ABSTRACT
The remarkable diversity of neurons in the nervous system is generated during development, when properties such as cell morphology, receptor profiles and neurotransmitter identities are specified. In order to gain a greater understanding of neurotransmitter specification we profiled the transcription state of cholinergic, GABAergic and glutamatergic neurons in vivo at three developmental time points. We identified 86 differentially expressed transcription factors that are uniquely enriched, or uniquely depleted, in a specific neurotransmitter type. Some transcription factors show a similar profile across development, others only show enrichment or depletion at specific developmental stages. Profiling of Acj6 (cholinergic enriched) and Ets65A (cholinergic depleted) binding sites in vivo reveals that they both directly bind the ChAT locus, in addition to a wide spectrum of other key neuronal differentiation genes. We also show that cholinergic enriched transcription factors are expressed in mostly non-overlapping populations in the adult brain, implying the absence of combinatorial regulation of neurotransmitter fate in this context. Furthermore, our data underlines that, similar to Caenorhabditis elegans, there are no simple transcription factor codes for neurotransmitter type specification.
This article has an associated First Person interview with the first author of the paper.
KEY WORDS: Targeted DamID, Neural development, Neurotransmitter, Specification, Transcription factors
Summary: Transcriptome profiling of cholinergic, GABAergic and glutamatergic neurons in Drosophila identified multiple transcription factors as potential regulators of neurotransmitter fate.
INTRODUCTION
The human brain is perhaps the most complex system known to mankind. It consists of approximately 85 billion neurons (Herculano-Houzel, 2016), which possess very diverse morphologies, neurotransmitter identities, electrical properties and preferences for synaptic partners. Understanding how this diversity is generated is one of the greatest challenges in biology and can only be achieved by identifying the underlying molecular mechanisms that determine these neuronal properties. Neurotransmitters allow neurons to communicate with each other, enabling organisms to sense, interpret and interact with their environment. Fast-acting neurotransmitters include acetylcholine and glutamate, which are, in general, excitatory, and GABA, which is inhibitory (Van Der Kloot and Robbins, 1959). The function of individual neurons depends on the specific types of neurotransmitters they produce, which in turn ensures proper information flow and can also influence the formation of neural circuits (Andreae and Burrone, 2018). Therefore, the proper specification of neurotransmitter fate is fundamental for nervous system development.
Model organism studies in Caenorhabditis elegans, mice and Drosophila have provided a wealth of information about factors and mechanisms involved in neurotransmitter specification. Comprehensive neurotransmitter maps (Hobert, 2016) and the description of terminal selector genes in C. elegans (Hobert, 2008) have provided important contributions to the field. These terminal selectors are transcription factors (or a transcription factor complex) that regulate the expression of a battery of terminal differentiation genes in the last phase of neuronal differentiation, and maintain the expression of these genes during the lifetime of a neuron (Hobert, 2008). For example, the C. elegans transcription factors ttx-3 and unc-86 act as terminal selectors in distinct cholinergic and serotonergic neuron populations, respectively (Zhang et al., 2014).
Cellular context is important for the action of these specifying factors, as misexpression of terminal selectors in other neuronal subtypes is often not sufficient to reprogram their fate (Duggan et al., 1998; Wenick and Hobert, 2004). The presence of co-factors, and likely the chromatin state, can also influence this plasticity (Altun-Gultekin et al., 2001; Patel and Hobert, 2017). Related to this, there appears to be little evidence for master regulators of cholinergic, GABAergic or glutamatergic fate (Konstantinides et al., 2018; Lacin et al., 2019; Serrano-Saiz et al., 2013). Rather, individual lineages, or subpopulations, utilise different transcription factors (or combinations of transcription factors) to specify the fast-acting neurotransmitter that they will utilise. Developmental context also plays a role in the mechanisms governing neurotransmitter specification. In Drosophila, early born embryonic neurons in a given lineage can use different neurotransmitters (Landgraf et al., 1997; Schmid et al., 1999). However, strikingly, each post-embryonic lineage only uses one neurotransmitter (Lacin et al., 2019), implying that specification occurs at the stem cell level during larval stages.
Neurotransmitter specification studies across different organisms have highlighted conserved mechanisms. A prominent example is the binding of the transcription factors AST-1 (C. elegans) and Etv1 (vertebrates) to a phylogenetically conserved DNA motif to specify dopaminergic fate (Flames and Hobert, 2009). Furthermore, orthologues acj6 (Drosophila), unc-86 (C. elegans) and Brn3A/POU4F1 (vertebrates) all have roles in cholinergic specification (Lee and Salvaterra, 2002; Serrano-Saiz et al., 2018; Zhang et al., 2014), while PITX2 (vertebrates) and unc-30 (C. elegans) both control GABAergic differentiation (Jin et al., 1994; Waite et al., 2011; Westmoreland et al., 2001).
In order to identify novel candidate genes, and investigate the dynamics of neurotransmitter specific transcription factors throughout development, we have performed cell specific profiling of RNA polymerase II occupancy, in vivo, in cholinergic, GABAergic and glutamatergic neurons of Drosophila. We identify 86 transcription factors that show differential expression between neurotransmitter types, in at least one developmental time point. There are both uniquely enriched and uniquely depleted transcription factors, and we show that acj6 (cholinergic enriched) and Ets65A (cholinergic depleted) both directly bind the choline acetyltransferase gene (ChAT) required for cholinergic fate.
RESULTS
Transcriptional profiling of neuronal types across development
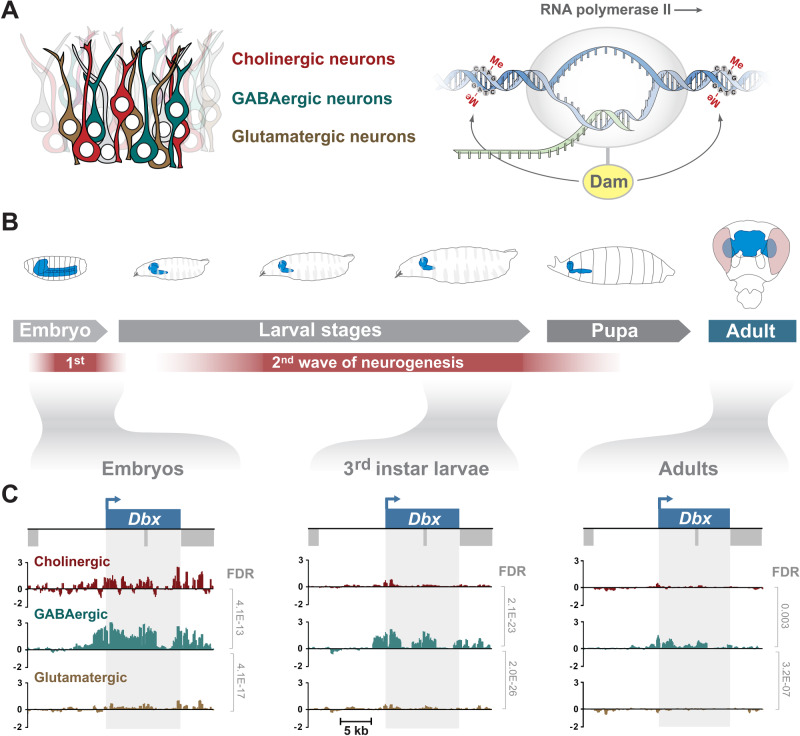
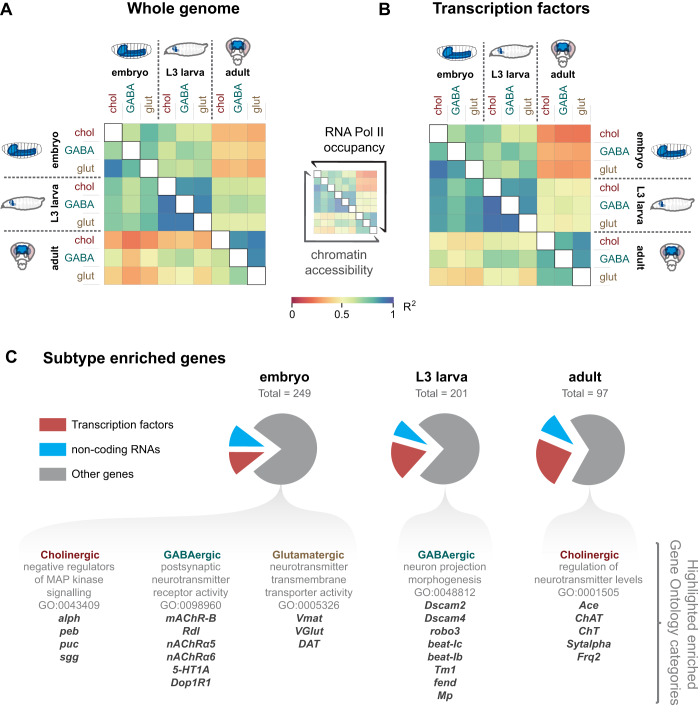
In order to investigate which genes participate in the specification of neuronal properties, namely, neurotransmitter choice, we applied the cell specific profiling technique Targeted DamID (TaDa). TaDa is based on DamID (van Steensel and Henikoff, 2000) and allows the profiling of protein–DNA interactions without the need for cell isolation, specific antibodies or fixation (Aughey et al., 2019; Southall et al., 2013). Transcriptional profiling is also possible with TaDa using the core subunit of RNA polymerase II (Pol II) (Southall et al., 2013). We have mapped the occupancy of Pol II in cholinergic, GABAergic and glutamatergic neurons, using specific GAL4 drivers that trap the expression of the genes ChAT (choline acetyltransferase), Gad1 (glutamic acid decarboxylase 1) and VGlut (vesicular glutamate transporter) (Diao et al., 2015). During Drosophila development, there are two neurogenic periods, the first to produce the larval nervous system, and the second to produce the adult nervous system. Therefore, to cover both developing stages and adult neurons, we profiled embryonic neurons, larval postembryonic neurons and adult neurons (see Fig. 1B). Windows of 20 h (embryo samples), and 24 h (third instar larvae and adult samples) were used for TaDa profiling and three replicates were performed for each experiment. The number of genes bound by Pol II ranged from 1170–1612 (see Table S1). To investigate the global differences in Pol II occupancy between neuronal types and developmental stages, we generated a correlation matrix (Fig. 2A). We found that the greatest variability is between developmental stages, rather than between cell types, with the adult brain data being more distinct from the embryonic and larval stages. When focusing on transcription factor genes, a similar pattern is evident (Fig. 2B). For each developmental stage, we identified uniquely enriched genes (i.e. genes enriched in one neurotransmitter compared to the other two neurotransmitter types) (Table S2). Encouragingly, a strong enrichment of Pol II occupancy is evident at ChAT, Gad1 and VGlut, the genes encoding the key enzymes involved in the biosynthesis of these neurotransmitters (Fig. S1). Transcription factors and non-coding RNAs make up a large proportion of all the enriched genes, at each developmental stage (Fig. 2C). In the adult, almost a quarter (23/97) of the enriched genes are transcription factors. Other enriched genes include the immunoglobulin domain containing beaten path (beat) and Down syndrome cell adhesion molecule (Dscam) genes, which play roles in axon guidance and dendrite self-avoidance (Pipes et al., 2001; Soba et al., 2007). Glutamatergic genes include twit and Dad, both of which are known to regulate synaptic homeostasis at the neuromuscular junction (Goold and Davis, 2007; Kim and Marques, 2012). Interestingly, there is an enriched expression of MAP kinase inhibitors in cholinergic neurons (Fig. 2C). Also, glutamatergic neurons express higher levels of the monoamine neurotransmitter related genes Vmat, DAT and Tdc2, while GABAergic neurons are enriched for serotonergic and dopaminergic receptors, relative to the other two fast-acting neurotransmitter types (Fig. 2C). Very few genes show enrichment across all developmental stages: five for cholinergic (ChAT, ChT, acj6, Mef2 and sosie), five for GABAergic (Gad1, Dbx, vg, CG13739 and CG14989) and two for glutamatergic (VGlut and oc) (Table S2). There is consistent enrichment of the GAL4-trapped genes (ChAT, Gad1 and VGlut) (Fig. S1) that provide type specific expression for the TaDa experiments.
Fig. 1.
Cell specific profiling of RNA Pol II occupancy in different neuronal subtypes throughout Drosophila development. (A) Profiling of RNA Pol II occupancy in cholinergic, GABAergic and glutamatergic neurons using TaDa. (B) Profiling windows cover embryonic nervous system development (5−25 h AEL), third instar larval nervous system development (24 h window before pupation) and the adult brain (heads from ∼3−4 day old adults after a 24 h expression window). Temporal restriction of Dam-Pol II expression was controlled using a temperature sensitive GAL80. (C) Bottom panels show an example of a transcription factor (Dbx) that is uniquely transcribed in GABergic neurons. Y-axis represent log2 ratios of Dam-Pol II over Dam-only. False discovery rate (FDR) values are shown for significant differences (<0.01).
Fig. 2.
Correlation of RNA Pol II occupancy and chromatin accessibility for neurotransmitter subtypes. (A) Correlation matrix for RNA Pol II signal (log2 over Dam-only for all genes) and chromatin accessibility (CATaDa) for all gene loci (extended 5 kb upstream and 2 kb downstream). (B) Correlation matrix for RNA Pol II and chromatin accessibility at annotated transcription factor genes. (C) Characterisation of subtype-enriched genes at each developmental stage. Examples of enriched GO term categories for the remaining genes are also included.
CATaDa, an adaption of TaDa, allows profiling of chromatin accessibility without the need for cell isolation using an untethered Dam protein (i.e. the control experiment in TaDa) (Fig. S2A) (Aughey et al., 2018). CATaDa reveals that, similar to RNA Pol II, global chromatin accessibility does not vary greatly between cell types (Fig. 2A,B) but shows more differences between developmental stages. Chromatin accessibility states of embryonic and larval neurons are more similar to each other than to those of adult neurons (Fig. 2A,B). When examining regions of the genome that display robust changes in chromatin accessibility (peaks that show >10 RPM differences across three consecutively methylated regions) during embryo development, only 37 GATC fragments (13 individual peaks) are identified, with 62% mapping to the loci of the three neurotransmitter synthesis genes (ChAT, Gad1 and VGlut) (Fig. S2B,C). This shows that across the population of neurons for each neurotransmitter type, major changes in accessibility are limited to genes involved in the respective neurotransmitter synthesis, with none of open regions directly corresponding to transcription factor loci. Differential accessibility is also present at sites outside of the gene and promoter for Gad1 and VGlut (yellow arrows in Fig. S2C). Weaker differences in accessibility are also observed at some of the differentially expressed transcription factor loci (Fig. S2D).
Identification of transcription factors uniquely enriched, or uniquely depleted in neurotransmitter types
Transcription factors play the major role in neurotransmitter specification and we have identified many with enriched Pol II occupancy in specific neurotransmitter types (Fig. 2A). Uniquely enriched transcription factors are candidates for activators of neurotransmitter identity and conversely, if there is depletion (or absence) of a transcription factor from only one type, they are candidates for repressors of neurotransmitter identity. For example, a hypothetical transcription factor that represses GABAergic fate would be present in both cholinergic and glutamatergic neurons but absent from GABAergic neurons.
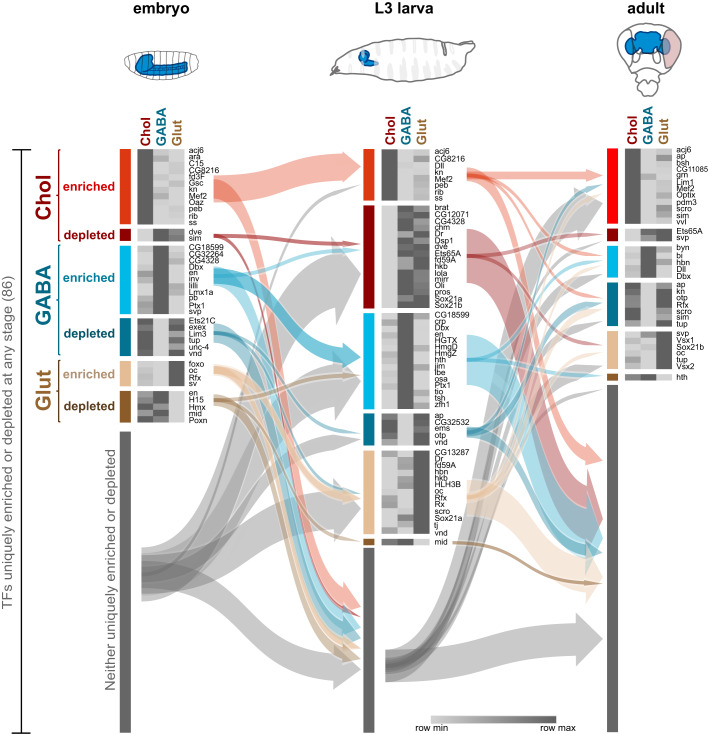
To investigate the expression pattern dynamics of both uniquely enriched and uniquely depleted transcription factors, we examined how their expression patterns transitioned across the stages of development (Fig. 3). We observe a great deal of flux between transcription factor expression in cell types and developmental stages. Many genes are enriched in one or two of the developmental stages. For example, kn, peb, rib and ss are cholinergic enriched in embryo and larva, but not in adults. Dll is an unusual case, as it is cholinergic enriched in larvae, however, switches to being GABAergic enriched in adults (Fig. 3; Fig. S3).
Fig. 3.
Transitions of uniquely enriched or depleted transcription factors during neural development. Transcription factors uniquely enriched or depleted in cholinergic, GABAergic and glutamatergic neurons. A total of 86 transcription factors are identified across all stages. Note that the arrows point to the group and not individual genes.
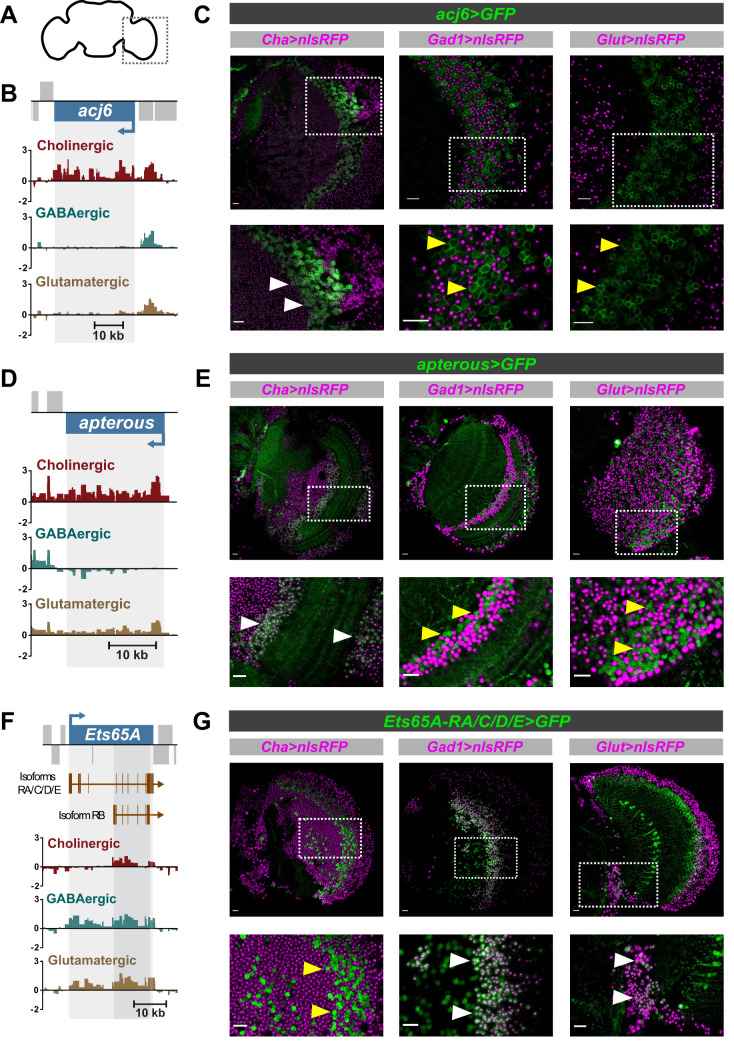
Exceptions to this are acj6 (cholinergic – see Fig. 4; Fig. S4), Dbx (GABAergic – see Fig. 1) and oc (glutamatergic), which are enriched in their respective neurotransmitter type throughout all stages. In support of our data, Acj6 is known to promote cholinergic fate in the peripheral nervous system (Lee and Salvaterra, 2002) and Dbx is important for the proper differentiation of a subset of GABAergic interneurons (Lacin et al., 2009). We checked the expression pattern of acj6 in adult brains, and as predicted by the transcriptomic data (Fig. 4B), we only found expression of acj6 in cholinergic neurons (Fig. 4C). We observed the same in larval brains, with the exception of some coexpression between glutamatergic neurons and acj6 (Fig. S4E). This agrees with the low-level signal observed in RNA Pol II occupancy plots for acj6 gene in third instar larva glutamatergic neurons (Fig. S4A).
Fig. 4.
Expression of acj6, apterous and Ets65A-RA/C/D/E in the adult brain. (A) Schematic of adult brain to show region of interest. (B) Pol II occupancy at acj6 in the adult brain. Y-axis represent log2 ratios of Dam-Pol II over Dam-only. FDR values are shown for significant differences (<0.01). (C) Expression pattern of acj6. White arrows show examples of colocalisation and yellow arrows absence of colocalisation. (D) Pol II occupancy at apterous. (E) Expression pattern of apterous in the adult brain. (F) Pol II occupancy at Ets65A in the adult brain. (G) Expression pattern of Ets65A-RA/C/D/E.
Candidate repressors of neurotransmitter fate (uniquely depleted transcription factors) also demonstrate dynamic changes in expression pattern across development (Fig. 3). Prominent examples are apterous (absent in GABAergic), and the longer transcript isoforms of Ets65A (absent in cholinergic) (Fig. 4D,F). We used genetic reporters to examine the expression pattern of apterous and Ets65A-RA/C/D/E in adult brains (Fig. 4E,G). In agreement with our data, the GABAergic reporter is absent in apterous positive cells, and the cholinergic reporter is absent in Ets65A-RA/C/D/E positive cells. We also observed an absence of apterous in larval GABAergic neurons (Fig. S5C,E), as predicted by the transcriptomic data (Fig. S5A). As for the longer transcripts of Ets65A-RA/C/D/E in larval neurons, we did identify their presence in a small number of cholinergic neurons (Fig. S6C,E), which could reflect the very low signal in the RNA Pol II occupancy plot (within the unique region of the long transcripts) (Fig. S6A).
We have identified transcription factors with potentially novel roles in regulating neurotransmitter identity. Therefore, we investigated candidate activators and candidate repressors for their potential to elicit pan-neural reprogramming of neurotransmitter identity. Pan-neural expression and RNAi knockdown of candidate activator transcription factors (Dbx, en, collier and CG4328) and candidate repressor transcription factors (ap, CG4328, Ets65A-RA and otp) during embryonic development, and larval stages did not result in any obvious changes in neurotransmitter expression patterns (Fig. S7).
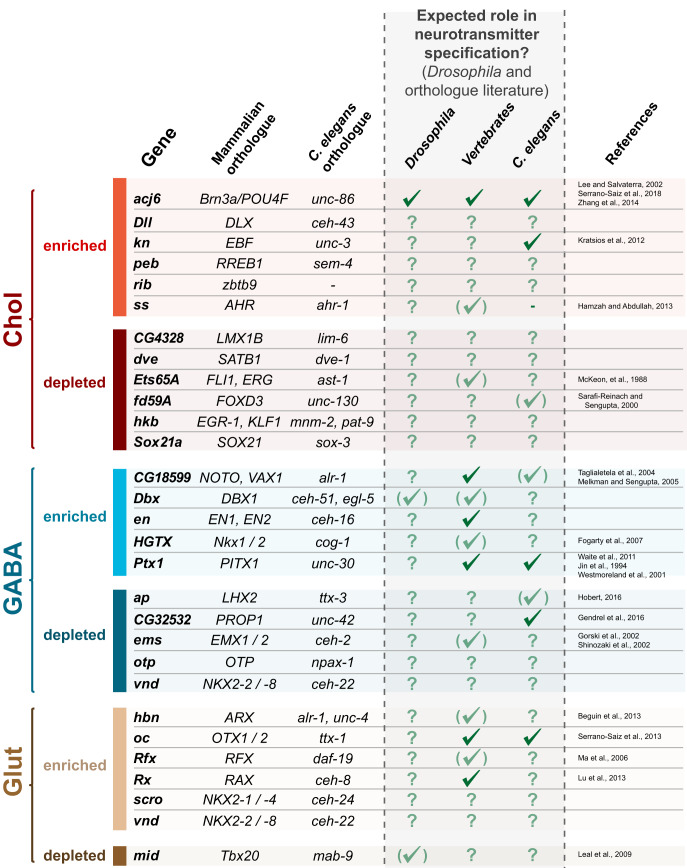
Focusing on candidate transcription factors demonstrating binary differences (clear on and off), we performed literature searches to examine whether they have been previously shown, or implicated in regulating neurotransmitter identity (Fig. 5). This included C. elegans and mouse orthologues, as much of the work in this field has utilised these model organisms. For example, the orthologues of cholinergic enriched acj6 (unc-86), GABAergic enriched Ptx1 (PITX1 and unc-30) and glutamatergic enriched oc (OTX1/2 and ttx-1) have all shown to have a role in promoting cholinergic, GABAergic and glutamatergic fate, respectively. However, there are many that have not been investigated in this context (38%), or that are only supported by indirect evidence (38%). These include Dll (DLX, ceh-43), sox21a (SOX21, sox-3), hbn (ARX, alr1, unc-4) and otp (OTP, npax-1). Given the strong conservation of neurotransmitter specification mechanisms, many of these newly highlighted factors provide promising research avenues for expanding our knowledge in this field.
Fig. 5.
Evidence for predicted roles of identified transcription factors and their orthologues. Strongly enriched or depleted transcription factors identified in developing larval brains. Uniquely enriched factors are predicted to be candidates that promote the respective neurotransmitter fate, whilst uniquely depleted factors are predicted to repress the neurotransmitter fate. A full tick indicates direct evidence that the transcription factor directly promotes or represses the neurotransmitter fate, whilst indirect supporting evidence is indicated by faded tick in brackets. A question mark signifies that nothing is currently known regarding neurotransmitter specification.
While non-coding ribosomal RNAs and tRNAs are transcribed by RNA polymerase I and III, micro RNAs (miRNAs) and long non-coding RNAs (lncRNAs) are primarily transcribed by Pol II. Our Dam-Pol II data identifies a set of differentially bound miRNAs and lncRNAs, between the neurotransmitter types (Fig. S8A). These include non-characterised lncRNAs and GABAergic enriched iab8, which is located in the Hox cluster and plays a role in the repression of abd-A (Gummalla et al., 2012). A small number of miRNAs were also identified, most notably, mir-87 (cholinergic), mir-184 (GABAergic) and mir-190 (glutamatergic), which are enriched during the developing states but not in the adult. Although annotated separately, mir-184 is embedded in CR44206 (Fig. S8B).
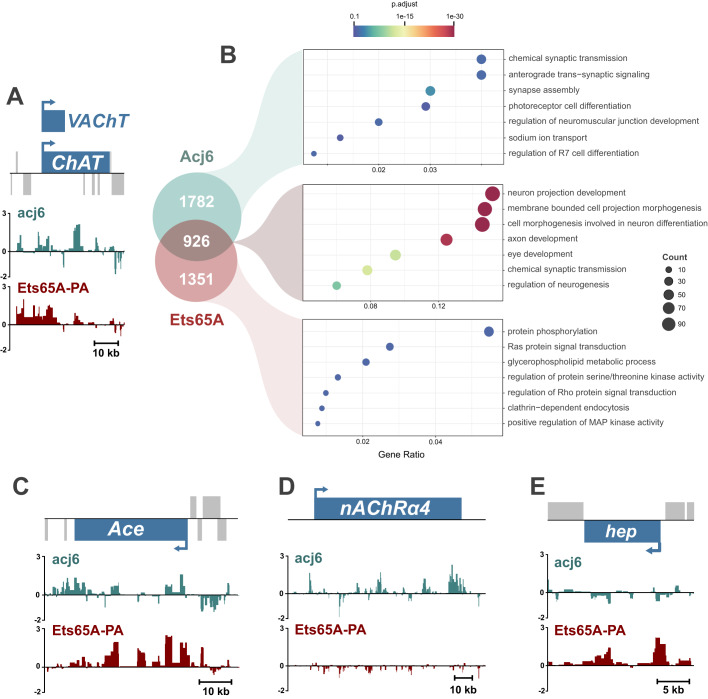
Acj6 and Ets65A-PA directly bind to ChAT and other key neuronal differentiation genes
Acj6 is enriched in cholinergic neurons (Fig. 3) and is known to promote cholinergic fate (Lee and Salvaterra, 2002). Acj6 can bind to specific sites upstream of ChAT in vitro (Lee and Salvaterra, 2002), however, the extent of Acj6 binding at the ChAT locus in vivo, and genome wide, is not known. In order to only profile the cells that endogenously express acj6, and therefore gain a more accurate readout of native Acj6 binding, we used an acj6 GAL4 line (Lai et al., 2008) to drive the expression of the Dam-acj6 transgene. Furthermore, we generated an Ets65A-RA/C/D/E MiMIC GAL4 trap line to investigate the in vivo binding of Ets65A-PA, with an interest to see whether, as a candidate cholinergic repressor, it could directly bind the ChAT locus. In the adult brain, both factors directly bind the ChAT locus (Fig. 6A). Acj6 binds at the upstream region studied by (Lee and Salvaterra, 2002), as well as strongly within intronic regions of ChAT. Ets65A-PA also binds at the same intronic region, however, it’s binding at the upstream region and transcriptional start site of ChAT is far more pronounced (Fig. 6A), which may reflect a different mode of regulation.
Fig. 6.
Acj6 and Ets65A-PA co-bind both ChAT and a whole suite of genes involved in neuronal differentiation. (A) Acj6 and Ets65-PA binding at ChAT (Y-axis represent log2 ratios of Dam-Pol II over Dam-only). (B) Enriched GO term categories for Acj6 and Ets65A-PA bound genes. (C−E) Binding at Ace, nAChRα4 and hep.
Acj6 and Ets65A-PA bind 2708 and 2277 genes, respectively, using a stringent false discovery rate (FDR) (FDR<0.0001) (Table S4). They co-bind 926 genes, which are highly enriched for nervous system genes, including genes involved in axon development [GO:0061564] and chemical synaptic transmission [GO:0007268] (Fig. 6B). While both factors bind the cholinergic signalling regulator gene Acetylcholine esterase (Ace) gene (Fig. 6C), Acj6 uniquely binds nicotinic Acetylcholine Receptor α4 (nAChRα4) (Fig. 6D), and Ets65A-RA binds multiple genes involved in MAP kinase signalling (e.g. hep, lic, Dsor and slpr) (Fig. 6E). Therefore, these factors have the potential to regulate not just a single neuronal property, but also a multitude of other genes that govern a wide spectrum of neuronal processes, such as their receptivity to extrinsic signals and synapse formation.
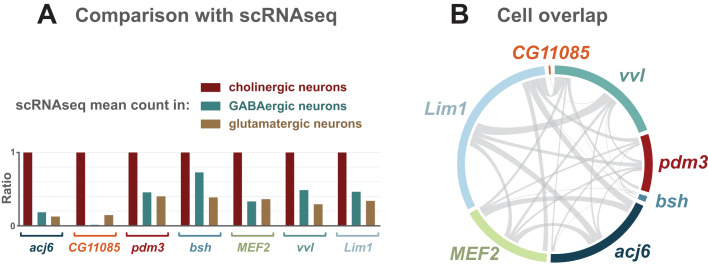
Enriched transcription factors are expressed in mostly non-overlapping populations
There are multiple transcription factors that show enriched expression in adult cholinergic neurons (Fig. 3). To investigate whether these factors are co-expressed within the cholinergic population, we mined single cell RNA-seq (scRNA-seq) data from adult brains (Davie et al., 2018). We find that the relative expression of the enriched factors, across the different neurotransmitter types, shows the same pattern, with enriched cholinergic factors also showing enrichment in the scRNAseq data (Fig. 7A). Due to the nature of scRNAseq data, we could then determine if the cholinergic cells expressing an enriched transcription factor also express other transcription factors identified as being enriched (Fig. 7B). Interestingly, there is relatively little overlap, demonstrating that these factors are expressed in distinct subpopulations of the cholinergic neurons in the adult brain.
Fig. 7.
Enriched transcription factors are expressed in mostly non-overlapping populations of adult cholinergic neurons. (A) Transcription factors identified as enriched in cholinergic neurons by TaDa are also enriched in scRNAseq data (adult brain). Mean counts are ratio normalised to the average count value in cholinergic neurons. (B) Circos plot displaying the overlap in cells expressing cholinergic enriched transcription factors.
DISCUSSION
Neurotransmitter identity is a key property of a neuron that needs to be tightly regulated in order to generate a properly functioning nervous system. Here we have investigated the dynamics and extent of transcription factor specificity in fast-acting neurotransmitter neuronal types in Drosophila. We profiled the transcription state of cholinergic, GABAergic and glutamatergic neurons in the developing and adult brain of Drosophila. We observe enriched Pol II occupancy at the relevant neurotransmitter synthesis genes (Fig. S1) and other genes associated with the activity of the specific types (Table S2). The monoamine neurotransmitter related genes Vmat, DAT and Tdc2 are enriched in glutamatergic neurons (Fig. 2C), which is not unprecedented, as monoamine populations can also be glutamatergic (Aguilar et al., 2017; Trudeau and El Mestikawy, 2018). Cholinergic, GABAergic, serotonergic and dopaminergic receptors are enriched in embryonic GABAergic neurons relative to the other two fast-acting neurotransmitter types (Fig. 2C), which correlates with GABAergic interneurons acting as integrative components of neural circuits. The enrichment of MAP kinase pathway regulators in cholinergic neurons is intriguing, suggesting that this signalling pathway may have a specific role in these neurons. This is supported by a recent study showing that MAP kinase signalling acts downstream of Gq-Rho signalling in C. elegans cholinergic neurons to control neuron activity and locomotion (Coleman et al., 2018).
Importantly, we have uncovered and highlighted transcription factors and non-coding RNAs differentially expressed between these types. Some of these are expected based on previous studies in Drosophila, including acj6 (cholinergic) (Lee and Salvaterra, 2002) and Dbx (GABAergic) (Lacin et al., 2009). Also, studies in other model organisms fit with our findings, for example, cholinergic enriched knot, whose orthologue, UNC-3 (C. elegans), is a terminal selector for cholinergic motor neuron differentiation (Kratsios et al., 2011). In addition, RFX, the vertebrate orthologue of Rfx, which we identified as glutamatergic enriched, can increase the expression of the neuronal glutamate transporter type 3 (Ma et al., 2006). However, we have identified many differentially expressed transcription factors that have not had their role studied with respect to neurotransmitter specification, or cases where there is supportive, but not direct, evidence for a role in neurotransmitter specification. For instance, vertebrate neuronal precursors expressing Nkx2.1 (HGTX orthologue) predominantly generate GABAergic interneurons (Fogarty et al., 2007), and a polyalanine expansion in ARX (hbn orthologue) causes remodelling and increased activity of glutamatergic neurons in vertebrates (Beguin et al., 2013). Acj6 is expressed in a subset of cholinergic neurons (Lee and Salvaterra, 2002) and Dbx in a subset of GABAergic neurons (Lacin et al., 2009). To the best of our knowledge, none of the enriched transcription factors we identified are expressed in all of the neurons of a particular neurotransmitter type. This highlights that, similar to C. elegans (Hobert, 2016), there are no simple transcription factor codes for neurotransmitter type specification in Drosophila.
Uniquely enriched factors are candidates for promoting a neurotransmitter fate, and we tested a number of them for their ability to reprogram neurons on a global scale in embryos (Fig. S7). No obvious changes were observed, however, this is not particularly surprising considering the importance of cellular context for the reprogramming of neuronal properties (Duggan et al., 1998; Wenick and Hobert, 2004). Successful reprograming may require intervention at a specific time point (e.g. at the progenitor stage), the co-expression of appropriate co-factors, and/or to exclusively target a neuronal subpopulation within each neurotransmitter type. Future work could investigate these factors in specific and relevant lineages, to shed light on important contextual information.
The majority of transcription factors identified as directly regulating neurotransmitter fate act in a positive manner, whereas only a handful of studies describe the role of repressors. Incoherent feedforward loops exist in C. elegans, where terminal selectors activate repressors, which feedback onto effector genes (for review, see Hobert, 2016). In vertebrates, both Neurogenin 2 and Tlx3 are required for the specification of certain glutamatergic populations but also act to repress GABAergic fate (Cheng et al., 2004; Schuurmans et al., 2004). Whether this is direct repression of Glutamic acid decarboxylase (Gad) genes (required for the synthesis of GABA), or indirectly, through another transcription factor, is unclear. We have identified several transcription factors that are expressed in two neurotransmitter types, but absent from the other. These include apterous (ap), Ets65A (long transcripts) and orthopedia (otp), which we hypothesise to be candidate repressors, given their absence from cells with a specific neurotransmitter identity. Our profiling of Ets65A-PA binding in vivo, reveals that it directly binds ChAT (Fig. 6A), and therefore has the potential to directly regulate cholinergic fate. Similar to the candidate activators, ectopic expression of these candidates did not show any obvious repression of the respective neurotransmitter genes (Fig. S7), however, again, this might be because they can only act as a repressor in specific contexts (e.g. when a co-repressor is present), or that they regulate genes associated with specific types but do not directly regulate neurotransmitter identity.
The development of single cell RNA-seq (scRNA-seq) technology has led to the profiling of several Drosophila tissues, including the whole adult brain (Davie et al., 2018), the central adult brain (Croset et al., 2018) and the adult optic lobes (Konstantinides et al., 2018). Here we mined the whole adult brain data (Davie et al., 2018) to compare and investigate the cholinergic enriched factors that we identified in adult brains. The enrichment of these transcription factors (compared to GABAergic and glutamatergic neurons) is also observed in the scRNAseq data (Fig. 7A). Furthermore, we discovered that the cholinergic cells that these factors are expressed in are almost non-overlapping (Fig. 7B). This is an intriguing finding, as it suggests that these factors, if they are indeed acting to promote/maintain cholinergic fate, they are not acting together in this context. This scenario maybe different during development, where specification is occurring, and it will be interesting to test this when high coverage scRNAseq data is available for the third instar larval brain. We observed more differentially expressed transcription factors in the L3 larval stage (58) compared to the embryo (40) or adults (33). This may reflect the existence of both the functioning larval nervous system (built during embryogenesis) and the developing adult nervous system at this stage (Fig. 3). While both the embryo and larval data are similar on a global scale, Pol II occupancy and chromatin accessibility in the adult brain is less correlated (Fig. 2). It is currently unclear whether this is due to adult VNCs being absent from the profiling experiments, or differences between immature and fully mature neurons, such as overall lower transcriptional activity in adults. We have previously shown that global chromatin accessibility distribution in adult neurons is distinct from larval neurons (Aughey et al., 2018), which may account for some of these differences.
Apart from the neurotransmitter synthesis genes, the chromatin accessibility of the different neuronal types, at a given stage, is surprisingly similar, as demonstrated in embryos (Fig. S2B). The enriched accessibility is not just restricted to the gene bodies of the neurotransmitter genes, and peaks are present upstream (Gad1) and downstream (VGlut) (Fig. S2C), which are likely enhancers. Accessibility at the ChAT gene is clearly higher in cholinergic neurons at the embryonic and adult stages, however, in third instar larvae, the difference is less pronounced (Fig. S2C). This could reflect increased plasticity at this stage, possibly linked to the dramatic remodelling of larval neurons during metamorphosis (for a review, see Yaniv and Schuldiner, 2016), or that this accessibility across the types is due to non-specific expression of the VAChT gene that overlaps with ChAT at its 5′ end. While a subset of transcription factors display obvious contrasts in Pol II occupancy, the same transcription factors have no observable, or minor, differences in accessibility (Fig. S2D). This could be due to transcription factors being expressed at relatively lower levels and/or that they are only expressed in a subset of the cells, therefore the difference is less prominent.
Evidence is emerging for the roles of miRNAs in generating neuronal diversity, including the differentiation of taste receptor neurons in worms (Chang et al., 2004; Johnston and Hobert, 2005) and dopaminergic neurons in vertebrates (Kim et al., 2007). Here, we found the enriched expression of mir-184 in GABAergic cells (Fig. S8B), which is intriguing, as mir-184 has been shown to downregulate GABRA3 (GABA-A receptor) mRNA (possibly indirectly) in vertebrate cell lines (Luo et al., 2017), and may be a mechanism to help prevent GABAergic neurons self-inhibiting. Furthermore, mir-87 has enriched RNA polymerase II occupancy in cholinergic neurons (Fig. S8A), and when mutated causes larval locomotion defects in Drosophila (Picao-Osorio et al., 2017).
Acj6 is expressed in adult cholinergic neurons (Fig. 4B,C) (Lee and Salvaterra, 2002), whilst Ets65A-PA is expressed in non-cholinergic adult neurons (Fig. 4F,G). However, despite this, they bind a large number of common target genes (Fig. 6). This includes 20% (101/493) of all genes annotated for a role in ‘neuron projection development’ (GO:0031175). This is quite striking, especially as this is in the adult, where there is virtually no neurogenesis or axonogenesis. However, this may reflect dendritic re-modelling processes, or a requirement of neurons to continuously express transcription factors, even after development, to maintain their fate. The acj6 orthologues, unc-86 and Brn3a are both required to maintain the fate of specific cholinergic populations (Serrano-Saiz et al., 2018), and transcriptional networks that specific Tv1/Tv4 neurons in Drosophila are also required to maintain them in the adult (Eade et al., 2012). Therefore, the binding of Acj6 and Ets65A-PA to developmental genes and ChAT in adult neurons could be required for the continued activation (and repression) of genes governing neuronal identity. MAP kinase signalling genes are enriched in cholinergic neurons (Fig. 2C) and Ets65A-PA specifically binds MAP kinase signalling genes (Fig. 6), making it tempting to speculate that Ets65A-PA acts to repress cholinergic specific genes such as ChAT and MAP kinase genes. These Acj6 and Ets65A-PA data also emphasise the diverse set of neuronal differentiation genes a single transcription factor could regulate.
The precise synthesis and utilisation of neurotransmitters ensures proper information flow and circuit function in the nervous system. The mechanisms of specification are lineage specific, predominantly through the action of transcription factors. Here we have provided further insights into the complement of different transcription factors that regulate neurotransmitter identity throughout development. Furthermore, we identified the genomic binding of a known activator, and a candidate repressor, of cholinergic fate in the adult, emphasising the broad spectrum of neural identity genes that they could be regulating outside of neurotransmitter use. Given the strong evidence for conserved mechanisms controlling neurotransmitter specification, these data will be a useful resource for not just researchers using Drosophila but other model systems too. Continued work to elucidate the mechanisms, co-factors and temporal windows in which these factors are acting will be fundamental in gaining a comprehensive understanding of neurotransmitter specification.
MATERIALS AND METHODS
Drosophila lines
Lines used in this study are as follows:
w; dvGlut-GAL4 [MI04979]/CyO act-GFP, (Bloomington #60312). w;; ChAT-GAL4 [MI04508] / TM3 act GFP, (Bloomington #60317). w;; Gad1-GAL4 [MI09277] / TM3 actin GFP (Diao et al., 2015). w[*]; Mi{Trojan-lexA:QFAD.2}VGlut[MI04979-TlexA:QFAD.2]/CyO, P{Dfd-GMR-nvYFP}2, (Bloomington #60314). w[*]; Mi{Trojan-lexA:QFAD.0}ChAT[MI04508-TlexA:QFAD.0]/TM6B, Tb[1], (Bloomington #60319). w[*]; Mi{Trojan-lexA:QFAD.2}Gad1[MI09277-TlexA:QFAD.2]/TM6B, Tb[1], (Bloomington, #60324) (all obtained from M. Landgraf).
UAS-LT3-NDam, tub-GAL80ts; UAS-LT3-NDam-RNA Pol II (from Andrea Brand). Ets65A-RA/C/D/E-GAL4 [MI07721] (this study). apterous-GAL4; UAS-GFP (from F Jiménez Díaz-Benjumea). acj6-GAL4-UAS-mCD8-GFP/FM7c; Pin/CyO (from DJ Luginbuhl) (Lai et al., 2008). elavG4;; Mi{PT-GFSTF.2}Gad [MI09277]/TM3 actin-GFP (Bloomington, #59304). elavG4;; Mi{PT-GFSTF.0}ChAT [MI04508]/TM3 actin-GFP (Bloomington, #60288).
UAS-Dbx (Bloomington, #56826). UAS-apterous (from F Jiménez Díaz-Benjumea), UAS-collier (from F Jiménez Díaz-Benjumea), UAS-engrailed [E9] (from Andrea Brand). UAS-otp (Fly ORF #F000016). UAS-CG4328 (FlyORF, #F0019111). UAS-Dbx sh RNAi attP40 (VDRC #330536). UAS-ap sh RNAi attP40 (VDRC #330463). UAS-Ets65A-RA RNAi attP2 (Bloomington #41682). UAS-Ets65A-RA attP2 (this study). yw, hs-Flp 1; +; Dr/TM6B. yw, hs-Flp 1; +; UAS-Ets65A-RA. AyGal4, UAS-mCD8-GFP/(CyO); Cha lexAQF, mCherry /TM6B.
Generation of Ets65A and acj6 Targeted DamID lines
Details and sequences of all primers used for generating constructs are shown in the Supplemental Material. pUAST-LT3-NDam-acj6-RF and pUAST-LT3-NDam-Ets65A-RA were generated by PCR amplifying acj6-RF and Ets65A-RA from an embryonic cDNA library. The resulting PCR products were cloned into pUAST-LT3-Dam plasmid (Southall et al., 2013) with NotI and XhoI sites, using Gibson assembly.
acj6-RF FW: CATCTCTGAAGAGGATCTGGCCGGCGCAGATCTGCGGCCGCTCATGACAATGTCGATGTATTCGACGACGG, acj6-RF RV:
GTCACACCACAGAAGTAAGGTTCCTTCACAAAGATCCTCTAGATCAGTATCCAAATCCCGCCGAACCG.
Ets65A-RA FW: CTGAAGAGGATCTGGCCGGCGCAGATCTGCGGCCGCTCATGTACGAGAACTCCTGTTCGTATCAGACG,
Ets65A-RA RV: ACAGAAGTAAGGTTCCTTCACAAAGATCCTCTAGATCATGCGTAGTGGGGATAGCTGCTC.
Generation of Ets65A-RA-GAL4 line
Ets65A-RA/C/D/E-GAL4 was generated by inserting a GAL4 trap cassette into the MI07721 MiMIC line (Bloomington #43913) using the triplet donor in vivo system described in (Diao et al., 2015).
Generation of UAS-Ets65A-RA line
pUAST-attB-Ets65A-RA was generated by PCR amplifying Ets65A-RA from an embryonic cDNA library. The resulting PCR product was cloned into pUAST-attB with NotI and XhoI sites, using Gibson assembly. Ets65A-FW CATCTCTGAAGAGGATCTGCGAGATCTGCGGATGTACGAGAACTCCTGTTCGTATCAGACGG Ets65A-RA RV GTTCCTTCACAAAGATCCTCTAGAGGTACCC TCATGCGTAGTGGGGATAGCTGCTCAG.
TaDa for RNA-Pol II mapping
Crosses producing larvae with the following genotypes were allowed to lay eggs over a minimum of 2 days at 25°C before timed collections were performed: tub-GAL80ts/+; UAS-LT3-NDam/ ChAT-GAL4MI04508. tub-GAL80ts/+; UAS-LT3-NDam-RNA Pol II/ ChAT-GAL4MI04508. tub-GAL80ts/+; UAS-LT3-NDam/ Gad1-GAL4 MI09277. tub-GAL80ts/+; UAS-LT3-NDam-RNA Pol II/ Gad1-GAL4 MI09277. tub-GAL80ts/ dvGlut-GAL4 MI04979; UAS-LT3-NDam/ +. tub-GAL80ts/ dvGlut-GAL4 MI04979; UAS-LT3-NDam-RNA Pol II/ +.
First instar larvae samples
Crosses of the right genotype were allowed to lay old eggs for 2 h at 25°C in fly cages. Wet yeast and two drops of 10% acetic acid were added to apple juice plates to promote egg laying. Then, egg laying was done for 5 h at 25°C, apple juice plates containing those embryos were transferred to 29°C (permissive temperature) for 20 h. After this time, first instar larvae were collected and stored in 1× PBS. Samples were flash-frozen in dry ice, and stored at −80°C until the appropriate amount of tissue was enough to start the experiment. No selection for the right genotype was done, and 20 µl worth of volume of tissue was used as a proxy to determine the appropriate amount of material for each replicate. Three replicates were done for each experiment. With this husbandry protocol, the collected first instar larvae were around 12 h after larvae hatching (ALH), just before the first larval neurons are being generated, then providing the transcriptome of embryonic neurogenesis.
Third instar larvae samples
Crosses of the right genotypes were allowed to lay eggs for 6 h at 25°C in fly food vials. These vials were then transferred to 18°C (restrictive temperature) for 7 days. They were then moved to 29°C (permissive temperature) for 24 h. Wandering stage larvae, around 96 h ALH, were selected with a GFP scope for the right genotype. Larvae were dissected in 1× PBS, leaving the anterior half of the larvae partly dissected, containing the CNS, but removing the gut and all the fat tissue. Samples were flash-frozen in dry ice, and stored at −80°C until the appropriate amount of tissue was enough to start the experiment. 100 partly dissected CNS were used for each replicate. Three replicates were done for each experiment.
Adult samples
Crosses of the right genotypes were allowed to lay eggs for 2 days at 18°C in fly food vials. Vials containing those eggs were kept at 18°C (restrictive temperature) until adult flies eclosed. They were then kept at 18°C for 5−10 days. After that, they were selected according for the right genotype, and transferred to 29°C (permissive temperature) for 24 h. Then, adult flies were flash-frozen in dry ice, and stored at −80°C. Around 50 fly heads were used for each replicate. Three replicates were done for each experiment.
When preparing the tissue to be used, neither larvae nor flies were sex sorted. It has been recently reported that transcriptomes from males and females, obtained with a cell specific driver combination expressed in neurons in the adult optic lobe, do not present major differences in their transcriptomes. Only a small number of genes, known sex-specific genes showed differences between sexes (Davis et al., 2018 preprint).
Our DamID protocol was based on Southall et al. (2013), and Marshall et al. (2016). Briefly, DNA was extracted using Qiagen DNeasy kit, and a minimum of 3 µg of DNA was precipitated for first instar larvae, 6 μg for third instar larvae, and 2.5 µg for adult samples. DNA was digested with DpnI overnight at 37°C. The next morning, 0.5 µl of DpnI was added for 1 h extra incubation, followed by DpnI heat inactivation (20 min, 80°C). Either Advantage cDNA polymerase, or Advantage 2 cDNA polymerase mix, 50×, Clontech, were used in PCR amplification. Enzymes Sau3AI or AlwI were used to remove DamID adaptors, from sonicated DNA.
Libraries were sequenced using Illumina HiSeq single-end 50 bp sequencing. Three replicates were performed for each experiment. A minimum of 25 million reads were obtained from the first instar larvae samples, 30 million reads from the third instar larvae, and 9 million reads from the adults' samples.
TaDa for identification of acj6 and Ets65A-RA binding sites
Crosses producing larvae with the following genotypes were allowed to lay eggs over a minimum of 2 days at 25°C:
acj6-GAL4-UAS-GFP; tub-GAL80ts/+; UAS-LT3-NDam/+. acj6-GAL4-UAS-GFP; tub-GAL80ts/+; UAS-LT3-NDam-acj6-RF/+. tub-GAL80ts/+; UAS-LT3-NDam/ Ets65A-RA-GAL4MI07721. tub-GAL80ts/+; UAS-LT3-NDam-Ets65A-RA/ Ets65A-RA-GAL4MI07721.
Crosses of the right genotypes were allowed to lay eggs for 2 days at 18°C in fly food vials. Vials containing those eggs were kept at 18°C (restrictive temperature) until adult flies eclosed. They were then kept at 18°C for around 10 days. Then, they were transferred to 29°C (permissive temperature) for 24 h, selected according for the right genotype, flash-frozen in dry ice, and stored at −80°C. A minimum of 150 fly heads were used for each replicate. Two replicates were done for each experiment.
The DamID protocol used for these samples is the same as described above, with minor changes, 6 µg of DNA were precipitated, Bioline Polymerase was used in the PCR amplification, and only AlwI was used to remove adaptors from sonicated DNA. Libraries were sequenced using Illumina HiSeq single-end 50 bp sequencing. Two replicates were acquired for each experiment. A minimum of 10 million reads were obtained from these samples.
TaDa data analysis
Sequencing data for TaDa and CATaDa were mapped back to release 6.03 of the Drosophila genome using a previously described pipeline (Aughey et al., 2018; Marshall and Brand, 2015). Transcribed genes (defined by Pol II occupancy) were identified using a Perl script described in Mundorf et al. (2019) based on one developed by Southall et al. (2013) (available at https://github.com/tonysouthall/Dam-RNA_POLII_analysis). Drosophila genome annotation release 6.11 was used, with 1% FDR and 0.2 log2 ratio thresholds. To compare data sets, log2 ratios were subtracted, in this case, producing three replicate comparison files (as three biological replicates were performed). These data were then analysed as described above to identify genes with significantly different Pol II occupancy. Due to the presence of negative log2 ratios in DamID experiments, these genes were filtered to check that any significantly enriched genes were also bound by Pol II in the experiment of interest (numerator data set). A gene list was generated from the transcript data using the values from the associated transcript with the most significant FDR. Correlation values (Fig. 2) were visualised using Morpheus (https://software.broadinstitute.org/morpheus/). The transition plot (Fig. 3) was generated in R using the transitionPlot function from the Gmisc R package (http://gforge.se/). Enrichment GO analysis was performed using the R package clusterProfiler (Yu et al., 2012).
For acj6 and Ets65A-RA TaDa, peaks were called and mapped to genes using a custom Perl program (available at https://github.com/tonysouthall/Peak_calling_DamID). In brief, a FDR was calculated for peaks (formed of two or more consecutive GATC fragments) for the individual replicates. Then each potential peak in the data was assigned a FDR. Any peaks with less than a 0.01% FDR were classified as significant. Significant peaks present in all replicates were used to form a final peak file. Any gene (genome release 6.11) within 5 kb of a peak (with no other genes in between) was identified as a potentially regulated gene.
For studying transcription factors specifically, we filtered the differentially expressed genes for known/predicted transcription factors using the FlyTF database (Pfreundt et al., 2010).
Extracting gene specific data from scRNAseq data
Data for specific genes were extracted from the adult scRNAseq matrix file (Davie et al., 2018) using the following Perl code:
#!usr/bin/perl #parse_scRNAseq_data use warnings;
my $file = ‘mtx_file.mtx’; #path to the scRNAseq matrix file
print “\nEnter gene number to extract for - see gene index file\n\n”; $genenum = < STDIN >; chomp $genenum; chomp $genenum;
open (OUTPUT, ‘> scRNAseq_data_for_gene’.“$genenum”.‘.txt’);
open my $fh, ‘<’, $file or die $!; while(<$fh>){@col = split(/\s/,$_); if($genenum == $col[0]) {print OUTPUT “$col[0]\t$col[1]\t$col[2]\n”;}} exit;
Cells with a transcript count (for the given gene) of less than three were excluded for further analysis.
Immunostaining and imaging
Third instar larval CNS or adult brains were dissected in 1x PBS. They were fixed in 4% formaldehyde (methanol free) 0.1% Triton X-100 PBS (PBST), for 30 min at room temperature. Samples were then rinsed twice with 0.1% PBST, and washed four times for 1 h with 0.1% PBST. 5% normal goat serum in 0.1% PBST was used as a blocking agent for 1 h at room temperature. Brains were then incubated overnight at 4°C with primary antibodies in 5% normal goat serum in 0.1% PBST. The primary antibodies used were: anti-chicken-GFP (Abcam #13970, 1:2000), and anti-rabbit-DsRed (Clontech #632496, 1:500). Brains were rinsed twice with 0.1% PBST, and washed four times with 0.1% PBST for 1 h. Secondary antibodies were diluted in 5% normal goat serum in 0.1% PBST and incubated with the brains for 1 h at room temperature. The secondary antibodies used were: anti-chicken-Alexa 488 (Thermo Fisher Scientific #A11039, 1:500), and anti-rabbit-Alexa 546 (Thermo Fisher Scientific #A11010, 1:500). Samples were then rinsed twice with 0.1% PBST, and washed four times for 1 h. Brains were mounted on glass cover slides in Vectashield (Vector laboratories). All incubations and washes were performed in a rotator. After dissection of first instar larvae CNS, they were placed in a polylisine coated microscope slide, where we performed all the incubations. Both experimental CNS and wild-type CNS were placed on the same slide. For all the immunostaining experiments, a minimum of five brains were dissected and visualised. Images were acquired using a Zeiss LSM 510 confocal microscope and edited using Fiji/ImageJ.
Supplementary Material
Acknowledgements
We would like to thank Matthias Landgraf, Eva Higginbotham, and Gabriel Aughey for feedback and advice on this project. We would like to thank Matthias Landgraf, Holly Ironfield, Benjamin White, Andrea Brand, Fernando J. Díaz-Benjumea, David John Luginbuhl, for providing fly stocks. For other fly stocks, we also thank Bloomington Drosophila Stock Center (NIH P40OD018537), the Vienna Drosophila Resource Center (VDRC, www.vdrc.at), and the FlyORF, Zurich ORFeome Project (https://flyorf.ch/).
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: A.E.G., T.D.S.; Methodology: A.E.G., T.D.S.; Software: T.D.S.; Validation: A.E.G., T.D.S.; Formal analysis: A.E.G., T.D.S.; Investigation: A.E.G., A.H., E.W., L.W.L., T.D.S.; Resources: A.E.G., A.H., E.W., T.D.S.; Data curation: A.E.G., T.D.S.; Writing - original draft: A.E.G., T.D.S.; Writing - review & editing: A.E.G., A.H., E.W., L.W.L., T.D.S.; Visualization: A.E.G., T.D.S.; Supervision: A.E.G., T.D.S.; Project administration: T.D.S.; Funding acquisition: T.D.S.
Funding
This work was funded by Wellcome Trust Investigator grant [104567/Z/14/Z to T.D.S.].
Data availability
All raw sequence files and processed files have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (accession number GSE139888).
Supplementary information
Supplementary information available online at http://bio.biologists.org/lookup/doi/10.1242/bio.052928.supplemental
References
- Aguilar J. I., Dunn M., Mingote S., Karam C. S., Farino Z. J., Sonders M. S., Choi S. J., Grygoruk A., Zhang Y., Cela C. et al. (2017). Neuronal depolarization drives increased dopamine synaptic vesicle loading via VGLUT. Neuron 95, 1074-1088.e1077. 10.1016/j.neuron.2017.07.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altun-Gultekin Z., Andachi Y., Tsalik E. L., Pilgrim D., Kohara Y. and Hobert O. (2001). A regulatory cascade of three homeobox genes, ceh-10, ttx-3 and ceh-23, controls cell fate specification of a defined interneuron class in C. elegans. Development 128, 1951-1969. [DOI] [PubMed] [Google Scholar]
- Andreae L. C. and Burrone J. (2018). The role of spontaneous neurotransmission in synapse and circuit development. J. Neurosci. Res. 96, 354-359. 10.1002/jnr.24154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aughey G. N., Estacio Gomez A., Thomson J., Yin H. and Southall T. D. (2018). CATaDa reveals global remodelling of chromatin accessibility during stem cell differentiation in vivo. Elife 7, e32341 10.7554/eLife.32341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aughey G. N., Cheetham S. W. and Southall T. D. (2019). DamID as a versatile tool for understanding gene regulation. Development 146, dev173666 10.1242/dev.173666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beguin S., Crepel V., Aniksztejn L., Becq H., Pelosi B., Pallesi-Pocachard E., Bouamrane L., Pasqualetti M., Kitamura K., Cardoso C. et al. (2013). An epilepsy-related ARX polyalanine expansion modifies glutamatergic neurons excitability and morphology without affecting GABAergic neurons development. Cereb. Cortex 23, 1484-1494. 10.1093/cercor/bhs138 [DOI] [PubMed] [Google Scholar]
- Chang S., Johnston R. J. Jr, Frøkjaer-Jensen C., Lockery S. and Hobert O. (2004). MicroRNAs act sequentially and asymmetrically to control chemosensory laterality in the nematode. Nature 430, 785-789. 10.1038/nature02752 [DOI] [PubMed] [Google Scholar]
- Cheng L., Arata A., Mizuguchi R., Qian Y., Karunaratne A., Gray P. A., Arata S., Shirasawa S., Bouchard M., Luo P. et al. (2004). Tlx3 and Tlx1 are post-mitotic selector genes determining glutamatergic over GABAergic cell fates. Nat. Neurosci. 7, 510-517. 10.1038/nn1221 [DOI] [PubMed] [Google Scholar]
- Coleman B., Topalidou I. and Ailion M. (2018). Modulation of Gq-Rho Signaling by the ERK MAPK Pathway Controls Locomotion in Caenorhabditis elegans. Genetics 209, 523-535. 10.1534/genetics.118.300977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croset V., Treiber C. D. and Waddell S. (2018). Cellular diversity in the Drosophila midbrain revealed by single-cell transcriptomics. Elife 7, e34550 10.7554/eLife.34550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davie K., Janssens J., Koldere D., De Waegeneer M., Pech U., Kreft L., Aibar S., Makhzami S., Christiaens V., Bravo González-Blas C. et al. (2018). A single-cell transcriptome atlas of the aging Drosophila brain. Cell 174, 982-998.e920. 10.1016/j.cell.2018.05.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis F. P., Nern A., Picard S., Reiser M. B., Rubin G. M., Eddy S. R. and Henry G. L. (2020). A genetic, genomic, and computational resource for exploring neural circuit function. Elife 9, e50901 10.7554/eLife.50901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao F., Ironfield H., Luan H., Diao F., Shropshire W. C., Ewer J., Marr E., Potter C. J., Landgraf M. and White B. H. (2015). Plug-and-play genetic access to drosophila cell types using exchangeable exon cassettes. Cell Rep 10, 1410-1421. 10.1016/j.celrep.2015.01.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggan A., Ma C. and Chalfie M. (1998). Regulation of touch receptor differentiation by the Caenorhabditis elegans mec-3 and unc-86 genes. Development 125, 4107-4119. [DOI] [PubMed] [Google Scholar]
- Eade K. T., Fancher H. A., Ridyard M. S. and Allan D. W. (2012). Developmental transcriptional networks are required to maintain neuronal subtype identity in the mature nervous system. PLoS Genet. 8, e1002501 10.1371/journal.pgen.1002501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flames N. and Hobert O. (2009). Gene regulatory logic of dopamine neuron differentiation. Nature 458, 885-889. 10.1038/nature07929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogarty M., Grist M., Gelman D., Marin O., Pachnis V. and Kessaris N. (2007). Spatial genetic patterning of the embryonic neuroepithelium generates GABAergic interneuron diversity in the adult cortex. J. Neurosci. 27, 10935-10946. 10.1523/JNEUROSCI.1629-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goold C. P. and Davis G. W. (2007). The BMP ligand Gbb gates the expression of synaptic homeostasis independent of synaptic growth control. Neuron 56, 109-123. 10.1016/j.neuron.2007.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gummalla M., Maeda R. K., Castro Alvarez J. J., Gyurkovics H., Singari S., Edwards K. A., Karch F. and Bender W. (2012). abd-A regulation by the iab-8 noncoding RNA. PLoS Genet. 8, e1002720 10.1371/journal.pgen.1002720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herculano-Houzel S. (2016). The Human Advantage: A New Understanding of How Our Brain Became Remarkable. Cambridge, Massachusetts: The MIT Press. [Google Scholar]
- Hobert O. (2008). Regulatory logic of neuronal diversity: terminal selector genes and selector motifs. Proc. Natl. Acad. Sci. USA 105, 20067-20071. 10.1073/pnas.0806070105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. (2016). A map of terminal regulators of neuronal identity in Caenorhabditis elegans. Wiley Interdiscip Rev. Dev. Biol. 5, 474-498. 10.1002/wdev.233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y., Hoskins R. and Horvitz H. R. (1994). Control of type-D GABAergic neuron differentiation by C. elegans UNC-30 homeodomain protein. Nature 372, 780-783. 10.1038/372780a0 [DOI] [PubMed] [Google Scholar]
- Johnston R. J. Jr and Hobert O. (2005). A novel C. elegans zinc finger transcription factor, lsy-2, required for the cell type-specific expression of the lsy-6 microRNA. Development 132, 5451-5460. 10.1242/dev.02163 [DOI] [PubMed] [Google Scholar]
- Kim N. C. and Marques G. (2012). The Ly6 neurotoxin-like molecule target of wit regulates spontaneous neurotransmitter release at the developing neuromuscular junction in Drosophila. Dev. Neurobiol. 72, 1541-1558. 10.1002/dneu.22021 [DOI] [PubMed] [Google Scholar]
- Kim J., Inoue K., Ishii J., Vanti W. B., Voronov S. V., Murchison E., Hannon G. and Abeliovich A. (2007). A MicroRNA feedback circuit in midbrain dopamine neurons. Science 317, 1220-1224. 10.1126/science.1140481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinides N., Kapuralin K., Fadil C., Barboza L., Satija R. and Desplan C. (2018). Phenotypic convergence: distinct transcription factors regulate common terminal features. Cell 174, 622-635.e613. 10.1016/j.cell.2018.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratsios P., Stolfi A., Levine M. and Hobert O. (2011). Coordinated regulation of cholinergic motor neuron traits through a conserved terminal selector gene. Nat. Neurosci. 15, 205-214. 10.1038/nn.2989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacin H., Zhu Y., Wilson B. A. and Skeath J. B. (2009). dbx mediates neuronal specification and differentiation through cross-repressive, lineage-specific interactions with eve and hb9. Development 136, 3257-3266. 10.1242/dev.037242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacin H., Chen H. M., Long X., Singer R. H., Lee T. and Truman J. W. (2019). Neurotransmitter identity is acquired in a lineage-restricted manner in the Drosophila CNS. Elife 8 10.7554/eLife.43701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai S. L., Awasaki T., Ito K. and Lee T. (2008). Clonal analysis of Drosophila antennal lobe neurons: diverse neuronal architectures in the lateral neuroblast lineage. Development 135, 2883-2893. 10.1242/dev.024380 [DOI] [PubMed] [Google Scholar]
- Landgraf M., Bossing T., Technau G. M. and Bate M. (1997). The origin, location, and projections of the embryonic abdominal motorneurons of Drosophila. J. Neurosci. 17, 9642-9655. 10.1523/JNEUROSCI.17-24-09642.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. H. and Salvaterra P. M. (2002). Abnormal chemosensory jump 6 is a positive transcriptional regulator of the cholinergic gene locus in Drosophila olfactory neurons. J. Neurosci. 22, 5291-5299. 10.1523/JNEUROSCI.22-13-05291.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Liu S. and Yao K. (2017). Transcriptome-wide investigation of mRNA/circRNA in miR-184 and its r.57c>u mutant type treatment of human lens epithelial cells. Mol. Ther. Nucleic Acids 7, 71-80. 10.1016/j.omtn.2017.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K., Zheng S. and Zuo Z. (2006). The transcription factor regulatory factor X1 increases the expression of neuronal glutamate transporter type 3. J. Biol. Chem. 281, 21250-21255. 10.1074/jbc.M600521200 [DOI] [PubMed] [Google Scholar]
- Marshall O. J. and Brand A. H. (2015). damidseq_pipeline: an automated pipeline for processing DamID sequencing datasets. Bioinformatics 31, 3371-3373. 10.1093/bioinformatics/btv386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall O. J., Southall T. D., Cheetham S. W. and Brand A. H. (2016). Cell-type-specific profiling of protein-DNA interactions without cell isolation using targeted DamID with next-generation sequencing. Nat. Protoc. 11, 1586-1598. 10.1038/nprot.2016.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundorf J., Donohoe C. D., McClure C. D., Southall T. D. and Uhlirova M. (2019). Ets21c governs tissue renewal, stress tolerance, and aging in the drosophila intestine. Cell Rep 27, 3019-3033.e3015. 10.1016/j.celrep.2019.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel T. and Hobert O. (2017). Coordinated control of terminal differentiation and restriction of cellular plasticity. Elife 6, e24100 10.7554/eLife.24100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfreundt U., James D. P., Tweedie S., Wilson D., Teichmann S. A. and Adryan B. (2010). FlyTF: improved annotation and enhanced functionality of the Drosophila transcription factor database. Nucleic Acids Res. 38, D443-D447. 10.1093/nar/gkp910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picao-Osorio J., Lago-Baldaia I., Patraquim P. and Alonso C. R. (2017). Pervasive behavioral effects of MicroRNA regulation in Drosophila. Genetics 206, 1535-1548. 10.1534/genetics.116.195776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipes G. C., Lin Q., Riley S. E. and Goodman C. S. (2001). The Beat generation: a multigene family encoding IgSF proteins related to the Beat axon guidance molecule in Drosophila. Development 128, 4545-4552. [DOI] [PubMed] [Google Scholar]
- Schmid A., Chiba A. and Doe C. Q. (1999). Clonal analysis of Drosophila embryonic neuroblasts: neural cell types, axon projections and muscle targets. Development 126, 4653-4689. [DOI] [PubMed] [Google Scholar]
- Schuurmans C., Armant O., Nieto M., Stenman J. M., Britz O., Klenin N., Brown C., Langevin L. M., Seibt J., Tang H. et al. (2004). Sequential phases of cortical specification involve Neurogenin-dependent and -independent pathways. EMBO J. 23, 2892-2902. 10.1038/sj.emboj.7600278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Saiz E., Poole R. J., Felton T., Zhang F., De La Cruz E. D. and Hobert O. (2013). Modular control of glutamatergic neuronal identity in C. elegans by distinct homeodomain proteins. Cell 155, 659-673. 10.1016/j.cell.2013.09.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Saiz E., Leyva-Diaz E., De La Cruz E. and Hobert O. (2018). BRN3-type POU homeobox genes maintain the identity of mature postmitotic neurons in nematodes and mice. Curr. Biol. 28, 2813-2823.e2. 10.1016/j.cub.2018.06.045 [DOI] [PubMed] [Google Scholar]
- Soba P., Zhu S., Emoto K., Younger S., Yang S. J., Yu H. H., Lee T., Jan L. Y. and Jan Y. N. (2007). Drosophila sensory neurons require Dscam for dendritic self-avoidance and proper dendritic field organization. Neuron 54, 403-416. 10.1016/j.neuron.2007.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southall T. D., Gold K. S., Egger B., Davidson C. M., Caygill E. E., Marshall O. J. and Brand A. H. (2013). Cell-type-specific profiling of gene expression and chromatin binding without cell isolation: assaying RNA Pol II occupancy in neural stem cells. Dev. Cell 26, 101-112. 10.1016/j.devcel.2013.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trudeau L. E. and El Mestikawy S. (2018). Glutamate cotransmission in cholinergic, GABAergic and monoamine systems: contrasts and commonalities. Front. Neural Circuits 12, 113 10.3389/fncir.2018.00113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Kloot W. G. and Robbins J. (1959). The effects of gamma-aminobutyric acid and picrotoxin on the junctional potential and the contraction of crayfish muscle. Experientia 15, 35-36. 10.1007/BF02157093 [DOI] [PubMed] [Google Scholar]
- van Steensel B. and Henikoff S. (2000). Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat. Biotechnol. 18, 424-428. 10.1038/74487 [DOI] [PubMed] [Google Scholar]
- Waite M. R., Skidmore J. M., Billi A. C., Martin J. F. and Martin D. M. (2011). GABAergic and glutamatergic identities of developing midbrain Pitx2 neurons. Dev. Dyn. 240, 333-346. 10.1002/dvdy.22532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenick A. S. and Hobert O. (2004). Genomic cis-regulatory architecture and trans-acting regulators of a single interneuron-specific gene battery in C. elegans. Dev. Cell 6, 757-770. 10.1016/j.devcel.2004.05.004 [DOI] [PubMed] [Google Scholar]
- Westmoreland J. J., McEwen J., Moore B. A., Jin Y. and Condie B. G. (2001). Conserved function of Caenorhabditis elegans UNC-30 and mouse Pitx2 in controlling GABAergic neuron differentiation. J. Neurosci. 21, 6810-6819. 10.1523/JNEUROSCI.21-17-06810.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaniv S. P. and Schuldiner O. (2016). A fly's view of neuronal remodeling. Wiley Interdiscip. Rev. Dev. Biol. 5, 618-635. 10.1002/wdev.241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L. G., Han Y. and He Q. Y. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284-287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Bhattacharya A., Nelson J. C., Abe N., Gordon P., Lloret-Fernandez C., Maicas M., Flames N., Mann R. S., Colon-Ramos D. A. et al. (2014). The LIM and POU homeobox genes ttx-3 and unc-86 act as terminal selectors in distinct cholinergic and serotonergic neuron types. Development 141, 422-435. 10.1242/dev.099721 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.