Abstract
Emerging evidence has demonstrated the feasibility of circulating miRNAs as robust non-invasive biomarkers for the diagnosis in colorectal cancer. The use of circulating miRNAs for the early detection of colorectal cancer (CRC) is of particular interest as it can offer a potential complementary approach to screening colonoscopy. However, the development of circulating miRNAs as “liquid biopsy” biomarkers for development into clinical screening tests has been hampered by several issues. In this article, we summarize the status of this field for the clinical utilization of miRNA biomarkers as liquid biopsies in colorectal cancer (CRC) and discuss their applications as screening tests for patients with colorectal adenoma (CRA) and CRC. Herein, we undertook a systematic search for citations in PubMed and the Cochrane Database from January 1, 2002 through December 31, 2017 as electronic sources for this study. All published studies were screened with no restriction on language, date, or country. We used database-specific combinations of the following index terms and text words, including: microRNA, colorectal cancer, serum, plasma, and exosomes. Based upon these searches, we summarize the progress and salient features of the current state of knowledge of miRNA diagnostic biomarkers in CRC, and focuses on the articles that attempt to optimize ideal methodologies to further advance their as liquid biopsies for clinical use. We conclude that the field of noncoding RNAs, particularly for the clinical use of miRNAs as liquid biopsy assays is maturing rapidly, and it is highly promising that these genomic signatures will likely be developed into clinically-viable tests for the early detection and clinical management of patients with colorectal cancer in the not so distant future.
Keywords: MicroRNA, diagnosis, biomarker, screening, colorectal cancer
1.1. INTRODUCTION
Colorectal cancer (CRC) is a significant healthcare problem worldwide and remains the second-leading cause of cancer-related deaths in the United States, with an estimated 50,000 deaths annually[1]. In spite of the progress made in terms of better strategies for screening and the development of targeted therapeutics for this malignancy, approximately 50% of CRC patients still die as a consequence of late detection of advanced disease with localized or distant metastases[2]. Most CRCs develop from premalignant colorectal adenomas (CRAs) that have accumulated mutations and epigenetic alterations in a stepwise fashion and eventually become malignant. Although current screening tests help reduce mortality, compliance with these approaches has been largely inadequate. Reasons for poor patient compliance include insufficient sensitivity and specificity of these screening tests, expense associated with these modalities, and their invasive nature[3]. These facts highlight and underscore a need for the identification and development of robust and inexpensive screening biomarkers that are minimally invasive, and facilitate earlier detection of CRC.
MicroRNAs (miRNAs) are an abundant class of small non-coding RNA molecules, 18 to 25 nucleotides in length, that regulate gene expression at the post-transcriptional level by promoting messenger RNA (mRNA) degradation or blocking their translation into functional proteins[4]. MiRNAs are frequently dysregulated during the pathogenesis of human cancers[5, 6], and are present in a wide variety of clinical specimens (including blood, saliva, urine and feces). They can be easily extracted from formalin-fixed paraffin-embedded tissue specimens, where these exist in a remarkably stable state. This has opened a new research frontier for the identification of clinically useful biomarkers in patients with cancer. However, the development of circulating miRNAs as clinical screening tests is currently hampered by several technical issues. Herein, we summarize the state of the art for the clinical utilization of miRNA biomarkers as liquid biopsies in CRC and discuss their applications as screening tests for patients with CRAs and CRCs.
1.2. METHODS
1.2.1. Data sources and literature search strategy
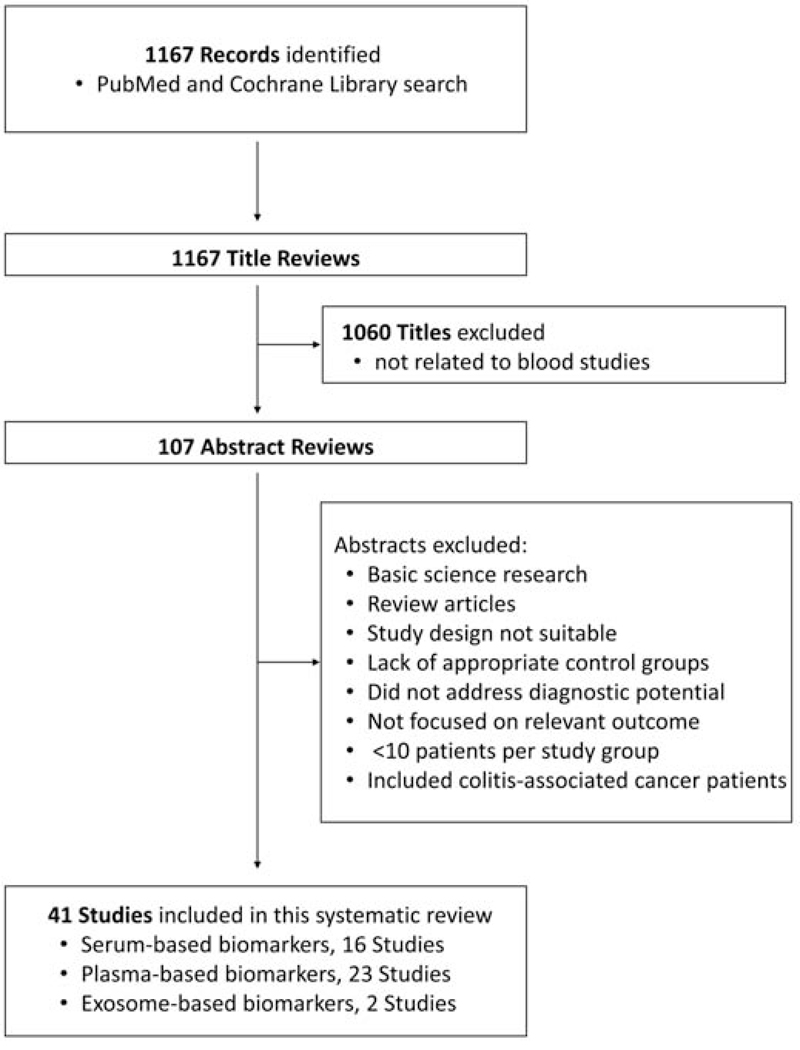
We searched for citations in PubMed and the Cochrane Database as electronic sources for this study (Figure 1). Each database was searched for entries from January 1, 2002 through December 31, 2017, and all published studies were screened with no restriction on language, date, or country. The year 2002 was chosen as the starting point because this was the first year in which miRNAs were implicated in cancer[5]. We used database-specific combinations of the following index terms and text words, including: microRNA, colorectal cancer, serum, plasma, and exosome. No search filter was applied for the study type.
Figure 1. Systematic literature search flow chart.
1.2.2. Study eligibility and quality assessment-inclusion and exclusion criteria
Two members of the research team (Y.T. and Y.O.) reviewed manuscript titles and abstracts. Studies were subjected to full-text review by both reviewers if one of the reviewers deemed it pertinent for inclusion. Keyword searches were run a second time before retrieving the final collection of articles. Finally, both reviewers discussed the reasons for initial inclusion or exclusion of all articles until a consensus was reached.
We were particularly interested in studies addressing diagnostic biomarker potential, minimally invasive biomarkers, extracellular miRNAs, and the clinical feasibility for use as screening biomarkers in colorectal neoplasia. Based on this approach, the inclusion criteria were: prospective or retrospective studies, adult human participants, miRNA studies, research on CRA and CRC, and English language. The exclusion criteria included the following: the description included only basic functional data, fewer than 10 patients in the study group, the sole description of prognostic biomarker potential, patients with colitis-associated cancer, case reports and series, editorials, literature reviews, and meta-analyses.
1.2.3. Data extraction and analysis
Study characteristics that were extracted from each study included the names of the target miRNAs, enrolled patient numbers (cases and controls), the study population (disease type), accuracy (sensitivity, specificity and area under the curve [AUC]), and the type of dysregulation (up- or down-regulated expression). Relevant findings were extracted by two members of this research team (Y.T. and Y.O.) and verified by the senior author (A.G.).
In characterizing research on the potential for a miRNA to serve as a diagnostic liquid biopsy for CRC patients, we sought to assess the quality of the studies included in our review. Both reviewers independently assessed the quality of studies using the Newcastle-Ottawa Scale[7]. This scale is comprised of eight specific criteria for evaluating the quality of observational cohort studies in terms of selection, comparability and outcome. Zero to seven stars were awarded for the methodologic quality of case selection, comparability of cohorts, and measurement outcomes. Studies received an overall quality rating of high, moderate, or low based on the Newcastle-Ottawa scale. An award of 6 or 7 stars was considered a high methodologic quality, while a score of ≤4 was considered low quality.
1.3. RESULTS
1.3.1. Systematic Review
The results of the systematic review are shown in Figure 1. A total of 1167 potential citations were identified initially through database retrieval. After screening titles and abstracts, we excluded 1060 articles that did not focus on blood-based biomarkers. We reviewed the full-text versions of the remaining 107 articles and excluded an additional 66 articles. Therefore, our analysis eventually included 41 articles in this systematic review (including 16 serum-based biomarkers; 23 plasma-based biomarkers; and 2 exosome-based biomarkers)
1.3.2. Potential use of miRNAs as liquid biopsy biomarkers
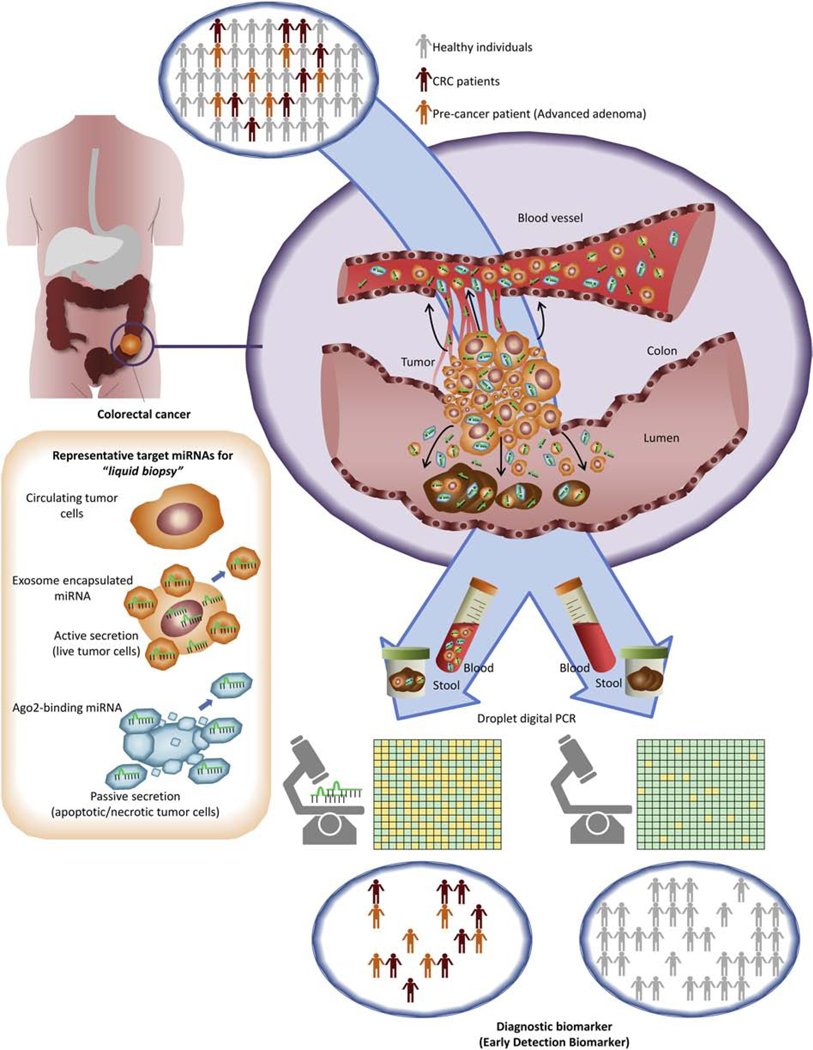
The discovery of extracellular miRNAs in biological fluids has opened a new era in the field of cancer liquid biopsy research (Figure 2). For the development of a clinically-meaningful biomarker, various steps taken during sample collection and subsequent long-term storage of specimens (months or years) are critical, and must be considered as potential sources of bias that can confound test results [8]. As a result, stability of the target molecule is absolutely essential for avoiding analytical variation between samples and independent laboratories – an important requisite for data reproducibility and universal adoption of specific biomarkers. The pioneering observation that miRNAs are quite resistant to RNase-mediated degradation and are largely preserved in biological bodily fluids was first made in 2008 by Chim and colleagues, wherein the investigators successfully identified placental miRNAs in maternal plasma at readily detectable concentrations[9]. Subsequently, it was revealed that several tumor-associated miRNAs could be detected in serum samples, and might offer a less invasive option for cancer diagnosis and prognosis in patients with diffuse B-cell lymphoma[10]. To further strengthen the argument favoring the stability of miRNAs in blood, a seminal study established the stability of endogenous miRNAs in human plasma and serum when specimens were exposed to various extreme experimental conditions[11]. MiRNAs were shown to remain stable for at least 24 hours at room temperature and even when subjected to eight freeze-thaw cycles. Based on these novel characteristics, accumulating studies have demonstrated that circulating miRNAs may serve as liquid biopsy biomarkers with relatively high sensitivity and specificity for the early detection of various gastrointestinal cancers, including esophageal, gastric, pancreatic, hepatobiliary, and colorectal cancer [12].
Figure 2. The biogenesis of extracellular miRNAs and their use as liquid biopsy biomarkers in CRC.
There are several opportunities for the development of an ideal screening test for CRC. The challenges for an optimal test include minimally invasive access to appropriate liquid specimens, a simple test with high sensitivity and specificity, and demonstrated improved performance over conventional screening tests (fecal immunochemical tests, fecal DNA, etc). Circulating miRs may provide this opportunity. After miRNA biogenesis in the nucleus and cytoplasm, miRNAs are released from tumor cells and are stable in extracellular spaces in various forms, such as exosomes, apoptotic bodies, shed microvesicles, high density lipoprotein particles, or Ago-2 bounded forms. The production of large numbers of miRNA molecules by tumor cells provides the best opportunity to detect asymptomatic cancer. MiRNA dysregulation measured in feces and blood might be exploited for use as non-invasive biomarkers to identify patients with CRAs and early CRCs.
1.3.3. Clinical feasibility of utilizing circulating miRNAs in blood as an early diagnostic tool in CRC
The evidence gathered thus far shows a remarkable degree of tissue specificity for aberrantly expressed miRNA in different types of human cancers. This finding, together with the fact that miRNAs exist in a highly stable manner in various body fluids, has invigorated a flurry of research interest in exploiting these small non-coding RNAs as promising bodily fluids-based, less-invasive, diagnostic biomarkers for colorectal neoplasia (Table 1 and 2). In various bodily fluids, feces have been widely used as non-invasive molecular screening biomarkers in CRC. Till date, several studies have attempted to identify the potential of fecal miRNAs as early diagnostic signatures in CRC[13–16]; however, in spite of all the positive development, compared with blood specimens, fecal miRNA suffer from several key limitations which must first be overcome. These shortcomings include: complexity of the fecal specimen, standardization of protocols for sample preparation and RNA extraction, and the use of appropriate normalization controls. Based on these complex characteristics of fecal specimens, a miRNA-based test using blood specimens seems more appropriate, as it can be deployed in a facile, easy-to-assess manner in the clinic, and relatively inexpensively. Accordingly, in this article, we focused on the diagnostic potential of circulating miRNAs in CRC, and the key studies are highlighted in the following sections.
Table 1.
Diagnostic miRNA markers using plasma specimens from patients with colorectal adenomas or CRCs
| miRNA |
Sample size |
Adenoma/CRC | Sensitivity (%) | Specificity (%) | AUC (95% CI) | Dysregulation | Quality assessment | References | |
|---|---|---|---|---|---|---|---|---|---|
| Cases | Control | ||||||||
| Plasma-based biomarker | |||||||||
| miR-17-3p | 90 | 50 | CRC | 64 | 70 | 0.72 (0.63–0.80) | Up-reulation | high | [18] |
| miR-18 | 78 | 86 | CRC | 73 | 79 | 0.8 | moderate | [53] | |
| miR-20a | 100 | 78 | CRC | 46 | 73.4 | 0.59 (0.51–0.67) | Up-regulation | moderate | [54] |
| miR-21 | 20 | 20 | CRC | 90 | 90 | 0.91 | Up-regulation | high | [27] |
| miR-21 | 29 | 29 | CRC | - | - | 0.65 | Up-regulation | moderate | [55] |
| miR-21 | 49 | 49 | CRC | 76.2 | 93.2 | 0.88 | Up-regulation | moderate | [56] |
| miR-24 | 111 | 130 | CRC | 78.4 | 83.9 | 0.84 (0.79–0.89) | Down-regulation | low | [57] |
| miR-26a-5p | 61 | 24 | CRC | - | - | 0.67 | Down-regulation | low | [58] |
| miR-29a | 100 | 59 | CRC | 69 | 89.1 | 0.84 (0.79–0.90) | Up-regulation | moderate | [19] |
| miR-29a | 37 | 59 | Adenoma | 62.2 | 84.7 | 0.77 (0.67–0.87) | Up-regulation | moderate | [19] |
| miR-34a | 37 | 20 | CRC | - | - | 0.71 (0.61–0.82) | Up-regulation | moderate | [59] |
| miR-92a | 90 | 50 | CRC | 89 | 70 | 0.89 (0.83–0.94) | Up-regulation | high | [18] |
| miR-92a | 100 | 59 | CRC | 84 | 71.2 | 0.84 (0.78–0.90) | Up-regulation | moderate | [19] |
| miR-92a | 37 | 59 | Adenoma | 64.9 | 81.4 | 0.75 (0.64–0.86) | Up-regulation | moderate | [19] |
| miR-96 | 187 | 47 | CRC | 65.4 | 73.3 | 0.74 (0.65–0.83) | Up-regulation | moderate | [60] |
| miR-106 | 100 | 78 | CRC | 74 | 44.4 | 0.61 (0.52–0.69) | Up-regulation | moderate | [54] |
| miR-142a-3p | 61 | 24 | CRC | - | - | 0.71 | Down-regulation | low | [58] |
| miR-150 | 37 | 20 | CRC | - | - | 0.68 (0.56–0.80) | Down-regulation | moderate | [59] |
| miR-183 | 118 | 61 | CRC | 73.7 | 88.5 | 0.83 (0.77–0.89) | Up-regulation | low | [61] |
| miR-200c | 78 | 86 | CRC | 64.1 | 73.3 | 0.75 | Up-regulation | moderate | [53] |
| miR-206 | 88 | 40 | CRC | - | - | 0.71 (0.61–0.80) | Up-regulation | low | [62] |
| miR-221 | 103 | 37 | CRC | 86 | 41 | 0.61 (0.49–0.72) | Up-regulation | low | [63] |
| miR-320a | 111 | 130 | CRC | 92.8 | 73.1 | 0.89 (0.85–0.93) | Down-regulation | low | [57] |
| miR-375 | 88 | 40 | CRC | 76.9 | 64.6 | 0.75 (0.65–0.84) | Down-regulation | low | [62] |
| miR-423–5p | 111 | 130 | CRC | 91.9 | 70.8 | 0.83 (0.78–0.89) | Down-regulation | low | [57] |
| miR-601 | 90 | 58 | CRC | 69.2 | 72.4 | 0.75 (0.67–0.83) | Down-regulation | low | [64] |
| miR-601 | 43 | 58 | Adenoma | 72.1 | 51.7 | 0.64 (0.53–0.75) | Down-regulation | low | [64] |
| miR-760 | 90 | 58 | CRC | 80.0 | 72.4 | 0.79 (0.71–0.86) | Down-regulation | low | [64] |
| miR-760 | 43 | 58 | Adenoma | 69.8 | 62.1 | 0.68 (0.58–0.79) | Down-regulation | low | [64] |
| miR-18a, -20a, -21, -29a, -92a, -106b, -133a, -143, -145 (panel) | 130 | 244 | CRC | - | - | 0.75 | Up-regulation | moderate | [65] |
| miR-15b, -17, -142-3p, -195, -331, -532, -532-3p, -652 (panel) | 25 | 38 | Adenoma | 88 | 64 | 0.87 | - | high | [66] |
| miR-7, -93, -409-3p (panel) | 22 | 27 | CRC | 82 | 89 | 0.90 (0.75–0.95) | - | moderate | [67] |
| miR-92a, -223 (panel) | 215 | 183 | CRC | 75.8 | 70.5 | 0.78 | Up-regulation | moderate | [68] |
| miR-19a, -19b, -15b (panel) | 42 | 53 | CRC | 78.6 | 79.3 | 0.84 (0.76–0.92) | Up-regulation | high | [69] |
| miR-21, miR-29c, miR-122, miR-192, miR-346, miR-372, miR-374a (panel) | 55 | 55 | CRC, Adenoma | 76–80 | 100 | 0.98(0.94–1.0) | - | high | [70] |
Table 2.
Diagnostic miRNA markers using serum specimen from patients with colorectal adenomas or CRCs
|
Serum-based biomarker |
|||||||||
| miR-17–3p | 70 | 70 | Early CRC | 83.6 | 72,9 | 0.81 (0.75–0.91) | Up-regulation | low | [71] |
| miR-21 | 186 | 53 | CRC | 91.9 | 81.1 | 0.92 (0.87–0.96) | Up-regulation | high | [26] |
| miR-21 | 43 | 53 | Adenoma | 81.1 | 76.7 | 0.81 (0.69–0.91) | Up-regulation | high | [26] |
| miR-21 | 200 | 80 | CRC | - | - | 0.8 | Up-regulation | low | [28] |
| miR-21 | 50 | 80 | Adenoma | - | - | 0.71 | Up-regulation | low | [28] |
| miR-21 | 40 | 40 | CRC | 77 | 78 | 0.87 | Up-regulation | low | [72] |
| miR-21 | 160 | 77 | Early CRC, Adenoma | - | - | 0.71 (0.64–0.77) | Up-regulation | high | [73] |
| miR-29a | 160 | 77 | Early CRC, Adenoma | - | - | 0.73 (0.66–0.79) | Up-regulation | high | [73] |
| miR-29b | 55 | 55 | CRC | - | - | - | Down-regulation | low | [74] |
| miR-92a | 200 | 80 | CRC | - | - | 0.77 | Up-regulation | low | [28] |
| miR-92a | 50 | 80 | Adenoma | - | - | 0.7 | Up-regulation | low | [28] |
| miR-103 | 84 | 32 | CRC | 55.9 | 75 | 0.66 | Up-regulation | low | [75] |
| miR-125b | 160 | 77 | Early CRC, Adenoma | - | - | 0.69 (0.62–0.77) | Up-regulation | high | [73] |
| miR-145 | 25 | 10 | CRC | - | - | 0.78 | Down-regulation | low | [76] |
| miR-155 | 146 | 60 | CRC | - | - | 0.78 | Up-regulation | low | [77] |
| miR-194 | 55 | 55 | CRC | - | - | - | Down-regulation | low | [74] |
| miR-210 | 218 | 102 | CRC | 74.6 | 73.5 | 0.82 | Up-regulation | low | [78] |
| miR-372 | - | - | Early CRC | 81.9 | 73.3 | 0.85 | Up-regulation | low | [79] |
| miR-720 | 84 | 32 | CRC | 58.3 | 56.3 | 0.63 | Up-regulation | low | [75] |
| miR-1290 | 56 | 57 | Adenoma | 46.4 | 91.2 | 0.72 | Up-regulation | High | [80] |
| miR-1290 | 211 | 57 | CRC | 70.1 | 91.2 | 0.83 | Up-regulation | high | [80] |
| miR-21, -31, -92a, -181b, -203, -let-7g (panel) | 113 | 89 | CRC | 96.4 | 88.1 | 0.92 | - | moderate | [81] |
| miR-23a-3p, -27a-3p, -142-5p, -376c-3p (panel) | 427 | 276 | CRC | 89 | 81 | 0.92 | - | moderate | [82] |
| miR-19a-3p, -21-5p, -425-5p | 169 | 121 | CRC | - | - | 0.78 (0.73–0.84) | Up-regulation | moderate | [83] |
The diagnostic potential for employing circulating miRNAs as liquid biopsies in CRC was first described by Chen et al in 2008[17]. This study demonstrated that a subset of 69 miRNAs was highly overexpressed in serum from CRC patients, but these miRNAs were virtually undetectable in sera from healthy volunteers. To further highlight the disease-specificity of this approach, the investigators reported that 14 of these miRNAs were exclusively upregulated in serum from CRC patients, but not in serum from patients with other types of cancer, primarily lung cancer. The first systematic and comprehensive analysis of blood-based CRC biomarkers was performed by Ng and coworkers[18]. In a discovery phase, these researchers performed miRNA expression profiling using tissue and plasma specimens from patients with CRC and healthy volunteers, and identified five candidate miRNAs that were later validated in a larger, independent cohort of plasma samples to highlight their potential application as diagnostic tools in CRC. There were three key findings inferred from this study. First, expression levels of miR-17–3p and miR-92a could discriminate patients with CRC from controls with great specificity and sensitivity (sensitivity, 64% & 89%; specificity, 70% & 70%; AUC, 0.72 & 0.89 respectively). Second, expression levels of both miRNAs were markedly lower in the pre vs. post-operative plasma specimens, suggesting that high levels of expression of these miRNAs were tumor-dependent. Third, miR-92a expression alone could discriminate patients with CRC from those with other types of gastrointestinal cancers, highlighting the potential for plasma miR-92 to serve as a novel diagnostic biomarker for CRC. A subsequent study explored the clinical usefulness of these two circulating miRNAs by including analysis of patients with CRAs - the precancerous lesions for CRC that represent a more pertinent target lesion for developing a CRC screening test[19]. This study showed that plasma levels of miR-29a and −92a could differentiate patients with CRAs and CRCs from healthy subjects, emphasizing the feasibility of these two miRNAs for use as potentially less invasive biomarkers for screening patients with colorectal neoplasia.
From a functional standpoint, miR-21 is one of the most well-established oncogenic miRNAs, and has been shown to participate in the multistep process of colorectal tumorigenesis, by not only regulation of MAPK pathway, but also by associating with WNT/β-Catenin signaling through targeting of PTEN, PDCD and DKK2[20–22]. Ma and coworkers crossed miR-21−/− mice with those lacking p53 (Trp53−/−), and demonstrated that Trp53−/−miR-21−/− mice developed tumors at a slightly later age. In addition, loss of miR-21 sensitizes transformed Trp53−/− cells to DNA-damage induced apoptosis through elevation of Pten expression [23]. In contrast, another group performed colitis-associated CRC model using azoxymethane and dextran sulfate sodium, and showed that miR-21−/− mice decreased in the size and numbers of tumors compared with control mice via increased expression of PDCD4 and modulation of nuclear factor (NF)-kB activation [24].
In addition to the oncogenic function of miR-21 from several basic researches, miR-21 has several distinct advantages favoring its development as an early detection biomarker for CRC. First, miR-21 is frequently up-regulated both in adenoma and cancer tissues compared to normal colonic epithelium. A miRNA microarray expression profiling study of tumor and paired normal tissues in a large cohort demonstrated that miR-21 expression was up-regulated significantly in adenomas compared with the matched adjacent normal mucosa, and demonstrated a stepwise increase in its expression in the adenoma-carcinoma sequence[6]. Second, state-of-the-art next generation sequencing techniques revealed that the absolute copy numbers of miR-21 in CRC tissues were significantly higher than other oncogenic miRNAs in cancer tissues[25]. Finally, miR-21 is a secreted miRNA that is often released by cancer cells and is present abundantly in plasma and serum, as evidenced from data reported by our group while analyzing supernatants from CRC cell lines and a large cohort of CRC-tissue and matched serum samples[26]. MiR-21 expression in cell supernatants significantly correlated with cell number and the duration of cell culture, while serum levels of miR-21 in CRC patients correlated positively with its expression status in the tumor tissues. Furthermore, levels of miR-21 expression robustly discriminated patients with CRAs and CRCs from healthy volunteers, suggesting that serum miR-21 may serve as a promising biomarker for screening patients with colorectal neoplasia. Analogous to serum, plasma levels of miR-21 may also have diagnostic potential in CRC[27]. In a study evaluating the expression levels of 380 miRNAs in primary cancer tissues and matched normal colonic tissue from 30 patients with CRC, the most dysregulated miRNAs were validated in an independent plasma test set from 40 patients with or without CRC. The results of this study established that plasma miR-21 could be used to identify patients with CRC from healthy volunteers with a very high sensitivity (90%) and specificity (90%). Likewise, another research group demonstrated the feasibility of using serum miR-21 levels as a diagnostic biomarker in serum samples from a cohort of 200 CRC patients, 50 advanced adenoma patients, and 80 healthy controls[28].
1.3.4. Several hurdles for developing this concept for clinical use
The evidence gathered thus far shows a remarkable degree of tissue specificity for aberrantly expressed miRNA in different types of human cancers. This finding, together with the fact that miRNAs are highly stable in various body fluids, has invigorated a flurry of research interest in exploiting these small non-coding RNAs as promising blood-based, less-invasive, diagnostic biomarkers for colorectal neoplasia. Although several studies to date have described the feasibility of utilizing miRNAs as potential diagnostic biomarkers in CRC (Table 1 and 2), there are several issues that must be considered for this concept to be developed for clinical use. Next, we focus on articles that attempt to elucidate each obstacle, and discuss the ideal methodology to further advance these research findings into actual clinical tests for early identification of colorectal neoplasia.
1.3.4.1 -. Protocol Standardization
For the development of meaningful biomarkers for clinical use, steps taken during sample collection and subsequent storage are critical, and must be considered as potential sources of bias that can confound test results[8]. The stability of target molecules is absolutely essential for avoiding analytical variation between samples and independent laboratories—an important requirement for data reproducibility and universal adoption of biomarkers. MiRNAs are stable in body fluids due to resistance to RNase activity[11, 17, 29], but studies have highlighted the importance of consistent pre-analytic preparation methods.
Glinge and colleagues performed a systematic analysis to evaluate the stability of miRNAs in blood under a variety of clinically relevant conditions, including the type of collection tubes, storage temperatures, physical disturbances, and serial freeze-thaw cycles[30].This study provides several key findings about the method of appropriate sample storage in clinical use of circulating miRNA test. First, the authors collected blood samples from healthy volunteers using different types of collection tubes and analyzed representative miRNAs (miR-1, miR-21, and miR-29b) to determine the impact of tube type on measurement outcomes. Tubes containing lithium-heparin were totally unsuitable for the detection of these three miRNAs, but collection tubes containing EDTA, citrate, or a serum separator did not alter the detectable levels of these miRNAs. There were subtle differences in circulating miRNA expression between various collection tubes and preparation techniques, indicating that samples should be collected using uniform protocols to avoid confounding the results. Second, levels of miR-1, miR-21 and miR-29b were stable for at least 24 hours in whole blood (some variability was seen in separated samples by 24 hours), but significant variations were detected in miR-21 and miR-29b in whole blood after 72 hours at room temperature due to the lysis of blood cells or platelets. Levels of miR-21 and miR-29b are significantly higher in buffy coats and red blood cells compared to plasma. Levels of miRs remained stable in whole blood up to 8 hours at room temperature before processing into plasma or serum. Plasma samples subjected to physical disturbance for 8 hours (rocker panels) showed reduced levels of miR-1 and miR-21, whereas just 1 hour of disturbance did not alter miR levels. The mechanistic basis of this is unclear. Third, long-time storage of EDTA-plasma samples at −80°C had no significant effect on miR-21 or miR-29b expression 9 months after freezing. However, miRNA expression was significantly increased in whole blood samples after freezing because of miRNAs released from cellular elements, particularly when there is hemolysis. In addition, miRNAs levels were decreased after 4 repetitive freeze-thaw cycles both in plasma and serum.
Combining these findings with previous data[11, 17, 31], the ideal protocol for sample collection could be described as follows: 1) use uniform collection tubes (but never lithium-heparin tubes); 2) whole blood may be preserved in EDTA or serum-separator tubes for up to 8 hours at room temperature during transportation to the clinical laboratory; 3) avoid storage of whole blood at −80°C; 4) after separation into plasma or serum fractions, store at −80°C promptly; 5) carefully separate the plasma or serum fractions to limit contamination from the cellular elements.
1.3.4.2 -. Establishment of direct quantification methods (extraction and standardization)
Sample collection, preparation and storage are major causes of technical variation in circulating miRNA biomarker studies. In addition, RNA extraction is more demanding than what is required in a protein-based assay. To date, most studies of circulating miRNAs use an endogenous or exogenous normalizer to address variations in the efficiency of RNA extraction. Although several studies have attempted to address the need for a normalization standard[32], there is as yet no consensus for the appropriate reference.
Recently, Zhao and coworkers demonstrated a direct quantification method that does not require RNA isolation to measure circulating miRNAs[33]. This study showed that treatment of 5 uL plasma aliquots with a denaturing buffer (Tween20, Tris, and EDTA) followed by heat treatment (75°C, 5 min) can efficiently release miRNAs for accurate measurement. The authors quantified let-7b and miR-92a expression in plasma specimens, and showed that this method was more accurate for analyzing circulating miRNAs than conventional RNA-based measurement methods. They used the direct quantification method to identify circulating miRNAs in patients with metastatic breast cancer, and showed that plasma miR-106a could be used to discriminate breast cancer patients with lymph node metastasis from those without it.
1.3.4.3 -. Quantification methods (relative expression vs absolute quantification)
Another challenge is the methodology for measuring miRNA levels. Quantitative real-time PCR (qPCR) is the standard method for this purpose, and most studies have quantitated circulating miRNA levels using the difference between cycle threshold (Ct) values of miRNAs of interest compared with those of endogenous or exogenous control miRNAs. One of the major limitations of using miRNAs in body fluids as liquid biopsy biomarkers is the selection of an endogenous control for normalizing the expression of target miRNAs. Unlike tissues, in which the target miRNA expression level can be normalized against RNU6B, U6, or miR-16, there is no consensus on the use of suitable reference genes to normalize the expression of target miRNAs from body fluids. Utilization of normalizers based on tissues is not advisable as it is difficult to exclude disease-specificity and other technical issues (e.g. sample collection, storage conditions, and importantly, hemolysis) that can inadvertently influence expression results[34]. Increasing numbers of studies have used the spike-in of synthetic, non-human mature miRNA from Caenorhabditis elegans such as cel-miR-39 and cel-miR-54 to normalize miRNA results. Although such an approach has some benefit in eliminating sample to sample variability in efficiency of RNA extraction and polymerase chain reaction amplification, this method still requires further refinement to account for differences that may occur during sample collection and storage. More recently, while performing genome-wide miRNA expression profiling on more than 500 serum samples, a study demonstrated that three miRNAs (miR-149–3p, miR-2861, and miR-4463) demonstrated stable expression across all samples, and hence may be adopted as more robust normalizers for serum miRNA expression profiling[32]. Although several studies have attempted to address the need for a normalization gene in evaluating the expression of circulating miRNAs[32], an appropriate reference gene or genes have not yet reached a consensus to serve as a normalizer in quantifying circulating miRNAs. Based on these backgrounds, the development of absolute quantification of miRs will be critical for inter-laboratory comparisons and standardization before such assays can be reliably used in clinical settings. Absolute quantification using qPCR with a calibrator curve relies on the accurate quantification of the number of copies of the calibrator, and accuracy problems may occur at several levels in the procedure. The challenges include: 1) initial quantification of the standard; 2) resuspension and preparation during serial dilutions; 3) reverse transcription; and 4) the amplification step.
PCR efficiency of pure synthetic calibrators may differ from clinical serum samples due to the presence of inhibitors[35]. Droplet digital PCR (ddPCR) may overcome this issue. This technology could provide absolute quantification of nucleic acids without the requirement of a standard curve, and estimate copy numbers of DNA and RNA targets with high accuracy compared to conventional qRT-PCR methods[36, 37]. A recent study directly compared conventional absolute qPCR method and ddPCR techniques for quantifying selected miRNAs (miR-21, miR-126, and let-7a) in serum. The authors quantified circulating miRNA levels by both methods using 85 serum specimens from 2 independent cohorts and demonstrated a significant positive correlation between ddPCR and qPCR in each miRNA. However, the coefficient of variation was similar or significantly smaller in ddPCR than qPCR for all miRNAs tested. In addition, expression levels of miR-126 and let-7a were significantly lower (2.4- and 3.9-fold, respectively) in ddPCR than in qPCR, and this discrepancy may be due to errors or imprecisions in the qPCR calibration curve, as described above. Reproducibility of ddPCR analysis was also successfully demonstrated using duplicate analysis or measuring at different times in this study. Although both techniques can be reliably used for quantification of circulating miRNAs, ddPCR technology showed similar or greater accuracy for absolute quantification of miRNA expression and may allow developing simple quantification protocols without calibrator curves to measure circulating miRNAs.
1.3.4.4 -. Circulating miR-containing exosomes in cancer patients
Considering that blood (serum/plasma) is one of the most easily accessible body fluids, it is most frequently used as diagnostic material for the development of screening biomarkers. Several studies have indicated that the critical issue is the use of standardized protocols using the same material (i.e., serum or plasma) for the development of circulating miRNAs into a universal screening test in clinical use.
Exosomes are microvesicles, 50–150 nm in diameter, and are actively secreted from various cell types, especially cancer cells[38]. Tumor-derived exosomes were first demonstrated in the peripheral circulation of patients with malignant disease in 1979[39, 40]. Emerging evidence has revealed that exosomal miRNAs released from cancer cells can be transported to other recipient cells within the tumor microenvironment—or to distant organs to enhance tumor progression and metastasis. In light of this feature, exosome-encapsulated miRNAs are gaining attention as potential of disease-specific diagnostic biomarkers in various types of cancer (Table 3)[41].
Table 3.
Diagnostic miRNA markers using exosome-containing specimens from patients with colorectal adenomas or CRCs
| Exosome-based biomarker | |||||||||
| let-7a | 88 | 11 | CRC | - | - | 0.67 | Up-regulation | moderate | [41] |
| miR-19a | 90 | 12 | CRC | - | - | - | Up-regulation | moderate | [84] |
| miR-21 | 88 | 11 | CRC | - | - | 0.8 | Up-regulation | moderate | [41] |
| miR-23a | 88 | 11 | CRC | - | - | 0.95 | Up-regulation | moderate | [41] |
| miR-92a | 90 | 12 | CRC | - | - | - | Up-regulation | moderate | [84] |
| miR-150 | 88 | 11 | CRC | - | - | 0.76 | Up-regulation | moderate | [41] |
| miR-223 | 88 | 11 | CRC | - | - | 0.72 | Up-regulation | moderate | [41] |
| miR-1229 | 88 | 11 | CRC | - | - | 0.61 | Up-regulation | moderate | [41] |
| miR-1246 | 88 | 11 | CRC | - | - | 0.95 | Up-regulation | moderate | [41] |
Uratani and colleagues compared the diagnostic accuracy of representative miRNA expression (miR-21, −29a, −92a, and −135b) between serum and exosomes from adenoma patients and healthy volunteers[42]. In serum, expression of miR-21, −29a, and −92a was significantly higher in patients with CRAs compared to healthy volunteers and significantly correlated with adenoma size and number in the colorectum. However, exosomal miR-21 and miR-29a levels in CRA patients were significantly higher than those of healthy volunteers, and only exosomal miR-21 significantly correlated with adenoma size and number. Furthermore, the diagnostic accuracy of these miRNAs is generally higher in serum compared to exosomes.. From the standpoint of a clinical test, extraction of total exosome requires a more complicated procedure and creates an additional hurdle for developing a uniform test for circulating miRNAs. Therefore, the merit to focus on total exosome looks still little as an early detection biomarker test in clinical use.
1.3.4.5 -. Cancer-specific miR-containing exosomes
The development of miRNA analysis from blood as a minimally invasive test for asymptomatic CRC is at hand. One would anticipate that the most powerful approach would be to utilize direct quantification of miRs with ddPCR on serum or plasma collected under standardized protocols. This approach can produce a simple test with high accuracy and specificity for the early detection of CRC patients.
The remaining challenge is the relative lack of disease-specificity for most of the circulating miRs in plasma or serum. To overcome this impediment, a novel methodology is based on the concept of cancer-specific exosomes, in spite of the above-mentioned technical challenges involved. Taylor and colleagues isolated exosomes that expressed the epithelial cell adhesion molecule (EpCAM) from sera of patients with ovarian and lung cancer, and detected the overlap in miRNA signatures between EpCAM-positive exosomes from sera and the corresponding tumor tissues[43]. Melo and colleagues identified glypican-1 as a specific marker of pancreatic cancer, and demonstrated that glypican-1(+)-expressing exosomes in the circulation could be used as early detection biomarkers to identify patients with this malignancy[44]. These studies are quite encouraging and there is a great degree of enthusiasm these approaches in combination with normalization of the protein marker and absolute quantification, which will provide additional measures to overcome the limitations and improve the sensitivity and specificity of diagnostic biomarkers for CRC.
1.4. Conclusion
Early diagnosis, especially detection of precancerous adenomas, is considered to be a key concept for improving patient survival in CRC management. Although colonoscopy is the most reliable method for early diagnosis of CRC, its very invasive nature and the associated high expense have hampered its worldwide adoption and compliance as a screening modality in average-risk populations. Currently available noninvasive screening methods, such as guaiac-based technologies and immunochemical fecal occult blood tests (gFOBT and FIT), result in a modest decrease in CRC-associated mortality [45–47]. However, larger screening studies have revealed the limitation of these tests such as their low sensitivity, especially with respect to detection of preneoplastic lesions [3, 48, 49]. Recently, two types of blood-based liquid biopsy assays, including the methylated SEPT9 and CancerSEEK have been proposed or adopted for CRC screening [50, 51]. Both assays were shown to be superior to fecal FIT in detecting CRC neoplasms. However, these approaches were also suboptimal for diagnosing patients with advanced adenomas because the amounts of circulating DNA load was extremely small in such early-stage, precancerous patients [52]. The feasibility of using miRNAs as liquid biopsy diagnostic biomarkers for cancer and precancerous lesions has become increasingly attractive as a large body of scientific evidence has accumulated regarding the functional roles of miRNAs in cancer development, as well as their stability and cancer-specific dysregulation in precancerous and cancerous tissues. Nonetheless, in spite of this enthusiasm, considerable work is required to overcome the technical challenges limiting the clinical application of miRNAs as early diagnostic biomarkers for CRC. Once these issues are resolved, prospective studies utilizing standardized assays in large population-based cross-sectional studies are needed to identify optimal miRNAs and marker panels that can be used for the early detection of colorectal polyps and cancers in clinical care.
Acknowledgments
Funding
The present work was supported by the CA72851, CA181572, CA184792, CA187956 and CA202797 grants from the National Cancer Institute, National Institute of Health; RP140784 from the Cancer Prevention Research Institute of Texas; grants from the Sammons Cancer Center and Baylor Foundation, as well as funds from the Baylor Scott & White Research Institute, Dallas, TX, USA awarded to AG. A part of this study was supported by a Grant in Aid for Scientific Research (26293301, 16K15612) from the Ministry of Education, Culture, Sports, Science, and Technology, Japan.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest: None of the authors have any potential conflicts to disclose.
REFERENCES
- [1].Siegel RL, Miller KD, Jemal A, Cancer statistics, 2016, CA: a cancer journal for clinicians, 66 (2016) 7–30. [DOI] [PubMed] [Google Scholar]
- [2].Figueredo A, Coombes ME, Mukherjee S, Adjuvant therapy for completely resected stage II colon cancer, The Cochrane database of systematic reviews, (2008) CD005390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Walsh JM, Terdiman JP, Colorectal cancer screening: clinical applications, JAMA : the journal of the American Medical Association, 289 (2003) 1297–1302. [DOI] [PubMed] [Google Scholar]
- [4].Bartel DP, MicroRNAs: target recognition and regulatory functions, Cell, 136 (2009) 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM, Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia, Proceedings of the National Academy of Sciences of the United States of America, 99 (2002) 15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, Liu CG, Calin GA, Croce CM, Harris CC, MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma, JAMA : the journal of the American Medical Association, 299 (2008) 425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wells GA, Shea B, O’Connell D, Peterson J, Welch V, Losos M, Tugwell P, The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. [Google Scholar]
- [8].Zou H, Harrington JJ, Klatt KK, Ahlquist DA, A sensitive method to quantify human long DNA in stool: relevance to colorectal cancer screening, Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology, 15 (2006) 1115–1119. [DOI] [PubMed] [Google Scholar]
- [9].Chim SS, Shing TK, Hung EC, Leung TY, Lau TK, Chiu RW, Lo YM, Detection and characterization of placental microRNAs in maternal plasma, Clinical chemistry, 54 (2008) 482–490. [DOI] [PubMed] [Google Scholar]
- [10].Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, Banham AH, Pezzella F, Boultwood J, Wainscoat JS, Hatton CS, Harris AL, Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma, British journal of haematology, 141 (2008) 672–675. [DOI] [PubMed] [Google Scholar]
- [11].Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M, Circulating microRNAs as stable blood-based markers for cancer detection, Proceedings of the National Academy of Sciences of the United States of America, 105 (2008) 10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Shigeyasu K, Toden S, Zumwalt TJ, Okugawa Y, Goel A, Emerging Role of MicroRNAs as Liquid Biopsy Biomarkers in Gastrointestinal Cancers, Clinical cancer research : an official journal of the American Association for Cancer Research, 23 (2017) 2391–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ahmed FE, Jeffries CD, Vos PW, Flake G, Nuovo GJ, Sinar DR, Naziri W, Marcuard SP, Diagnostic microRNA markers for screening sporadic human colon cancer and active ulcerative colitis in stool and tissue, Cancer genomics & proteomics, 6 (2009) 281–295. [PubMed] [Google Scholar]
- [14].Link A, Balaguer F, Shen Y, Nagasaka T, Lozano JJ, Boland CR, Goel A, Fecal MicroRNAs as novel biomarkers for colon cancer screening, Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology, 19 (2010) 1766–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Koga Y, Yasunaga M, Takahashi A, Kuroda J, Moriya Y, Akasu T, Fujita S, Yamamoto S, Baba H, Matsumura Y, MicroRNA expression profiling of exfoliated colonocytes isolated from feces for colorectal cancer screening, Cancer prevention research (Philadelphia, Pa.), 3 (2010) 1435–1442. [DOI] [PubMed] [Google Scholar]
- [16].Koga Y, Yamazaki N, Yamamoto Y, Yamamoto S, Saito N, Kakugawa Y, Otake Y, Matsumoto M, Matsumura Y, Fecal miR-106a is a useful marker for colorectal cancer patients with false-negative results in immunochemical fecal occult blood test, Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology, 22 (2013) 1844–1852. [DOI] [PubMed] [Google Scholar]
- [17].Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, Li Q, Li X, Wang W, Zhang Y, Wang J, Jiang X, Xiang Y, Xu C, Zheng P, Zhang J, Li R, Zhang H, Shang X, Gong T, Ning G, Wang J, Zen K, Zhang J, Zhang CY, Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases, Cell research, 18 (2008) 997–1006. [DOI] [PubMed] [Google Scholar]
- [18].Ng EK, Chong WW, Jin H, Lam EK, Shin VY, Yu J, Poon TC, Ng SS, Sung JJ, Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening, Gut, 58 (2009) 1375–1381. [DOI] [PubMed] [Google Scholar]
- [19].Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X, Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer, International journal of cancer. Journal international du cancer, 127 (2010) 118–126. [DOI] [PubMed] [Google Scholar]
- [20].Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T, MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer, Gastroenterology, 133 (2007) 647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Talotta F, Cimmino A, Matarazzo MR, Casalino L, De Vita G, D’Esposito M, Di Lauro R, Verde P, An autoregulatory loop mediated by miR-21 and PDCD4 controls the AP-1 activity in RAS transformation, Oncogene, 28 (2009) 73–84. [DOI] [PubMed] [Google Scholar]
- [22].Kawakita A, Yanamoto S, Yamada SI, Naruse T, Takahashi H, Kawasaki G, Umeda M, MicroRNA-21 Promotes Oral Cancer Invasion via the Wnt/beta-Catenin Pathway by Targeting DKK2, Pathol Oncol Res, (2013). [DOI] [PubMed] [Google Scholar]
- [23].Ma X, Choudhury SN, Hua X, Dai Z, Li Y, Interaction of the oncogenic miR-21 microRNA and the p53 tumor suppressor pathway, Carcinogenesis, 34 (2013) 1216–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Shi C, Yang Y, Xia Y, Okugawa Y, Yang J, Liang Y, Chen H, Zhang P, Wang F, Han H, Wu W, Gao R, Gasche C, Qin H, Ma Y, Goel A, Novel evidence for an oncogenic role of microRNA-21 in colitis-associated colorectal cancer, Gut, 65 (2016) 1470–1481. [DOI] [PubMed] [Google Scholar]
- [25].Schee K, Lorenz S, Worren MM, Gunther CC, Holden M, Hovig E, Fodstad O, Meza-Zepeda LA, Flatmark K, Deep Sequencing the MicroRNA Transcriptome in Colorectal Cancer, PloS one, 8 (2013) e66165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Toiyama Y, Takahashi M, Hur K, Nagasaka T, Tanaka K, Inoue Y, Kusunoki M, Boland CR, Goel A, Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer, Journal of the National Cancer Institute, 105 (2013) 849–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kanaan Z, Rai SN, Eichenberger MR, Roberts H, Keskey B, Pan J, Galandiuk S, Plasma miR-21: a potential diagnostic marker of colorectal cancer, Annals of surgery, 256 (2012) 544–551. [DOI] [PubMed] [Google Scholar]
- [28].Liu GH, Zhou ZG, Chen R, Wang MJ, Zhou B, Li Y, Sun XF, Serum miR-21 and miR-92a as biomarkers in the diagnosis and prognosis of colorectal cancer, Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine, 34 (2013) 2175–2181. [DOI] [PubMed] [Google Scholar]
- [29].Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, Benjamin H, Kushnir M, Cholakh H, Melamed N, Bentwich Z, Hod M, Goren Y, Chajut A, Serum microRNAs are promising novel biomarkers, PloS one, 3 (2008) e3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Glinge C, Clauss S, Boddum K, Jabbari R, Jabbari J, Risgaard B, Tomsits P, Hildebrand B, Kaab S, Wakili R, Jespersen T, Tfelt-Hansen J, Stability of Circulating Blood-Based MicroRNAs - Pre-Analytic Methodological Considerations, PloS one, 12 (2017) e0167969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Turchinovich A, Weiz L, Langheinz A, Burwinkel B, Characterization of extracellular circulating microRNA, Nucleic acids research, 39 (2011) 7223–7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Shimomura A, Shiino S, Kawauchi J, Takizawa S, Sakamoto H, Matsuzaki J, Ono M, Takeshita F, Niida S, Shimizu C, Fujiwara Y, Kinoshita T, Tamura K, Ochiya T, Novel combination of serum microRNA for detecting breast cancer in the early stage, Cancer science, 107 (2016) 326–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhao Q, Deng S, Wang G, Liu C, Meng L, Qiao S, Shen L, Zhang Y, Lu J, Li W, Zhang Y, Wang M, Pestell RG, Liang C, Yu Z, A direct quantification method for measuring plasma MicroRNAs identified potential biomarkers for detecting metastatic breast cancer, Oncotarget, 7 (2016) 21865–21874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kirschner MB, Kao SC, Edelman JJ, Armstrong NJ, Vallely MP, van Zandwijk N, Reid G, Haemolysis during sample preparation alters microRNA content of plasma, PloS one, 6 (2011) e24145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Corbisier P, Pinheiro L, Mazoua S, Kortekaas AM, Chung PY, Gerganova T, Roebben G, Emons H, Emslie K, DNA copy number concentration measured by digital and droplet digital quantitative PCR using certified reference materials, Analytical and bioanalytical chemistry, 407 (2015) 1831–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Pinheiro LB, Coleman VA, Hindson CM, Herrmann J, Hindson BJ, Bhat S, Emslie KR, Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification, Analytical chemistry, 84 (2012) 1003–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Huggett JF, Foy CA, Benes V, Emslie K, Garson JA, Haynes R, Hellemans J, Kubista M, Mueller RD, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT, Bustin SA, The digital MIQE guidelines: Minimum Information for Publication of Quantitative Digital PCR Experiments, Clinical chemistry, 59 (2013) 892–902. [DOI] [PubMed] [Google Scholar]
- [38].Huber V, Fais S, Iero M, Lugini L, Canese P, Squarcina P, Zaccheddu A, Colone M, Arancia G, Gentile M, Seregni E, Valenti R, Ballabio G, Belli F, Leo E, Parmiani G, Rivoltini L, Human colorectal cancer cells induce T-cell death through release of proapoptotic microvesicles: role in immune escape, Gastroenterology, 128 (2005) 1796–1804. [DOI] [PubMed] [Google Scholar]
- [39].Taylor DD, Homesley HD, Doellgast GJ, Binding of specific peroxidase-labeled antibody to placental-type phosphatase on tumor-derived membrane fragments, Cancer research, 40 (1980) 4064–4069. [PubMed] [Google Scholar]
- [40].Taylor DD, Doellgast GJ, Quantitation of peroxidase-antibody binding to membrane fragments using column chromatography, Analytical biochemistry, 98 (1979) 53–59. [DOI] [PubMed] [Google Scholar]
- [41].Ogata-Kawata H, Izumiya M, Kurioka D, Honma Y, Yamada Y, Furuta K, Gunji T, Ohta H, Okamoto H, Sonoda H, Watanabe M, Nakagama H, Yokota J, Kohno T, Tsuchiya N, Circulating exosomal microRNAs as biomarkers of colon cancer, PloS one, 9 (2014) e92921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Uratani R, Toiyama Y, Kitajima T, Kawamura M, Hiro J, Kobayashi M, Tanaka K, Inoue Y, Mohri Y, Mori T, Kato T, Goel A, Kusunoki M, Diagnostic Potential of Cell-Free and Exosomal MicroRNAs in the Identification of Patients with High-Risk Colorectal Adenomas, PloS one, 11 (2016) e0160722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Taylor DD, Gercel-Taylor C, MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer, Gynecologic oncology, 110 (2008) 13–21. [DOI] [PubMed] [Google Scholar]
- [44].Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, LeBleu VS, Mittendorf EA, Weitz J, Rahbari N, Reissfelder C, Pilarsky C, Fraga MF, Piwnica-Worms D, Kalluri R, Glypican-1 identifies cancer exosomes and detects early pancreatic cancer, Nature, 523 (2015) 177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Levine JS, Screening and surveillance for colorectal neoplasia: uncertainties of colonoscopic management, Polskie Archiwum Medycyny Wewnetrznej, 118 (2008) 302–306. [PubMed] [Google Scholar]
- [46].Dominic OG, McGarrity T, Dignan M, Lengerich EJ, American College of Gastroenterology Guidelines for Colorectal Cancer Screening 2008, The American journal of gastroenterology, 104 (2009) 2626–2627; author reply 2628–2629. [DOI] [PubMed] [Google Scholar]
- [47].Whitlock EP, Lin JS, Liles E, Beil TL, Fu R, Screening for colorectal cancer: a targeted, updated systematic review for the U.S. Preventive Services Task Force, Annals of internal medicine, 149 (2008) 638–658. [DOI] [PubMed] [Google Scholar]
- [48].Ahlquist DA, Sargent DJ, Loprinzi CL, Levin TR, Rex DK, Ahnen DJ, Knigge K, Lance MP, Burgart LJ, Hamilton SR, Allison JE, Lawson MJ, Devens ME, Harrington JJ, Hillman SL, Stool DNA and occult blood testing for screen detection of colorectal neoplasia, Annals of internal medicine, 149 (2008) 441–450, w481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Morikawa T, Kato J, Yamaji Y, Wada R, Mitsushima T, Shiratori Y, A comparison of the immunochemical fecal occult blood test and total colonoscopy in the asymptomatic population, Gastroenterology, 129 (2005) 422–428. [DOI] [PubMed] [Google Scholar]
- [50].Lofton-Day C, Model F, Devos T, Tetzner R, Distler J, Schuster M, Song X, Lesche R, Liebenberg V, Ebert M, Molnar B, Grutzmann R, Pilarsky C, Sledziewski A, DNA methylation biomarkers for blood-based colorectal cancer screening, Clin Chem, 54 (2008) 414–423. [DOI] [PubMed] [Google Scholar]
- [51].Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, Douville C, Javed AA, Wong F, Mattox A, Hruban RH, Wolfgang CL, Goggins MG, Dal Molin M, Wang TL, Roden R, Klein AP, Ptak J, Dobbyn L, Schaefer J, Silliman N, Popoli M, Vogelstein JT, Browne JD, Schoen RE, Brand RE, Tie J, Gibbs P, Wong HL, Mansfield AS, Jen J, Hanash SM, Falconi M, Allen PJ, Zhou S, Bettegowda C, Diaz LA Jr., Tomasetti C, Kinzler KW, Vogelstein B, Lennon AM, Papadopoulos N, Detection and localization of surgically resectable cancers with a multi-analyte blood test, Science (New York, N.Y.), 359 (2018) 926–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Jin P, Kang Q, Wang X, Yang L, Yu Y, Li N, He YQ, Han X, Hang J, Zhang J, Song L, Han Y, Sheng JQ, Performance of a second generation methylated SEPT9 test in detecting colorectal neoplasm, J Gastroenterol Hepatol, (2014). [DOI] [PubMed] [Google Scholar]
- [53].Zhang GJ, Zhou T, Liu ZL, Tian HP, Xia SS, Plasma miR-200c and miR-18a as potential biomarkers for the detection of colorectal carcinoma, Molecular and clinical oncology, 1 (2013) 379–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Chen WY, Zhao XJ, Yu ZF, Hu FL, Liu YP, Cui BB, Dong XS, Zhao YS, The potential of plasma miRNAs for diagnosis and risk estimation of colorectal cancer, International journal of clinical and experimental pathology, 8 (2015) 7092–7101. [PMC free article] [PubMed] [Google Scholar]
- [55].Zanutto S, Pizzamiglio S, Ghilotti M, Bertan C, Ravagnani F, Perrone F, Leo E, Pilotti S, Verderio P, Gariboldi M, Pierotti MA, Circulating miR-378 in plasma: a reliable, haemolysis-independent biomarker for colorectal cancer, British journal of cancer, 110 (2014) 1001–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Du M, Liu S, Gu D, Wang Q, Zhu L, Kang M, Shi D, Chu H, Tong N, Chen J, Adams TS, Zhang Z, Wang M, Clinical potential role of circulating microRNAs in early diagnosis of colorectal cancer patients, Carcinogenesis, 35 (2014) 2723–2730. [DOI] [PubMed] [Google Scholar]
- [57].Fang Z, Tang J, Bai Y, Lin H, You H, Jin H, Lin L, You P, Li J, Dai Z, Liang X, Su Y, Hu Q, Wang F, Zhang ZY, Plasma levels of microRNA-24, microRNA-320a, and microRNA-423–5p are potential biomarkers for colorectal carcinoma, Journal of experimental & clinical cancer research : CR, 34 (2015) 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Ghanbari R, Mosakhani N, Asadi J, Nouraee N, Mowla SJ, Yazdani Y, Mohamadkhani A, Poustchi H, Knuutila S, Malekzadeh R, Downregulation of Plasma MiR-142–3p and MiR-26a-5p in Patients With Colorectal Carcinoma, Iranian journal of cancer prevention, 8 (2015) e2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Aherne ST, Madden SF, Hughes DJ, Pardini B, Naccarati A, Levy M, Vodicka P, Neary P, Dowling P, Clynes M, Circulating miRNAs miR-34a and miR-150 associated with colorectal cancer progression, BMC cancer, 15 (2015) 329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Sun Y, Liu Y, Cogdell D, Calin GA, Sun B, Kopetz S, Hamilton SR, Zhang W, Examining plasma microRNA markers for colorectal cancer at different stages, Oncotarget, 7 (2016) 11434–11449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Yuan D, Li K, Zhu K, Yan R, Dang C, Plasma miR-183 predicts recurrence and prognosis in patients with colorectal cancer, Cancer biology & therapy, 16 (2015) 268–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Xu L, Li M, Wang M, Yan D, Feng G, An G, The expression of microRNA-375 in plasma and tissue is matched in human colorectal cancer, BMC cancer, 14 (2014) 714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Pu XX, Huang GL, Guo HQ, Guo CC, Li H, Ye S, Ling S, Jiang L, Tian Y, Lin TY, Circulating miR-221 directly amplified from plasma is a potential diagnostic and prognostic marker of colorectal cancer and is correlated with p53 expression, Journal of gastroenterology and hepatology, 25 (2010) 1674–1680. [DOI] [PubMed] [Google Scholar]
- [64].Wang Q, Huang Z, Ni S, Xiao X, Xu Q, Wang L, Huang D, Tan C, Sheng W, Du X , Plasma miR-601 and miR-760 are novel biomarkers for the early detection of colorectal cancer, PloS one, 7 (2012) e44398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Luo X, Stock C, Burwinkel B, Brenner H, Identification and evaluation of plasma microRNAs for early detection of colorectal cancer, PloS one, 8 (2013) e62880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Kanaan Z, Roberts H, Eichenberger MR, Billeter A, Ocheretner G, Pan J, Rai SN, Jorden J, Williford A, Galandiuk S, A plasma microRNA panel for detection of colorectal adenomas: a step toward more precise screening for colorectal cancer, Annals of surgery, 258 (2013) 400–408. [DOI] [PubMed] [Google Scholar]
- [67].Wang S, Xiang J, Li Z, Lu S, Hu J, Gao X, Yu L, Wang L, Wang J, Wu Y, Chen Z, Zhu H, A plasma microRNA panel for early detection of colorectal cancer, International journal of cancer. Journal international du cancer, 136 (2015) 152–161. [DOI] [PubMed] [Google Scholar]
- [68].Chang PY, Chen CC, Chang YS, Tsai WS, You JF, Lin GP, Chen TW, Chen JS, Chan EC, MicroRNA-223 and microRNA-92a in stool and plasma samples act as complementary biomarkers to increase colorectal cancer detection, Oncotarget, 7 (2016) 10663–10675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Giraldez MD, Lozano JJ, Ramirez G, Hijona E, Bujanda L, Castells A, Gironella M, Circulating microRNAs as biomarkers of colorectal cancer: results from a genome-wide profiling and validation study, Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association, 11 (2013) 681–688.e683. [DOI] [PubMed] [Google Scholar]
- [70].Carter JV, Roberts HL, Pan J, Rice JD, Burton JF, Galbraith NJ, Eichenberger MR, Jorden J, Deveaux P, Farmer R, Williford A, Kanaan Z, Rai SN, Galandiuk S, A Highly Predictive Model for Diagnosis of Colorectal Neoplasms Using Plasma MicroRNA: Improving Specificity and Sensitivity, Annals of surgery, 264 (2016) 575–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Zhu J, Dong H, Zhang Q, Zhang S, Combined assays for serum carcinoembryonic antigen and microRNA-17–3p offer improved diagnostic potential for stage I/II colon cancer, Molecular and clinical oncology, 3 (2015) 1315–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Basati G, Emami Razavi A, Abdi S, Mirzaei A, Elevated level of microRNA-21 in the serum of patients with colorectal cancer, Medical oncology (Northwood, London, England), 31 (2014) 205. [DOI] [PubMed] [Google Scholar]
- [73].Yamada A, Horimatsu T, Okugawa Y, Nishida N, Honjo H, Ida H, Kou T, Kusaka T, Sasaki Y, Yagi M, Higurashi T, Yukawa N, Amanuma Y, Kikuchi O, Muto M, Ueno Y, Nakajima A, Chiba T, Boland CR, Goel A, Serum miR-21, miR-29a, and miR-125b Are Promising Biomarkers for the Early Detection of Colorectal Neoplasia, Clinical cancer research : an official journal of the American Association for Cancer Research, 21 (2015) 4234–4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Basati G, Razavi AE, Pakzad I, Malayeri FA, Circulating levels of the miRNAs, miR-194, and miR-29b, as clinically useful biomarkers for colorectal cancer, Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine, 37 (2016) 1781–1788. [DOI] [PubMed] [Google Scholar]
- [75].Nonaka R, Miyake Y, Hata T, Kagawa Y, Kato T, Osawa H, Nishimura J, Ikenaga M, Murata K, Uemura M, Okuzaki D, Takemasa I, Mizushima T, Yamamoto H, Doki Y, Mori M, Circulating miR-103 and miR-720 as novel serum biomarkers for patients with colorectal cancer, International journal of oncology, 47 (2015) 1097–1102. [DOI] [PubMed] [Google Scholar]
- [76].Ramzy I, Hasaballah M, Marzaban R, Shaker O, Soliman ZA, Evaluation of microRNAs-29a, 92a and 145 in colorectal carcinoma as candidate diagnostic markers: An Egyptian pilot study, Clinics and research in hepatology and gastroenterology, 39 (2015) 508–515. [DOI] [PubMed] [Google Scholar]
- [77].Lv ZC, Fan YS, Chen HB, Zhao DW , Investigation of microRNA-155 as a serum diagnostic and prognostic biomarker for colorectal cancer, Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine, 36 (2015) 1619–1625. [DOI] [PubMed] [Google Scholar]
- [78].Wang W, Qu A, Liu W, Liu Y, Zheng G, Du L, Zhang X, Yang Y, Wang C, Chen X, Circulating miR-210 as a diagnostic and prognostic biomarker for colorectal cancer, European journal of cancer care, (2016). [DOI] [PubMed] [Google Scholar]
- [79].Yu J, Jin L, Jiang L, Gao L, Zhou J, Hu Y, Li W, Zhi Q, Zhu X, Serum miR-372 is a Diagnostic and Prognostic Biomarker in Patients with Early Colorectal Cancer, Anti-cancer agents in medicinal chemistry, 16 (2016) 424–431. [DOI] [PubMed] [Google Scholar]
- [80].Imaoka H, Toiyama Y, Fujikawa H, Hiro J, Saigusa S, Tanaka K, Inoue Y, Mohri Y, Mori T, Kato T, Toden S, Goel A, Kusunoki M, Circulating microRNA-1290 as a novel diagnostic and prognostic biomarker in human colorectal cancer, Annals of oncology : official journal of the European Society for Medical Oncology / ESMO, (2016). [DOI] [PubMed] [Google Scholar]
- [81].Wang J, Huang SK, Zhao M, Yang M, Zhong JL, Gu YY, Peng H, Che YQ, Huang CZ, Identification of a circulating microRNA signature for colorectal cancer detection, PloS one, 9 (2014) e87451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Vychytilova-Faltejskova P, Radova L, Sachlova M, Kosarova Z, Slaba K, Fabian P, Grolich T, Prochazka V, Kala Z, Svoboda M, Kiss I, Vyzula R, Slaby O, Serum-based microRNA signatures in early diagnosis and prognosis prediction of colon cancer, Carcinogenesis, 37 (2016) 941–950. [DOI] [PubMed] [Google Scholar]
- [83].Zhu M, Huang Z, Zhu D, Zhou X, Shan X, Qi LW, Wu L, Cheng W, Zhu J, Zhang L, Zhang H, Chen Y, Zhu W, Wang T, Liu P, A panel of microRNA signature in serum for colorectal cancer diagnosis, Oncotarget, 8 (2017) 17081–17091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Matsumura T, Sugimachi K, Iinuma H, Takahashi Y, Kurashige J, Sawada G, Ueda M, Uchi R, Ueo H, Takano Y, Shinden Y, Eguchi H, Yamamoto H, Doki Y, Mori M, Ochiya T, Mimori K, Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer, British journal of cancer, 113 (2015) 275–281. [DOI] [PMC free article] [PubMed] [Google Scholar]