Abstract
Melanomas that have histopathologic features that overlap with those of Spitz nevus are referred to as spitzoid melanomas. However, the diagnostic concept is used inconsistently and genomic analyses suggest it is a heterogeneous category. Spitz tumors, the spectrum of melanocytic neoplasms extending from Spitz nevi to their malignant counterpart Spitz melanoma, are defined in the 2018 WHO classification of skin tumors by the presence of specific genetic alterations such a kinase fusions or HRAS mutations. It is unclear what fraction of ‘spitzoid melanomas’ defined solely by their histopathologic features belong to the category of Spitz melanoma or to other melanoma subtypes. We assembled a cohort of 25 spitzoid melanomas diagnosed at a single institution over an eight-year period and performed high coverage DNA sequencing of 480 cancer related genes. Transcriptome wide RNA sequencing was performed for select cases. Only 9 cases (36%) had genetic alterations characteristic of Spitz melanoma, including HRAS mutation or fusion involving BRAF, ALK, NTRK1, or MAP3K8. The remaining cases were divided into those with a MAPK activating mutation and those without. Both Spitz melanoma and spitzoid melanomas in which a MAPK activating mutation could not be identified tended to occur in younger patients on skin with little solar elastosis, infrequently harbored TERT promoter mutations, and had a lower burden of pathogenic mutations than spitzoid melanomas with non-Spitz MAPK activating mutations. The MAPK activating mutations identified affected non-V600 residues of BRAF as well as NRAS, MAP2K1/2, NF1 and KIT while BRAF V600 mutations, the most common mutations in melanomas of the WHO low-CSD category, were entirely absent.
While the ‘spitzoid melanomas’ comprising our cohort were enriched for bona fide Spitz melanomas, the majority of melanomas fell outside of the genetically defined category of Spitz melanomas, indicating that histomorphology is an unreliable predictor of Spitz lineage.
Introduction
Spitz nevi are composed of epithelioid to spindled melanocytes with abundant homogenous cytoplasm and large vesicular nuclei, and often display epidermal hyperplasia, clefts around and between melanocytes, Kamino bodies, and maturation with descent. They can have limited scatter of melanocytes into the upper levels of the epidermis1. They were first described by Sophie Spitz in 1948 as melanomas of childhood2. Clinically, they are most commonly encountered as rapidly growing lesions in children but can occur at any age1,3. Spitz tumor refers to the spectrum of melanocytic tumors with histopathologic features of Spitz nevus that ranges from benign to malignant. They may be challenging to classify as benign or malignant because histopathologic features that are commonly taken as indicators of malignancy such as nuclear atypia, scatter of melanocytes in the upper epidermis, poor maturation within the dermis, deep extension, and deep dermal mitoses are not uncommonly seen in Spitz tumors with benign biologic behavior4.
Recently, a broad spectrum of MAPK activating alterations have been identified in Spitz nevi that differ from those in other types of melanocytic nevi. These include activating point mutations in HRAS (often with copy number gain of mutant HRAS) and rearrangements involving the serine/threonine kinases BRAF and MAP3K8 or the receptor tyrosine kinases (RTKs) ROS1, ALK, NTRK1, NTRK3, RET, MET, and MERTK5–13 and occur in a mutually exclusive fashion with only one mutation in a given Spitz nevus. Despite comprehensive genetic profiling of many Spitz nevi for other oncogenes and tumor suppressors, additional concomitant oncogenic drivers have not been identified, suggesting that these mutations alone are capable of initiating the clonal expansion that results in a Spitz nevus. Early studies indicated that the histomorphologic features of Spitz nevi correspond with their initiating mutations to some degree14–16. Thus, the diverse spectrum of initiating mutations in Spitz tumors and the resulting variation in the clinical phenotype likely contributes to the diagnostic difficulty of Spitz tumors. In contrast, the initiating mutations in common acquired nevi are much more homogeneous with over 80% harboring BRAF V600E mutations17,18. While there have been a number of iterations of criteria to distinguish Spitz nevi from melanoma19, even their authors acknowledge their limitations.
The term spitzoid melanoma refers to histopathologically malignant tumors comprised of large epithelioid melanocytes resembling those found in Spitz nevi. Spitzoid melanoma is thus a purely morphology-based diagnosis, and prior studies have shown that many tumors classified as such have genetic hallmarks of melanomas of skin with low-cumulative sun-induced damage (low-CSD melanomas), such as BRAF and NRAS mutations and a high mutation burden with a UV signature. As Spitz nevi do not show activating BRAF or NRAS point mutations, spitzoid melanomas with these mutations are not genetically related to Spitz nevi. In one study, fusions involving BRAF or RTKs were identified in 41% of Spitz nevi, but only 13% of spitzoid melanomas7. In another study of six pediatric spitzoid melanomas, kinase fusions typical of Spitz nevi were identified in 5 cases20. Activating BRAF or NRAS mutations, which are typical of other melanomas were identified in 20–86% of spitzoid melanomas across various studies21–24. De Forno et al. did not identify HRAS mutation, found in a subset of Spitz tumors, in a cohort of 21 spitzoid melanomas and on this basis argued that spitzoid melanomas are not related to HRAS-mutated Spitz nevi25. Lazova et al. performed exome sequencing of 27 spitzoid melanomas and BRAF V600 mutations were found in 37% and NRAS mutations in 15% of cases. Only one case (4%) had an HRAS mutation indicative of a Spitz lineage24. Gene fusions are not detectable by exome sequencing and thus could not be analyzed in this study. The majority of cases in this series also had a mutation burden comparable to low- and high-CSD melanomas.
Based on the observation that a considerable portion of spitzoid melanomas have the genetic hallmarks of other types of cutaneous melanomas, the 2018 WHO classification of skin tumors introduced the concept of Spitz melanoma (malignant Spitz tumor) as a melanoma subtype that not only has the morphologic features of Spitz tumors, but also has their genetic hallmarks, namely the distinct spectrum of MAPK activating alterations described above26. This is likely an important distinction, as clinical evidence suggests that tumors at the malignant end of the spectrum of Spitz tumors have a better prognosis than other types of cutaneous melanomas. This applies particularly to children, in whom spitzoid tumors with malignant histopathologic features less commonly metastasize with lethal outcome as compared to conventional melanomas27. Therefore, we have designated spitzoid melanomas in children as “spitzoid melanoma, childhood type” in our practice and provide a comment that although they have histopathologic features of malignancy, their risk for recurrence and death appears lower than for conventional (or ‘adult-type’) cutaneous melanoma.
Here we investigate what fraction of spitzoid melanomas in our practice fall into the newly defined entity Spitz melanoma, and analyze their clinical, histomorphologic and genetic features.
Materials and Methods
Case Selection and Histopathologic analysis
After approval from the institutional review board at the University of California, San Francisco, we searched our archives for melanomas diagnosed from 2010–2017 and annotated as having spitzoid features. Each case was re-reviewed by three pathologists (S.P., S.R., I.Y.) and only cases that were determined to have spitzoid cytomorphologic features by consensus and had archival tissue available for molecular analysis were included. Pathologic features such epidermal effacement and hyperplasia, ulceration, Kamino body formation, dermal mitoses, and cytology/pleomorphism were recorded for each case. Pigmentation was graded on a 0–4 scale as previously described28. Similarly, the extent of solar elastosis was graded on a previously developed scale29. Clinical follow-up data was obtained from a combination of chart review and patient outreach.
DNA and RNA Extraction
Hematoxylin and eosin stained sections were used to guide tumor microdissection from formalin-fixed paraffin embedded (FFPE) sections of 10 μm in thickness. DNA and RNA extraction was performed using the QIAGEN Allprep DNA/RNA FFPE extraction kit (QIAGEN, Germantown, MD, p/n 80204).
DNA Sequencing and Analysis
50–250 ng of DNA was prepared for sequencing using the KAPA Hyper Prep Kit (KAPA Biosciences, Wilmington, MA p/n KK8504) according to the manufacturer’s instructions. Custom-designed bait libraries were used to target the coding regions of 480 genes (Supplementary Table 1). Select introns of kinases rearranged in melanocytic tumors (ALK, BRAF, MET, NTRK1, NTRK3, RET, ROS1, PRKCA) and the promoter region of TERT were also targeted. Sequencing was performed as paired-end 100bp reads on a HiSeq2500 (Illumina).
Sequence reads were mapped to the human genome (hg19) using the Burrows-Wheeler aligner (BWA)30. Recalibration of reads and variant calling were performed using the Genome Analysis Toolkit (GATK)31 and FreeBayes32. Quality metrics were determined using Picard33 (Supplementary Table 2). Three samples with coverage below 25x tumor-specific coverage were excluded from the study. Variant annotation was performed with Annovar34. Structural variant detection was performed with Delly35 and reviewed by visual inspection in the Integrative Genomics Viewer36. Resulting fusion transcripts were predicted by joining the exons upstream and downstream from the genomic breakpoint. Genomic copy number assessment was performed with CNVkit37 which uses off-target reads with adjustment of copy ratios for tumor fraction as estimated by THetA38.
We reviewed all alterations annotated as pathogenic or likely pathogenic in ClinVar39. We excluded variants with greater than 1% prevalence in the NHLBI GO Exome Sequencing Project40 or 1000 Genomes Project41 from further review as they likely represent benign germline polymorphisms. We classified truncating mutations in known tumor suppressors as pathogenic. For genes in which a cancer related pathogenic alteration was identified, we manually annotated all remaining variants of that gene as pathogenic, likely pathogenic, uncertain significance, and likely benign by literature review, using various databases as annotation resources (OMIM42, MyCancerGenome43). We determined the number of pathogenic copy number alterations in each case. Shallow deletions of a tumor suppressor accompanied by pathogenic alteration of that tumor suppressor was considered a pathogenic copy number alteration. Deep deletions of tumor suppressors were counted as two pathogenic alterations. Copy number increase of an oncogene was counted as one pathogenic alteration, whereas amplification was counted as two pathogenic alterations. We also counted copy loss of chromosome 9p without mutation in the remaining allele of CDKN2A as a pathogenic alteration, given the importance of this tumor suppressor in melanoma and reports of gene silencing by promoter hypermethylation.
RNA Sequencing and Analysis
For the nine cases in which a pathogenic MAPK activating alteration was not identified by DNA sequencing, whole transcriptome RNA sequencing was performed. 170–400 ng of total RNA was prepared for sequencing using the KAPA stranded RNA-seq kit (KAPA Biosciences, Wilmington, MA, p/n KK8400) according to the manufacturer’s instructions. Subsequently, exome capture was carried out using NimbleGen SeqCap EZ MedExome target enrichment probes (Roche, Basel, Switzerland, p/n 07681330001) following the manufacturer’s protocol. Sequencing was performed on a NovaSeq 6000 system (Illumina Inc, San Diego, CA) with 150-bp paired-end reads. Sequence reads were mapped to the reference human genome (hg19) using STAR44 and fusions were identified by FusionCatcher45 and verified by manual review.
Statistical Analyses
A genetic dataset for cutaneous melanoma provided by the Cancer Genome Atlas (Skin Cutaneous melanoma, TCGA)46 was downloaded from cBioPortal47,48, providing mutational data for 448 cases of cutaneous melanomas. Previous studies have identified kinase fusions in the TCGA49,50. The prevalence of mutations in the TCGA dataset was compared to that of our cohort by Fisher’s exact test.
Pearson correlation was calculated between age, number of pathogenic mutations, and extent of solar elastosis (converting the scale from Landi et al. which ranges from 0 to 3+ to an numeric scale ranging from 1–11). The statistical significance of the differences between each group was calculated by two-sided t-test.
Results
A total of 25 patients with spitzoid melanoma were included in this study (see Table 1 for summary of clinical and histologic features). For 19 patients, we received biopsies in formalin, and for the remaining six patients we received slides sent in consultation. Their average age was 43.8 years (range 4–80 years), including six patients 16 years of age or younger. Males and females were evenly represented. The spitzoid melanomas most commonly involved the extremities (64%, 16/25) and less commonly the head and neck (20%, 5/25) or the trunk (16%, 4/25). The average Breslow thickness was 2.1 mm (range 0.4 – 8.4 mm) and the mean diameter was 5.9 mm (range 2.2 – 9.8 mm). It should be noted that some melanomas were transected at the base of the biopsy, so the Breslow thickness could be underestimated.
Table 1.
Clinical and Histopathologic features of Spitzoid Melanomas.
| CASE | AGE | SEX | SITE | EPIDERMAL HYPERPLASIA | EPIDERMAL EFFACEMENT | ULCERATION | KAMINO BODIES | PIGMENTATION (SCORE 0–4) | SOLAR ELASTOSIS | DERMAL MITOSES (PER MM2) | CYTOLOGY | PLEOMORPHISM | BRESLOW DEPTH (MM) | DIAMETER (MM) | CLINICAL FOLLOW-UP |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 4 | M | LOWER LEG | MILD | NO | NO | NO | 1 | 0 | 2 | EPITHELIOID | MODERATE TO SEVERE | 6 | 2.9 | |
| 2 | 11 | F | RIGHT ARM | YES | NO | NO | NO | 3 | 0 | 1 | EPITHELIOID, SPINDLED | MODERATE | 1.8 | 7.5 | |
| 3 | 11 | F | LEFT EAR | YES | YES | NO | NO | 1 | 0 | 3 | EPITHELIOID, SPINDLED | MILD TO MODERATE | 1.1 | ≥2.2 | No recurrence at 34 months |
| 4 | 13 | F | LEFT LATERAL CALF | NO | NO | NO | NO | 3 | 0 | 0 | EPITHELIOID, SPINDLED | MILD | N/A | N/A | |
| 5 | 13 | F | LEFT CHEEK | YES | NO | NO | NO | 0 | 0 | 2 | EPITHELIOID, SPINDLED | MILD | 8.4 | 9.2 | |
| 6 | 16 | M | LEFT POSTERIOR UPPER EAR RIM | YES | YES | NO | NO | 2 | 0+ | 1 | EPITHELIOID, SPINDLED | MILD | 1.3 | ≥3.3 | |
| 7 | 23 | F | LEFT MEDIAL THIGH | YES | YES | NO | NO | 1 | 0 | 3 | EPITHELIOID, SPINDLED | MODERATE | 2.2 | ≥8.9 | |
| 8 | 26 | F | LEFT CALF | YES | YES | NO | NO | 1 | 1 | 1 | EPITHELIOID, SPINDLED | MILD TO MODERATE | 1.2 | 5.3 | |
| 9 | 27 | M | RIGHT BUTTOCK | YES | NO | NO | NO | 2 | 0 | 0 | EPITHELIOID, SPINDLED | MILD | 0.6 | 5.9 | No recurrence at 48 months |
| 10 | 34 | M | RIGHT LATERAL LEG | YES | NO | NO | YES | 3 | 0+ | 2 | EPITHELIOID | MODERATE TO SEVERE | 1.3 | 5.5 | No recurrence at 36 months |
| 11 | 39 | M | RIGHT THIGH | YES | NO | NO | NO | 0 | 2 | 0 | EPITHELIOID | SEVERE | 3.7 | 6.9 | |
| 12 | 47 | F | LEFT KNEE | YES | YES | NO | NO | 0 | 0+ | 8 | SPINDLED | MILD | 3.8 | 9.9 | No recurrence at 31 months |
| 13 | 49 | M | RIGHT LOWER LEG | YES | NO | NO | YES | 4 | 1 | 0 | EPITHELIOID | SEVERE | 0.7 | 5.5 | Died after 60 months (unknown reason) |
| 14 | 50 | F | LEFT LEG | NO | NO | NO | NO | 0 | 1 | 2 | EPITHELIOID, SPINDLED | MODERATE | 1.3 | ≥5.7 | |
| 15 | 52 | F | RIGHT PROXIMAL FOREARM | YES | YES | NO | NO | 1 | 2 | 0 | EPITHELIOID | MODERATE | 0.4 | 6.7 | |
| 16 | 55 | M | LEFT LATERAL BACK | YES | NO | NO | NO | 0 | 1+ | 0 | EPITHELIOID, SPINDLED | MODERATE | 0.7 | 7.9 | |
| 17 | 57 | M | RIGHT CHEST | YES | YES | YES | NO | 4 | 1 | 1 | EPITHELIOID | SEVERE | 0.9 | ≥8.5 | |
| 18 | 61 | M | RIGHT THIGH | YES | YES | NO | NO | 1 | 2− | 2 | EPITHELIOID, SPINDLED | MODERATE | 1.9 | 4.4 | No recurrence at 36 months, sentinel node negative |
| 19 | 68 | M | LEFT FOREARM | YES | YES | NO | NO | 0 | 2+ | 5 | EPITHELIOID | SEVERE | 1.9 | ≥5.7 | No recurrence at 57 months |
| 20 | 68 | M | RIGHT CHEEK | YES | YES | YES | NO | 1 | 3 | 1 | EPITHELIOID, SPINDLED | MODERATE | 1.9 | 4.4 | No recurrence at 34 months |
| 21 | 69 | M | LEFT POSTERIOR SHOULDER | YES | YES | NO | NO | 4 | 1+ | 0 | EPITHELIOID, SPINDLED | MILD TO MODERATE | 1.1 | 5.5 | Multiple cutaneous recurrences beginning 26 months after diagnosis |
| 22 | 70 | F | RIGHT ANTERIOR THIGH | YES | NO | NO | NO | 4 | 1+ | 0 | EPITHELIOID, SPINDLED | MODERATE | 0.5 | 5.8 | No recurrence at 54 months |
| 23 | 75 | F | RIGHT TRICEPS | NO | NO | NO | NO | 3 | 2− | 2 | SPINDLED | MILD | 1.1 | 3.8 | |
| 24 | 78 | F | LEFT LATERAL THIGH | NO | YES | NO | NO | 2 | 1− | 4 | EPITHELIOID | MODERATE | 1.1 | 2.6 | No recurrence at 13 months |
| 25 | 80 | F | RIGHT CHEEK | NO | YES | NO | NO | 2 | 3 | 9 | EPITHELIOID | MODERATE | 5.2 | 9.8 | Local lymph node metastasis detected after 1 month, underwent left neck dissection; No recurrence after 60 months |
The majority of tumors (n=23, 92%) were predominantly composed of large epithelioid melanocytes with abundant cytoplasm with a minority of spindled melanocytes. Two cases (8%) were composed exclusively of spindled melanocytes. Cytoplasmic melanin was identified in 19 cases (76%), with 8 tumors (32%) demonstrating strong (grade 3–4) pigmentation. Epidermal hyperplasia was present in 80% of cases. Kamino bodies were uncommon, identified in only two (8%) cases.
Spitz melanomas
Two spitzoid melanomas contained HRAS hotspot mutations with gain of the mutant allele (8%). One occurred on the leg of a 50-year-old woman and there was also loss of chromosome 9 on which CDKN2A resides (case 14). In this case, p16 expression was absent by immunohistochemistry, suggesting inactivation of the remaining allele of CDKN2A. The other melanoma occurred on the arm of a 75 year-old-woman, and in addition to the HRAS mutation there was a three-codon deletion in MAP2K1 (p.102_104del), a known gain-of-function mutation51,52 with a copy number gain of the mutant allele, a hemizygous truncating mutation in ARID1A, an amplification of NOTCH2, and homozygous deletion of CDKN2A (case 23). Both HRAS-mutated Spitz melanomas were predominantly intradermal with a broad, horizontal silhouette and haphazardly arranged, thickened collagen bundles (Figure 1), similar to HRAS-mutated Spitz nevi5.
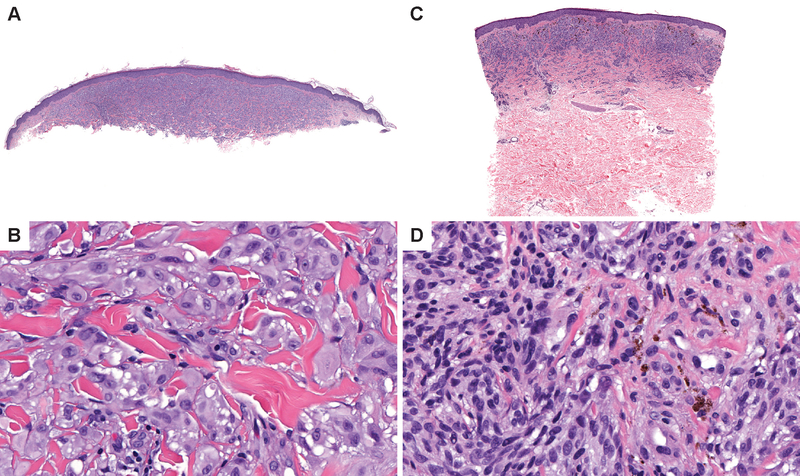
Figure 1. Spitz melanomas with HRAS mutations share features with HRAS mutant Spitz nevi.
(A) Low power view of case 14 which contained an HRASQ61R mutation. The melanoma is predominantly intradermal and has a broad distribution. (B) On higher power view, the melanocytes are epithelioid and pleomorphic and distributed as single cells and elongated nests in between dense sclerotic collagen bundles. Occasional intra-nuclear inclusions are noted. (C) Low power view of case 23 which contained an HRASG12D mutation. The melanoma is predominantly intradermal and broader than it is deep. (D) On higher power, superficial melanocytes are irregularly pigmented and deeper within the dermis. Dermal melanocytes have scant cytoplasm and hyperchromatic nuclei.
BRAF fusions were identified in two cases (8%). Case 12 occurred on the knee of a 47 year old woman and had an NRF1-BRAF fusion, similar to those reported in pleomorphic xanthoastrocytoma53, a TERT promoter hotspot mutation54,55, and a hemizygous PTENG165R mutation56 (Figure 2). Case 4 (included in a previous study of BRAF fusions in melanocytic tumors6) harbored a SOX6-BRAF fusion with focal amplification of the fusion gene. While additional pathogenic point mutations were not identified, there were multiple copy number aberrations, including gains of distal chromosome 1p, chromosomes 2, 12, 13, and 15 as well as copy number neutral loss of heterozygosity of chromosome 3.
Figure 2. Histopathology of Spitz melanomas with BRAF fusions.
(A) and (B) Case 12-- NRF1-BRAF fusion. Sections show a highly cellular tumor with a thin overlying epidermis. The growth pattern is predominantly syncytial and the lesion is composed of epithelioid and spindled melanocytes with vesicular nuclei. (C) and (D) Case 4-- SOX6-BRAF fusion with amplification of the fusion gene. The melanocytes show vesicular nuclei and prominent nucleoli with heterogeneous pigment. There is mild epidermal hyperplasia.
Rearrangements affected the receptor tyrosine kinases (RTKs) ALK in two cases (8%) and NTRK1 in one case (4%). Case 7 occurred on the thigh of a 23 year-old-female and contained a DCTN1-ALK fusion with loss of chromosome 9 and increased dermal mitoses (3/mm2) (Figure 3). Case 10 occurred on the leg of a 34 year-old-man and harbored a fusion of the 3’ portion of ALK to an intergenic region of chromosome 11q, with a gain of the distal portion of ALK encoding the kinase domain, suggesting that a complex rearrangement resulted in an ALK kinase fusion as the oncogene in this case. The tumors with ALK rearrangements demonstrated fascicular nests of melanocytes as characteristic of Spitz nevi with ALK fusions14,15. A TPM3-NTRK1 fusion was present in one case along with gain of chromosome 17 and loss of chromosome 5p upstream of TERT suggestive of a genomic rearrangement which altered TERT promoter or enhancer sequences, a mechanism of telomerase reactivation previously described in neuroblastoma57. This tumor contained rosette-like structures within the dermal component (Figure 4), a feature that is more common in Spitz nevi with NTRK1 fusion16.
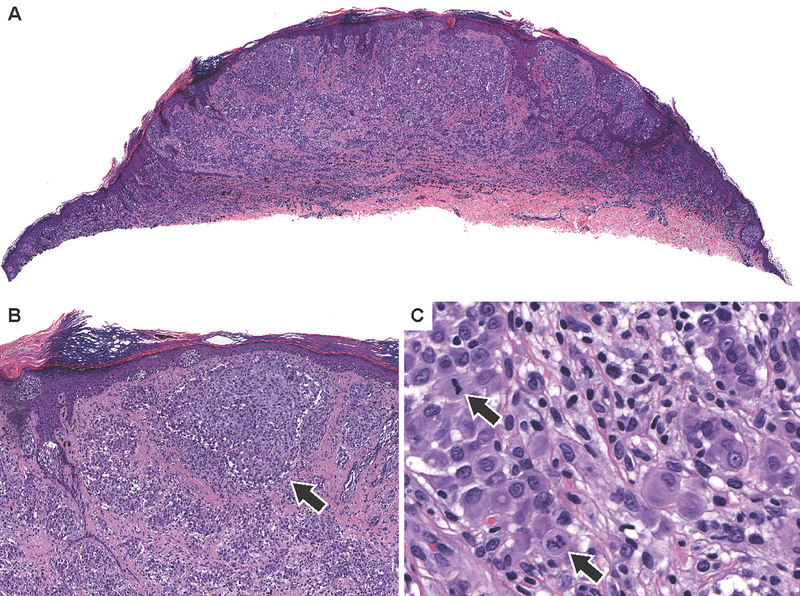
Figure 3. Spitz melanoma with ALK fusion.
(A) Low power view of case 7 with a DCTN1-ALK fusion. As is characteristic in melanomas with ALK fusions, there are large vertically oriented nests in the superficial dermis. (B) Medium power view shows fusiform melanocytes arrayed in irregularly sized dermal nests with discohesive melanocytes (arrow). (C) High power view shows epithelioid melanocytes and multiple dermal mitoses.
Figure 4. Rosette-like structures in Spitz melanoma with NTRK1 fusion.
Case 3--TPM3-NTRK fusion. (A) Low power view demonstrates a dome shaped lesion with mild epidermal thinning. (B) High power shows epithelioid to spindled melanocytes with rosette-like structures (arrows in B), a feature that can be seen in Spitz tumors with NTRK1 fusions. (C) Mitotic figures in the deep dermis are easily recognized.
Rearrangements of MAP3K8 were identified in 2 cases by RNA sequencing (8%). Case 2 demonstrated a ZFP36L1-MAP3K8 fusion and occurred on the right arm of an 11-year-old girl and was notable for pigment rich epithelioid and spindled cells with some adnexal extension. Case 11 contained a MAP3K8-SVIL fusion and presented on the right thigh of a 55-year-old man and contained severely pleomorphic epithelioid cells with mild epidermal hyperplasia. Both cases also demonstrated bi-allelic loss of CDKN2A without other pathogenic alterations.
While HRAS mutations and BRAF or RTK fusions were identified in the Cancer Genome Atlas (TCGA) skin cutaneous melanoma (SKCM) cohort46,49, these alterations occurred at greater prevalence in our cohort (P=0.0479 for HRAS mutation, P=0.00041 for fusions of BRAF, NTRK1, or ALK). The difference in prevalence of MAP3K8 fusion was not statistically significant (P=0.0942).
Mutations activating the MAP-kinase pathway were present in half of spitzoid melanomas without Spitz nevus associated mutations
Altogether, nine (36%) of the spitzoid melanomas were classified as Spitz melanoma due to the presence of an oncogenic alterations typical of Spitz tumors. Other MAP-kinase pathway activating alterations not associated with Spitz nevi were identified in another nine (36%) spitzoid melanomas. Interestingly, we did not identify any BRAFV600 mutations in our cohort. BRAFV600E is common in melanomas without chronic sun-induced damage (non-CSD) and was significantly underrepresented in our spitzoid melanomas as compared to the TCGA SKCM cohort (P=1.3*10^−6). This underrepresentation was not due to excluding cases that were positive by BRAF V600E immunohistochemistry. We identified a BRAF D594G mutation in a single case (case 19), which is a class 3 BRAF mutation that is considered weakly activating58. This tumor had a concomitant activating MAP2K2F57Y mutation59–61. MAP2K2, also known as MEK2, acts directly downstream of BRAF in the MAPK pathway, and the MAP2K2 mutation likely cooperates with the class 3 BRAF mutation. This tumor also had a hotspot mutation in the promoter of TERT and an activating EZH2Y646N mutation62. Histopathologically, this tumor contained large epithelioid cells with a high mitotic index and severe nuclear pleomorphism (Figure 5).
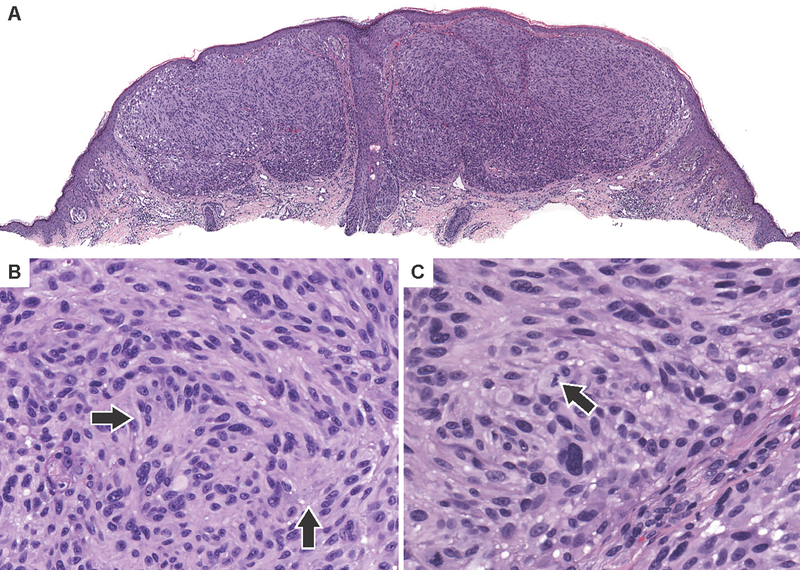
Figure 5. Spitzoid melanoma with a class 3 BRAF mutation.
(A) Low power view of case 19 which harbors BRAFD594G mutation along with EZH2Y646S, BLMW803*, MAP2K2F57Y and TERT promoter mutations. The neoplasm is predominantly dermal and is composed of coalescing nests and sheets of large epithelioid melanocytes with a focally dense infiltrate of lymphocytes. (B)Medium power shows a predominantly nested architecture with numerous admixed lymphocytes. (C) Nuclei are pleomorphic with prominent nucleoli and brisk mitotic activity (arrows).
NRAS hotspot mutations were identified in five of cases (20%), at a similar prevalence as in the SKCM cohort of TCGA. It included a melanoma on the helix of the ear of a 16-year-old boy (case 6, Figure 6) with an NRASQ61H mutation that underwent secondary amplification as the sole detected pathogenic mutation. In case 25, the NRASQ61H mutation occurred in combination with an activating CTNNB1G34I mutation, a TERT promoter mutation, and IDH1R132C and NFKBIEG41E mutations63–65. This spitzoid melanoma also demonstrated a minor small cell component, which did not demonstrate the strong and uniform beta-catenin staining indicative of CTNNB1 mutation observed in the majority of the melanoma. This finding suggests that the CTNNB1 mutation arose later in melanoma progression resulting in the epithelioid cytomorphology present in the majority of the melanoma (Supplementary Figure 1).
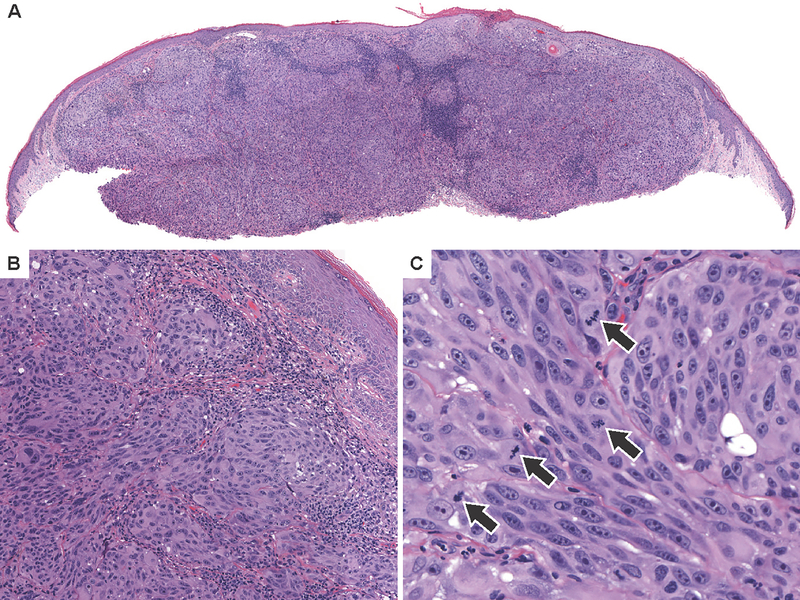
Figure 6. Spitzoid melanomas with NRAS mutation.
(A) Low power view of case 6 which harbors NRASQ61H with high level focused amplification the mutant allele shows a dome shaped compound tumor with epidermal hyperplasia and clefting around nests of melanocytes in the superficial dermis. (B) The lesion is composed of epithelioid melanocytes with hyperchromatic nuclei, growing as nests and fascicles distributed between thickened sclerotic collagen bundles. (C) Whole genome copy number profile demonstrates NRAS amplification as well as loss of chromosome 1p distal to NRAS and gain of chromosome 1q. (D) Low power view of case 25—which harbors NRASQ61K, CTNNB1G34I, IDH1R132C, and NFKBIE and TERT promoter mutations shows a nodular tumor that extends to the deep dermis. (E) The lesion is predominantly composed of large epithelioid melanocytes with focal areas of small more nevoid cells, predominantly near the dermal-epidermal junction. (F) Many dermal mitoses are present (arrows).
Bi-allelic loss of NF1 occurred in two (8%) cases, in combination with NRASG12D in one case (case 17). Activating KITL576P and MAP2K1E203K mutations were identified in the absence of additional MAPK pathway activating mutations in one case each66,67.
In the remaining seven (28%) of cases, no mutations in MAP-kinase pathway were identified. This included an unpigmented epithelioid cell tumor with a CRTC1-TRIM11 fusion, typical for a new class of tumors that bear resemblance to clear cell sarcoma68, on the cheek of a 13-year-old girl (case 5, Supplementary Figure 2).
Subclasses of spitzoid melanoma
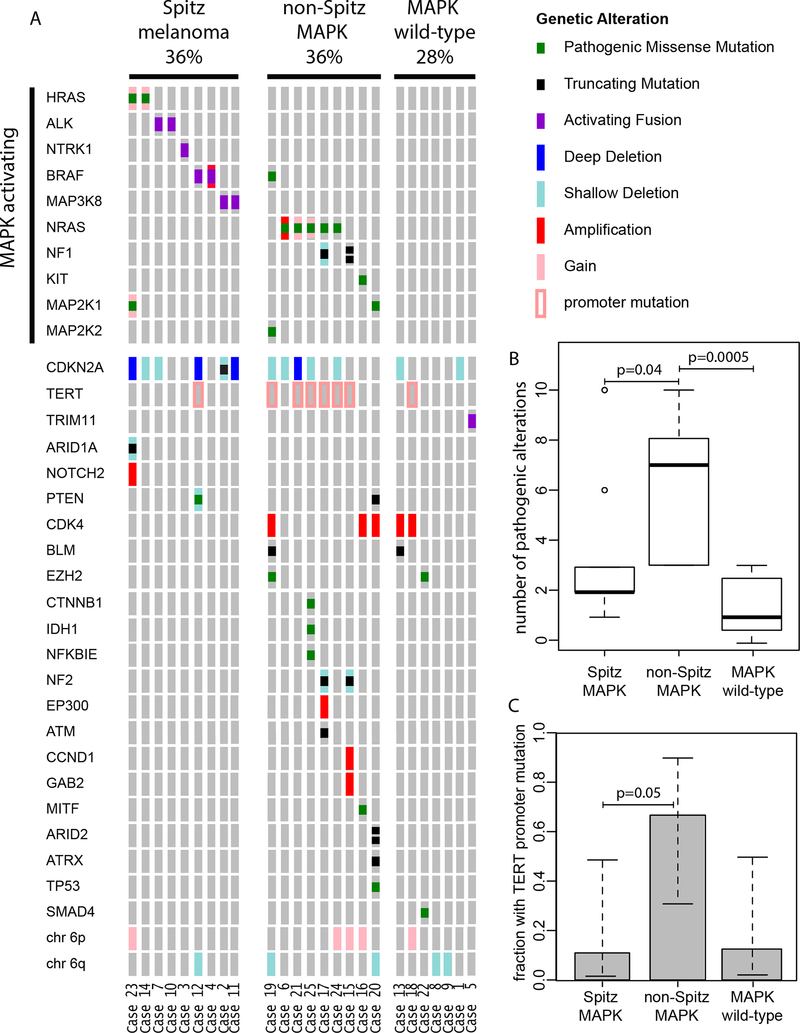
A summary of the pathogenic mutations identified in our series is presented in Figure 7A. We identified three genetic subcategories in our series: 1) Spitz melanoma, 2) spitzoid melanoma without identified MAPK activating mutations and 3) spitzoid melanoma non-Spitz MAPK activating mutations. The latter tended to occur in older patients and arose in skin with higher degrees of solar elastosis and had more pathogenic alterations (Figure 7B, Supplemental Figure 3).
Figure 7. Genetic alterations in spitzoid melanomas.
(A) Oncoplot highlighting the genetic landscape of spitzoid melanomas in our cohort, stratified by those with classical Spitz drivers, non-Spitz MAPK drivers, and those without an identifiable MAPK mutation. (B) Number of pathogenic mutations in each subset. (C) Proportion of cases with TERT promoter mutations. Note that those tumors with non-Spitz MAPK drivers demonstrated an increase in both the number of pathogenic mutations as well as TERT promoter mutations.
Mutations in the promoter of TERT are present in ~70% of cutaneous melanomas and are associated with worse prognosis in both conventional and spitzoid melanoma69,70. Hotspot TERT promoter alterations were identified in 8 spitzoid melanomas (32%) and were more common in spitzoid melanomas with a non-Spitz nevus MAPK activating mutation (Figure 7C).
Inactivation of the tumor suppressor CDKN2A is a major driver in melanoma, and homozygous deletion of CDKN2A may be associated with poor outcome in atypical Spitz tumors69,71. Bi-allelic loss of function of CDKN2A was identified in 5 (20%) spitzoid melanomas. In four cases, bi-allelic loss occurred by homozygous deletion of CDKN2A, and in the remaining case, there was a truncating mutation of CDKN2A with loss of the wild-type allele. Four of nine (44%) Spitz melanomas demonstrated bi-allelic inactivation of CDKN2A.
Clinical Follow-Up
We were able to obtain clinical follow up data for 12 patients, 11 of whom were alive at the time of this study (Table 1, mean follow up 44 months). One patient died 60 months after excision from unknown causes. One patient (case 21) had multiple cutaneous recurrences beginning 26 months after excision. Another patient had a positive sentinel lymph node biopsy that was followed by completion lymphadenectomy with no record of relapse during follow-up. Nine patients were treated with simple excision all of which had an uneventful follow-up period.
Discussion
The genetic alterations in our series of 25 spitzoid melanomas differed significantly from those reported for cutaneous melanomas. BRAF V600 mutations found in the majority of low-CSD cutaneous melanomas were completely absent, indicating that spitzoid melanomas do not typically arise from common acquired nevi. Instead we found an increased incidence (30%) of oncogenic alterations typical for Spitz tumors such as HRAS mutation and activating fusions BRAF, MAP3K8, ALK, and NTRK1.
The Spitz melanomas with HRAS mutation or ALK or NTRK1 kinase fusion retained the some of the distinct histopathologic features associated with Spitz nevi with these mutations. Two of the Spitz melanomas had genetic features that indicate malignancy in low-and high- CSD melanoma (Cases 12 and 23). Two additional Spitz melanomas had bi-allelic inactivation of CDKN2A. Case 3 with an NTRK1 fusion, but without other definitely pathogenic mutations demonstrated copy number loss upstream of TERT, suggesting a structural rearrangement. Similar rearrangements reactivate TERT in neuroblastoma and have been reported in lethal Spitz melanoma in the absence of a TERT promoter mutation13. By design, our targeted assay cannot detect structural rearrangements far upstream of TERT and therefore we may have missed similar alterations in other cases.
Most of our spitzoid melanomas were not classifiable as Spitz melanomas based on their genetic alterations. We further subdivided these ‘non-Spitz’ spitzoid melanomas into those without any identifiable MAP-kinase pathway mutations and those with MAPK pathway mutation(s) not associated with Spitz nevi such as in NRAS, BRAF, KIT, NF1, and or MAP2K1/2.
The melanomas in the latter of these two groups occurred in older patients, with more solar elastosis in the adjacent dermis and had a higher number of pathogenic mutations than the rest of our spitzoid melanomas, suggesting that they have a higher somatic mutation burden due to UV exposure. One notable exception was a spitzoid melanoma with focused high-level amplification of mutant NRAS that occurred on the ear of a teenager and demonstrated minimal solar elastosis and no additional pathogenic alterations. Focused high-level amplification of mutant NRAS has been described in spitzoid tumors on the ear of children72. Perhaps secondary amplification of mutant NRAS leads to spitzoid cytomorphologic features by a mechanism similar to HRAS mutation.
The different histopathologic features of conventional and Spitz nevi likely reflect differences in the oncogenic signaling outputs of their initiating mutations. Melanocytes of a conventional nevus may acquire secondary genetic alterations that lead to additional proliferation and a change in the cytomorphology of the neoplastic melanocytes. For example, when a melanocyte of a conventional nevus, which was initiated by a BRAF (or rarely RAS) mutation, acquires an activating CTNNB1 mutation in addition to the mutation which initiated the nevus, a secondary clonal expansion of melanocytes characterized by increased cell size and pigmentation results in a component of deep penetrating nevus (melanocytoma)52. Bi-allelic loss of BAP1 in a melanocyte of a conventional nevus results in a secondary proliferation of epithelioid melanocytes with distinctive spitzoid cytomorphology73–76. Similarly, bi-allelic loss of PRKAR1A leads to development of a pigmented epithelioid melanocytoma within a conventional nevus77. While we identified a CTNNB1 mutation in one spitzoid melanoma, we did not identify any pathogenic BAP1 mutations in our series. This may indicate that cutaneous melanomas with BAP1 loss are rare and represent only a small fraction of spitzoid melanoma, or that the spitzoid cytomorphologic features of melanocytic tumors with BAP1 loss are not retained during malignant progression. Intriguingly, two spitzoid melanomas harbored activating EZH2 mutations. Loss of BAP1 has been shown to lead to increased EZH2 activity and EZH2-dependent transformation78. Perhaps mutational activation of EZH2 is functionally similar to loss of BAP1 and also results in a spitzoid phenotype.
Most spitzoid melanomas without identified MAP-kinase pathway mutations occurred in young patients with little solar elastosis, similar to Spitz melanomas. Their genomes did not contain many deletions and amplifications suggesting they are distinct from “triple wild-type” melanomas in the TCGA that characteristically contain many copy number aberrations and structural rearrangements46. In one case we identified a CRTC1-TRIM11 fusion which is not predicted to affect MAPK pathway signaling and has been recently described in dermal melanocytic tumors composed of epithelioid and spindled cells with minimal pigmentation that resemble clear cell sarcoma68. This case lacked melanin (pigmentation score of 0) and was characterized by epithelioid to spindled melanocytes with vesicular chromatin and a single prominent nucleolus, findings very characteristic of clear cell sarcoma.
Interestingly, our findings differ from those of the study performed by Lazova et al., in which the authors found a high genetic similarity between spitzoid and conventional melanomas with ~40% of the spitzoid melanomas in their series having a BRAF V600 mutation79. Perhaps the younger age of patients in our study accounts in part for the differences as the average age in our study was 45 years, whereas the average age in the study by Lazova et al. was 64 years. More likely, the disparate results may reflect different histopathologic criteria for spitzoid melanoma across institutions.
Our findings suggest that multiple evolutionary pathways can lead to spitzoid melanoma, and that it is not a homogeneous category. Given the genomic diversity of Spitz melanoma, and indeed the likelihood that spitzoid melanomas without Spitz initiating oncogenes are also genomically diverse, the failure of histopathologic criteria to distinguish all Spitz nevi from all spitzoid melanomas is not surprising. Diagnostic criteria likely need to be developed for each molecular sub-lineage (i.e. ALK-fused Spitz tumors, HRAS mutant Spitz tumors) and may need to include molecular as well as histopathologic findings. Future studies will aim to determine differences in clinical behavior for Spitz melanoma as compared to low- or high-CSD melanoma and the impact of specific initiating mutation, additional pathogenic mutations, and mutation burden on clinical outcomes.
Supplementary Material
Acknowledgements
We thank Connie Jang and Yvonne Lee for their assistance in organizing and obtaining the cases. Shyam Raghavan was funded in part by the American Society of Dermatopathology Mentorship Award, and Sandra Peternel was supported by a grant from the European Academy of Dermatology and Venereology (RF-2017-17). This work was supported by the National Cancer Institute at the National Institutes of Health (grant number 1R35CA220481).
Footnotes
Disclosure/Conflict of Interest
The authors declare no relevant disclosures or conflicts of interest.
References
- 1.Barnhill RL. The Spitzoid lesion: rethinking Spitz tumors, atypical variants, “Spitzoid melanoma” and risk assessment. Mod Pathol Off J U S Can Acad Pathol Inc 2006;19 Suppl 2:S21–33. [DOI] [PubMed] [Google Scholar]
- 2.Spitz S Melanomas of Childhood. Am J Pathol 1948;24:591–609. [PMC free article] [PubMed] [Google Scholar]
- 3.Dika E, Fanti PA, Fiorentino M, Capizzi E, Neri I, Piraccini BM, et al. Spitzoid tumors in children and adults: a comparative clinical, pathological, and cytogenetic analysis. Melanoma Res 2015;25:295–301. [DOI] [PubMed] [Google Scholar]
- 4.Gerami P, Busam K, Cochran A, Cook MG, Duncan LM, Elder DE, et al. Histomorphologic assessment and interobserver diagnostic reproducibility of atypical spitzoid melanocytic neoplasms with long-term follow-up. Am J Surg Pathol 2014;38:934–940. [DOI] [PubMed] [Google Scholar]
- 5.Bastian BC, LeBoit PE, Pinkel D. Mutations and copy number increase of HRAS in Spitz nevi with distinctive histopathological features. Am J Pathol 2000;157:967–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Botton T, Yeh I, Nelson T, Vemula SS, Sparatta A, Garrido MC, et al. Recurrent BRAF kinase fusions in melanocytic tumors offer an opportunity for targeted therapy. Pigment Cell Melanoma Res 2013;26:845–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiesner T, He J, Yelensky R, Esteve-Puig R, Botton T, Yeh I, et al. Kinase fusions are frequent in Spitz tumours and spitzoid melanomas. Nat Commun 2014;5:3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeh I, Botton T, Talevich E, Shain AH, Sparatta AJ, de la Fouchardiere A, et al. Activating MET kinase rearrangements in melanoma and Spitz tumours. Nat Commun 2015;6:7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yeh I, Tee MK, Botton T, Shain AH, Sparatta AJ, Gagnon A, et al. NTRK3 kinase fusions in Spitz tumours. J Pathol 2016;240:282–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang L, Busam KJ, Benayed R, Cimera R, Wang J, Denley R, et al. Identification of NTRK3 Fusions in Childhood Melanocytic Neoplasms. J Mol Diagn JMD 2017;19:387–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.VandenBoom T, Quan VL, Zhang B, Garfield EM, Kong BY, Isales MC, et al. Genomic Fusions in Pigmented Spindle Cell Nevus of Reed: Am J Surg Pathol 2018;:1. [DOI] [PubMed] [Google Scholar]
- 12.Quan VL, Zhang B, Mohan LS, Shi K, Isales MC, Panah E, et al. Activating Structural Alterations In MAPK Genes Are Distinct Genetic Drivers In a Unique Subgroup Of Spitzoid Neoplasms. Am J Surg Pathol 2019. doi: 10.1097/PAS.0000000000001213. [DOI] [PubMed] [Google Scholar]
- 13.Newman S, Fan L, Pribnow A, Silkov A, Rice SV, Lee S, et al. Clinical genome sequencing uncovers potentially targetable truncations and fusions of MAP3K8 in spitzoid and other melanomas. Nat Med 2019;:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Busam KJ, Kutzner H, Cerroni L, Wiesner T. Clinical and pathologic findings of Spitz nevi and atypical Spitz tumors with ALK fusions. Am J Surg Pathol 2014;38:925–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeh I, de la Fouchardiere A, Pissaloux D, Mully TW, Garrido MC, Vemula SS, et al. Clinical, histopathologic, and genomic features of Spitz tumors with ALK fusions. Am J Surg Pathol 2015;39:581–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amin SM, Haugh AM, Lee CY, Zhang B, Bubley JA, Merkel EA, et al. A Comparison of Morphologic and Molecular Features of BRAF, ALK, and NTRK1 Fusion Spitzoid Neoplasms. Am J Surg Pathol 2017;41:491–498. [DOI] [PubMed] [Google Scholar]
- 17.Pollock PM, Harper UL, Hansen KS, Yudt LM, Stark M, Robbins CM, et al. High frequency of BRAF mutations in nevi. Nat Genet 2003;33:19–20. [DOI] [PubMed] [Google Scholar]
- 18.Yeh I, von Deimling A, Bastian BC. Clonal BRAF mutations in melanocytic nevi and initiating role of BRAF in melanocytic neoplasia. J Natl Cancer Inst 2013;105:917–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ackerman AB. Spitz’s Nevus: Reassessment Critical, Revision Radical. New York: Ardor Scribendi, 2007. [Google Scholar]
- 20.Wu G, Barnhill RL, Lee S, Li Y, Shao Y, Easton J, et al. The landscape of fusion transcripts in spitzoid melanoma and biologically indeterminate spitzoid tumors by RNA sequencing. Mod Pathol 2016. doi: 10.1038/modpathol.2016.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Dijk MCRF, Bernsen MR, Ruiter DJ. Analysis of mutations in B-RAF, N-RAS, and H-RAS genes in the differential diagnosis of Spitz nevus and spitzoid melanoma. Am J Surg Pathol 2005;29:1145–1151. [DOI] [PubMed] [Google Scholar]
- 22.Fullen DR, Poynter JN, Lowe L, Su LD, Elder JT, Nair RP, et al. BRAF and NRAS mutations in spitzoid melanocytic lesions. Mod Pathol Off J U S Can Acad Pathol Inc 2006;19:1324–1332. [DOI] [PubMed] [Google Scholar]
- 23.Da Forno PD, Pringle JH, Fletcher A, Bamford M, Su L, Potter L, et al. BRAF, NRAS and HRAS mutations in spitzoid tumours and their possible pathogenetic significance. Br J Dermatol 2009;161:364–372. [DOI] [PubMed] [Google Scholar]
- 24.Lazova R, Pornputtapong N, Halaban R, Bosenberg M, Bai Y, Chai H, et al. Spitz nevi and Spitzoid melanomas: exome sequencing and comparison with conventional melanocytic nevi and melanomas. Mod Pathol 2017. doi: 10.1038/modpathol.2016.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Da Forno PD, Pringle JH, Fletcher A, Bamford M, Su L, Potter L, et al. BRAF, NRAS and HRAS mutations in spitzoid tumours and their possible pathogenetic significance. Br J Dermatol 2009;161:364–372. [DOI] [PubMed] [Google Scholar]
- 26.Elder DE, Massi D, Scolyer R, Willemze R. WHO Classification of Skin Tumours, 4th Edition. Lyon, France: IARC Press, 2018. [Google Scholar]
- 27.Paradela S, Fonseca E, Pita-Fernández S, Prieto V g. Spitzoid and non-spitzoid melanoma in children. A prognostic comparative study. J Eur Acad Dermatol Venereol 2013;27:1214–1221. [DOI] [PubMed] [Google Scholar]
- 28.Viros A, Fridlyand J, Bauer J, Lasithiotakis K, Garbe C, Pinkel D, et al. Improving melanoma classification by integrating genetic and morphologic features. PLoS Med 2008;5:e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landi MT, Bauer J, Pfeiffer RM, Elder DE, Hulley B, Minghetti P, et al. MC1R Germline Variants Confer Risk for BRAF-Mutant Melanoma. Science 2006;313:521–522. [DOI] [PubMed] [Google Scholar]
- 30.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinforma Oxf Engl 2009;25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garrison, Gabor Erik M. Haplotype-based variant detection from short-read sequencing. arXiv 2012;1207.3907 [q-bio.GN].
- 33.Broad Institute. Picard. Available from: http://broadinstitute.github.io/picard/.
- 34.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164–e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rausch T, Zichner T, Schlattl A, Stütz AM, Benes V, Korbel JO. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 2012;28:i333–i339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 2013;14:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing. PLOS Comput Biol 2016;12:e1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oesper L, Satas G, Raphael BJ. Quantifying tumor heterogeneity in whole-genome and whole-exome sequencing data. Bioinformatics 2014;30:3532–3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res 2016;44:D862–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.NHLBI GO Exome Sequencing Project (ESP). Exome Variant Server. Seattle, WA: Available from: http://evs.gs.washington.edu/EVS/. [Google Scholar]
- 41.Consortium T 1000 GP. A global reference for human genetic variation. Nature 2015;526:nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Online Mendelian Inheritance in Man, OMM.McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University; (Baltimore, MD: ). Available from: https://omim.org. [Google Scholar]
- 43.My Cancer Genome. Available from: https://www.mycancergenome.org.
- 44.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nicorici D, Şatalan M, Edgren H, Kangaspeska S, Murumägi A, Kallioniemi O, et al. FusionCatcher – a tool for finding somatic fusion genes in paired-end RNA-sequencing data. bioRxiv 2014;:011650. [Google Scholar]
- 46.Akbani R, Akdemir KC, Aksoy BA, Albert M, Ally A, Amin SB, et al. Genomic Classification of Cutaneous Melanoma. Cell 2015;161:1681–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci Signal 2013;6:pl1–pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov 2012;2:401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stransky N, Cerami E, Schalm S, Kim JL, Lengauer C. The landscape of kinase fusions in cancer. Nat Commun 2014;5. doi: 10.1038/ncomms5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lehmann BD, Shaver TM, Johnson DB, Li Z, Gonzalez-Ericsson PI, Sanchez V, et al. Identification of targetable recurrent MAP3K8 rearrangements in melanomas lacking known driver mutations. Mol Cancer Res MCR 2019. doi: 10.1158/1541-7786.MCR-19-0257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chakraborty R, Hampton OA, Shen X, Simko SJ, Shih A, Abhyankar H, et al. Mutually exclusive recurrent somatic mutations in MAP2K1 and BRAF support a central role for ERK activation in LCH pathogenesis. Blood 2014;124:3007–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yeh I, Lang UE, Durieux E, Tee MK, Jorapur A, Shain AH, et al. Combined activation of MAP kinase pathway and β-catenin signaling cause deep penetrating nevi. Nat Commun 2017;8:644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Phillips JJ, Gong H, Chen K, Joseph NM, van Ziffle J, Jin L-W, et al. Activating NRF1-BRAF and ATG7-RAF1 fusions in anaplastic pleomorphic xanthoastrocytoma without BRAF p.V600E mutation. Acta Neuropathol (Berl) 2016;132:757–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA. Highly recurrent TERT promoter mutations in human melanoma. Science 2013;339:957–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, et al. TERT promoter mutations in familial and sporadic melanoma. Science 2013;339:959–961. [DOI] [PubMed] [Google Scholar]
- 56.Han S-Y, Kato H, Kato S, Suzuki T, Shibata H, Ishii S, et al. Functional Evaluation of PTEN Missense Mutations Using in Vitro Phosphoinositide Phosphatase Assay. Cancer Res 2000;60:3147–3151. [PubMed] [Google Scholar]
- 57.Valentijn LJ, Koster J, Zwijnenburg DA, Hasselt NE, van Sluis P, Volckmann R, et al. TERT rearrangements are frequent in neuroblastoma and identify aggressive tumors. Nat Genet 2015;47:1411–1414. [DOI] [PubMed] [Google Scholar]
- 58.Yao Z, Yaeger R, Rodrik-Outmezguine VS, Tao A, Torres NM, Chang MT, et al. Tumours with class 3 BRAF mutants are sensitive to the inhibition of activated RAS. Nature 2017;548:234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nyström A-M, Ekvall S, Berglund E, Björkqvist M, Braathen G, Duchen K, et al. Noonan and cardio-facio-cutaneous syndromes: two clinically and genetically overlapping disorders. J Med Genet 2008;45:500–506. [DOI] [PubMed] [Google Scholar]
- 60.Senawong T, Phuchareon J, Ohara O, McCormick F, Rauen KA, Tetsu O. Germline mutations of MEK in cardio-facio-cutaneous syndrome are sensitive to MEK and RAF inhibition: implications for therapeutic options. Hum Mol Genet 2008;17:419–430. [DOI] [PubMed] [Google Scholar]
- 61.Gos M, Smigiel R, Kaczan T, Landowska A, Abramowicz A, Sasiadek M, et al. MAP2K2 mutation as a cause of cardio-facio-cutaneous syndrome in an infant with a severe and fatal course of the disease. Am J Med Genet A 2018;176:1670–1674. [DOI] [PubMed] [Google Scholar]
- 62.Souroullas GP, Jeck WR, Parker JS, Simon JM, Liu J-Y, Paulk J, et al. An oncogenic Ezh2 mutation induces tumors through global redistribution of histone 3 lysine 27 trimethylation. Nat Med 2016;22:632–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Parsons DW, Jones S, Zhang X, Lin JC-H, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science 2008;321:1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science 2009;324:261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shain AH, Garrido M, Botton T, Talevich E, Yeh I, Sanborn JZ, et al. Exome sequencing of desmoplastic melanoma identifies recurrent NFKBIE promoter mutations and diverse activating mutations in the MAPK pathway. Nat Genet 2015;47:1194–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Curtin JA, Busam K, Pinkel D, Bastian BC. Somatic activation of KIT in distinct subtypes of melanoma. J Clin Oncol 2006;24:4340–4346. [DOI] [PubMed] [Google Scholar]
- 67.Nikolaev SI, Rimoldi D, Iseli C, Valsesia A, Robyr D, Gehrig C, et al. Exome sequencing identifies recurrent somatic MAP2K1 and MAP2K2 mutations in melanoma. Nat Genet 2012;44:133–139. [DOI] [PubMed] [Google Scholar]
- 68.Cellier L, Perron E, Pissaloux D, Karanian M, Haddad V, Alberti L, et al. Cutaneous Melanocytoma With CRTC1-TRIM11 Fusion: Report of 5 Cases Resembling Clear Cell Sarcoma. Am J Surg Pathol 2018;42:382–391. [DOI] [PubMed] [Google Scholar]
- 69.Lee S, Barnhill RL, Dummer R, Dalton J, Wu J, Pappo A, et al. TERT Promoter Mutations Are Predictive of Aggressive Clinical Behavior in Patients with Spitzoid Melanocytic Neoplasms. Sci Rep 2015;5:11200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Griewank KG, Murali R, Puig-Butille JA, Schilling B, Livingstone E, Potrony M, et al. TERT Promoter Mutation Status as an Independent Prognostic Factor in Cutaneous Melanoma. J Natl Cancer Inst 2014;106:dju246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gerami P, Scolyer RA, Xu X, Elder DE, Abraham RM, Fullen D, et al. Risk assessment for atypical spitzoid melanocytic neoplasms using FISH to identify chromosomal copy number aberrations. Am J Surg Pathol 2013;37:676–684. [DOI] [PubMed] [Google Scholar]
- 72.Dubruc E, Balme B, Dijoud F, Disant F, Thomas L, Wang Q, et al. Mutated and amplified NRAS in a subset of cutaneous melanocytic lesions with dermal spitzoid morphology: report of two pediatric cases located on the ear. J Cutan Pathol 2014;41:866–872. [DOI] [PubMed] [Google Scholar]
- 73.Wiesner T, Murali R, Fried I, Cerroni L, Busam K, Kutzner H, et al. A Distinct Subset of Atypical Spitz Tumors is Characterized by BRAF Mutation and Loss of BAP1 Expression. Am J Surg Pathol 2012;36:818–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Busam KJ, Sung J, Wiesner T, von Deimling A, Jungbluth A. Combined BRAF(V600E)-positive melanocytic lesions with large epithelioid cells lacking BAP1 expression and conventional nevomelanocytes. Am J Surg Pathol 2013;37:193–199. [DOI] [PubMed] [Google Scholar]
- 75.Yeh I, Mully TW, Wiesner T, Vemula SS, Mirza SA, Sparatta AJ, et al. Ambiguous melanocytic tumors with loss of 3p21. Am J Surg Pathol 2014;38:1088–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vilain RE, McCarthy SW, Thompson JF, Scolyer RA. BAP1-inactivated spitzoid naevi. Am J Surg Pathol 2015;39:722. [DOI] [PubMed] [Google Scholar]
- 77.Cohen JN, Joseph NM, North JP, Onodera C, Zembowicz A, LeBoit PE. Genomic Analysis of Pigmented Epithelioid Melanocytomas Reveals Recurrent Alterations in PRKAR1A, and PRKCA Genes. Am J Surg Pathol 2017;41:1333–1346. [DOI] [PubMed] [Google Scholar]
- 78.LaFave LM, Béguelin W, Koche R, Teater M, Spitzer B, Chramiec A, et al. Loss of BAP1 function leads to EZH2-dependent transformation. Nat Med 2015;21:1344–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lazova R, Pornputtapong N, Halaban R, Bosenberg M, Bai Y, Chai H, et al. Spitz nevi and Spitzoid melanomas: exome sequencing and comparison with conventional melanocytic nevi and melanomas. Mod Pathol 2017. doi: 10.1038/modpathol.2016.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.