The clonal evolution of metastatic colorectal cancer reveals novel insights into cancer biology.
Abstract
Tumor heterogeneity and evolution drive treatment resistance in metastatic colorectal cancer (mCRC). Patient-derived xenografts (PDXs) can model mCRC biology; however, their ability to accurately mimic human tumor heterogeneity is unclear. Current genomic studies in mCRC have limited scope and lack matched PDXs. Therefore, the landscape of tumor heterogeneity and its impact on the evolution of metastasis and PDXs remain undefined. We performed whole-genome, deep exome, and targeted validation sequencing of multiple primary regions, matched distant metastases, and PDXs from 11 patients with mCRC. We observed intricate clonal heterogeneity and evolution affecting metastasis dissemination and PDX clonal selection. Metastasis formation followed both monoclonal and polyclonal seeding models. In four cases, metastasis-seeding clones were not identified in any primary region, consistent with a metastasis-seeding-metastasis model. PDXs underrepresented the subclonal heterogeneity of parental tumors. These suggest that single sample tumor sequencing and current PDX models may be insufficient to guide precision medicine.
INTRODUCTION
Colorectal cancer (CRC) is the most common gastrointestinal malignancy and is a leading cause of cancer-related death. Since the initial description of the “adenoma-carcinoma” initiation of primary CRC (1), the genomic landscape of primary CRC has been detailed by large-scale sequencing studies (2).
The genomic characterization of metastatic CRC (mCRC) includes studies using exome or targeted sequencing of cancer-related genes with limited breadth and depth of coverage, some of which directly compare metastases to their matched primary tumors (3–8). These studies have allowed us to begin to understand the clonal progression and evolution of metastases; however, notable limitations exist. First, exome- and gene panel–targeted sequencing have limited breadth of coverage that primarily targets the coding regions that represent ~2% of the genome, and therefore, they are unable to identify the majority of passenger mutations, which are mostly in the noncoding genome. However, whole-genome sequencing allows for the detection of many more mutations, which improves cancer cellular prevalence estimates. Second, most studies only evaluated one biopsy per tumor, thus largely underestimated the intratumor heterogeneity, particularly in the primary tumors. To be able to understand the evolution of metastases, comprehensively evaluating the primary tumor is essential, as metastasis-seeding clones could be present at low frequency in the primary tumors and not captured in single region sequencing. Because of these limitations in current studies, critical questions on mCRC metastatic evolution have not been comprehensively addressed: Do metastases arise from single or multiple cells? Do metastases occur after multiple clonal expansions in the primary? Do metastases seed other metastases?
The practice of using human tumors grown in immunosuppressed mice [referred to as patient-derived xenografts (PDXs)] has been widely used to model cancer biology, predict treatment response, and delineate mechanisms of therapy resistance (https://pdmr.cancer.gov/) (9). However, it is not clear how accurately PDX models capture the genomic heterogeneity observed in human tumors, creating doubt regarding their utility for precision medicine applications (10–12). There exists no study in mCRC that evaluated the clonal heterogeneity and evolution of PDXs in conjunction with their matched primary and metastatic tumors from the same patients.
We prospectively collected matched normal, primary, and metastatic tumor tissues from patients with mCRC and developed PDXs from this cohort, allowing us to deeply explore the landscape of tumor heterogeneity and clonal evolution. Here, we report on the comprehensive genomic profiling of 102 samples (11 matched normal and 91 unique patient and PDX tumor samples) from 11 patients with mCRC. These results describe complex intratumor heterogeneity and its impact on the formation and evolution of metastasis and the relationship of PDXs to patient tumors, which have important clinical implications.
RESULTS
Somatic mutations in mCRC
We sequenced 91 unique tumor specimens (58 multiregion primary tumor samples, 24 metastatic samples, and 9 PDX samples) and 11 matched normal samples from 11 patients (CRC1 to CRC11) with mCRC (Fig. 1 and table S1). All patients were microsatellite stable. Eight patients received chemotherapy before primary or metastatic tumor resection (Fig. 1). The initially resected primary tumor region and all metastases were sequenced using both whole-genome and exome sequencing, and PDXs were sequenced using exome sequencing for the discovery of somatic mutations. Subsequently, all samples, including 47 remaining multiregion primary samples, were then subjected to a targeted validation sequencing of 152 cancer genes and >100,000 somatic mutations chosen in the discovery phase (Fig. 1).
Fig. 1. Overview of patient cohort, samples, and study design.
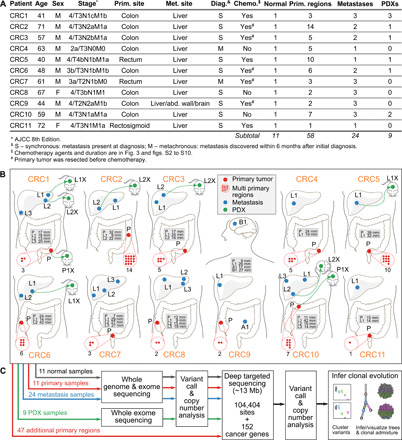
(A) Summary of the clinical data. (B) Locations of the primary tumors and distant metastasis sites for all 11 patients. Sample names are prefixed by a letter representing the site of tumor (P, primary; L, liver metastasis; A, abdominal wall metastasis; and B, brain metastasis). Tumor diameters are in the gray boxes. The number of red dots inside red dashed circles indicates the total primary regions for the corresponding primary tumors. Several primary and metastasis samples were implanted into immunodeficient mice yielding PDX tumors (named with suffix letter X with a green dotted arrow linked to the parental patient sample). (C) Sequencing and clonal evolution analysis.
Analysis of whole-genome, exome, and targeted validation sequencing data identified ~11,000 somatic mutations per patient, including single nucleotide variants (SNVs) and indels (fig. S1). The most frequently mutated genes were genes known in CRC including APC, TP53, KRAS, PIK3CA, TCF7L2, SMAD3, and SMAD4 (1, 2). Frequent copy number gain was observed in IRS2, EGFR, and MYC, while frequent copy number loss was observed in TP53, SMAD4, TCF4, and NRAS. Direct comparison of mutational profiles of matched primary and metastatic samples revealed very few cancer-related genes mutated in the metastasis that were not seen in any of the primary regions evaluated in the same patient (fig. S1 and Fig. 2).
Fig. 2. Clonal evolution in mCRC: From primary to metastasis and patient-derived xenograft.
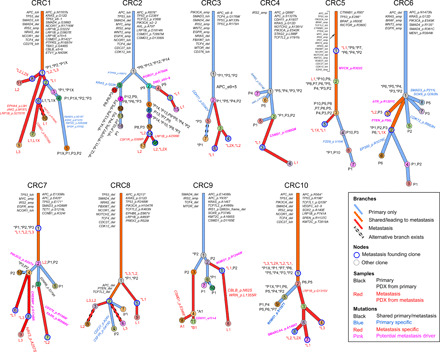
Each patient’s clonal history is presented by a tree whose nodes represent clones; branches represent evolution paths (length scaled by the square root of number of clonal marker mutations). Branches are labeled with potential driver mutations, and clone nodes are labeled with samples where the clones are found (with nonzero cellular fraction). A star (*) next to a sample indicates that the clone is the founding clone of the sample. Sample names are prefixed by a letter representing the site of tumor (P, primary; L, liver metastasis; A, abdominal wall metastasis; and B, brain metastasis). Suffix X indicates PDX. Clones marked as “alternative branch exists” were those predicted by ClonEvol to have an alternative position that does not change seeding model (see the Supplementary Figures of individual patients). Patient CRC11 was excluded from clonal evolution analysis because of low-quality primary sample.
To characterize metastatic progression, we reconstructed the clonal evolution of the tumors within individual patients and found that metastasis was often established after the accumulation of a large number of mutations and acquisition of many driver events (Fig. 2 and data S1), consistent with previous studies (3, 7). On average, the numbers of clonal marker variants that we were able to assign were ~1507 (~58 coding) for the founding clones and ~155 (~7.3 coding) for the subclones (data S3). Genes commonly mutated before metastasis establishment included APC, TP53, KRAS, PIK3CA, TCF7L2, and SMAD4. To further delineate potential metastasis drivers, we traced subclones leading to metastasis (i.e., those detected in the primary tumor that seeded metastasis). We found several mutations as clonal marker variants of clones that metastasized, including those targeting PTEN, PIK3CG, ROBO1, DMD, CDH11, and LRP1B (Fig. 2; pink mutations).
Intra- and intertumor heterogeneity is substantial and is reflected in metastatic progression
An understanding of the intra- and intertumor heterogeneity of mCRC and how it is reflected in the evolution and establishment of distant metastases may have implications in the response to treatment and subsequent development of resistance. We sampled all available metastatic sites and profiled all areas of the primary tumors where high-quality unique tissue cores could be extracted, yielding as many as 14 regions from a single primary tumor (Fig. 1). In general, we sequenced higher numbers of primary regions from bigger tumors (Pearson correlation, ~0.8; Fig. 1). We observed substantial intratumor heterogeneity with metastatic progression. All unique specimens carried only a subset (~10 to 70%; mean, 25%) of total clones identified in all samples from a given patient (Fig. 3A). Primary and metastasis-specific subclones were found in all patients (Fig. 2). Notably, the metastasis-founding subclones were not detected in any primary regions we studied in four cases (CRC3, fig. S2; CRC4, fig. S3; CRC8 clone 5, fig. S4; and CRC9 clone 4, fig. S5) and were detected in only a subset of primary regions in six cases (CRC2, Fig. 3; CRC5, fig. S6; CRC6, fig. S7; CRC7, fig. S8; CRC9 clone 5, fig. S5; and CRC1, fig. S9). In the case with the highest number of primary regions (CRC2), two metastasis-seeding subclones were found with low cellular fractions in fewer than 3 of the 14 primary regions (Fig. 3). Subclone 4 (salmon color) seeding metastasis L1 was found in only one primary region (P9 at 5%). Subclone 10 (brown color) seeding metastasis L2 was found in only two primary regions (P3 at 3.5% and P8 at 3%; Fig. 3). While mutations in APC, TP53, KRAS, PIK3CA, TCF7L2, and SMAD4 were often clonal, mutations in SMAD3, CDH11, CSMD1, EP300, and ROBO1 were usually subclonal (figs. S1 and S2).
Fig. 3. CRC has substantial heterogeneity reflected in metastatic progression and establishment of PDXs.
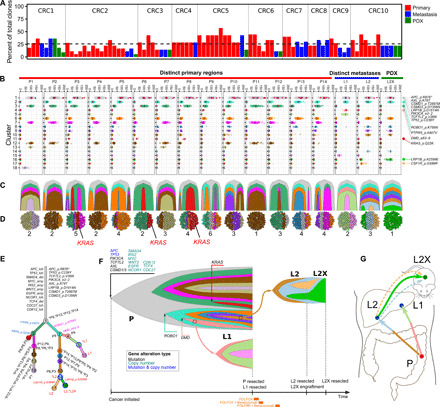
(A) Percentage of total patient cancer clones detected in individual samples. (B to G) Clonal heterogeneity and evolution in patient CRC2. (B) Clustering of variants displaying purity-corrected cancer cellular fraction (CCF) across 14 primary regions, 2 metastases, and a PDX. Black bars, mean CCFs; red dots, nonsilent mutations in cancer genes (details on the far right). (C) Fishplot of clonal evolution and (D) clonal admixture of individual samples. (E) Clonal evolution tree with branch length scaled by the square root of the number of clonal marker variants. (F) Fishplot of the clonal evolution across all samples (time not to scale; sample acquisitions at the right end). Treatments are presented at the bottom. Primary tumor is presented as a combination of all primary regions. Arrows indicate metastasis seeding. Cancer genes, whose somatic alterations are clonal markers of a clone, are indicated with arrows pointing to the tips of the fishplot corresponding to the clone. (G) Anatomic representation of tumor location and metastatic progression. Arrows represent seeding clones between samples/sites. Dashed arrows represent clones regressed in PDX. Colors are matched throughout panels (B to G).
The clonal evolution of individual patients highlights heterogeneous subclonal mutations found in genes likely contributing to metastasis or treatment resistance: KRAS (13, 14) (CRC2), PTEN (15) (CRC6), and PIK3CG (16) (CRC7). Most notably, a subclonal KRAS Q22K mutation was present in only 3 of 14 primary regions from CRC2 and was not detected by clinical genomic testing of the standard-of-care clinical biopsy (Fig. 3). The KRAS Q22K mutation is known to activate KRAS and increase RAS–GTP (guanosine triphosphate) levels (17), a mechanism of resistance to EGFR inhibitors (13). This suggests that rare subclones could carry clinically relevant mutations that may only be detected by extensive, multiregion genomic profiling. In addition, a PTEN F56L mutation in patient CRC6 was a marker of the founding clone of the PDX derived from a metastasis, but it was subclonal in the primary tumor. It may therefore be a driver of the metastatic subclones found in the primary tumor (fig. S7) (15).
While most chromosomal aberrations were early clonal events (data S2), we also found subclonal copy number alterations including amplification of EGFR (CRC9 and CRC10) and WNT2 (CRC7 and CRC9) and deletion of FBXW7 (CRC1), APC, PTEN, TCF7L2, TET1 (CRC8), and NRAS (CRC9) (fig. S1). Notably, we found that patient CRC8 had large deletions involving chromosome 5 (carrying APC) and chromosome 10 (carrying PTEN and TCF7L2) that were among the subclonal events that occurred independent of the subclone that seeded first liver metastasis L1 (fig. S4B). The mutant alleles of APC and TCF7L2 were lost, and the subclone carrying the losses of APC, PTEN, and TCF7L2 seeded two liver metastases L2 and L3. This suggests that metastasis can develop from cells carrying monoallelic inactivation of APC contrasting with biallelic inactivation of APC in other APC-positive patients (fig. S1). Similar to the PTEN mutation in CRC6, loss of PTEN in CRC8 may have contributed to clonal expansion and metastasis.
Distant metastases can arise via polyclonal seeding
Single-cell seeding has classically been viewed as the primary model of metastasis dissemination in cancer (18). However, there is growing evidence that metastasis seeding may involve or require the cooperation of multiple cells that either represent a single clone (monoclonal seeding) or multiple distinct clones (polyclonal seeding) (19, 20). Our data provide the ability to find rare metastasis-seeding subclones present in primary tumors. We identified two cases (CRC5 and CRC7) whose metastases were seeded by multiple distinct clones from the primary tumor (Fig. 4 and figs. S6 and S8). We observed at least one subclone with marker variants that were present at subclonal levels in both the primary and metastasis. This supports polyclonal metastasis seeding by the subclones and one of their ancestor clones that was also found to be present in both the primary and metastasis. In CRC5, the founding clone (1) and two subclones (2 and 3) in the primary tumor were found to seed the liver metastasis (fig. S6). The metastasis-seeding subclones were not present in the first primary region (P1) but were present in all the subsequently studied primary regions (fig. S6). In CRC7, at least three clones, including the founding clone (1) and two subclones (2 and 4), were identified in metastases. Those clones were involved in the establishment of three liver metastases: two were seeded by multiple clones/cells, and one was seeded by a single clone/cell (fig. S8). In this case, the metastasis-seeding clones were present in only a subset of the primary regions (fig. S8). We also observed two cases where the metastasis-seeding clones were present at the subclonal levels only in the metastatic samples (Fig. 4), suggesting polyclonal metastasis seeding from one metastasis to another metastasis. In summary, metastatic biology and progression have been thought to follow a single-cell seeding mechanism (a monoclonal model). This model was supported in seven of our cases. However, polyclonal metastasis seeding was detected in four cases (CRC5, CRC7, CRC8, and CRC10).
Fig. 4. Hepatic metastases from CRC may arise from polyclonal seeding from the primary tumor.
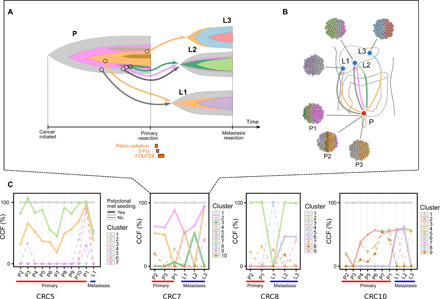
(A) Time-based fishplot presentation of clonal evolution from the initiation of cancer (far left) through metastatic progression in patient CRC7. Cancer genes whose mutation is a clonal marker of a clone are indicated with arrows pointing to the tip of the corresponding clone’s fishplot. (B) Anatomic representation of tumor progression and metastatic progression, with the clonal subpopulation of each sample shown. These data support the initiation of metastases by two distinct clones from the primary P, in L1 and L2 of this patient. Radiation and chemotherapy do not alter the general scheme of seeding models (i.e., change from monoclonal to polyclonal model). (C) The median purity-corrected CCF of the variant clusters identified in patients CRC5, CRC7, CRC8, and CRC10. A polyclonal model is supported by the clusters whose CCF is present at subclonal levels in both primary and metastasis samples of these patients.
Model of metastasis-seeding-metastasis
Recent progress in understanding metastatic progression has highlighted several mechanisms that allow a metastasis-seeding clone to leave the primary tumor (“seed”) and grow in a distant organ where it finds an appropriate “soil” (21). However, it is not clear whether the seed or its evolved progeny are able to leave the soil again, allowing metastasis to seed another metastasis via hematogenous spread (22). Our clonal evolution models revealed four cases (CRC3, CRC8, CRC9, and CRC10) where the clonal marker variants of the metastasis-seeding clones were not detected in any of the primary regions (Fig. 5 and figs. S4 and S10) but were observed in another metastasis. This observation raises a possibility that those mutations arose in the metastasis itself, consistent with a metastasis-seeding-metastasis model.
Fig. 5. Metastasis-seeding-metastasis models in mCRC.
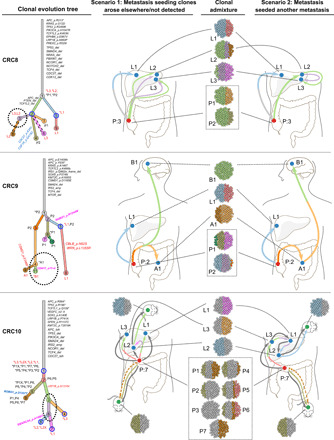
In patient CRC8 (top), clone 3 (purple, dotted circle) was found at subclonal frequency in metastases L2 and L3 but was absent in all primary regions. In patient CRC9 (middle), B1 founding clone 11 (green, dotted circle) arose from clone 4 (light orange, dotted circle) in A1, and both were absent in all primary regions. In patient CRC10 (bottom), clones 3 (light green, dotted circle) and 6 (purple, dotted circle) led to establishment of metastases L1 and L2, respectively, but were not involved in the evolution of any primary regions. These results suggest that many metastases developed not from the primary tumor but from other metastases.
In CRC8, clonal marker variants of subclone 3 were present in two distinct metastatic sites in the right lobe of the liver (L2 and L3, respectively). None of the three primary regions contained mutations found in the metastasis-shared subclone, suggesting the shared clone and its parent were involved in the seeding between the two liver metastases (Fig. 5 and fig. S4). In CRC10, none of the seven primary regions captured either of the two metastasis-seeding subclones between liver metastases L1 and L2 and between L1 and L3 (Fig. 5 and fig. S10). In CRC9, a subclone shared between the abdominal wall metastasis and brain metastasis was not detected elsewhere (Fig. 5B and fig. S5). The marker variants of this clone were found to be present at similar levels in the brain metastasis, indicating a monoclonal metastasis-seeding-metastasis mechanism. In CRC3, two liver metastases L1 and L2 shared variants (clone 3, green color) that were not detected in all primary regions, also supporting a monoclonal metastasis-seeding-metastasis model (fig. S2).
PDX models underestimate the heterogeneity of parental tumors
Our comprehensive genomic profiling and clonal evolution models allowed for extensive characterization of PDXs and comparison to their parental (patient) tumors. Furthermore, our PDX generation from subcutaneous injection of single-cell suspensions is expected to reduce geographic selection bias. We observed substantial subclonal skewing in xenografts compared with their matched parental samples (Fig. 6). In four of nine xenografts, the dominant parental tumor clones were underrepresented, while minor parental subclones became dominant (CRC2, CRC10/P1X, and CRC1/P1X,L2X). Only two xenografts contained all of the subclones found in the parental tumor, but in these cases, the parental sample contained a small number of subclones (CRC3 L2/1 clone and CRC10 L2/2 clones). Six other xenografts lost between 20 and 67% of clones compared with their parental tumors (Fig. 6).
Fig. 6. Clonal architecture and evolution of PDXs in CRC.
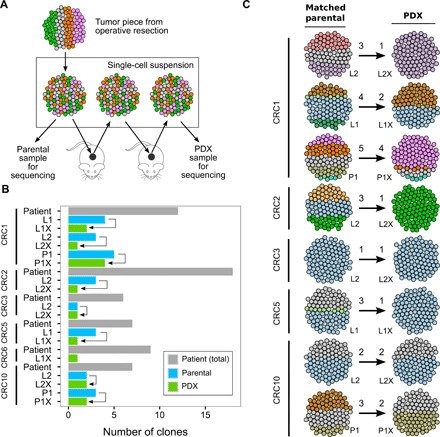
(A) Experimental procedure for tumor harvesting and PDX development via single-cell suspension. All samples were derived from tumor tissue resected at the time of surgery. Engraftment procedures uniformly involve creation of single-cell suspension at each step of harvest and subsequent PDX development to maintain the clonal diversity of each sample. All PDX samples were collected after two passages when the PDXs were considered established. (B) Summary of the clones seen in each PDX compared with matched parental tumor sample and number of clones observed in patient. (C) Clonal selection from parental tumor to its matched PDX. Numbers above the arrows represent the number of clones present in each sample. These data demonstrate that most PDXs fail to accurately recapitulate the clonal architecture of their parental tumors.
Mutations in well-characterized CRC genes (such as APC, TP53, and TCF7L2) were among the initiating events for several tumors and were concordant between the parental sample and PDX tumors, as expected. However, we also found subclonal mutations that were underrepresented in the PDX compared with the parental tumor in ROBO1 (CRC10, fig. S10), SMAD3, and KMT2C (CRC1, fig. S9). Additional comparison of PDX tumors, multiregion primary, and metastasis tumors revealed subclonal mutations with implications for treatment that were present in nonparental patient tumors but absent in the PDX. For example, in patient CRC2, a subclonal KRAS Q22K mutation present in only 3 of 14 primary regions was not recapitulated in the PDX (Figs. 3 and 6). Together, these observations demonstrate that the clonal heterogeneity of CRC tumors is not accurately represented by PDX models.
DISCUSSION
We used comprehensive genome sequencing to study clonal evolution in mCRC and defined the relationship between PDXs and patient tumors. Our reconstruction of the clonal evolution of patients with mCRC highlights complex intra- and intertumor heterogeneity reflected in metastasis dissemination and xenograft clonal selection. This may have direct clinical implications, as the failure of cancer therapies is clearly related to tumor heterogeneity and the constant, adaptive evolution of human tumors in the context of treatment response and resistance.
Treatment decisions in clinical oncology are largely based on the principle that metastases are seeded by cells from the tumor of origin (23). Metastatic biology and progression have also been thought to follow a single-cell seeding mechanism (a monoclonal model). This model was supported in seven cases. However, polyclonal metastasis seeding was detected in four cases (CRC5, CRC7, CRC8, and CRC10). This result is consistent with the earlier analyses using exome data (5, 24). Earlier study also found distinct origins between lymph node and distant metastases in primary tumors (4), supporting our model of primary seeding metastasis. Moreover, our whole-genome data allowed us to reconstruct more detailed metastasis seeding models that involved subclones established late after multiple clonal expansions, consistent with an earlier smaller study using single-cell sequencing (7). Clearly, metastasis is a complex process that may require the cooperation of multiple cells from different subclones or occur in multiple rounds of seeding that involve distinct clones. These findings suggest the need for new approaches to target the interactions of multiple cells and/or clones to inhibit the establishment and progression of metastases.
Our multiregion and whole-genome analysis allowed us to extensively assess the intratumor heterogeneity of the primary tumor and how it is reflected in metastasis. Compared with earlier exome or targeted sequencing studies (4, 6, 25), our analysis revealed substantial intratumor heterogeneity in the primary tumor affecting the metastasis dissemination. We frequently observed that metastases were seeded by rare subclones from the primary tumor. Potential metastatic clones do not necessarily represent the most abundant subpopulations in the primary tumor; instead, some metastasis-prone subclones may have a survival advantage in the bloodstream or may have a growth advantage in specific distant organs. Notably, metastasis-seeding clones are often seen in only a small subset of primary regions as a result of spatial heterogeneity in the primary tumor. This poses a challenge to design effective personalized therapies using a single or limited number of biopsies collected at a tumor site. Circulating tumor cells and/or DNA could potentially complement tumor biopsies by providing genomic signatures of subclones traveling between distant tumors (26).
As the first study to assay xenograft tumors together with matched primary regions and distant metastases, we showed that intratumor heterogeneity coupled with clonal selection in PDX models could result in a limited understanding of a patient’s entire cancer using single biopsy and xeno-engraftment experiments. This is a reflection of the inherent heterogeneity in developing PDXs and other model systems based on a small biopsy or operative specimen used for research taken from a larger tumor that results in a model that does not contain all of the cellular components of a complex tumor. This issue is even more complex due to the patient tumor microenvironment, which is increasingly recognized to influence clinical outcomes (27) but not modeled in PDXs. Furthermore, distinct tumor clones could demonstrate various survival capabilities in novel microenvironments when introduced to mouse models. Higher-resolution single-cell analyses of tumors corroborate this research as substantial diversity, and differential treatment responses were observed in patient-derived organoids (PDOs) developed from single CRC cells from one biopsy (12). Another study found that PDOs recapitulated the characteristics of the parental tumors from which they were derived, including responsiveness to a panel of drugs (11). While only a subset of clones is often represented in PDXs and PDOs, these models could still mimic treatment response of the parental tumor and may cause these different observations between studies. However, mutations in targetable or treatment-resistant markers (such as KRAS and PTEN) could be easily missed in a single biopsy, leading to missed opportunities for timely and effective therapy. Furthermore, with the success of immunotherapies in the treatment of solid tumors (28), ensuring that tumor biopsies recapitulate the complete clonal and mutational spectrum of a tumor (including all potential neoantigens) may prove to be very important.
Last, our analysis suggests that metastases may seed other metastases. Although there is a possibility that the seeding clones found in multiple metastases were not detectable in the primary due to unsequenced regions (those used for diagnostics or having low-quality material) or low clonal frequency, we have made an attempt to exhaust the primary tumors by sequencing all available tumor regions with high-quality DNA. Our results, although remaining to be validated, are indeed consistent with previous studies in circulating tumor cells. Circulating tumor cells can be seen in both patients with localized and metastatic diseases and are often more abundant in metastatic patients (29). Mutations specific to metastasis can also be detected in the blood (30). Together, this suggests that tumor cells are released into the blood from both primary and metastatic sites and, therefore, are capable of traveling to distant organs. The ability of metastases to seed other metastases has implications for clinical decision-making in patients with metastatic cancer, where directed therapy to a site of metastasis could affect subsequent progression.
These data provide insights into several aspects of CRC, including treatment response, the development of resistance, metastatic progression, and preclinical cancer modeling. The characterization of the complex clonal evolution from primary tumor to metastases may have direct implications for the treatment of patients with CRC.
MATERIALS AND METHODS
Patient cohort and sample acquisition
We evaluated matched normal, primary, and metastasis samples from 11 patients (CRC1 to CRC11) with mCRC undergoing treatment at the Alvin J. Siteman Cancer Center at the Washington University School of Medicine in St. Louis, MO, USA. This included a total of 102 samples including 91 unique tumor specimens (58 multiregion primary tumor samples, 24 metastatic samples, and 9 PDX samples) and 11 matched normal samples (10 from the blood and 1 from uninvolved colon) (Fig. 1 and table S1). Our protocol allowed the retrieval and pathological evaluation of the archived primary tumors to select for regions with high tumor content and high isolated DNA quality for multiregion sequencing (see the Supplementary Methods). This yielded various numbers of primary regions per patient depending on tissue quality (Fig. 1). All patients provided informed consent under an institutional review board–approved protocol.
Patient-derived xenografts
PDXs were grown from nine tumors (two primary and seven metastatic) from six patients for two generations of passaging in nonobese diabetic/severe combined immunodeficient mice (NOD/SCID). This followed our recently published protocol (see also the Supplementary Methods) (31).
Multiregion whole-genome, exome, and targeted sequencing
The initially resected primary tumor region corresponding to the clinical specimen evaluated on standard pathology, all metastasis, and xenograft samples were sequenced for the discovery of somatic mutations. The initial primary region and all metastatic samples were sequenced using both whole-genome (~67× tumor mean coverage) and exome (~215× tumor mean coverage) sequencing. PDX samples were subjected to exome sequencing. All samples, including 47 remaining multiregion primary samples, were then subjected to targeted validation sequencing (~251× tumor mean coverage) of 152 cancer genes and >100,000 somatic mutations (detected in the discovery phase) covering all nonsilent coding SNVs, small insertions/deletions (indels), and diploid heterozygous noncoding SNVs (Fig. 1, data S1, and Supplementary Methods). All sequencing data were deposited to the database of genotypes and phenotypes (dbGap) under the accession phs001722.
Reconstruction of clonal evolution
Heterozygous mutations validated in targeted sequencing were first clustered on the basis of their variant allele fraction using sciClone (32) to identify the founding clones and subclones that were subsequently analyzed using ClonEvol (33) to infer clonal evolution models (Fig. 1). Further details regarding patients, PDXs, and materials and methods are provided in the Supplementary Materials.
Supplementary Material
Acknowledgments
We thank all the patients who participated in this research. Funding: R.C.F., C.A.Ma., and E.R.M. received funding from The Alvin J. Siteman Cancer Center Siteman Investment Program, The Foundation for Barnes-Jewish Hospital Cancer Frontier Fund, the National Cancer Institute Cancer Center Support Grant P30 CA091842, and the Barnard Trust. R.C.F. also received funding from the Washington University PDX Development and Trial Center (U54CA224083), the American Surgical Association Foundation Fellowship, the American Cancer Society Institutional Review Grant, the Society of Surgical Oncology James Ewing Foundation Clinical Investigator Award, the Sidney Kimmel Translational Science Scholar Award, and the David Riebel Cancer Research Fund. Funding was also provided for C.A.Ma. in part by a Research Scholar Grant (130878-RSG-17-058-01-RMC) from the American Cancer Society, NIH R21 (R21CA185983-01), and NIH CTSA (UL1 TR002345). B.A.K., J.G.G., and M.S.S. were all supported by the Washington University Surgical Oncology Training Grant (T32 CA009621). T.J.L. was supported by R35 CA197561. B.S.W. received funding from the Barnes-Jewish Hospital Foundation. We thank the Siteman Tissue Procurement Core and the core grant/services of the Washington University Digestive Diseases Research Core Center (P30 DK052574) for supporting this work. Author contributions: H.X.D., C.A.Ma., and R.C.F. oversaw the experimental design, data generation, data analysis, and interpretation of the results. B.A.K., J.G.G., M.S.S., S.P.G., B.D.G., W.G.H., S.M.S., D.C.L., K.H.L., A.C.L., and R.C.F. developed the patient cohort, obtained tissues, and contributed to sample processing and obtaining of clinical data. J.G.G. developed xenograft models. C.C.F. and R.S.F. oversaw library construction, targeted enrichment, and DNA sequencing. H.X.D., B.S.W., J.Z., C.R.C., C.A.Mi., D.E.L., J.R.W., and M.G. performed data analysis and contributed to methods and algorithm development. E.R.M., R.K.W., T.J.L., C.A.Ma., and R.C.F. conceived the study concept and supervised the study. H.X.D., C.A.Ma., and R.C.F. wrote the manuscript, which was reviewed by all authors. Competing interests: All authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. Sequencing data in this study are available in the dbGap database (accession phs001722).
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/24/eaay9691/DC1
REFERENCES AND NOTES
- 1.Fearon E. R., Vogelstein B., A genetic model for colorectal tumorigenesis. Cell 61, 759–767 (1990). [DOI] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Network , Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brannon A. R., Vakiani E., Sylvester B. E., Scott S. N., McDermott G., Shah R. H., Kania K., Viale A., Oschwald D. M., Vacic V., Emde A.-K., Cercek A., Yaeger R., Kemeny N. E., Saltz L. B., Shia J., D’Angelica M. I., Weiser M. R., Solit D. B., Berger M. F., Comparative sequencing analysis reveals high genomic concordance between matched primary and metastatic colorectal cancer lesions. Genome Biol. 15, 454 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Naxerova K., Reiter J. G., Brachtel E., Lennerz J. K., van de Wetering M., Rowan A., Cai T., Clevers H., Swanton C., Nowak M. A., Elledge S. J., Jain R. K., Origins of lymphatic and distant metastases in human colorectal cancer. Science 357, 55–60 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei Q., Ye Z., Zhong X., Li L., Wang C., Myers R. E., Palazzo J. P., Fortuna D., Yan A., Waldman S. A., Chen X., Posey J. A., Basu-Mallick A., Jiang B. H., Hou L., Shu J., Sun Y., Xing J., Li B., Yang H., Multiregion whole-exome sequencing of matched primary and metastatic tumors revealed genomic heterogeneity and suggested polyclonal seeding in colorectal cancer metastasis. Ann. Oncol. 28, 2135–2141 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim T.-M., Jung S.-H., An C. H., Lee S. H., Baek I.-P., Kim M. S., Park S.-W., Rhee J.-K., Lee S.-H., Chung Y.-J., Subclonal genomic architectures of primary and metastatic colorectal cancer based on intratumoral genetic heterogeneity. Clin. Cancer Res. 21, 4461–4472 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Leung M. L., Davis A., Gao R., Casasent A., Wang Y., Sei E., Vilar E., Maru D., Kopetz S., Navin N. E., Single-cell DNA sequencing reveals a late-dissemination model in metastatic colorectal cancer. Genome Res. 27, 1287–1299 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu Z., Ding J., Ma Z., Sun R., Seoane J. A., Scott Shaffer J., Suarez C. J., Berghoff A. S., Cremolini C., Falcone A., Loupakis F., Birner P., Preusser M., Lenz H.-J., Curtis C., Quantitative evidence for early metastatic seeding in colorectal cancer. Nat. Genet. 51, 1113–1122 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stewart E., Federico S. M., Chen X., Shelat A. A., Bradley C., Gordon B., Karlstrom A., Twarog N. R., Clay M. R., Bahrami A., Freeman B. B. III, Xu B., Zhou X., Wu J., Honnell V., Ocarz M., Blankenship K., Dapper J., Mardis E. R., Wilson R. K., Downing J., Zhang J., Easton J., Pappo A., Dyer M. A., Orthotopic patient-derived xenografts of paediatric solid tumours. Nature 549, 96–100 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding L., Ellis M. J., Li S., Larson D. E., Chen K., Wallis J. W., Harris C. C., McLellan M. D., Fulton R. S., Fulton L. L., Abbott R. M., Hoog J., Dooling D. J., Koboldt D. C., Schmidt H., Kalicki J., Zhang Q., Chen L., Lin L., Wendl M. C., McMichael J. F., Magrini V. J., Cook L., McGrath S. D., Vickery T. L., Appelbaum E., Deschryver K., Davies S., Guintoli T., Lin L., Crowder R., Tao Y., Snider J. E., Smith S. M., Dukes A. F., Sanderson G. E., Pohl C. S., Delehaunty K. D., Fronick C. C., Pape K. A., Reed J. S., Robinson J. S., Hodges J. S., Schierding W., Dees N. D., Shen D., Locke D. P., Wiechert M. E., Eldred J. M., Peck J. B., Oberkfell B. J., Lolofie J. T., Du F., Hawkins A. E., O’Laughlin M. D., Bernard K. E., Cunningham M., Elliott G., Mason M. D., Thompson D. M., Ivanovich J. L., Goodfellow P. J., Perou C. M., Weinstock G. M., Aft R., Watson M., Ley T. J., Wilson R. K., Mardis E. R., Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 464, 999–1005 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vlachogiannis G., Hedayat S., Vatsiou A., Jamin Y., Fernández-Mateos J., Khan K., Lampis A., Eason K., Huntingford I., Burke R., Rata M., Koh D.-M., Tunariu N., Collins D., Hulkki-Wilson S., Ragulan C., Spiteri I., Moorcraft S. Y., Chau I., Rao S., Watkins D., Fotiadis N., Bali M., Darvish-Damavandi M., Lote H., Eltahir Z., Smyth E. C., Begum R., Clarke P. A., Hahne J. C., Dowsett M., de Bono J., Workman P., Sadanandam A., Fassan M., Sansom O. J., Eccles S., Starling N., Braconi C., Sottoriva A., Robinson S. P., Cunningham D., Valeri N., Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 359, 920–926 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roerink S. F., Sasaki N., Lee-Six H., Young M. D., Alexandrov L. B., Behjati S., Mitchell T. J., Grossmann S., Lightfoot H., Egan D. A., Pronk A., Smakman N., van Gorp J., Anderson E., Gamble S. J., Alder C., van de Wetering M., Campbell P. J., Stratton M. R., Clevers H., Intra-tumour diversification in colorectal cancer at the single-cell level. Nature 556, 457–462 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Misale S., Yaeger R., Hobor S., Scala E., Janakiraman M., Liska D., Valtorta E., Schiavo R., Buscarino M., Siravegna G., Bencardino K., Cercek A., Chen C.-T., Veronese S., Zanon C., Sartore-Bianchi A., Gambacorta M., Gallicchio M., Vakiani E., Boscaro V., Medico E., Weiser M., Siena S., Di Nicolantonio F., Solit D., Bardelli A., Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 486, 532–536 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boutin A. T., Liao W.-T., Wang M., Hwang S. S., Karpinets T. V., Cheung H., Chu G. C., Jiang S., Hu J., Chang K., Vilar E., Song X., Zhang J., Kopetz S., Futreal A., Wang Y. A., Kwong L. N., DePinho R. A., Oncogenic Kras drives invasion and maintains metastases in colorectal cancer. Genes Dev. 31, 370–382 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Roock W., De Vriendt V., Normanno N., Ciardiello F., Tejpar S., KRAS, BRAF, PIK3CA, and PTEN mutations: Implications for targeted therapies in metastatic colorectal cancer. Lancet Oncol. 12, 594–603 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Semba S., Itoh N., Ito M., Youssef E. M., Harada M., Moriya T., Kimura W., Yamakawa M., Down-regulation of PIK3CG, a catalytic subunit of phosphatidylinositol 3-OH kinase, by CpG hypermethylation in human colorectal carcinoma. Clin. Cancer Res. 8, 3824–3831 (2002). [PubMed] [Google Scholar]
- 17.Janakiraman M., Vakiani E., Zeng Z., Pratilas C. A., Taylor B. S., Chitale D., Halilovic E., Wilson M., Huberman K., Ricarte Filho J. C., Persaud Y., Levine D. A., Fagin J. A., Jhanwar S. C., Mariadason J. M., Lash A., Ladanyi M., Saltz L. B., Heguy A., Paty P. B., Solit D. B., Genomic and biological characterization of exon 4 KRAS mutations in human cancer. Cancer Res. 70, 5901–5911 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Talmadge J. E., Fidler I. J., AACR centennial series: The biology of cancer metastasis: Historical perspective. Cancer Res. 70, 5649–5669 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aceto N., Bardia A., Miyamoto D. T., Donaldson M. C., Wittner B. S., Spencer J. A., Yu M., Pely A., Engstrom A., Zhu H., Brannigan B. W., Kapur R., Stott S. L., Shioda T., Ramaswamy S., Ting D. T., Lin C. P., Toner M., Haber D. A., Maheswaran S., Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 158, 1110–1122 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheung K. J., Padmanaban V., Silvestri V., Schipper K., Cohen J. D., Fairchild A. N., Gorin M. A., Verdone J. E., Pienta K. J., Bader J. S., Ewald A. J., Polyclonal breast cancer metastases arise from collective dissemination of keratin 14-expressing tumor cell clusters. Proc. Natl. Acad. Sci. U.S.A. 113, E854–E863 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fidler I. J., The pathogenesis of cancer metastasis: The “seed and soil” hypothesis revisited. Nat. Rev. Cancer 3, 453–458 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Micalizzi D. S., Maheswaran S., Haber D. A., A conduit to metastasis: Circulating tumor cell biology. Genes Dev. 31, 1827–1840 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanahan D., Weinberg R. A., Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Ulintz P. J., Greenson J. K., Wu R., Fearon E. R., Hardiman K. M., Lymph node metastases in colon cancer are polyclonal. Clin. Cancer Res. 24, 2214–2224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goswami R. S., Patel K. P., Singh R. R., Meric-Bernstam F., Kopetz S., Subbiah V., Alvarez R. H., Davies M. A., Jabbar K. J., Roy Chowdhuri S., Lazar A. J., Medeiros L. J., Broaddus R. R., Luthra R., Routbort M. J., Hotspot mutation panel testing reveals clonal evolution in a study of 265 paired primary and metastatic tumors. Clin. Cancer Res. 21, 2644–2651 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pantel K., Alix-Panabières C., Liquid biopsy and minimal residual disease - latest advances and implications for cure. Nat. Rev. Clin. Oncol. 16, 409–424 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Woolston A., Khan K., Spain G., Barber L. J., Griffiths B., Gonzalez-Exposito R., Hornsteiner L., Punta M., Patil Y., Newey A., Mansukhani S., Davies M. N., Furness A., Sclafani F., Peckitt C., Jiménez M., Kouvelakis K., Ranftl R., Begum R., Rana I., Thomas J., Bryant A., Quezada S., Wotherspoon A., Khan N., Fotiadis N., Marafioti T., Powles T., Lise S., Calvo F., Guettler S., von Loga K., Rao S., Watkins D., Starling N., Chau I., Sadanandam A., Cunningham D., Gerlinger M., Genomic and transcriptomic determinants of therapy resistance and immune landscape evolution during anti-EGFR treatment in colorectal cancer. Cancer Cell 36, 35–50.e9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ott P. A., Bang Y.-J., Piha-Paul S. A., Razak A. R. A., Bennouna J., Soria J.-C., Rugo H. S., Cohen R. B., O’Neil B. H., Mehnert J. M., Lopez J., Doi T., van Brummelen E. M. J., Cristescu R., Yang P., Emancipator K., Stein K., Ayers M., Joe A. K., Lunceford J. K., T-cell-inflamed gene-expression profile, programmed death ligand 1 expression, and tumor mutational burden predict efficacy in patients treated with pembrolizumab across 20 cancers: KEYNOTE-028. J. Clin. Oncol. 37, 318–327 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Bettegowda C., Sausen M., Leary R. J., Kinde I., Wang Y., Agrawal N., Bartlett B. R., Wang H., Luber B., Alani R. M., Antonarakis E. S., Azad N. S., Bardelli A., Brem H., Cameron J. L., Lee C. C., Fecher L. A., Gallia G. L., Gibbs P., Le D., Giuntoli R. L., Goggins M., Hogarty M. D., Holdhoff M., Hong S. M., Jiao Y., Juhl H. H., Kim J. J., Siravegna G., Laheru D. A., Lauricella C., Lim M., Lipson E. J., Marie S. K., Netto G. J., Oliner K. S., Olivi A., Olsson L., Riggins G. J., Sartore-Bianchi A., Schmidt K., Shih l.-M., Oba-Shinjo S. M., Siena S., Theodorescu D., Tie J., Harkins T. T., Veronese S., Wang T. L., Weingart J. D., Wolfgang C. L., Wood L. D., Xing D., Hruban R. H., Wu J., Allen P. J., Schmidt C. M., Choti M. A., Velculescu V. E., Kinzler K. W., Vogelstein B., Papadopoulos N., Diaz L. A. Jr., Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 6, 224ra24 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tie J., Wang Y., Tomasetti C., Li L., Springer S., Kinde I., Silliman N., Tacey M., Wong H.-L., Christie M., Kosmider S., Skinner I., Wong R., Steel M., Tran B., Desai J., Jones I., Haydon A., Hayes T., Price T. J., Strausberg R. L., Diaz L. A. Jr., Papadopoulos N., Kinzler K. W., Vogelstein B., Gibbs P., Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci. Transl. Med. 8, 346ra92 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grossman J. G., Nywening T. M., Belt B. A., Panni R. Z., Krasnick B. A., DeNardo D. G., Hawkins W. G., Goedegebuure S. P., Linehan D. C., Fields R. C., Recruitment of CCR2+ tumor associated macrophage to sites of liver metastasis confers a poor prognosis in human colorectal cancer. Oncoimmunology 7, e1470729 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller C. A., White B. S., Dees N. D., Griffith M., Welch J. S., Griffith O. L., Vij R., Tomasson M. H., Graubert T. A., Walter M. J., Ellis M. J., Schierding W., DiPersio J. F., Ley T. J., Mardis E. R., Wilson R. K., Ding L., SciClone: Inferring clonal architecture and tracking the spatial and temporal patterns of tumor evolution. PLOS Comput. Biol. 10, e1003665 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dang H. X., White B. S., Foltz S. M., Miller C. A., Luo J., Fields R. C., Maher C. A., ClonEvol: Clonal ordering and visualization in cancer sequencing. Ann. Oncol. 28, 3076–3082 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kandoth C., McLellan M. D., Vandin F., Ye K., Niu B., Lu C., Xie M., Zhang Q., McMichael J. F., Wyczalkowski M. A., Leiserson M. D. M., Miller C. A., Welch J. S., Walter M. J., Wendl M. C., Ley T. J., Wilson R. K., Raphael B. J., Ding L., Mutational landscape and significance across 12 major cancer types. Nature 502, 333–339 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Griffith M., Griffith O. L., Smith S. M., Ramu A., Callaway M. B., Brummett A. M., Kiwala M. J., Coffman A. C., Regier A. A., Oberkfell B. J., Sanderson G. E., Mooney T. P., Nutter N. G., Belter E. A., Du F., Long R. L., Abbott T. E., Ferguson I. T., Morton D. L., Burnett M. M., Weible J. V., Peck J. B., Dukes A., McMichael J. F., Lolofie J. T., Derickson B. R., Hundal J., Skidmore Z. L., Ainscough B. J., Dees N. D., Schierding W. S., Kandoth C., Kim K. H., Lu C., Harris C. C., Maher N., Maher C. A., Magrini V. J., Abbott B. S., Chen K., Clark E., Das I., Fan X., Hawkins A. E., Hepler T. G., Wylie T. N., Leonard S. M., Schroeder W. E., Shi X., Carmichael L. K., Weil M. R., Wohlstadter R. W., Stiehr G., McLellan M. D., Pohl C. S., Miller C. A., Koboldt D. C., Walker J. R., Eldred J. M., Larson D. E., Dooling D. J., Ding L., Mardis E. R., Wilson R. K., Genome modeling system: A knowledge management platform for genomics. PLOS Comput. Biol. 11, e1004274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuilman T., Velds A., Kemper K., Ranzani M., Bombardelli L., Hoogstraat M., Nevedomskaya E., Xu G., de Ruiter J., Lolkema M. P., Ylstra B., Jonkers J., Rottenberg S., Wessels L. F., Adams D. J., Peeper D. S., Krijgsman O., CopywriteR: DNA copy number detection from off-target sequence data. Genome Biol. 16, 49 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Griffith M., Miller C. A., Griffith O. L., Krysiak K., Skidmore Z. L., Ramu A., Walker J. R., Dang H. X., Trani L., Larson D. E., Demeter R. T., Wendl M. C., McMichael J. F., Austin R. E., Magrini V., McGrath S. D., Ly A., Kulkarni S., Cordes M. G., Fronick C. C., Fulton R. S., Maher C. A., Ding L., Klco J. M., Mardis E. R., Ley T. J., Wilson R. K., Optimizing cancer genome sequencing and analysis. Cell Syst. 1, 210–223 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller C. A., McMichael J., Dang H. X., Maher C. A., Ding L., Ley T. J., Mardis E. R., Wilson R. K., Visualizing tumor evolution with the fishplot package for R. BMC Genomics 17, 880 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/24/eaay9691/DC1


