ABSTRACT
Basal bodies (BBs) are microtubule-based organelles that act as a template for and stabilize cilia at the cell surface. Centrins ubiquitously associate with BBs and function in BB assembly, maturation and stability. Human POC5 (hPOC5) is a highly conserved centrin-binding protein that binds centrins through Sfi1p-like repeats and is required for building full-length, mature centrioles. Here, we use the BB-rich cytoskeleton of Tetrahymena thermophila to characterize Poc5 BB functions. Tetrahymena Poc5 (TtPoc5) uniquely incorporates into assembling BBs and is then removed from mature BBs prior to ciliogenesis. Complete genomic knockout of TtPOC5 leads to a significantly increased production of BBs, yet a markedly reduced ciliary density, both of which are rescued by reintroduction of TtPoc5. A second Tetrahymena POC5-like gene, SFR1, is similarly implicated in modulating BB production. When TtPOC5 and SFR1 are co-deleted, cell viability is compromised and BB overproduction is exacerbated. Overproduced BBs display defective transition zone formation and a diminished capacity for ciliogenesis. This study uncovers a requirement for Poc5 in building mature BBs, providing a possible functional link between hPOC5 mutations and impaired cilia.
This article has an associated First Person interview with the first author of the paper.
KEY WORDS: Basal body, Centriole, Centrin, Poc5, Tetrahymena, Electron tomography
Summary: Loss of Tetrahymena thermophila Poc5 reveals an important role for this centrin-binding protein in basal body maturation, which also impacts basal body production and ciliogenesis.
INTRODUCTION
Centrioles and basal bodies (BBs) are evolutionarily ancient microtubule-based organelles that nucleate and organize microtubules into specific arrangements for fundamental cellular processes (Carvalho-Santos et al., 2011; Hodges et al., 2010; Kobayashi and Dynlacht, 2011). During cell division, microtubule arrays emanating from centriole-comprising centrosomes function to assemble and organize the mitotic spindle for cellular control of chromosome segregation. BBs are structurally similar to centrioles, and in some cell types centrioles and BBs are interconverted as a function of the cell cycle; however, BBs are distinctly required for templating, orienting, and anchoring cilia and flagella at the cell surface. Depending on the cell type, cilia and flagella have central roles in diverse mechanical and sensory functions, including generating directional fluid flow for cell motility and normal left–right axis determination, as well as coordinating several important signal transduction pathways (Ishikawa and Marshall, 2011; Nachury and Mick, 2019; Nonaka et al., 1998). In humans, cilia are present on almost all cell types, and ciliary dysfunction is associated with complex disorders known as ciliopathies, which display a broad spectrum of clinical features reflective of the ubiquity and functional diversity of cilia (Badano et al., 2006; Waters and Beales, 2011). Furthermore, investigating ciliopathies has solidified the importance of the cilium-associated BB for ciliary function, as many genetic mutations underlying ciliopathies encode defective BB proteins as well as proteins that reside and function in both BBs and centrioles (Reiter and Leroux, 2017).
The characteristic cylindrical structure of centrioles and BBs is primarily composed of a nine-fold radially symmetric arrangement of specialized triplet microtubules. The definitive nine-fold internal symmetry is imparted during early assembly by the proximal cartwheel structure, which consists of nine spokes radiating from a central hub, with the tip of each spoke typically containing a set of triplet microtubules (Guichard et al., 2018; Hirono, 2014; Kilburn et al., 2007). As centrioles and BBs mature, these structures become polarized, with an apparent transition from triplet-to-doublet microtubules occurring at the distal end, as well as the variable acquisition of subdistal and distal appendages (Pearson, 2014; Tanos et al., 2013). Doublet microtubules of the ciliary axoneme originate from the distal end of a mature BB, in a region known as the transition zone (TZ), which is an important subdomain for initiating the compartmentalization of the cilium during early assembly and maintaining a distinct ciliary protein composition (Garcia-Gonzalo and Reiter, 2017; Gonçalves and Pelletier, 2017). Thus, a mature TZ is required for ciliary assembly and maintenance of ciliary function, and a significant number of ciliopathy-associated proteins localize to and function in the TZ (Reiter and Leroux, 2017).
Centriole and BB duplication is a tightly regulated process generally coupled with the cell cycle, to ensure that constant numbers of these structures are maintained after cell division for specific cellular requirements (Fırat-Karalar and Stearns, 2014; Gönczy, 2012; Pearson and Winey, 2009). To maintain constant centriole and BB numbers, duplication occurs only once per cell cycle, and new assembly is typically limited to only one site on a preexisting structure. This assembly process differs in multiciliated cells, where the requisite near-simultaneous assembly of BBs occurs during differentiation and proceeds by multiple BBs forming on a preexisting template BB as well as by de novo assembly around a deuterosome (Brooks and Wallingford, 2014; Dawe et al., 2007; Yan et al., 2016). Notably, numerous studies in diverse model systems have elucidated a strong evolutionary conservation of core molecules that regulate duplication of these structures, including molecular assembly factors with shared functions across assembly pathways (Rodrigues-Martins et al., 2008; Strnad and Gönczy, 2008; Vladar and Stearns, 2007).
Centrins are small Ca2+-binding proteins that are widely conserved in microtubule-organizing centers including the yeast spindle pole body (yeast centrosome), centrioles and BBs, where enrichment in centrioles and BBs is found at both the site of new assembly and in the distal portion of these structures (Baum et al., 1986; Kilburn et al., 2007; Laoukili et al., 2000; Paoletti et al., 1996; Stemm-Wolf et al., 2005; Vonderfecht et al., 2012). While the role of centrins in centrosome duplication is less understood, centrins have key functions that contribute to BB duplication and maintenance, including roles in BB assembly, maturation, separation and stability (Koblenz et al., 2003; Ruiz et al., 2005; Stemm-Wolf et al., 2005; Vonderfecht et al., 2012, 2011). Furthermore, defective ciliary assembly and function, as well as an array of ciliopathy-like phenotypes, have been observed in mouse and zebrafish studies of centrin depletion (Delaval et al., 2011; Ying et al., 2019, 2014). Notably, centrins also perform non-centrosomal functions, including roles in DNA damage repair, mRNA export and fibroblast growth factor-mediated signaling (Dantas et al., 2011; Nishi et al., 2005; Shi et al., 2015).
An interaction between centrin and centrosomal Sfi1p is required for duplication of the yeast spindle pole body, and in mammalian cells, the mammalian ortholog SFI1 promotes centriole duplication (Kilmartin, 2003; Kodani et al., 2019). Structural studies of Sfi1 uncovered centrin-binding repeats (CBRs) that each contain a conserved sequence motif, Ax7LLx3F/Lx2WK/R, that directly binds the C-terminus of centrin (Kilmartin, 2003; Li et al., 2006; Martinez-Sanz et al., 2010, 2006). This conserved motif enabled the identification of centrin-binding proteins across eukaryotes that contain centrioles and/or BBs, including five in humans (Azimzadeh et al., 2009; Eisen et al., 2006; Gogendeau et al., 2007; Heydeck et al., 2016; Kilmartin, 2003; Stemm-Wolf et al., 2013). The centrin-binding protein, Poc5, was identified previously in the Chlamydomonas reinhardtii centriole proteome and characterized as an ancestral, core centriolar protein based on broad evolutionary conservation among eukaryotes (Hodges et al., 2010; Keller et al., 2009). Furthermore, human POC5 (hPOC5) was found to be enriched in the distal portion of human centrioles, where it has an essential role in centriole elongation and maturation (Azimzadeh et al., 2009). This role for hPOC5 is notable because molecular mechanisms that contribute to building a full-length centriole/BB remain poorly understood, especially compared with the mechanistic and molecular understanding of early assembly (Chang et al., 2016; Chen et al., 2017; Comartin et al., 2013; Keller et al., 2009; Schmidt et al., 2009). Furthermore, a truncating mutation in hPOC5 was recently implicated in an inherited form of retinal degeneration, retinitis pigmentosa, characterized by progressive loss of photoreceptors, and Poc5 has been found to colocalize with centrin in the connecting cilium of zebrafish photoreceptors, where it is important for normal retinal development and function (Weisz Hubshman et al., 2018; Wheway et al., 2014). Additionally, hPOC5 mutations associated with adolescent idiopathic scoliosis lead to mislocalization of hPOC5, impaired cell cycle progression and shorter cilia (Hassan et al., 2019).
The ciliate, Tetrahymena thermophila, has a microtubule-based cytoskeleton containing roughly 750 BBs organized along cortical rows and within the feeding structure of the cell, the oral apparatus (Bayless et al., 2015). Previous Tetrahymena studies have provided fundamental insight on the multiple BB functions of centrins (Kilburn et al., 2007; Stemm-Wolf et al., 2005; Vonderfecht et al., 2012, 2011). Furthermore, members of the large family of 13 centrin-binding proteins in Tetrahymena (Sfr1–Sfr13) each localize distinctly to subsets of cortical row and/or oral apparatus BBs, as well as to specific regions within BBs overlapping with known patterns of centrin localization (Heydeck et al., 2016; Stemm-Wolf et al., 2013). In addition, centrin-binding proteins have diverse roles in BBs, where Sfr1 and Sfr13 function in modulating BB production and in separating/stabilizing BBs, respectively. Taken together, these findings suggest that centrin-binding proteins may collectively provide the precise spatiotemporal coordination necessary for centrin to perform multiple BB functions. Despite the strong evolutionary conservation of Poc5 in centrioles and BBs, this study provides the first functional characterization of Poc5 in BBs. This investigation reveals a dynamic BB localization pattern for Tetrahymena Poc5 (TtPoc5) and an essential role for Poc5 in building a mature BB capable of nucleating a cilium.
RESULTS
Identification of TtPoc5 through evolutionarily conserved Poc5 domains
The hallmark feature of centriole and BB-associated centrin-binding proteins is the presence of a variable number of CBRs that each contain a conserved sequence motif, Ax7LLx3F/Lx2WK/R (Kilmartin, 2003; Li et al., 2006). Despite the expanded number of centrin-binding proteins in Tetrahymena, mining the Tetrahymena genome using solely this conserved sequence motif within CBRs was not a sufficient method for identifying TtPoc5. This was primarily due to limited sequence conservation between centrin-binding proteins, as exemplified by moderate overall sequence conservation (mean 16% identity, 29% similarity) even within the Poc5 family (Azimzadeh et al., 2009). Despite limited overall sequence conservation, Poc5 orthologs can be differentiated from other centrin-binding proteins because they contain a 21-amino-acid (AA) signature sequence motif, known as the Poc5 box, that is both highly conserved (mean 57% identity, 81% similarity) and uniquely found only in Poc5 orthologs (Azimzadeh et al., 2009). Using the full protein sequence of hPOC5 to search the Tetrahymena genome, a Poc5 ortholog was identified that shares characteristic Poc5 domain organization and sequence conservation with hPOC5 [overall identities, 68/291 (23%); positives, 135/291 (46%)] (Fig. 1A). Similar to hPOC5, TtPoc5 (TTHERM_00079160) has three CBRs organized as an isolated N-terminal CBR (CBR1) and two CBRs in tandem (CBR2,3) (Fig. 1A,B), as well as predicted coiled-coil domains on both sides of the tandem CBR repeats (green regions in Fig. 1A) (Azimzadeh et al., 2009). Notably, the defining residues of the conserved sequence motif between the three CBRs of hPOC5 and TtPoc5 are primarily either conserved or their hydrophobicity is retained despite amino acid differences, suggesting that the functional centrin-binding property of TtPoc5 CBRs is likely intact (denoted by asterisks in Fig. 1B, highlighted in Fig. 1C). Additionally, TtPoc5 contains the characteristic Poc5 box, which exhibits a high degree of shared sequence conservation with hPOC5 [overall identities, 15/21 (71%); positives, 17/21 (81%)] (Fig. 1B) and with select Poc5 orthologs (Fig. 1D) (Azimzadeh et al., 2009).
Fig. 1.
TtPoc5 contains evolutionarily conserved Poc5 domains. (A) Schematics showing conserved organization of centrin-binding repeats (CBRs; gray boxes), predicted coiled-coil domains (green regions) and the Poc5 box (yellow box) in hPOC5 and TtPoc5. (B) Sequence alignment within the region containing an isolated CBR (CBR1), tandem CBRs (CBR2,3), and the Poc5 box. Identical residues are shaded in black and similar residues are shaded in gray. Asterisks denote conserved centrin-binding sequence motif (Ax7LLx3F/Lx2WK/R) residues. (C) Sequence logos showing the amino acid composition within 18-amino-acid sequence motifs (positions 6–23) of hPOC5 and TtPoc5 CBRs plus five upstream amino acids. Black residues indicate the high proportion of hydrophobic AAs at conserved positions (marked with asterisks). Hydrophilic residues are indicated in blue and neutral residues are indicated in green. (D) Multiple sequence alignment of the highly conserved 21-amino-acid Poc5 box across selected Poc5 orthologs.
TtPoc5 localizes exclusively to assembling BBs
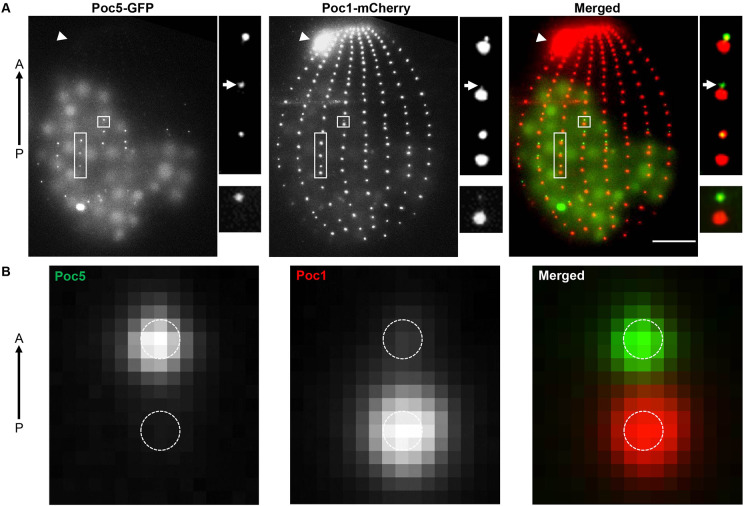
Tetrahymena cells have a highly organized cytoskeleton with hundreds of BBs maintained along cortical rows as well as in the BB-comprised oral apparatus (Bayless et al., 2015). To determine whether TtPoc5 localizes to BBs, Tetrahymena cells containing endogenously tagged Poc5–GFP and the BB-specific marker, Poc1–mCherry, were imaged for colocalization (Heydeck et al., 2016; Pearson et al., 2009; Stemm-Wolf et al., 2013). Endogenous TtPoc5 only colocalized with Poc1 in a subset of cortical row BBs (highlighted with boxes in Fig. 2A) and was notably absent from the oral apparatus (arrowheads in Fig. 2A). Notably, live imaging of Tetrahymena cells in the GFP channel captured autofluorescence in large cell vacuoles that was not reflective of the Poc5–GFP signal (Fig. 2A).
Fig. 2.
Endogenously tagged Poc5–GFP localizes to assembling BBs. (A) Live-cell imaging of C-terminally tagged Poc5–GFP relative to Poc1–mCherry (BB marker) with the anterior (A)–posterior (P) axis indicated with an arrow. Poc5–GFP localizes to a subset of cortical row BBs (highlighted with boxes) but is not detected in the mature oral apparatus (labeled with arrowheads). Scale bar: 10 µm. Upper right panels are representative regions of cortical rows containing three BB pairs. Poc5–GFP localizes to the anterior (assembling) BBs and is absent in the posterior (mature) BBs. Poc5 incorporation can precede that of Poc1 (labeled with arrows). Small (1.1 µm×1.2 µm) panels show the representative BB pair used for image averaging in B. (B) Image averaging of Poc5–GFP and Poc1–mCherry signals across 58 cortical row BB pairs reveals Poc5–GFP localizes exclusively at assembling BBs. The BB scaffolds are approximated with 200 nm diameter dashed circles based on the center of the Poc1–mCherry signal, reflecting the average diameter of a Tetrahymena BB.
To dissect further the TtPoc5 BB localization pattern, analysis focused on characterizing the Poc5–GFP signal relative to Poc1–mCherry in cortical row BBs. In Tetrahymena, new BBs assemble anteriorly from existing mature BBs in cortical rows and can be identified in closely adjoining BB pairs (Bayless et al., 2015). Endogenously tagged Poc1–mCherry was used for this study because it slowly incorporates into assembling BBs and gradually accumulates during BB maturation, therefore differentiating assembling from mature BBs (Pearson et al., 2009). Within a representative region of a cortical row containing three BB pairs, Poc5–GFP resided in assembling BBs and was not detected in mature BBs, marked by prominent Poc1–mCherry signal (Fig. 2A). Image averaging of Poc5–GFP and Poc1–mCherry signals across 58 BB pairs (a representative BB pair is shown in Fig. 2A) consistently detected TtPoc5 exclusively at the assembling BB (Fig. 2B). Notably, the timing of TtPoc5 incorporation into assembling BBs appeared to precede that of the slowly incorporated Poc1, since Poc5–GFP was found in assembling BBs devoid of Poc1–mCherry (arrows in Fig. 2A). Furthermore, the absence of TtPoc5 in mature BBs may signify why Poc5–GFP was not observed in the oral apparatus BBs (Fig. 2A), which contain primarily mature BBs, as indicated by enrichment of Poc1–mCherry.
Dynamic incorporation and removal of TtPoc5 in BBs
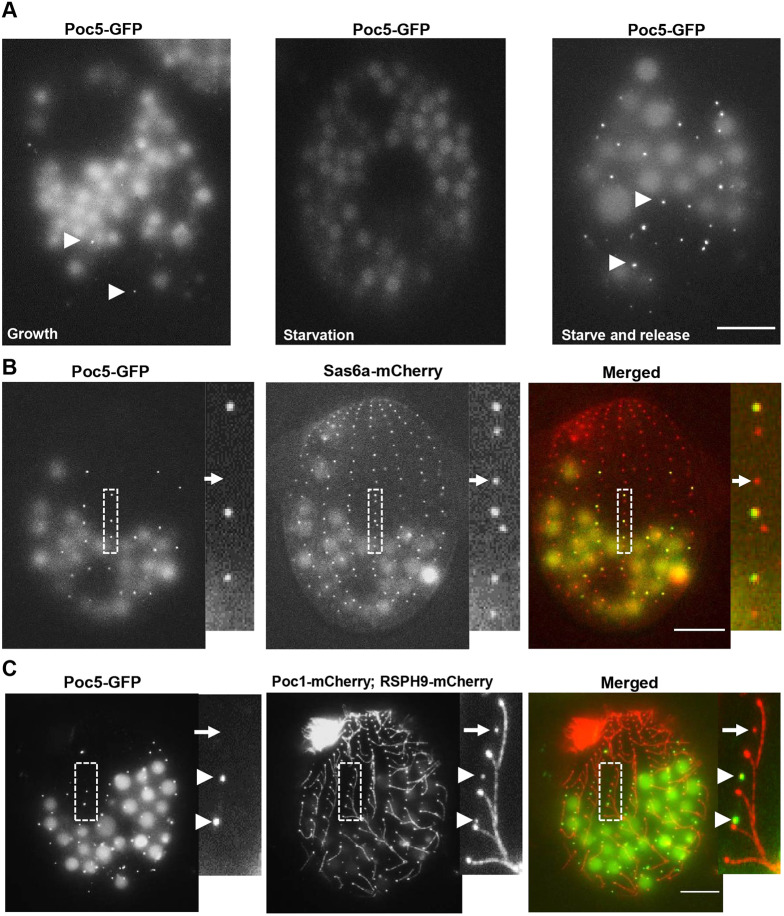
To verify that TtPoc5 localizes preferentially to assembling BBs, Tetrahymena cells with endogenously tagged Poc5–GFP were cultured in three conditions that elicit different cellular responses to BB assembly and maintenance: growth medium, starvation medium and starvation medium followed by release into growth medium (Fig. 3A). During logarithmic growth, Tetrahymena BBs are stabilized for the entirety of the cell cycle and BB assembly is not synchronous, thus, the majority of cortical row BBs at any given time are mature, with a minimal number of newly assembled BBs (Bayless et al., 2015; Galati et al., 2015). In growth, Poc5–GFP was only detected in a small proportion of cortical row BBs (arrowheads in Fig. 3A), which was consistent with TtPoc5 exclusively residing in assembling BBs (Fig. 2A,B). When Tetrahymena cells are shifted from growth medium into starvation medium, cells arrest in G1 preventing new BB assembly (Pearson et al., 2009). Under medium starvation for 24 h when only mature BBs persisted, the Poc5–GFP signal was not detected in any cortical row BBs (Fig. 3A). Furthermore, when starved cells are shifted back into growth medium this releases cells from cell cycle arrest (starve and release) and correspondingly initiates a synchronous wave of new BB assembly (Pearson et al., 2009). In contrast to the depleted Poc5–GFP signal in starved cells, there was a marked enrichment of Poc5–GFP-positive BBs during synchronized BB assembly in starved and released cells (arrowheads in Fig. 3A).
Fig. 3.
TtPoc5 is enriched during BB assembly and removed prior to cilia formation. (A) Live-cell imaging of endogenously tagged, C-terminal Poc5–GFP during logarithmic growth, in starvation and after release back into growth medium (starve and release). During logarithmic growth, BB assembly occurs throughout the cell cycle and low levels of Poc5–GFP signal are detected at a single moment (indicated with arrowheads). In starvation, cells are arrested in G1 preventing new BB assembly and Poc5–GFP is not detectable. Overnight starvation followed by release into growth medium (stimulating new BB assembly) enriches the Poc5–GFP signal (marked with arrowheads). (B,C) Live-cell imaging of starved and released cells with representative sections of cortical rows (marked with dashed boxes) expanded in the smaller panels. (B) Endogenous Poc5–GFP was co-expressed with Sas6a–mCherry (early marker of BB cartwheel structure) to assess the timing of TtPoc5 BB incorporation. Sas6a–mCherry labels all BBs whereas Poc5–GFP is only at the anterior BB in a pair. The Poc5–GFP signal always coincides with the Sas6a–mCherry signal, therefore TtPoc5 BB incorporation does not precede that of Sas6a. (C) Endogenous Poc5–GFP was co-expressed with Poc1–mCherry (BB marker) and RSPH9–mCherry (evenly labels ciliary axonemes) to assess the timing of Poc5 removal from maturing BBs. Poc5–GFP removal precedes cilia formation since Poc5–GFP-positive BBs are not ciliated (indicated with arrowheads) and maturing, non-ciliated BBs with detectable Poc1–mCherry (highlighted with an arrow) can be devoid of Poc5-GFP signal. Scale bars: 10 µm.
To better understand the timing of TtPoc5 BB incorporation relative to early assembly, cells co-expressing Poc5–GFP and Sas6a–mCherry were used since Sas6a is a critical component of the early-forming BB cartwheel structure that acts as a template for the nine-fold symmetry of BBs (Culver et al., 2009). In Tetrahymena, the cartwheel is a stable structure that persists from early BB assembly through maturation, therefore Sas6a–mCherry evenly labeled all cortical row BBs (Fig. 3B) (Bayless et al., 2015). To increase the number of Poc5–GFP-positive BBs for this analysis, cells were starved and released. Poc5–GFP localized exclusively to the assembling BBs within BB pairs, and Poc5–GFP-positive BBs were always marked with Sas6a–mCherry (Fig. 3B). Given that Poc5–GFP-positive BBs were never devoid of Sas6a–mCherry, TtPoc5 BB incorporation preceded that of Poc1 (Fig. 2A) but did not precede early cartwheel formation (Fig. 3B). Notably, Sas6a–mCherry-positive BBs lacking any detectable Poc5–GFP were also observed (arrows in Fig. 3B), which are likely mature BBs where TtPoc5 has been removed.
To elucidate the timing of TtPoc5 BB removal relative to BB maturation and the onset of cilia formation, starved and released cells were used that co-expressed Poc5–GFP, Poc1–mCherry (a BB marker) and a tagged Tetrahymena ortholog of the ciliary-specific protein, radial spoke head 9 (RSPH9–mCherry; Yang et al., 2006). As seen in Fig. 3C, RSPH9–mCherry evenly labeled ciliary axonemes along cortical rows as well as in the mature oral apparatus, and there was no detected signal overlap with Poc5–GFP in BBs. Within a representative region of a cortical row (boxed area in Fig. 3C), Poc5–GFP colocalized with Poc1–mCherry in anteriorly positioned BBs of pairs (arrowheads in Fig. 3C) but was not observed in mature BBs with prominent RSPH9–mCherry-labeled cilia. The absence of TtPoc5 in ciliated BBs suggested that TtPoc5 removal from BBs was either prior to or prompted by the onset of cilia formation (arrowheads in Fig. 3C). In Tetrahymena, not all mature BBs are ciliated, providing an intermediate stage between BB maturation/stabilization and cilia formation for further dissection of this timing (Bayless et al., 2012; Nanney, 1975, 1971). As shown in Fig. 3C, mature nonciliated Poc1–mCherry-positive BBs were present that lacked Poc5–GFP signal (denoted with arrows), indicating that TtPoc5 removal occurred prior to the onset of cilia formation. Collectively, TtPoc5 is transiently incorporated into assembling BBs and is then removed prior to BB maturation, suggesting that TtPoc5 potentially functions during this dynamic stage of BB development, which is similar to the known function of hPOC5 in centriole elongation/maturation (Azimzadeh et al., 2009).
Loss of TtPoc5 leads to overproduced cortical row BBs and a reduction of cilia
To determine the BB function(s) of TtPoc5, a complete genomic knockout (KO) strain was generated by replacing the TtPOC5 open reading frame (ORF) through homologous recombination with a high efficiency, codon-optimized NEO2 cassette (coNEO2) built for usage in Tetrahymena and used for positive selection of transformants (Fig. 4A) (Mochizuki, 2008). The TtPOC5 KO strain was validated by PCR using isolated genomic DNA from wild-type (WT) control and poc5Δ cells (Fig. 4B). RT-PCR using isolated RNA from WT and poc5Δ cells further confirmed that poc5Δ cells lacked TtPOC5, revealing expression of coNEO2 only in poc5Δ cells and expression of TtPOC5 only in WT cells (Fig. S1).
Fig. 4.
poc5Δ cells are viable and display overproduced BBs but fewer cilia. (A) Schematic outlining the homologous recombination strategy for generating a complete micronuclear knockout of TtPOC5, where the TtPOC5 ORF is replaced with a codon-optimized NEO2 (coNEO2) cassette for drug selection. The red arrowheads indicate the primers used to amplify PCR products specific to WT POC5 and coNEO2. (B) PCR confirming success of the knockout strategy, with only wild-type (WT) cells containing the TtPOC5 ORF and only poc5Δ cells containing the coNEO2 cassette. Sizes of select marker (M) bands are displayed in kilobase pairs. (C) Growth curves for WT and poc5Δ cells grown at either 30°C or 37°C for 8 h in SPP medium. Cell density (cells/ml) measurements are gathered at the initiation of the experiment (cultures starting at 0.5×105 cells/ml), and 4 and 8 h after initiation. Results are mean±s.d. for n=3 analyzed samples per strain and incubation temperature. (D) Representative WT and poc5Δ cells stained with an antibody against Cen1 (labels BBs) after incubation at either room temperature (RT), 30°C or 37°C. Loss of TtPoc5 does not disrupt the organization/orientation of cortical row BBs. The bottom panel shows a representative poc5Δ rescue (Rescue) cell at 30°C with no CdCl2 induction (N.I.) due to ‘leakiness’ of the MTT1 promoter, where TtPOC5 is reincorporated in poc5Δ cells through transformed MTT1pr-GFP-Poc5. Scale bars: 10 µm. (E) BB density quantification (average BBs/10 µm) for WT and poc5Δ cells at RT, 30°C and 37°C. Significant BB overproduction is observed in poc5Δ cells at all tested growth temperatures and is rescued with MTT1pr-GFP–Poc5 to near WT levels at 30°C. Results are mean±s.e.m. for n=300 total counts (five counts per cell across 20 cells, in triplicate) per condition. ***P<0.001 (Student's t-test). (F) Representative 10 µm sections of cortical rows labeled with an antibody against polyglutamylated tubulin (labels ciliary axonemes and BBs). (G) Ciliary density quantification (average number of cilia per 10 µm) for WT and poc5Δ cells at 30°C reveals significantly reduced ciliary density in poc5Δ cells that is rescued with TtPOC5 reincorporation. Results are mean±s.e.m. for n=100 total counts per condition. ***P<0.001; n.s., not significant (Student's t-test).
TtPOC5 is not essential for cell viability given that poc5Δ cells persist indefinitely; and through time course analysis, we determined that poc5Δ cells exhibited similar growth rates to WT cells at both an optimal growth temperature for Tetrahymena (30°C) and at a restrictive temperature (37°C) used to identify temperature-sensitive mutants (Fig. 4C). From a gross morphological standpoint, poc5Δ cells grown at room-temperature (RT), 30°C and 37°C displayed properly oriented BBs organized along cortical rows, as well as morphologically normal oral apparatuses (Fig. 4D). Since BB orientation was maintained in poc5Δ cells, the spatial organization of cortical row BBs in poc5Δ and WT cells was assessed by quantifying the BB density (average BBs/10 µm) along cortical rows (Fig. 4E). Measuring BB density in growing Tetrahymena cells is a powerful method for identifying disrupted BB homeostasis since the number of cortical row BBs is maintained at a nearly constant number (Frankel, 2008, 1980; Nanney, 1971, 1968, 1966). At all tested growth temperatures (RT, 30°C and 37°C), poc5Δ cells had a significantly greater cortical row BB density (8.0, 7.8, 8.0 BBs/10 µm, respectively) than WT cells (6.9, 6.8, 6.9 BBs/10 µm, respectively), suggesting that TtPoc5 potentially functions to inhibit BB overproduction or that the loss of TtPoc5 elicits a compensatory cellular response leading to BB overproduction (Fig. 4E).
To determine that this observed BB overproduction was a direct result of loss of TtPoc5, a rescue strain was generated that reintroduced TtPOC5 in poc5Δ cells through transformation of a CdCl2-inducible rescuing construct, MTT1pr-GFP-Poc5 (Fig. 4D). To assess rescue, the BB density was quantified in cells grown at 30°C with no added CdCl2 (no induction; N.I.), due to the established ‘leakiness’ of the MTT1 promoter (Heydeck et al., 2016; Shang et al., 2002) and because MTT1pr-mCherry–Poc5 exogenously expressed with N.I. in a WT background localized to cortical row BBs in a similar manner to endogenous Poc5–GFP (Fig. 2A; Fig. S2). Furthermore, CdCl2-induced MTT1pr-mCherry–Poc5 overexpression led to an aberrant TtPoc5 BB localization pattern and formation of elongated Poc5-positive fibers observed previously with overexpression of Poc5 (arrows in Fig. S2) (Dantas et al., 2013). As seen in Fig. 4D and quantified in Fig. 4E, poc5Δ rescue cells grown at 30°C with N.I. exhibited a reduced BB density (7.1 BBs/10 µm) to near WT levels (6.8 BBs/10 µm) upon TtPOC5 reincorporation, demonstrating that TtPoc5 has a direct function in modulating Tetrahymena BB production. Interestingly, despite the cortical row BB overproduction seen with loss of TtPoc5, poc5Δ cells had fewer ciliated BBs than WT cells (Fig. 4F,G). For this analysis, the ciliary density (average number of cilia per 10 µm) was examined in WT, poc5Δ and poc5Δ rescue cells (N.I.) grown at 30°C with an antibody against polyglutamylation, which marks microtubule glutamylation in both BBs and cilia (Bayless et al., 2016; Wolff et al., 1992). As shown in Fig. 4F and quantified in Fig. 4G, poc5Δ cells nucleated on average 2.5 cilia/10 µm along cortical rows compared with 3.5 cilia/10 µm in WT cells, and TtPOC5 reincorporation in poc5Δ rescue cells grown at 30°C with N.I. led to a significantly increased ciliary density (3.3 cilia/10 µm) to near WT levels. Collectively, phenotypic analysis of poc5Δ cells elucidated a function for TtPoc5 in cortical row BB production and uncovered a diminished capacity in poc5Δ cells to maintain WT ciliary density levels despite BB overproduction.
Loss of TtPoc5 and Sfr1 results in exacerbated BB overproduction and cell death
A potentially antagonistic role for TtPoc5 in modulating BB production is interestingly not unique for Tetrahymena centrin-binding proteins, where the previously characterized loss of Sfr1 led to a significantly greater BB density (7.9 BBs/10 µm) than in WT cells (6.98 BBs/10 µm) (Heydeck et al., 2016). Beyond the phenotypic overlap in poc5Δ and sfr1Δ cells, the BLAST search that identified TtPoc5 through sequence conservation with hPOC5 (described in Fig. 1) identified Sfr1 as the second orthologous hit to hPOC5 [overall identities, 40/172 (23%); positives, 74/172 (43%)]. This shared sequence conservation between hPOC5, TtPoc5 and Sfr1 primarily stemmed from a CBR organization that appears to be distinctive to Poc5 orthologs, with these proteins typically containing three CBRs organized as an isolated CBR and two CBRs in tandem (Fig. 1A) (Azimzadeh et al., 2009; Heydeck et al., 2016; Stemm-Wolf et al., 2013). Notably, despite apparent BB overproduction in both poc5Δ and sfr1Δ cells, loss of Sfr1 did not lead to the reduced ciliary density (Fig. S3) observed with loss of Poc5 (Fig. 4F,G). Furthermore, the highly conserved Poc5 box that is a hallmark of Poc5 orthologs (Fig. 1D) is notably absent in Sfr1, and therefore TtPoc5 was considered the primary Tetrahymena Poc5 ortholog (Azimzadeh et al., 2009; Heydeck et al., 2016; Stemm-Wolf et al., 2013).
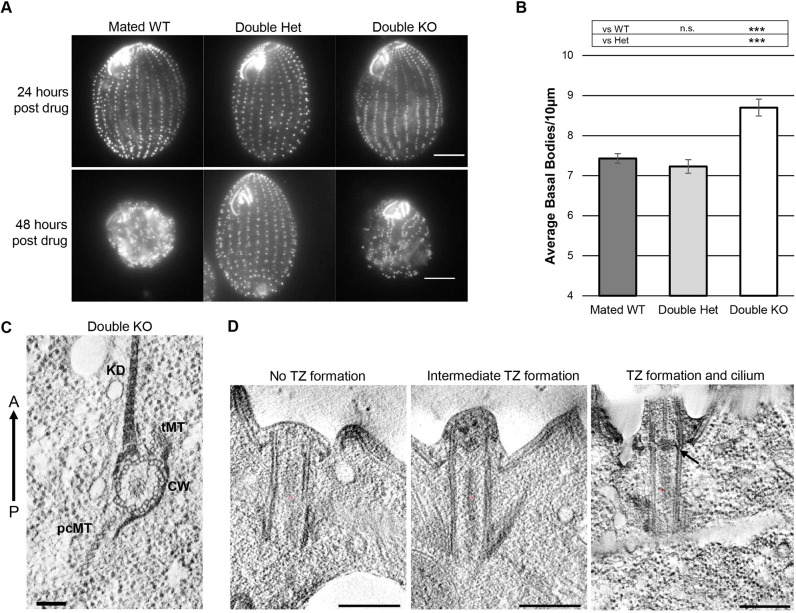
Given the potential functional redundancy between TtPoc5 and Sfr1, double KO cells lacking both TtPOC5 and SFR1 (poc5Δ;sfr1Δ cells) were generated using established methods (Hai et al., 2000; Heydeck et al., 2016). Briefly, heterokaryon strains with germline deletion of TtPOC5 or SFR1 were crossed, and drug-selection followed by PCR confirmation produced heterokaryon cells with germline deletion of both loci (Fig. S4). These cells were then mated together and drug-selected to generate poc5Δ;sfr1Δ cells with homozygous deletion of both loci in both nuclei (germline and somatic). Unlike poc5Δ and sfr1Δ single KO cells that are viable and persist indefinitely, poc5Δ;sfr1Δ cells were inviable following 48 h of drug selection at 30°C, suggesting that the overlapping BB function of TtPoc5 and Sfr1 was vital for Tetrahymena cell viability (Fig. 5A) (Heydeck et al., 2016). To test whether the underlying poc5Δ;sfr1Δ cell lethality was due to the unsuccessful transmission of the drug-resistance neomycin cassettes, heterokaryon cells were also mated in parallel with WT cells to generate a strain heterozygous for both TtPOC5 and SFR1, resulting in viable cells at 48 h post drug treatment (Fig. 5A) (Cassidy-Hanley, 2012; Hai et al., 2000). Furthermore, WT cells lacking drug-resistance cassettes were mated together to test the efficacy of drug selection, resulting in incomplete drug selection by 24 h post drug treatment but cell death by 48 h of drug treatment (Fig. 5A). Thus, poc5Δ;sfr1Δ cell death by 48 h of drug treatment was due to the combined loss of TtPoc5 and Sfr1, revealing an essential function for these Tetrahymena centrin-binding proteins that was obscured in poc5Δ and sfr1Δ single KO cells due to some functional redundancy (Heydeck et al., 2016).
Fig. 5.
Loss of both TtPoc5 and Sfr1 results in cell lethality and exacerbated overproduction of defective BBs. A previously characterized centrin-binding protein, Sfr1, is the second best hPOC5 ortholog in Tetrahymena genome BLAST search, despite lacking the Poc5 box (Heydeck et al., 2016). Beyond sequence conservation, Sfr1 has a similar role in modulating BB production, prompting the generation of double knockout (poc5Δ;sfr1Δ) cells using established methods (Hai et al., 2000). (A) Representative control mated WT, double heterozygous (Het), and double knockout (KO) cells stained with anti-Cen1 antibody after 24 h and 48 h of drug selection following mating. Double KO cell death within 48 h post drug selection is not due to a lack of drug resistance since double heterozygous cells are viable after 48 h in drug. Drug selection is effective since mated WT cells that do not carry drug resistance cassettes die after 48 h in drug. Scale bars: 10 µm. (B) BB density quantification (average BBs/10 µm) at 24 h. Double KO cells have significantly higher BB density relative to mated WT and double heterozygous cells. Results are mean±s.e.m. for n=90 total counts (three counts per cell across 15 cells, in two biological replicates). ***P<0.001; n.s., not sginificant (Student's t-test). (C,D) Electron tomography of double KO BBs. (C) Cross-sectional view showing normal Tetrahymena BB accessory structures [kinetodesmal fiber (KD), transverse microtubules (tMT), post ciliary microtubules (pcMT)] and intact proximal cartwheel (CW). The anterior (A)-posterior (P) axis is indicated with an arrow. Scale bar: 100 nm. (D) Longitudinal views showing BBs that do not template cilia and have varying degrees of transition zone (TZ) formation (left and middle panels). Right panel shows a BB with a templated cilium and a morphologically normal TZ (arrow indicates electron dense axosome). See Movie 1. Scale bars: 100 nm.
To investigate the functional consequences of loss of both TtPoc5 and Sfr1 on BB production, phenotypic analysis of BB density was conducted after 24 h of drug treatment at 30°C, when mated WT, double heterozygous, and poc5Δ;sfr1Δ cells persisted (Fig. 5A). Notably, mated WT cells were analyzed in parallel to control for BB rearrangement after mating and a potentially increased BB density from requisite BB assembly during cell division (Pearson and Winey, 2009). After 24 h of drug treatment, poc5Δ;sfr1Δ cells maintained properly oriented cortical row BBs and morphologically normal oral apparatuses, similar to what was observed in poc5Δ or sfr1Δ single KO cells (Fig. 4D) (Heydeck et al., 2016). As shown in Fig. 5A and quantified in Fig. 5B, the BB density in poc5Δ;sfr1Δ cells was significantly exacerbated (8.7 BBs/10 µm) compared with double heterozygous (7.2 BBs/10 µm) and mated WT cells (7.4 BBs/10 µm), while there was a no significant difference in BB density between double heterozygous and mated WT cells. Furthermore, the BB density in poc5Δ;sfr1Δ cells (8.7 BBs/10 µm) exceeded what was measured in poc5Δ cells grown at 30°C (7.8 BBs/10 µm) (Fig. 4E) and in sfr1Δ cells (7.9 BBs/10 µm) (Heydeck et al., 2016). Collectively, the increased cortical row BB overproduction in poc5Δ;sfr1Δ cells revealed functionally redundant roles for TtPoc5 and Sfr1 in BB production. Thus, poc5Δ;sfr1Δ cell death may result from an inability to compensate for a greatly increased level of BB production and consequently BB-dense cortical rows, which supports the importance of retaining a typical, highly regulated BB number in Tetrahymena (Frankel, 2008, 1980; Nanney, 1971, 1968, 1966).
Given that poc5Δ;sfr1Δ cells died by 48 h of drug treatment despite an abundance of BBs after 24 h (Fig. 5A,B), an ultrastructural examination using electron microscopy (EM) was conducted to elucidate potential BB defects driving poc5Δ;sfr1Δ cell lethality (Fig. 5C,D). From cross-sectional views of poc5Δ;sfr1Δ cortical row BBs, it was apparent that the proximal cartwheel structures comprising radially symmetric arrays of triplet microtubules were intact (Fig. 5C) (Allen, 1969; Bayless et al., 2015; Hirono, 2014; Kilburn et al., 2007). Additionally, BB accessory structures that aid in properly orienting and positioning BBs along cortical rows were also intact, which was not unexpected given the retained BB orientation in poc5Δ;sfr1Δ cells (Fig. 5C) (Bayless et al., 2015; Meehl et al., 2016). Contrary to what was seen on examining the proximal end of poc5Δ;sfr1Δ BBs, longitudinal views captured BBs docked at the cell surface with varying degrees of TZ formation at the distal end (WT comparison found in Giddings et al., 2010), suggesting that BB maturation was delayed or impaired upon loss of both TtPoc5 and Sfr1 (Fig. 5D; see Movie 1). Consequently, poc5Δ;sfr1Δ BBs with aberrant TZs did not template cilia at the cell surface (Fig. 5D) and lacked distinct TZ/axonemal features, including a transition from triplet-to-doublet microtubules, the electron-dense axosome, and the central pair of microtubules emanating from the axosome through the ciliary axoneme (Fig. S5; see Movies 2, 3) (Bayless et al., 2015; Meehl et al., 2016; Sattler and Staehelin, 1974). Notably, BBs that templated cilia and exhibited morphologically normal TZs were also observed through EM (arrow marks an axosome in Fig. 5D), which might represent inherited BBs that formed prior to the loss of TtPoc5 and Sfr1 or reveal that there could be a dysregulated timing/delay of BB maturation in poc5Δ;sfr1Δ cells. By disrupting the timing of BB maturation and impairing TZ formation, the cortical row BB overproduction observed in poc5Δ;sfr1Δ cells was likely a compensatory response that resulted in an increased number of immature BBs, a diminished capacity to form cilia and an inability to survive by 48 h.
DISCUSSION
In Tetrahymena, the tightly controlled timing and positioning of BB assembly, combined with the stabilization of BBs throughout the cell cycle, establishes a highly organized cytoskeleton with hundreds of BBs maintained along cortical rows and in the oral apparatus (Bayless et al., 2015). This intricately patterned and BB-rich cytoskeleton was previously exploited for characterizing the essential BB functions of centrin, which broadly encompass roles in BB assembly, orientation, separation and stability (Kilburn et al., 2007; Stemm-Wolf et al., 2005; Vonderfecht et al., 2012, 2011). Here, TtPoc5 is uniquely incorporated into assembling cortical row BBs and is then removed from mature BBs prior to the onset of cilia formation. Tetrahymena cells lacking TtPOC5 are viable yet overproduce cortical row BBs and, despite this overproduction of BBs, exhibit a significantly reduced number of cilia along cortical rows. Upon reintroduction of TtPoc5, the cortical row BB overproduction and reduced ciliary density seen with loss of TtPoc5 were both rescued. Interestingly, Tetrahymena has two POC5-like genes, TtPOC5 and SFR1, which show similarity within their CBRs, and each encodes BB-specific proteins with roles in modulating cortical row BB production (Heydeck et al., 2016). When TtPOC5 and SFR1 are co-deleted, Tetrahymena cells are no longer viable and display exacerbated levels of cortical row BB overproduction. Furthermore, underlying BB maturation defects are apparent with varying degrees of distal TZ formation in these cells, leading to a consequently diminished capacity to nucleate a cilium. This study uncovers a specific requirement for Poc5 in building the distal portion of BBs, which corresponds with the distinct timing of TtPoc5 BB localization and may signify an added importance of centrin reported at the BB distal end (Kilburn et al., 2007). By revealing this functional redundancy, the dysregulation of BB number seen with loss of TtPoc5 and/or Sfr1 is likely a cellular response in Tetrahymena to compensate for delayed or defective BB maturation. Additionally, the essential role for TtPoc5 in Tetrahymena BB maturation provides a possible functional explanation for the observed association between hPOC5 mutations and impaired cilia (Hassan et al., 2019; Oliazadeh et al., 2017; Patten et al., 2015; Weisz Hubshman et al., 2018).
TtPoc5 is exclusively present in assembling BBs
In a previous analysis of hPOC5, this highly conserved centrin-binding protein was found to be recruited to procentrioles in the G2 phase of the cell cycle as procentrioles elongate and mature (Azimzadeh et al., 2009). Furthermore, hPOC5 was observed in both the daughter and mother centrioles, similar to what is seen for centrin, indicating that once hPOC5 is recruited to procentrioles it is then stably incorporated. In comparison with hPOC5, this study finds endogenous TtPoc5 exclusively in assembling BBs after the initial cartwheel structure is formed and removal of TtPoc5 from mature BBs precedes ciliary assembly (Figs 2 and 3). This dynamic incorporation and removal of TtPoc5 relative to BB production has not been observed in previous analyses of other centrin-binding proteins, suggesting that the precise timing of TtPoc5 BB localization and function is important. Accordingly, overexpressing TtPoc5 using an inducible promoter appears to disrupt this critical timing, leading to the formation of elongated Poc5-positive fibers and an aberrant BB localization pattern with TtPoc5 residing in all cortical row BBs (Fig. S2). Interestingly, overexpression of Sfr1 did not produce Sfr1-positive fibers and Sfr1 was found to be stably present in all cortical row BBs as well as in the mature BBs within the oral apparatus (Heydeck et al., 2016). However, the functional significance and extent of this discrepancy in BB localization patterns is not currently understood primarily because of the technical challenges of endogenously tagging Sfr1, precluding a direct comparison with endogenous TtPoc5 (Heydeck et al., 2016). Given the stable presence of centrin in both mature BBs and centrioles, furthering our understanding of the molecular underpinnings driving Poc5 removal from mature BBs (and not mature centrioles), as well as illuminating the functional importance of Sfr1 residing in all BBs, will both be interesting to address in future research.
Role of TtPoc5 in BB production and Tetrahymena BB number constancy
Beyond the established role for hPOC5 in centriole elongation/maturation, a Poc5 BB function has not been previously described (Azimzadeh et al., 2009; Chang et al., 2016; Chen et al., 2017; Comartin et al., 2013). In this study of TtPoc5 BB function, poc5Δ cells have a significantly increased cortical row BB density (Fig. 4E). This increase in BB number through BB overproduction is notable because Tetrahymena cells have an intrinsic capability to maintain a nearly constant total number of cortical row BBs by altering the spatial organization of BBs through changes in the BB density and/or the overall number of cortical rows (Frankel, 2008, 1980; Nanney, 1971, 1968, 1966). The mechanism underlying the regulation of BB number constancy in Tetrahymena has not been elucidated; however, Sfr1 was similarly implicated in modulating cortical row BB production after observing more densely packed, supernumerary cortical rows in sfr1Δ cells (Heydeck et al., 2016). The absence of both TtPoc5 and Sfr1 leads to a greater increase in BB number (Fig. 5B), which has not been seen with genetic deletions of other characterized centrin-binding proteins, and uniquely implicates Poc5 regulating BB number. Notably, future studies are needed to address the significance of centrin-binding for Poc5 BB function, as well as the functional relevance of non-centrin interactors, including known ciliary and cytoskeleton components that bind hPOC5 (Hassan et al., 2019). Furthermore, the highly conserved Poc5 box does not have a reported function, thus, this additional domain found only in Poc5 orthologs may specify Poc5 BB function.
Although most metazoan cell types lack the cortical organization of BBs that is characteristic of ciliates, there is a strong evolutionary conservation of molecules that tightly control both BB and centriole duplication irrespective of cortical organization (Firat-Karalar et al., 2014; Pearson and Winey, 2009; Rodrigues-Martins et al., 2008). Furthermore, previous studies have established that centrosome amplification and the underlying deregulation of centriole number are widespread features of many human cancers (Chan, 2011; Marteil et al., 2018; Wong et al., 2015). Using a diverse panel of 60 human cancer cell lines derived from nine distinct tissues (the NCI-60 panel), significant centrosome amplification was evident in all tissue types and within half of the analyzed cell lines (Marteil et al., 2018). Furthermore, there was marked variability in the percentage of cells with supernumerary centrioles, indicating that cancer cell lines possess distinct centriole numbers and levels of centrosome amplification. Interestingly, hPOC5 is capable of binding both human CETN2 and human CETN3, which have roles in promoting and antagonizing centrosome duplication, respectively (Sawant et al., 2015). Thus, depletion of CETN3 in HeLa cells results in a centriole overproduction, suggesting that centrins are important for modulating centriole number, and this bidirectional contribution to centrosome duplication may be impaired in some human cancers. Collectively, these findings suggest that conserved mechanisms and/or signaling cues that are important for maintaining both centriole and BB number may exist, of which, Poc5 and centrins are good candidates for directing this process in metazoans.
Requirement for Poc5 in BB maturation
The recruitment of hPOC5 to nascent centrioles occurs after initial assembly and coincides with building the centriole distal end, where hPOC5 and centrin colocalize (Azimzadeh et al., 2009; Paoletti et al., 1996). Consistent with this localization pattern, hPOC5 is essential for centriole elongation/maturation but is not required for the initiation of procentriole assembly. In this study, cortical row BB production is significantly heightened in Tetrahymena cells lacking both POC5 genes, and cross-sections of the BB ultrastructure reveal intact proximal cartwheel structures, indicating that Poc5 is not critical for initiating BB assembly (Fig. 5C). In contrast to the BB proximal end, cortical row BBs docked at or near the cell surface display variable TZ formation at the distal end, illuminating an important role for Poc5 in Tetrahymena BB maturation and a shared function for Poc5 in building the distal end of both BBs and centrioles (Fig. 5D) (Azimzadeh et al., 2009). Furthermore, a role for Poc5 in BB maturation provides a functional explanation for the dynamic removal of TtPoc5 from mature BBs, suggesting that TtPoc5 is removed after completed formation of the BB distal end. Notably, a previous study in Chlamydomonas found concentrated centrin at the BB distal end and centrin-depletion uncovered a similar requirement for centrin in BB maturation, indicating that centrin and Poc5 may cooperate in this important process (Koblenz et al., 2003). Interestingly, centrin-depleted Chlamydomonas cells with BB maturation defects also contained aberrant BB numbers, including BB overproduction, suggesting that delayed and/or impaired BB development is functionally connected with dysregulation of BB number. Thus, the overproduction of cortical row BBs in Tetrahymena cells lacking TtPoc5 and Sfr1 is likely to be a mechanism to compensate for delayed and/or defective BB maturation rather than a novel role for Poc5 in antagonizing BB production. The functional extent of this compensatory mechanism requires further investigation, especially how BB production is impacted in response to distinct changes in the rate of BB biogenesis. Finally, cancer cells frequently lose precise control over centriole length, and the recurrent feature of centriole size deregulation is associated with persistent centriole amplification (Marteil et al., 2018). Together, these findings may have uncovered a conserved cellular response that requires further examination, in which BBs and centrioles are overproduced when development of these structures, but not initial assembly, is compromised.
Impact on ciliogenesis with loss of Poc5
Whereas hPOC5-depletion leads to impaired centriole maturation and subsequently defective cell cycle progression, the combined loss of TtPoc5 and Sfr1 in Tetrahymena results in immature cortical row BBs that consequently do not template cilia at the cell surface (Fig. 5D) (Azimzadeh et al., 2009). Additionally, structural features found within the distal TZ and ciliary axoneme, such as a triplet-to-doublet MT transition and a central MT pair, are not evident in cross-sections of the BB ultrastructure (Fig. S5) (Meehl et al., 2016). Taken together, this delayed and/or failed ciliary assembly is not unexpected, since a mature TZ serves to compartmentalize the cilium and is required for ciliary assembly and maintained ciliary function (Czarnecki and Shah, 2012). Similarly, flagella are frequently absent in centrin-depleted Chlamydomonas cells as a result of delayed BB development and improper BB maturation (Koblenz et al., 2003). Notably, this role for Poc5 in BB maturation may provide a functional explanation for ciliary defects associated with hPOC5 mutations, including shorter cilia observed with overexpression of a hPOC5 variant associated with adolescent idiopathic scoliosis (Hassan et al., 2019; Patten et al., 2015; Weisz Hubshman et al., 2018; Xu et al., 2018). In addition, Poc5 and centrin colocalize in the connecting cilium of photoreceptors, which corresponds structurally to the TZ and is vital for retinal function (Weisz Hubshman et al., 2018; Wheway et al., 2014; Ying et al., 2019). Although the function of Poc5 in the connecting cilium is not understood, a hPOC5 mutation is associated with a form of retinal degeneration hallmarked by progressive loss of photoreceptors (Weisz Hubshman et al., 2018). Interestingly, a recent study of mouse centrins uncovered an important cooperative function between CETN2 and CETN3 in stabilizing the connecting cilium, suggesting that a role for Poc5 and centrins in BB maturation may translate to maturation of the analogous connecting cilium (Ying et al., 2019).
Tetrahymena cells are not viable when TtPOC5 and SFR1 are co-deleted, yet cells lacking either POC5 gene persist indefinitely despite the presence of overproduced cortical row BBs. This suggests that the lethality observed in cells lacking both POC5 genes is likely a direct consequence of delayed and/or failed ciliogenesis rather than an overproduction of BBs, given that the hundreds of cilia on the surface of Tetrahymena cells have essential roles for survival, including locomotory and feeding functions (Bayless et al., 2015; Gaertig et al., 2013). Interestingly, a co-occurrence of overproduced BBs and a reduced ciliary density is apparent in viable cells lacking TtPoc5 alone; this indicates that further examination is needed to understand whether the degree by which cilia numbers are reduced, potentially in combination with exacerbated BB overproduction, underlies cell death when TtPoc5 and SFR1 are absent. Additionally, loss of Sfr1 does not alter ciliary density (Fig. S3), suggesting that TtPoc5 and Sfr1 have functional overlap that modulates BB production, but as their differing BB localization patterns indicate, TtPoc5 and Sfr1 do not appear to be functionally redundant. Finally, loss of TtPoc5 and Sfr1 may result in mis-localization of Tetrahymena Cen1 and/or Cen2 at the BB distal end, where proper localization and function of Cen1 is essential for cell viability (Stemm-Wolf et al., 2005; Vonderfecht et al., 2012). Thus, increasing our understanding of the role of centrin in this process may be critical for understanding the full extent of Poc5 BB function. In summary, Tetrahymena has two paralogous POC5 genes that are required for BB maturation, highlighting a requirement for Poc5 in building the distal end of both BBs and centrioles. In the absence of Poc5, Tetrahymena cells elicit a compensatory response to defective BB maturation that leads to BB overproduction and a consequential increase in the typically constant BB number. These abundant, immature BBs lack an ability to nucleate a cilium, illuminating the importance of Poc5 in ciliary assembly and potentially revealing the functional implications of hPOC5 mutations on ciliary function.
MATERIALS AND METHODS
T. thermophila strains and culture media
A WT strain derived from the progeny of a cross between B2086 and CU428 was used as the control for assaying the growth rate of the poc5Δ strain and for phenotypic analysis of the poc5Δ and poc5Δ rescue strains. A WT strain derived from the progeny of a cross between CU427 and SB1969 was the control comparison for phenotypic analysis of the poc5Δ;sfr1Δ strain (all parental WT strains were from the Tetrahymena Stock Center, Cornell University, Ithaca, NY). Cells were grown either at room-temperature (RT), 30°C or 37°C as indicated, in 2% super proteose peptose (SPP) medium (Orias et al., 2000) to mid-log phase (∼3×105 cells/ml). Cell density measurements were determined on a Z2 Coulter Counter (Beckman Coulter, Brea, CA). For starvation (arresting cells in G1), cells were grown to mid-log phase in SPP medium, washed in 10 mM Tris-HCl pH 7.4, and then resuspended in 10 mM Tris-HCl (pH 7.4) for overnight incubation at 30°C. For ‘starve and release’ experiments, cells were grown to mid-log phase in SPP medium, washed and resuspended in 10 mM Tris-HCl pH 7.4 for overnight incubation at 30°C, and then released back into SPP medium for 5 h (to stimulate synchronous BB assembly). For experiments with the cadmium-inducible MTT1 promoter (Shang et al., 2002), cells were incubated overnight at 30°C in SPP medium either containing no CdCl2 or 0.5 µg/ml CdCl2 at 30°C, as indicated in Fig. 4 and Fig. S2.
Identification of TtPoc5 and sequence comparison of Poc5 orthologs
Tetrahymena Poc5 (TtPoc5; TTHERM_00079160) was identified as the reciprocal best BLAST hit of full-length human POC5 (hPOC5) (Azimzadeh et al., 2009) in the Tetrahymena genome (Eisen et al., 2006). Sequence conservation was assessed by calculating the number of amino acid identities and positives. An identity is when the same amino acid is present at a given position in a sequence alignment. A positive is when a different amino acid is present at a given position, but the physico-chemical properties of the amino acids are preserved. Of note, a prior evolutionary analysis of centriolar proteins used a previous alias of the same TtPoc5 ortholog, 5.m00513 (Hodges et al., 2010). The sequence alignment of hPOC5 and TtPoc5 was performed using ClustalW and the amino acid box shading was performed using BoxShade; both of these tools are available through the ExPASy Bioinformatics Resource Portal (https://www.expasy.org/genomics). Detection of predicted coiled-coil domains in TtPoc5 and hPOC5 was through the DeepCoil tool (Ludwiczak et al., 2019). Sequence logos were generated by WebLogo3 (Crooks et al., 2004; Schneider and Stephens, 1990) to visualize the amino acid composition of the conserved sequence motifs (the canonical motif is Ax7LLx3F/Lx2WK/R) found within the three centrin-binding repeats (CBRs) of hPOC5 and TtPoc5 (Azimzadeh et al., 2009; Kilmartin, 2003). The 18-amino-acid sequence motifs span positions 6–23, with five additional amino acids upstream of the motifs (positions 1–5) included in the sequence logo. The size of the residue correlates with conservation, and the color of the residue signifies the type of amino acid (hydrophobic is indicated in black, hydrophilic is blue and neutral is green). The multiple sequence alignment of the highly conserved Poc5 box across select eukaryotes was performed using ClustalW and BoxShade. Select Poc5 orthologs are as follows: human (NP_001092741), mouse (NP_080449), zebrafish (XP_691080), Chlamydomonas reinhardtii (Protein Id:9798, v4.0 through the Joint Genome Institute), and Paramecium tetraurelia (GSPATT00029936001, through the Paramecium Genome Database).
Plasmids and strain construction
The endogenous C-terminal GFP fusion used to localize TtPoc5 was made in the pGFP-LAP-NEO2 plasmid (based on the p4T2-1 vector; Gaertig et al., 1994), which provides integration into the endogenous TtPOC5 locus and expression of Poc5–GFP under control of the endogenous promoter. Briefly, 1 kb of DNA from the 3′ end of TtPOC5 (immediately upstream of the stop codon) was cloned adjacent to the GFP–LAP tag, and 1 kb of 3′ sequence downstream of the stop codon was cloned into the vector flanking the NEO2 drug selection marker (conferring paromomycin resistance). A p4T21-MTT1pr-mCherry-Poc5 plasmid was created for TtPoc5 overexpression experiments, which provides integration into the endogenous TtPOC5 locus and expression of the N-terminal mCherry–Poc5 fusion was under control of the cadmium-inducible MTT1 promoter (Shang et al., 2002).
The poc5Δ rescue strain was generated by creating a TtPOC5 rescue construct (pBS-MTT1pr-GFP-Poc5) that integrated into the RPL29 locus and was under control of the cadmium-inducible MTT1 promoter (Shang et al., 2002; Winey et al., 2012). This exogenous N-terminal GFP fusion was made by first cloning TtPOC5 into the pENTR4 Dual Selection Entry Vector (Invitrogen, Carlsbad, CA) and then, using the Gateway cloning system (Invitrogen, Carlsbad, CA), TtPOC5 was subcloned into the pBS-MTT1pr-GFP-gtw vector (conferring cycloheximide resistance).
Sequence-confirmed constructs were linearized and transformed into the macronucleus of WT cells by biolistic bombardment using a PDS-1000 particle bombarder (Bio-Rad, Hercules, CA) (Bruns and Cassidy-Hanley, 2000), except for pBS-MTT-GFP-POC5 (the poc5Δ rescue construct), which was transformed into the macronucleus of poc5Δ cells. For colocalization studies, cells expressing endogenous Poc5-GFP were transformed with endogenous Poc1–mCherry (Pearson et al., 2009), Sas6a–mCherry (Culver et al., 2009), and/or RSPH9–mCherry (a gift from Chad Pearson, Department of Cell and Developmental Biology, University of Colorado School of Medicine, Aurora, CO). In cells co-expressing Poc5–GFP, Poc1–mCherry and RSPH9–mCherry, endogenous Poc1–mCherry was drug-selected with blasticidin while RSPH9–mCherry was drug-selected with cycloheximide (to allow for independent drug selection of each mCherry gene fusion).
Generation of the poc5Δ and poc5Δ;sfr1Δ strains
The poc5Δ strain was generated by replacing the TtPOC5 open reading frame with a high efficiency, codon-optimized NEO2 (coNEO2) gene built for usage in Tetrahymena (Hai et al., 2000; Mochizuki, 2008). Targeted homologous recombination of the TtPOC5 locus was achieved by using a knockout cassette with coNEO2 flanked by 1 kb of DNA upstream of the TtPOC5 start codon and 1 kb of DNA downstream of the TtPOC5 stop codon. For micronuclear transformation, mating strains B2086/CrNeo and CU428/CrNeo were used, which both contain a NEO gene in their macronucleus with two frameshift mutations to prevent DNA elimination of the selectable marker (Mochizuki et al., 2002; Yao et al., 2003). During conjugation, the TtPOC5-knockout construct was initially transformed into the germline micronucleus using biolistic bombardment of DNA-coated gold particles, as previously described (Bruns and Cassidy-Hanley, 2000; Hai et al., 2000). Following biolistic bombardment, cells were incubated overnight in 10 mM Tris-HCl pH 7.4 to finish conjugation, and then introduced to SPP medium for paromomycin drug-selection of positive transformants. Two different mating types of micronuclear knockout heterokaryons were generated through star crosses, as previously described (Hai et al., 2000). Single mating pairs were isolated from a mating between the two micronuclear knockout heterokaryon strains, and progeny were confirmed by PCR and RT-PCR for complete loss of TtPOC5 (poc5Δ cells). Primers used were: (1) for PCR validation: WT TtPOC5 forward, 5′-ATGAATTCAAATAAGAATCAACCAAAGAAGAAA-3′ and WT TtPOC5 reverse, 5′-TTTTTTGGTAGTTGTTGTTTTTGTTATTGC-3′; coNEO2 forward (located in the POC5 5′ UTR, upstream of the integrated coNEO2 cassette), 5′-ATTAATAACATTGCTGATGCTTTT-3′ and coNEO2 reverse (located within the coNEO2 cassette), 5′-GATTAATTACCTTCTAATAATTTGAAATAATTAATCC-3′; and (2) for RT-PCR: RNA was isolated from WT and poc5Δ cells using the RNeasy Mini and QIAshredder kits (Qiagen, Hilden, Germany). cDNA was generated using the Superscript III One-Step RT-PCR system (Invitrogen, Carlsbad, CA). Standard PCR followed with WT TtPOC5 forward, 5′-CCAAAGAAGAAAACATTACTTGATGACTTCAATG-3′ and WT TtPOC5 reverse, 5′-GGTAGTTGTTGTTTTTGTTATTGCTTTAGATGG-3′; and coNEO2 forward, 5′-ATTAATAACATTGCTGATGCTTTT-3′, and coNEO2 reverse, 5′-GAAGACGATAGAAGGCGATACG-3′.
For generation of the poc5Δ;sfr1Δ strain, micronuclear knockout heterokaryon strains of two different mating types were initially generated with germline micronuclei homozygous for both coNEO2 in the TtPOC5 locus and NEO2 in the SFR1 locus (Heydeck et al., 2016). Genotyping heterokaryons was performed by PCR since both cassettes conferred paromomycin drug resistance. PCR confirmation used: coNEO2 forward: 5′-ATTAATAACATTGCTGATGCTTTT-3′, coNEO2 reverse: 5′-GATTAATTACCTTCTAATAATTTGAAATAATTAATCC-3′, NEO2 forward: 5′-AATCTACTAATTTGCTTTATTTTTCATAAGC-3′, and NEO2 reverse: 5′-TCCATACTTTGAAGATATCAAGC-3′. An equal number of starved cells for each validated double heterokaryon strain were mixed together for mating overnight, and 24 h after the initiation of mating an equal volume of 2× SPP medium was added. Cells were allowed to recover from mating in 2× SPP for 4 h before adding paromomycin (200 µg/ml), this was considered the 0-h timepoint for the poc5Δ;sfr1Δ double knockout (with BB density measurements at 24 and 48 h after paromomycin addition). In parallel to the double knockout mating, two control matings were performed as described with the double knockout cells: WT×WT (CU427×SB1969) and WT x heterokaryons (to generate cells that are heterozygous for TtPOC5 and SFR1).
Fluorescence imaging
Images were collected using a Nikon Eclipse Ti inverted microscope (Nikon USA, Melville, NY) with a CFI Plan Apo VC 60× H NA 1.4 objective and a charge-coupled device (CCD) CoolSNAP HQ2 camera (Teledyne Photometrics, Tucson, AZ). Metamorph Imaging Software (Molecular Devices, San Jose, CA) was used for image acquisition, with all images acquired at RT.
For live-cell imaging, cells were grown in 2% SPP medium, washed in 10 mM Tris-HCl pH 7.4, pelleted, and placed on microscope slides (VWR, Radnor, PA). For immunofluorescence, cells were fixed using 3% formaldehyde and fixed cells were placed on poly-L-lysine-coated multi-well slides (Polysciences Inc., Warrington, PA). Cells were then blocked for 1 h using phosphate-buffed saline (PBS)+1% bovine serum albumin (BSA) before overnight incubation at 4°C in primary antibody, followed by a 2-h RT incubation in secondary antibody. All primary and secondary antibodies were diluted in PBS+1% BSA. Primary antibodies used were against TtCen1 at 1:1000 (Stemm-Wolf et al., 2005) to label BBs, and glutamylated tubulin at 1:1000 (GT335 by AdipoGen, San Diego, CA) to label ciliary axonemes (antibody also labels BBs) (Wolff et al., 1992). Secondary antibodies used: goat anti-mouse-IgG or goat anti-rabbit-IgG Alexa Fluor 488 both at 1:1000 (Invitrogen, Carlsbad, CA). Cells were mounted in Citifluor mounting medium (Ted Pella Inc., Redding, CA).
In cells co-expressing Poc5–GFP and Poc1–mCherry, image averaging of GFP and mCherry signals was performed using ImageJ (National Institutes of Health, Bethesda, MD) across 58 BB pairs. Images of Poc5–GFP and Poc1–mCherry BB signals were placed in a stack and the sum fluorescence of the stack was measured to create an averaged fluorescence image. The BB scaffolds were approximated using 200 nm diameter dashed circles, based on the average diameter of a Tetrahymena BB and the positioning of the center of the Poc1–mCherry signal.
Electron microscopy
Cells were prepared for electron tomography as described previously (Giddings et al., 2010; Meehl et al., 2009). Briefly, cells were gently spun into 15% dextran (average molecular mass 9000–11,000 Da; Sigma-Aldrich, St Louis, MO) with 5% BSA in 2% SPP medium. A small volume of the cell pellet was transferred to a sample holder and high-pressure frozen using a Wohlwend Compact 02 high pressure freezer (Technotrade International, Manchester, NH). The frozen cells were freeze substituted in 0.25% glutaraldehyde and 0.1% uranyl acetate in acetone and then embedded in Lowicryl HM20 resin. Serial thick (250–300 nm) sections were cut using a Leica UCT ultramicrotome, and section ribbons were collected on Formvar-coated copper slot grids. The grids were poststained with 2% aqueous uranyl acetate followed by Reynold's lead citrate. 15 nm gold beads (BBI Solutions, Crumlin, UK) were affixed to both sides of the grid to serve as markers for subsequent tilt series alignment. Serial thin (80 nm) sections were collected and imaged to evaluate overall basal body structure and cortical row organization. Dual-axis tilt series of Tetrahymena cells were collected on a Tecnai F30 intermediate voltage electron microscope (Thermo Fisher Scientific, Waltham, MA). Images were collected every one degree over a ±60° range using the SerialEM acquisition program (Mastronarde, 2005) with a Gatan Oneview camera at 1.2 nm pixel. Serial section tomograms of Tetrahymena BBs were generated using the IMOD software package (Giddings et al., 2010; Kremer et al., 1996; Mastronarde, 1997). In total, five tomograms from the double KO and one serial tomogram from WT were reconstructed.
Calculation of growth curves
Growth curves for the poc5Δ strain were calculated from cells grown in 2% SPP medium at 30°C and 37°C over the course of 8 h. Using a Z2 Coulter Counter (Beckman Coulter, Brea, CA), cell density (cells/ml) measurements were taken at the 0-h (with all cultures starting at 0.5×105 cells/ml), 4-h, and 8-h time points. The final growth curves for WT and poc5Δ cells were based on averaging cell density measurements from three independent experiments. The WT strain used to calculate growth curves (derived from the progeny of B2086xCU428) was the same WT strain used for phenotypic analysis of the poc5Δ strain.
Quantification of cortical row BB and ciliary density
The BB density (average BBs/10 µm) along cortical rows was quantified for WT and poc5Δ cells at RT, 30°C and 37°C. BBs were labeled with the anti-TtCen1 antibody, and ImageJ (National Institutes of Health, Bethesda, MD) was used to measure 10 µm regions along five separate cortical rows per cell (measurements on the side of the cell containing the oral apparatus and on the opposite side of the cell), across 20 cells in triplicate experiments. The final BB density was based on 300 total counts per condition. The same method was used to quantify the BB density for poc5Δ rescue and poc5Δ;sfr1Δ cells, but cells were only analyzed after growth in 2% SPP medium at 30°C. Quantification of the ciliary density (average cilia/10 µm) was quantified for WT, poc5Δ, sfr1Δ, and poc5Δ rescue cells at 30°C. Notably, variable ciliary density measurements were observed across experiments, but a reduced ciliary density in poc5Δ cells compared with WT cells was a consistent finding. Ciliary axonemes (and BBs) were labeled with the anti-glutamylated tubulin (GT335) antibody, and ImageJ was used to measure 10 µm regions on both sides of the cell (similar to measuring the BB density), with the average ciliary density based on 100 total counts per strain.
Supplementary Material
Acknowledgements
We thank Chad Pearson for generously gifting the RSPH9–mCherry plasmid and Janet B. Meehl for her critical guidance of the electron microscopy work. Electron microscopy was performed at the Boulder Electron Microscopy Core Facility, University of Colorado, Boulder.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: W.H., B.A.B., A.J.S.-W., M.W.; Methodology: W.H., B.A.B., A.J.S.-W., E.T.O., M.N.; Validation: W.H., B.A.B., M.N.; Formal analysis: W.H., B.A.B., E.T.O., A.S.F., M.W.; Investigation: W.H., B.A.B., M.N., E.T.O., A.S.F., M.N.; Data curation: W.H., B.A.B., M.N., E.T.O., A.S.F.; Writing - original draft: W.H., M.W.; Writing - review & editing: W.H., B.A.B., A.J.S.-W., E.T.O., A.S.F., M.W.; Visualization: W.H., B.A.B., A.S.F., E.T.O.; Supervision: W.H., B.A.B., M.W.; Project administration: M.W.; Funding acquisition: M.W.
Funding
The work was supported by the National Institutes of Health, initially under NIH RO1 GM074746 and completed under NIH RO1 GM127571 (both awarded to M.W.). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.240838.supplemental
Peer review history
The peer review history is available online at https://jcs.biologists.org/lookup/doi/10.1242/jcs.240838.reviewer-comments.pdf
References
- Allen R. D. (1969). The morphogenesis of basal bodies and accessory structures of the cortex of the ciliated protozoan Tetrahymena pyriformis. J. Cell Biol. 40, 716-733. 10.1083/jcb.40.3.716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimzadeh J., Hergert P., Delouvée A., Euteneuer U., Formstecher E., Khodjakov A. and Bornens M. (2009). hPOC5 is a centrin-binding protein required for assembly of full-length centrioles. J. Cell Biol. 185, 101-114. 10.1083/jcb.200808082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badano J. L., Mitsuma N., Beales P. L. and Katsanis N. (2006). The ciliopathies: an emerging class of human genetic disorders. Annu. Rev. Genomics Hum. Genet. 7, 125-148. 10.1146/annurev.genom.7.080505.115610 [DOI] [PubMed] [Google Scholar]
- Baum P., Furlong C. and Byers B. (1986). Yeast gene required for spindle pole body duplication: homology of its product with Ca2+-binding proteins. Proc. Natl. Acad. Sci. USA 83, 5512-5516. 10.1073/pnas.83.15.5512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayless B. A., Giddings T. H. Jr., Winey M. and Pearson C. G (2012). Bld10/Cep135 stabilizes basal bodies to resist cilia-generated forces. Mol. Biol. Cell 23, 4820-4832. 10.1091/mbc.e12-08-0577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayless B. A., Galati D. F. and Pearson C. G. (2015). Tetrahymena basal bodies. Cilia 5, 1 10.1186/s13630-016-0022-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayless B. A., Galati D. F., Junker A. D., Backer C. B., Gaertig J. and Pearson C. G. (2016). Asymmetrically localized proteins stabilize basal bodies against ciliary beating forces. J. Cell Biol. 215, 457-466. 10.1083/jcb.201604135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks E. R. and Wallingford J. B. (2014). Multiciliated cells: a review. Curr. Biol. CB 24, R973-R982. 10.1016/j.cub.2014.08.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns P. J. and Cassidy-Hanley D. (2000). Biolistic transformation of macro- and micronuclei. Methods Cell Biol. 62, 501-512. 10.1016/S0091-679X(08)61553-8 [DOI] [PubMed] [Google Scholar]
- Carvalho-Santos Z., Azimzadeh J., Pereira-Leal J. B. and Bettencourt-Dias M. (2011). Evolution: tracing the origins of centrioles, cilia, and flagella. J. Cell Biol. 194, 165-175. 10.1083/jcb.201011152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy-Hanley D. M. (2012). Tetrahymena in the laboratory: strain resources, methods for culture, maintenance, and storage. Methods Cell Biol. 109, 237-276. 10.1016/B978-0-12-385967-9.00008-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J. Y. (2011). A clinical overview of centrosome amplification in human cancers. Int. J. Biol. Sci. 7, 1122-1144. 10.7150/ijbs.7.1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.-W., Hsu W.-B., Tsai J.-J., Tang C.-J. C. and Tang T. K. (2016). CEP295 interacts with microtubules and is required for centriole elongation. J. Cell Sci. 129, 2501-2513. 10.1242/jcs.186338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.-Y., Wu C.-T., Tang C.-J. C., Lin Y.-N., Wang W.-J. and Tang T. K. (2017). Human microcephaly protein RTTN interacts with STIL and is required to build full-length centrioles. Nat. Commun. 8, 247 10.1038/s41467-017-00305-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comartin D., Gupta G. D., Fussner E., Coyaud É, Hasegan M., Archinti M., Cheung S. W. T., Pinchev D., Lawo S., Raught B., et al. (2013). CEP120 and SPICE1 cooperate with CPAP in centriole elongation. Curr. Biol. 23, 1360-1366. 10.1016/j.cub.2013.06.002 [DOI] [PubMed] [Google Scholar]
- Crooks G. E., Hon G., Chandonia J.-M. and Brenner S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188-1190. 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver B. P., Meehl J. B., Giddings T. H. and Winey M. (2009). The two SAS-6 homologs in Tetrahymena thermophila have distinct functions in basal body assembly. Mol. Biol. Cell 20, 1865-1877. 10.1091/mbc.e08-08-0838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarnecki P. G. and Shah J. V. (2012). The ciliary transition zone: from morphology and molecules to medicine. Trends Cell Biol. 22, 201-210. 10.1016/j.tcb.2012.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantas T. J., Wang Y., Lalor P., Dockery P. and Morrison C. G. (2011). Defective nucleotide excision repair with normal centrosome structures and functions in the absence of all vertebrate centrins. J. Cell Biol. 193, 307-318. 10.1083/jcb.201012093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantas T. J., Daly O. M., Conroy P. C., Tomas M., Wang Y., Lalor P., Dockery P., Ferrando-May E. and Morrison C. G. (2013). Calcium-binding capacity of centrin2 is required for linear POC5 assembly but not for nucleotide excision repair. PLoS ONE 8, e68487 10.1371/journal.pone.0068487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe H. R., Farr H. and Gull K. (2007). Centriole/basal body morphogenesis and migration during ciliogenesis in animal cells. J. Cell Sci. 120, 7-15. 10.1242/jcs.03305 [DOI] [PubMed] [Google Scholar]
- Delaval B., Covassin L., Lawson N. D. and Doxsey S. (2011). Centrin depletion causes cyst formation and other ciliopathy-related phenotypes in zebrafish. Cell Cycle Georget. Tex. 10, 3964-3972. 10.4161/cc.10.22.18150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen J. A., Coyne R. S., Wu M., Wu D., Thiagarajan M., Wortman J. R., Badger J. H., Ren Q., Amedeo P., Jones K. M., et al. (2006). Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 4, e286 10.1371/journal.pbio.0040286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fırat-Karalar E. N. and Stearns T. (2014). The centriole duplication cycle. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 369, 20130460 10.1098/rstb.2013.0460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firat-Karalar E. N., Rauniyar N., Yates J. R. III and Stearns T. (2014). Proximity interactions among centrosome components identify regulators of centriole duplication. Curr. Biol. CB 24, 664-670. 10.1016/j.cub.2014.01.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel J. (1980). Propagation of cortical differences in tetrahymena. Genetics 94, 607-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel J. (2008). What do genic mutations tell us about the structural patterning of a complex single-celled organism? Eukaryot. Cell 7, 1617-1639. 10.1128/EC.00161-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaertig J., Gu L., Hai B. and Gorovsky M. A. (1994). High frequency vector-mediated transformation and gene replacement in Tetrahymena. Nucleic Acids Res. 22, 5391-5398. 10.1093/nar/22.24.5391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaertig J., Wloga D., Vasudevan K. K., Guha M. and Dentler W. (2013). Discovery and functional evaluation of ciliary proteins in Tetrahymena thermophila. Methods Enzymol. 525, 265-284. 10.1016/B978-0-12-397944-5.00013-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galati D. F., Abuin D. S., Tauber G. A., Pham A. T. and Pearson C. G. (2015). Automated image analysis reveals the dynamic 3-dimensional organization of multi-ciliary arrays. Biol. Open 5, 20-31. 10.1242/bio.014951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalo F. R. and Reiter J. F. (2017). Open sesame: how transition fibers and the transition zone control ciliary composition. Cold Spring Harb. Perspect. Biol. 9, a028134 10.1101/cshperspect.a028134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giddings T. H. Jr., Meehl J. B., Pearson C. G. and Winey M (2010). Electron tomography and immuno-labeling of Tetrahymena thermophila basal bodies. Methods Cell Biol. 96, 117-141. 10.1016/S0091-679X(10)96006-8 [DOI] [PubMed] [Google Scholar]
- Gogendeau D., Beisson J., de Loubresse N. G., Le Caer J.-P., Ruiz F., Cohen J., Sperling L., Koll F. and Klotz C. (2007). An Sfi1p-like centrin-binding protein mediates centrin-based Ca2+ -dependent contractility in Paramecium tetraurelia. Eukaryot. Cell 6, 1992-2000. 10.1128/EC.00197-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves J. and Pelletier L. (2017). The ciliary transition zone: finding the pieces and assembling the gate. Mol. Cells 40, 243-253. 10.14348/molcells.2017.0054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P. (2012). Towards a molecular architecture of centriole assembly. Nat. Rev. Mol. Cell Biol. 13, 425-435. 10.1038/nrm3373 [DOI] [PubMed] [Google Scholar]
- Guichard P., Hamel V. and Gönczy P. (2018). The rise of the cartwheel: seeding the centriole organelle. BioEssays 40, 1700241 10.1002/bies.201700241 [DOI] [PubMed] [Google Scholar]
- Hai B., Gaertig J. and Gorovsky M. A. (2000). Knockout heterokaryons enable facile mutagenic analysis of essential genes in Tetrahymena. Methods Cell Biol. 62, 513-531. 10.1016/S0091-679X(08)61554-X [DOI] [PubMed] [Google Scholar]
- Hassan A., Parent S., Mathieu H., Zaouter C., Molidperee S., Bagu E. T., Barchi S., Villemure I., Patten S. A. and Moldovan F. (2019). Adolescent idiopathic scoliosis associated POC5 mutation impairs cell cycle, cilia length and centrosome protein interactions. PLoS ONE 14, e0213269 10.1371/journal.pone.0213269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heydeck W., Stemm-Wolf A. J., Knop J., Poh C. C. and Winey M. (2016). Sfr1, a tetrahymena thermophila Sfi1 repeat protein, modulates the production of cortical row basal bodies. mSphere 1 10.1128/mSphere.00257-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirono M. (2014). Cartwheel assembly. Philos. Trans. R. Soc. B Biol. Sci. 369, 20130458 10.1098/rstb.2013.0458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges M. E., Scheumann N., Wickstead B., Langdale J. A. and Gull K. (2010). Reconstructing the evolutionary history of the centriole from protein components. J. Cell Sci. 123, 1407-1413. 10.1242/jcs.064873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H. and Marshall W. F. (2011). Ciliogenesis: building the cell's antenna. Nat. Rev. Mol. Cell Biol. 12, 222-234. 10.1038/nrm3085 [DOI] [PubMed] [Google Scholar]
- Keller L. C., Geimer S., Romijn E., Yates J III, Zamora I. and Marshall W. F. (2009). Molecular architecture of the centriole proteome: the conserved WD40 domain protein POC1 is required for centriole duplication and length control. Mol. Biol. Cell 20, 1150-1166. 10.1091/mbc.e08-06-0619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilburn C. L., Pearson C. G., Romijn E. P., Meehl J. B., Giddings T. H. Jr., Culver B. P., Yates J. R III and Winey M. (2007). New Tetrahymena basal body protein components identify basal body domain structure. J. Cell Biol. 178, 905-912. 10.1083/jcb.200703109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin J. V. (2003). Sfi1p has conserved centrin-binding sites and an essential function in budding yeast spindle pole body duplication. J. Cell Biol. 162, 1211-1221. 10.1083/jcb.200307064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T. and Dynlacht B. D. (2011). Regulating the transition from centriole to basal body. J. Cell Biol. 193, 435-444. 10.1083/jcb.201101005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koblenz B., Schoppmeier J., Grunow A. and Lechtreck K.-F. (2003). Centrin deficiency in Chlamydomonas causes defects in basal body replication, segregation and maturation. J. Cell Sci. 116, 2635-2646. 10.1242/jcs.00497 [DOI] [PubMed] [Google Scholar]
- Kodani A., Moyer T., Chen A., Holland A., Walsh C. A. and Reiter J. F. (2019). SFI1 promotes centriole duplication by recruiting USP9X to stabilize the microcephaly protein STIL. J. Cell Biol. 218, 2185-2197. 10.1083/jcb.201803041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer J. R., Mastronarde D. N. and McIntosh J. R. (1996). Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71-76. 10.1006/jsbi.1996.0013 [DOI] [PubMed] [Google Scholar]
- Laoukili J., Perret E., Middendorp S., Houcine O., Guennou C., Marano F., Bornens M. and Tournier F. (2000). Differential expression and cellular distribution of centrin isoforms during human ciliated cell differentiation in vitro. J. Cell Sci. 113, 1355-1364. [DOI] [PubMed] [Google Scholar]
- Li S., Sandercock A. M., Conduit P., Robinson C. V., Williams R. L. and Kilmartin J. V. (2006). Structural role of Sfi1p-centrin filaments in budding yeast spindle pole body duplication. J. Cell Biol. 173, 867-877. 10.1083/jcb.200603153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwiczak J., Winski A., Szczepaniak K., Alva V. and Dunin-Horkawicz S. (2019). DeepCoil-a fast and accurate prediction of coiled-coil domains in protein sequences. Bioinforma. Oxf. Engl. 35, 2790-2795. 10.1093/bioinformatics/bty1062 [DOI] [PubMed] [Google Scholar]
- Marteil G., Guerrero A., Vieira A. F., de Almeida B. P., Machado P., Mendonça S., Mesquita M., Villarreal B., Fonseca I., Francia M. E., et al. (2018). Over-elongation of centrioles in cancer promotes centriole amplification and chromosome missegregation. Nat. Commun. 9, 1258 10.1038/s41467-018-03641-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Sanz J., Yang A., Blouquit Y., Duchambon P., Assairi L. and Craescu C. T. (2006). Binding of human centrin 2 to the centrosomal protein hSfi1. FEBS J. 273, 4504-4515. 10.1111/j.1742-4658.2006.05456.x [DOI] [PubMed] [Google Scholar]
- Martinez-Sanz J., Kateb F., Assairi L., Blouquit Y., Bodenhausen G., Abergel D., Mouawad L. and Craescu C. T. (2010). Structure, dynamics and thermodynamics of the human centrin 2/hSfi1 complex. J. Mol. Biol. 395, 191-204. 10.1016/j.jmb.2009.10.041 [DOI] [PubMed] [Google Scholar]
- Mastronarde D. N. (1997). Dual-axis tomography: an approach with alignment methods that preserve resolution. J. Struct. Biol. 120, 343-352. 10.1006/jsbi.1997.3919 [DOI] [PubMed] [Google Scholar]
- Mastronarde D. N. (2005). Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36-51. 10.1016/j.jsb.2005.07.007 [DOI] [PubMed] [Google Scholar]
- Meehl J. B., Giddings T. H. and Winey M. (2009). High pressure freezing, electron microscopy, and immuno-electron microscopy of Tetrahymena thermophila basal bodies. Methods Mol. Biol. Clifton NJ 586, 227-241. 10.1007/978-1-60761-376-3_12 [DOI] [PubMed] [Google Scholar]
- Meehl J. B., Bayless B. A., Giddings T. H., Pearson C. G. and Winey M. (2016). Tetrahymena Poc1 ensures proper intertriplet microtubule linkages to maintain basal body integrity. Mol. Biol. Cell 27, 2394-2403. 10.1091/mbc.e16-03-0165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K. (2008). High efficiency transformation of Tetrahymena using a codon-optimized neomycin resistance gene. Gene 425, 79-83. 10.1016/j.gene.2008.08.007 [DOI] [PubMed] [Google Scholar]
- Mochizuki K., Fine N. A., Fujisawa T. and Gorovsky M. A. (2002). Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in tetrahymena. Cell 110, 689-699. 10.1016/S0092-8674(02)00909-1 [DOI] [PubMed] [Google Scholar]
- Nachury M. V. and Mick D. U. (2019). Establishing and regulating the composition of cilia for signal transduction. Nat. Rev. Mol. Cell Biol. 20, 389 10.1038/s41580-019-0116-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanney D. L. (1975). Patterns of basal body addition in ciliary rows in Tetrahymena. J. Cell Biol. 65, 503-512. 10.1083/jcb.65.3.503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanney D. L. (1971). The pattern of replication of cortical units in Tetrahymena. Dev. Biol. 26, 296-305. 10.1016/0012-1606(71)90128-X [DOI] [PubMed] [Google Scholar]
- Nanney D. L. (1966). Corticotype transmission in Tetrahymena. Genetics 54, 955-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanney D. L. (1968). Cortical patterns in cellular morphogenesis. Differences in cortical patterns in ciliates may be hereditary, but independent of genic differences. Science 160, 496-502. 10.1126/science.160.3827.496 [DOI] [PubMed] [Google Scholar]
- Nishi R., Okuda Y., Watanabe E., Mori T., Iwai S., Masutani C., Sugasawa K. and Hanaoka F. (2005). Centrin 2 stimulates nucleotide excision repair by interacting with xeroderma pigmentosum group C protein. Mol. Cell. Biol. 25, 5664-5674. 10.1128/MCB.25.13.5664-5674.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka S., Tanaka Y., Okada Y., Takeda S., Harada A., Kanai Y., Kido M. and Hirokawa N. (1998). Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell 95, 829-837. 10.1016/S0092-8674(00)81705-5 [DOI] [PubMed] [Google Scholar]
- Oliazadeh N., Gorman K. F., Eveleigh R., Bourque G. and Moreau A. (2017). Identification of elongated primary cilia with impaired mechanotransduction in idiopathic scoliosis patients. Sci. Rep. 7, 44260 10.1038/srep44260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orias E., Hamilton E. P. and Orias J. D. (2000). Tetrahymena as a laboratory organism: useful strains, cell culture, and cell line maintenance. Methods Cell Biol. 62, 189-211. 10.1016/S0091-679X(08)61530-7 [DOI] [PubMed] [Google Scholar]
- Paoletti A., Moudjou M., Paintrand M., Salisbury J. L. and Bornens M. (1996). Most of centrin in animal cells is not centrosome-associated and centrosomal centrin is confined to the distal lumen of centrioles. J. Cell Sci. 109, 3089-3102. [DOI] [PubMed] [Google Scholar]
- Patten S. A., Margaritte-Jeannin P., Bernard J.-C., Alix E., Labalme A., Besson A., Girard S. L., Fendri K., Fraisse N., Biot B., et al. (2015). Functional variants of POC5 identified in patients with idiopathic scoliosis. J. Clin. Invest. 125, 1124-1128. 10.1172/JCI77262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson C. G. (2014). Choosing sides – asymmetric centriole and basal body assembly. J. Cell Sci. 127, 2803-2810. 10.1242/jcs.151761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson C. G. and Winey M. (2009). Basal body assembly in ciliates: the power of numbers. Traffic Cph. Den. 10, 461-471. 10.1111/j.1600-0854.2009.00885.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson C. G., Osborn D. P. S., Giddings T. H. Jr., Beales P. L. and Winey M. (2009). Basal body stability and ciliogenesis requires the conserved component Poc1. J. Cell Biol. 187, 905-920. 10.1083/jcb.200908019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter J. F. and Leroux M. R. (2017). Genes and molecular pathways underpinning ciliopathies. Nat. Rev. Mol. Cell Biol. 18, 533-547. 10.1038/nrm.2017.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues-Martins A., Riparbelli M., Callaini G., Glover D. M. and Bettencourt-Dias M. (2008). From centriole biogenesis to cellular function: centrioles are essential for cell division at critical developmental stages. Cell Cycle Georget. Tex 7, 11-16. 10.4161/cc.7.1.5226 [DOI] [PubMed] [Google Scholar]
- Ruiz F., Garreau de Loubresse N., Klotz C., Beisson J. and Koll F. (2005). Centrin deficiency in Paramecium affects the geometry of basal-body duplication. Curr. Biol. CB 15, 2097-2106. 10.1016/j.cub.2005.11.038 [DOI] [PubMed] [Google Scholar]
- Sattler C. A. and Staehelin L. A. (1974). Ciliary membrane differentiations in Tetrahymena pyriformis. Tetrahymena has four types of cilia. J. Cell Biol. 62, 473-490. 10.1083/jcb.62.2.473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawant D. B., Majumder S., Perkins J. L., Yang C.-H., Eyers P. A. and Fisk H. A. (2015). Centrin 3 is an inhibitor of centrosomal Mps1 and antagonizes centrin 2 function. Mol. Biol. Cell 26, 3741-3753. 10.1091/mbc.E14-07-1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T. I., Kleylein-Sohn J., Westendorf J., Le Clech M., Lavoie S. B., Stierhof Y.-D. and Nigg E. A. (2009). Control of centriole length by CPAP and CP110. Curr. Biol. CB 19, 1005-1011. 10.1016/j.cub.2009.05.016 [DOI] [PubMed] [Google Scholar]
- Schneider T. D. and Stephens R. M. (1990). Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18, 6097-6100. 10.1093/nar/18.20.6097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y., Song X., Bowen J., Corstanje R., Gao Y., Gaertig J. and Gorovsky M. A. (2002). A robust inducible-repressible promoter greatly facilitates gene knockouts, conditional expression, and overexpression of homologous and heterologous genes in Tetrahymena thermophila. Proc. Natl. Acad. Sci. USA 99, 3734-3739. 10.1073/pnas.052016199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Zhao Y., Vonderfecht T., Winey M. and Klymkowsky M. W. (2015). Centrin-2 (Cetn2) mediated regulation of FGF/FGFR gene expression in Xenopus. Sci. Rep 5, 10283 10.1038/srep10283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemm-Wolf A. J., Morgan G., Giddings T. H. Jr., White E. A., Marchione R., McDonald H. B. and Winey M (2005). Basal body duplication and maintenance require one member of the Tetrahymena thermophila centrin gene family. Mol. Biol. Cell 16, 3606-3619. 10.1091/mbc.e04-10-0919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemm-Wolf A. J., Meehl J. B. and Winey M. (2013). Sfr13, a member of a large family of asymmetrically localized Sfi1-repeat proteins, is important for basal body separation and stability in Tetrahymena thermophila. J. Cell Sci. 126, 1659-1671. 10.1242/jcs.120238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad P. and Gönczy P. (2008). Mechanisms of procentriole formation. Trends Cell Biol. 18, 389-396. 10.1016/j.tcb.2008.06.004 [DOI] [PubMed] [Google Scholar]
- Tanos B. E., Yang H.-J., Soni R., Wang W.-J., Macaluso F. P., Asara J. M. and Tsou M.-F. B. (2013). Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes Dev. 27, 163-168. 10.1101/gad.207043.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladar E. K. and Stearns T. (2007). Molecular characterization of centriole assembly in ciliated epithelial cells. J. Cell Biol. 178, 31-42. 10.1083/jcb.200703064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonderfecht T., Stemm-Wolf A. J., Hendershott M., Giddings T. H. Jr., Meehl J. B. and Winey M (2011). The two domains of centrin have distinct basal body functions in Tetrahymena. Mol. Biol. Cell 22, 2221-2234. 10.1091/mbc.e11-02-0151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonderfecht T., Cookson M. W., Giddings T. H., Clarissa C. and Winey M. (2012). The two human centrin homologues have similar but distinct functions at Tetrahymena basal bodies. Mol. Biol. Cell 23, 4766-4777. 10.1091/mbc.e12-06-0454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters A. M. and Beales P. L. (2011). Ciliopathies: an expanding disease spectrum. Pediatr. Nephrol. Berl. Ger 26, 1039-1056. 10.1007/s00467-010-1731-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisz Hubshman M., Broekman S., van Wijk E., Cremers F., Abu-Diab A., Khateb S., Tzur S., Lagovsky I., Smirin-Yosef P., Sharon D. et al. (2018). Whole-exome sequencing reveals POC5 as a novel gene associated with autosomal recessive retinitis pigmentosa. Hum. Mol. Genet. 27, 614-624. 10.1093/hmg/ddx428 [DOI] [PubMed] [Google Scholar]
- Wheway G., Parry D. A. and Johnson C. A. (2014). The role of primary cilia in the development and disease of the retina. Organogenesis 10, 69-85. 10.4161/org.26710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M., Stemm-Wolf A. J., Giddings T. H. and Pearson C. G. (2012). Cytological analysis of Tetrahymena thermophila. Methods Cell Biol. 109, 357-378. 10.1016/B978-0-12-385967-9.00013-X [DOI] [PubMed] [Google Scholar]
- Wolff A., de Néchaud B., Chillet D., Mazarguil H., Desbruyères E., Audebert S., Eddé B., Gros F. and Denoulet P. (1992). Distribution of glutamylated alpha and beta-tubulin in mouse tissues using a specific monoclonal antibody, GT335. Eur. J. Cell Biol. 59, 425-432. [PubMed] [Google Scholar]
- Wong Y. L., Anzola J. V., Davis R. L., Yoon M., Motamedi A., Kroll A., Seo C. P., Hsia J. E., Kim S. K., Mitchell J. W., et al. (2015). Reversible centriole depletion with an inhibitor of Polo-like kinase 4. Science 348, 1155-1160. 10.1126/science.aaa5111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Sheng F., Xia C., Li Y., Feng Z., Qiu Y. and Zhu Z. (2018). Common variant of POC5 is associated with the susceptibility of adolescent idiopathic scoliosis. Spine 43, E683-E688. 10.1097/BRS.0000000000002490 [DOI] [PubMed] [Google Scholar]
- Yan X., Zhao H. and Zhu X. (2016). Production of Basal Bodies in bulk for dense multicilia formation. F1000Research 5, 1533 10.12688/f1000research.8469.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P., Diener D. R., Yang C., Kohno T., Pazour G. J., Dienes J. M., Agrin N. S., King S. M., Sale W. S., Kamiya R., et al. (2006). Radial spoke proteins of Chlamydomonas flagella. J. Cell Sci. 119, 1165-1174. 10.1242/jcs.02811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M.-C., Fuller P. and Xi X. (2003). Programmed DNA deletion as an RNA-guided system of genome defense. Science 300, 1581-1584. 10.1126/science.1084737 [DOI] [PubMed] [Google Scholar]
- Ying G., Avasthi P., Irwin M., Gerstner C. D., Frederick J. M., Lucero M. T. and Baehr W. (2014). Centrin 2 is required for mouse olfactory ciliary trafficking and development of ependymal cilia planar polarity. J. Neurosci. Off. J. Soc. Neurosci. 34, 6377-6388. 10.1523/JNEUROSCI.0067-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying G., Frederick J. M. and Baehr W. (2019). Deletion of both centrin 2 (CETN2) and CETN3 destabilizes the distal connecting cilium of mouse photoreceptors. J. Biol. Chem. 294, 3957-3973. 10.1074/jbc.RA118.006371 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.