Brown JM, Hansen K, Rocha Marques M, Aho RJ, Grishina I, Hurtado R, Herzlinger D, Ferretti E, et al. Pbx loss in cranial neural crest, unlike in epithelium, results in cleft palate only and a broader midface. J Anat. 2018;233:222–242. doi: 10.1111/joa.12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Longobardi E, Penkov D, Mateos D, De Florian G, Torres M, Blasi F. Biochemistry of the tale transcription factors PREP, MEIS, and PBX in vertebrates. Dev Dyn. 2014;243:59–75. doi: 10.1002/dvdy.24016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nourse J, Mellentin JD, Galili N, Wilkinson J, Stanbridge E, Smith SD, Cleary ML. Chromosomal translocation t(1;19) results in synthesis of a homeobox fusion mRNA that codes for a potential chimeric transcription factor. Cell. 1990;60:535–545. doi: 10.1016/0092-8674(90)90657-z. [DOI] [PubMed] [Google Scholar]
- 12.Monica K, Galili N, Nourse J, Saltman D, Cleary ML. PBX2 and PBX3, new homeobox genes with extensive homology to the human proto-oncogene PBX1. Mol Cell Biol. 1991;11:6149–6157. doi: 10.1128/mcb.11.12.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pöpperl H, Rikhof H, Chang H, Haffter P, Kimmel CB, Moens CB. Lazarus is a novel pbx gene that globally mediates hox gene function in zebrafish. Mol Cell. 2000;6:255–267. doi: 10.1016/s1097-2765(00)00027-7. [DOI] [PubMed] [Google Scholar]
- 14.Heidet L, Moriniére V, Henry C, De Tomasi L, Reilly ML, Humbert C, Alibeu O, Fourrage C, Bole-Feysot C, Nitschké P, et al. Targeted exome sequencing identifies PBX1 as involved in monogenic congenital anomalies of the Kidney and urinary tract. J Am Soc Nephrol. 2017;28:2901–2914. doi: 10.1681/ASN.2017010043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Villaescusa JC, Li B, Toledo EM, Rivetti di Val Cervo P, Yang S, Stott SR, Kaiser K, Islam S, Gyllborg D, Laguna-Goya R, et al. A PBX1 transcriptional network controls dopaminergic neuron development and is impaired in Parkinson's disease. EMBO J. 2016;35:1963–1978. doi: 10.15252/embj.201593725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu B, Tian X, Zhang L, Feng R. Hematopoietic PBX-interaction protein promotes breast cancer sensitivity to paclitaxel through a microtubule-dependent mechanism. DNA Cell Biol. 2016;35:740–745. doi: 10.1089/dna.2016.3318. [DOI] [PubMed] [Google Scholar]
- 17.Ma YY, Zhang Y, Mou XZ, Liu ZC, Ru GQ, Li E. High level of homeobox A9 and PBX homeobox 3 expression in gastric cancer correlates with poor prognosis. Oncol Lett. 2017;14:5883–5889. doi: 10.3892/ol.2017.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Errico MC, Felicetti F, Bottero L, Mattia G, Boe A, Felli N, Petrini M, Bellenghi M, Pandha HS, Calvaruso M, et al. The abrogation of the HOXB7/PBX2 complex induces apoptosis in melanoma through the miR-221&222-c-FOS pathway. Int J Cancer. 2013;133:879–892. doi: 10.1002/ijc.28097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han HB, Gu J, Ji DB, Li ZW, Zhang Y, Zhao W, Wang LM, Zhang ZQ. PBX3 promotes migration and invasion of colorectal cancer cells via activation of MAPK/ERK signaling pathway. World J Gastroenterol. 2014;20:18260–18270. doi: 10.3748/wjg.v20.i48.18260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jung JG, Shih IM, Park JT, Gerry E, Kim TH, Ayhan A, Handschuh K, Davidson B, Fader AN, Selleri L, Wang TL. Ovarian cancer chemoresistance relies on the stem cell reprogramming factor PBX1. Cancer Res. 2016;76:6351–6361. doi: 10.1158/0008-5472.CAN-16-0980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu X, Cai N, Bao Z, You Y, Ji J, Liu N. Silencing Pre-B-cell leukemia homeobox 3 decreases the proliferation of human glioma cells in vitro and in vivo. J Neurooncol. 2017;135:453–463. doi: 10.1007/s11060-017-2603-9. [DOI] [PubMed] [Google Scholar]
- 22.Aspland SE, Bendall HH, Murre C. The role of E2A-PBX1 in leukemogenesis. Oncogene. 2001;20:5708–5717. doi: 10.1038/sj.onc.1204592. [DOI] [PubMed] [Google Scholar]
- 23.Rosales-Aviña JA, Torres-Flores J, Aguilar-Lemarroy A, Gurrola-Díaz C, Hernández-Flores G, Ortiz-Lazareno PC, Lerma-Díaz JM, de Celis R, González-Ramella Ó, Barrera-Chaires E, et al. MEIS1, PREP1, and PBX4 are differentially expressed in acute lymphoblastic leukemia: Association of MEIS1 expression with higher proliferation and chemotherapy resistance. J Exp Clin Cancer Res. 2011;30(112) doi: 10.1186/1756-9966-30-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu X, Bao Z, Liu Y, Jiang K, Zhi T, Wang D, Fan L, Liu N, Ji J. PBX3/MEK/ERK1/2/LIN28/let-7b positive feedback loop enhances mesenchymal phenotype to promote glioblastoma migration and invasion. J Exp Clin Cancer Res. 2018;37(158) doi: 10.1186/s13046-018-0841-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramberg H, Grytli HH, Nygård S, Wang W, Ögren O, Zhao S, Løvf M, Katz B, Skotheim RI, Bjartell A, et al. PBX3 is a putative biomarker of aggressive prostate cancer. Int J Cancer. 2016;139:1810–1820. doi: 10.1002/ijc.30220. [DOI] [PubMed] [Google Scholar]
- 26.Alharbi RA, Pandha HS, Simpson GR, Pettengell R, Poterlowicz K, Thompson A, Harrington K, El-Tanani M, Morgan R. Inhibition of HOX/PBX dimer formation leads to necroptosis in acute myeloid leukemia cells. Oncotarget. 2017;8:89566–89579. doi: 10.18632/oncotarget.20023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramberg H, Alshbib A, Berge V, Svindland A, Taskén KA. Regulation of PBX3 expression by androgen and Let-7d in prostate cancer. Mol Cancer. 2011;10(50) doi: 10.1186/1476-4598-10-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dardaei L, Longobardi E, Blasi F. Prep1 and Meis1 competition for Pbx1 binding regulates protein stability and tumorigenesis. Proc Natl Acad Sci USA. 2014;111:E896–E905. doi: 10.1073/pnas.1321200111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ando H, Natsume A, Senga T, Watanabe R, Ito I, Ohno M, Iwami K, Ohka F, Motomura K, Kinjo S, et al. Peptide-based inhibition of the HOXA9/PBX interaction retards the growth of human meningioma. Cancer Chemother Pharmacol. 2014;73:53–60. doi: 10.1007/s00280-013-2316-5. [DOI] [PubMed] [Google Scholar]
- 30.Lin HY, Zeng Liang YK, Wei XL, Chen CF. GATA3 and TRPS1 are distinct biomarkers and prognostic factors in breast cancer: Database mining for GATA family members in malignancies. Oncotarget. 2017;8:34750–34761. doi: 10.18632/oncotarget.16160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin Z, Xu L, Zhang L, Zhao M, Li D, Ye L, Ma Y, Ren S, Yu H, Wang D, et al. Interleukin enhancer binding factor 2 is a prognostic biomarker for breast cancer that also predicts neoadjuvant chemotherapy responses. Am J Transl Res. 2018;10:1677–1689. [PMC free article] [PubMed] [Google Scholar]
- 32.Lánczky A, Nagy Á, Bottai G, Munkácsy G, Szabó A, Santarpia L, Győrffy B. miRpower: A web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res Treat. 2016;160:439–446. doi: 10.1007/s10549-016-4013-7. [DOI] [PubMed] [Google Scholar]
- 33.Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, Chen H, Omeroglu G, Meterissian S, Omeroglu A, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 34.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 35.Beck S, Sommer P, dos Santos Silva E, Blin N, Gött P. Hepatocyte nuclear factor 3 (winged helix domain) activates trefoil factor gene TFF1 through a binding motif adjacent to the TATAA box. DNA Cell Biol. 1999;18:157–164. doi: 10.1089/104454999315547. [DOI] [PubMed] [Google Scholar]
- 36.Bernardo GM, Lozada KL, Miedler JD, Harburg G, Hewitt SC, Mosley JD, Godwin AK, Korach KS, Visvader JE, Kaestner KH, et al. FOXA1 is an essential determinant of ERalpha expression and mammary ductal morphogenesis. Development. 2010;137:2045–2054. doi: 10.1242/dev.043299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Farmer P, Bonnefoi H, Becette V, Tubiana-Hulin M, Fumoleau P, Larsimont D, Macgrogan G, Bergh J, Cameron D, Goldstein D, et al. Identification of molecular apocrine breast tumours by microarray analysis. Oncogene. 2005;24:4660–4671. doi: 10.1038/sj.onc.1208561. [DOI] [PubMed] [Google Scholar]
- 38.Guo F, Kuo YF, Shih YCT, Giordano SH, Berenson AB. Trends in breast cancer mortality by stage at diagnosis among young women in the United States. Cancer. 2018;124:3500–3509. doi: 10.1002/cncr.31638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferlay J, Colombet M, Soerjomataram I, Dyba T, Randi G, Bettio M, Gavin A, Visser O, Bray F. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries and 25 major cancers in 2018. Eur J Cancer. 2018;103:356–387. doi: 10.1016/j.ejca.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 40.Baeyens-Fernández JA, Molina-Portillo E, Pollan M, Rodríguez-Barranco M, Del Moral R, Arribas-Mir L, Sánchez-Cantalejo Ramírez E, Sánchez MJ. Trends in incidence, mortality and survival in women with breast cancer from 1985 to 2012 in Granada, Spain: A population-based study. BMC Cancer. 2018;18(781) doi: 10.1186/s12885-018-4682-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dard A, Reboulet J, Jia Y, Bleicher F, Duffraisse M, Vanaker JM, Forcet C, Merabet S. Human HOX proteins use diverse and context-dependent motifs to interact with TALE class cofactors. Cell Rep. 2018;22:3058–3071. doi: 10.1016/j.celrep.2018.02.070. [DOI] [PubMed] [Google Scholar]
- 42.Roux M, Zaffran S. Hox genes in cardiovascular development and diseases. J Dev Biol. 2016;4(pii: E14) doi: 10.3390/jdb4020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parker HJ, Piccinelli P, Sauka-Spengler T, Bronner M, Elgar G. Ancient Pbx-Hox signatures define hundreds of vertebrate developmental enhancers. BMC Genomics. 2011;12(637) doi: 10.1186/1471-2164-12-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duque-Afonso J, Feng J, Scherer F, Lin CH, Wong SH, Wang Z, Iwasaki M, Cleary ML. Comparative genomics reveals multistep pathogenesis of E2A-PBX1 acute lymphoblastic leukemia. J Clin Invest. 2015;125:3667–3680. doi: 10.1172/JCI81158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duque-Afonso J, Lin CH, Han K, Wei MC, Feng J, Kurzer JH, Schneidawind C, Wong SH, Bassik MC, Cleary ML. E2A-PBX1 remodels oncogenic signaling networks in B-cell precursor acute lymphoid leukemia. Cancer Res. 2016;76:6937–6949. doi: 10.1158/0008-5472.CAN-16-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Y, Xu X, Lin P, He Y, Zhang Y, Cao B, Zhang Z, Sethi G, Liu J, Zhou X, Mao X. Inhibition of the deubiquitinase USP9x induces pre-B cell homeobox 1 (PBX1) degradation and thereby stimulates prostate cancer cell apoptosis. J Biol Chem. 2019;294:4572–4582. doi: 10.1074/jbc.RA118.006057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu C, Li H, Zhang L, Jia T, Duan L, Lu C. MicroRNA-1915-3p prevents the apoptosis of lung cancer cells by downregulating DRG2 and PBX2. Mol Med Rep. 2016;13:505–512. doi: 10.3892/mmr.2015.4565. [DOI] [PubMed] [Google Scholar]
- 48.Qiu Y, Wang ZL, Jin SQ, Pu YF, Toyosawa S, Aozasa K, Morii E. Expression level of pre-B-cell leukemia transcription factor 2 (PBX2) as a prognostic marker for gingival squamous cell carcinoma. J Zhejiang Univ Sci B. 2012;13:168–175. doi: 10.1631/jzus.B1100077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qiu Y, Morii E, Tomita Y, Zhang B, Matsumura A, Kitaichi M, Okumura M, Aozasa K. Prognostic significance of pre B cell leukemia transcription factor 2 (PBX2) expression in non-small cell lung carcinoma. Cancer Sci. 2009;100:1198–1209. doi: 10.1111/j.1349-7006.2009.01156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qiu Z, Wang X, Shi Y, Da M. miR-129-5p suppresses proliferation, migration, and induces apoptosis in pancreatic cancer cells by targeting PBX3. Acta Biochim Biophys Sin. 2019;(Shanghai 51):997–1007. doi: 10.1093/abbs/gmz096. [DOI] [PubMed] [Google Scholar]
- 51.Li YS, Zou Y, Dai DQ. MicroRNA-320a suppresses tumor progression by targeting PBX3 in gastric cancer and is downregulated by DNA methylation. World J Gastrointest Oncol. 2019;11:842–856. doi: 10.4251/wjgo.v11.i10.842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li H, Wang J, Xu F, Wang L, Sun G, Wang J, Yang Y. By downregulating PBX3, miR-526b suppresses the epithelial-mesenchymal transition process in cervical cancer cells. Future Oncol. 2019;15:1577–1591. doi: 10.2217/fon-2018-0575. [DOI] [PubMed] [Google Scholar]
- 53.Garcia-Cuellar MP, Steger J, Füller E, Hetzner K, Slany RK. Pbx3 and Meis1 cooperate through multiple mechanisms to support Hox-induced murine leukemia. Haematologica. 2015;100:905–913. doi: 10.3324/haematol.2015.124032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lamprecht S, Kaller M, Schmidt EM, Blaj C, Schiergens TS, Engel J, Jung A, Hermeking H, Grünewald TGP, Kirchner T, Horst D. PBX3 is part of an EMT regulatory network and indicates poor outcome in colorectal cancer. Clin Cancer Res. 2018;24:1974–1986. doi: 10.1158/1078-0432.CCR-17-2572. [DOI] [PubMed] [Google Scholar]
- 55.Li H, Sun G, Liu C, Wang J, Jing R, Wang J, Zhao X, Xu X, Yang Y. PBX3 is associated with proliferation and poor prognosis in patients with cervical cancer. Onco Targets Ther. 2017;10:5685–5694. doi: 10.2147/OTT.S150139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Magnani L, Ballantyne EB, Zhang X, Lupien M. PBX1 genomic pioneer function drives ERalpha signaling underlying progression in breast cancer. PLoS Genet. 2011;7(e1002368) doi: 10.1371/journal.pgen.1002368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Magnani L, Patten DK, Nguyen VT, Hong SP, Steel JH, Patel N, Lombardo Y, Faronato M, Gomes AR, Woodley L, et al. The pioneer factor PBX1 is a novel driver of metastatic progression in ERα-positive breast cancer. Oncotarget. 2015;6:21878–21891. doi: 10.18632/oncotarget.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Costa RH, Grayson DR, Darnell JE Jr. Multiple hepatocyte-enriched nuclear factors function in the regulation of transthyretin and alpha 1-antitrypsin genes. Mol Cell Biol. 1989;9:1415–1425. doi: 10.1128/mcb.9.4.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hu Q, Luo Z, Xu T, Zhang JY, Zhu Y, Chen WX, Zhong SL, Zhao JH, Tang JH. FOXA1: A promising prognostic marker in breast cancer. Asian Pac J Cancer Prev. 2014;15:11–16. doi: 10.7314/apjcp.2014.15.1.11. [DOI] [PubMed] [Google Scholar]
- 60.Yamaguchi N, Shibazaki M, Yamada C, Anzai E, Morii M, Nakayama Y, Kuga T, Hashimoto Y, Tomonaga T, Yamaguchi N. Tyrosine phosphorylation of the pioneer transcription factor FoxA1 promotes activation of estrogen signaling. J Cell Biochem. 2017;118:1453–1461. doi: 10.1002/jcb.25804. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.