Abstract
Objectives
In 2017, AMP, ASCO and CAP jointly published the first formalized classification system for the interpretation and reporting of sequence variants in cancer. The challenges of incorporating new variant interpretation guidelines into existing, validated workflows have likely hampered adoption and implementation in labs with classification methods in place. Ambiguity in assigning clinical significance across guidelines is grounded in differential weighting of evidence used in variant assessment. Therefore, we undertook an internal process-improvement exercise to correlate the two classification schemes using historical laboratory data.
Design and methods
Existing clinical variant assignments from 40 consecutive oncology cases comprising 150 somatic variants were re-assessed according to the 2017 AMP/ASCO/CAP scheme. Approximately 50% of these were cancers of the gynecologic tract.
Results
Our laboratory-developed (GPS) classifications for ‘actionable’ variants and variants of uncertain clinical significance mapped consistently with the AMP/ASCO/CAP Tiers I-III. The majority of Level 1 variants were reclassified to Tier I (21/25; 84%) while all Level 2 and Level 4 variants were assigned to Tier II (9/9; 100%) and Tier III (17/17; 100%), respectively. The greatest variability was seen for GPS Level 3 variants, which was strongly influenced by TP53 interpretations. Ultimately, we found that most GPS Level 3 variants were classified as Tier III (77/99; 77.8%).
Conclusions
Our internally developed 5-level classifications mapped consistently with the proposed AMP/ASCO/CAP 4-Tiered system. As a result of this analysis, we can provide a framework for other labs considering a similar transition to the 2017 AMP/ASCO/CAP guidelines and a rationale for explaining specific discrepancies.
Keywords: Somatic variant classification, Clinical sequencing, Variant guidelines
Graphical abstract
1. Introduction
In 2011, the Genomics and Pathology Services (GPS) clinical laboratory at Washington University School of Medicine implemented guidelines for the interpretation and reporting of sequence variants identified through clinical next-generation sequencing (NGS) analysis. The NGS assays offered by GPS include tumor mutational profiling, diagnostic testing for disorders of somatic mosaicism, and whole exome-based gene panels for diagnosis of cardiac and renal diseases, and immunodeficiency (congenital neutropenia) with a suspected genetic component. In practice, multiple lines of evidence are evaluated in order to determine the reported clinical classifications. These sources, reviewed in Ref. [1], include, but are not limited to allelic fraction within population databases (e.g., ExAC, gnomAD), literature review (e.g., OMIM, ClinVar, PubMed), and in silico prediction tools based on evolutionary conservation and tertiary protein domain structure (e.g., PolyPhen, SIFT, GERP++). For oncology, additional variant level analyses utilize specific databases, including COSMIC, cBioPortal, and CIViC.
In combination with guidelines and recommendations from professional organizations (e.g. NCCN), our clinical NGS laboratory developed a strategy to classify somatic variants identified in tumor-only mutational profiling for patients with cancer [2]. The overall goal of the classification scheme was to highlight the most important and thus ‘actionable’ variants to the oncologist. As a result, five levels were chosen for stratification and are briefly described here. Level 1 are those variants deemed to be of diagnostic, prognostic, and/or therapeutic value in the cancer type of the patient. Level 2 is reserved for variants similar in scope to Level 1 but apply to a cancer type distinct from the one the patient presents with. This distinction allows the treating physician to evaluate interventions, including off-label drug usage that may prove to be relevant. Level 3 refers to variants that have been previously reported in cancer or another genetic disease, but without compelling evidence to predict its clinical significance or relevance to the patient’s disease. Level 4 denotes variants of uncertain clinical significance, which includes novel (previously unreported) variants and variants for which there is insufficient evidence to support a pathogenic contribution. Finally, Level 5 refers to well-documented polymorphisms occurring at a frequency greater than 1.0% within any population or subpopulation.
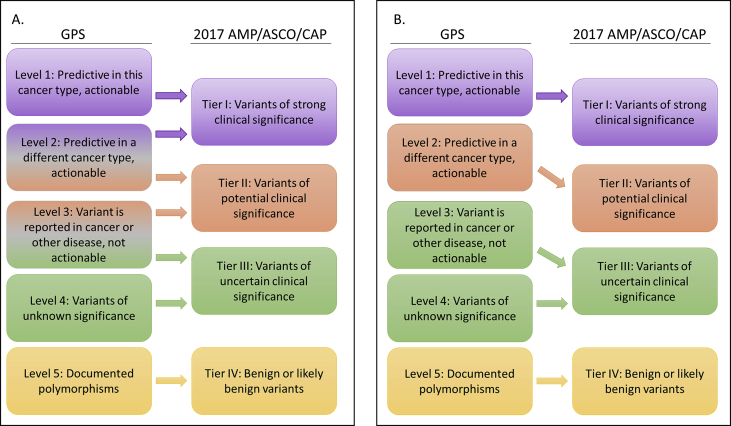
It was not until 2017, that AMP, ASCO and CAP jointly published the first formalized a clinical classification system for the interpretation and reporting of sequence variants in cancer [1]. This classification scheme was predicated on a 4-Tiered system, with significant weight placed upon published evidence for making stratification decisions. These current recommendations are briefly summarized here and compared with our current GPS-developed classification system. Tier I includes variants of strong clinical significance based upon therapeutic, prognostic and diagnostic criteria. Two distinct levels of evidence provide support for this classification: Level A identifies FDA-approved disease-specific therapies as well as professional guidelines whereas Level B is reserved for well-powered studies with expert consensus that have not yet entered professional recommendations. As such, the AMP/ASCO/CAP Tier I and the GPS Level 1 are well aligned. As a general theme, while our established GPS classification assignments are not based upon a formal structure for the strength of the evidence levels, scientific rigor is considered in determining the actionable nature of the variant under evaluation. Tier II classifies variants similar to Tier I, with the allowance that levels of evidence are less stringent. Thus, this tier describes variants of potential clinical significance in terms of their diagnostic, therapeutic and prognostic attributes. For example, evidence Level C includes off-label use of FDA-approved therapies and multiple small studies, whereas Level D is reserved for those variants for which there is only preclinical data or case reports. Tier III refers to variants of uncertain clinical significance (VUS), which maps directly to the GPS Level 4; the AMP/ASCO/CAP Tier IV describes benign or likely benign variants, corresponding to GPS Level 5.
The challenges of adopting new guidelines in variant interpretation are multi-fold and are likely encountered by labs like ours that have implemented classification schemes prior to the publication of the 2017 guidelines. Primarily, it is unclear if one can easily correlate the new classification hierarchy to the current one, especially with Tiers I and II (Fig. 1A). The difficulty in mapping assignments across guidelines stems from differential weighting of criteria used to inform the process. Secondly, comparing historical results with contemporary ones may prove difficult for longitudinal quality assessment and process improvement activities. Lastly, a certain degree of subjectivity in variant assessment among clinical variant scientists precludes the development of a linear, and thus automated, transition between classification guidelines. With these challenges in mind, we developed a method to correlate the two classification schemes using historical de-identified variant data.
Fig. 1.
(A) Predicted and (B) observed transition of current (GPS) classification of variants to the 2017 guidelines proposed by AMP/ASCO/CAP. (A) Gradient colors (for Level 2 and Level 3) represent the uncertainty of re-assignment based upon differences in the classification criteria. (B) The differences in mapping variants compared to panel A reflect the data collected as described within the text. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2. Methods
We specifically developed the following approach to better gauge the impact of replacing classification methods from our current, established GPS scheme to the 2017 AMP/ASCO/CAP guidelines. A series of 150 somatic variants previously assigned to the top 4 GPS levels (Level 1, 2, 3 and 4) identified in 40 solid tumor and hematologic cancer cases were selected for a retrospective analysis. Inclusion criteria for this study is that all variants were identified from consecutive cases over a 6-month period (June through Dec 2017). This was performed by a single qualified pathologist and ultimately reviewed by a team of clinical variant scientists including three experienced pathologists at Washington University formally trained in clinical variant interpretation. Prior interpretive information available at the time of clinical reporting along with further evaluation of the clinical case allowed these variants to be scored according to the 2017 AMP/ASCO/CAP guidelines. The variants in this report were classified with the evidence available in May 2019. In our subset of data, evidence at that time did not change the classifications previously applied to the GPS system. The institutional review board of Washington University School of Medicine deemed this study as not fitting the definition of human subjects research; thus, no IRB protocol was required.
3. Results and discussion
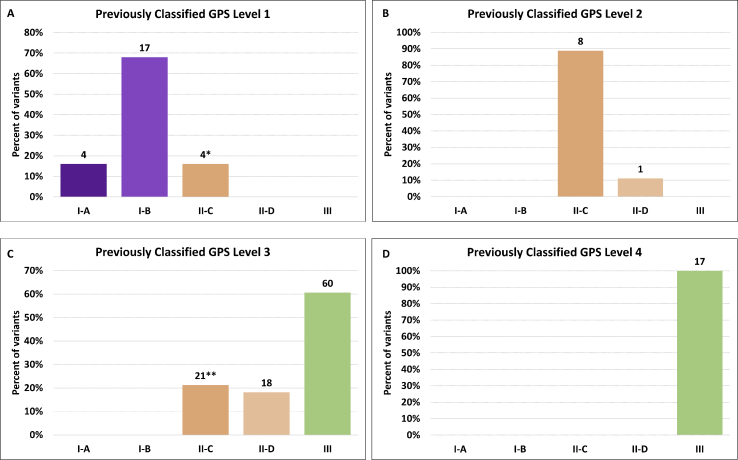
Variants for the mapping exercise were selected from previously reported oncology cases involving the following cancer types: cervix (10), ovarian (6), endometrial (4), hematopoietic/myeloid (4), thyroid (2), gastrointestinal tract (2), head and neck (2), unknown primary (2), and one case each representing lung, breast, pancreas, sarcoma, as well as a rhabdoid tumor and a fibroma. Of note, these cases were skewed toward cancers of the gynecologic tract (50%) and so the variants under investigation would potentially be overrepresented by those prevalent in these tumors. Across this group of tumors, GPS-reported variants included 25 at Level 1, 9 at Level 2, 99 at Level 3, and 17 at Level 4. Noticeably, the most common variant classification reported was Level 3. As suggested above, this level represents the greatest variability in correlating variant assignment to the 2017 guidelines, allowing us to provide a meaningful comparison.
Focusing initially on Level 1 (Fig. 2A), we found that a majority of variants were reclassified as Tier I-A or Tier I–B (21/25; 84%) based on the clinical presentation and existing published evidence. Most of these were Tier I–B, representing the stricter criteria of evidence required to support a level Tier I-A classification. Of the 4 variants identified as Tier II-C, arising largely from a lack of evidence to support a higher classification, 2 were PIK3CA variants. As of May 2019, the FDA approved therapies for HER2-negative, ER/PR-positive breast carcinoma include alpelisib, a PI3Kα inhibitor for use in patients with PIK3CA mutations [3]. Off-label FDA use for other tumor types clearly placed these variants in Tier II-C, although prior interpretations of these variants as prognostic for certain tumor types placed them into the GPS Level 1. Such discrepancies are rare, but point out limitations in transitioning variant level assignments. Our 9 GPS Level 2-assigned variants (Fig. 2B) as all mapped to AMP/ASCO/CAP Tier II, with all but one given an evidence level of C.
Fig. 2.
Reassignments of variants by level. Data represent the 40 tumor types and 150 total variants assessed. The percent of variants reclassified into the respective Tier is shown with actual values above the individual measurements in the bar graph. GPS classification levels shown for each graph are as described in Fig. 1. ∗2 Level 1 to II-C variants were in PIK3CA in cervical cancer (see text). ∗∗17 of 21 Level 3 to II-C variants were in TP53 (see text).
The greatest variability, as predicted, was observed for our diverse GPS Level 3 variants (Fig. 2C). A majority (60/99; 60.6%) of these were classified as AMP/ASCO/CAP Tier III (VUS). This highlights the utility of the AMP/ASCO/CAP guidelines, since many GPS Level 3 variants are thus classified based upon inclusion in publicly-accessible variant databases, where detailed annotations are often lacking. In other words, that a variant has been previously identified does not mean it has escaped the VUS classification. The onus then resides on the clinical variant scientist to report potentially interesting or actionable VUS, with the consequence that these variants will not come to the immediate attention of oncologists using laboratories that do not report or provide interpretations for these VUS. Thus, adoption the 2017 guidelines has the potential to decrease uncertainty and improve clarity the final clinical report, but with the potential cost of ineffective communication of VUS that may nonetheless have important implications for a particular case (e.g., a patient for whom clinical trial eligibility criteria include unspecified variants identified in specific genes).
Interestingly, 17 of 21 (81%) GPS Level 3 variants mapped to AMP/ASCO/CAP Tier II-C were variants in TP53. There is a conflicting body of literature on the actionability of TP53 variants, based largely upon the type of cancer under review and not limited to a specific variant. For example, TP53 can be a biomarker of poor therapeutic response in AML [4] but not in other cancer types. Additionally, therapeutic approaches have been proposed to inhibit dominant-negative functions of mutant p53 or rescue p53 loss of function in a variety of tumor types. In breast cancer, TP53 mutations have been used as a marker of responsiveness to depirubicin-cyclophosphamide [5]. In non-small cell lung cancer, TP53 has been used as a marker of responsiveness to carboplatin/gemcitabine [6]. In chemotherapy-refractory metastatic colorectal carcinoma patients treated with cetuximab, TP53 is a predictor of better clinical outcomes [7]. Finally, there are ongoing clinical trials targeting either wild type or mutant TP53 suggesting use as positive and negative selection strategy (i.e. inclusion or exclusion criteria). However, factoring in the weight of the evidence and lack of preclinical trials, a conservative interpretation places these variants more appropriately into Tier III, thus increasing the number of Level 3 variants reclassified as Tier III at 77.8% (77/99). Finally, all 17 Level 4 variants were correctly assigned to Tier III as variants of unknown clinical significance (Fig. 2D). While we did not explicitly interrogate variants at Level 5, a preliminary mapping indicates that all would map directly into the AMP/ASCO/CAP Tier IV, as expected. In the absence of paired normal tissue for comparison, these include well-documented polymorphisms occurring at a population frequency greater than 1.0%. However, we have recently shown that even this cutoff may be too high [8]. A summary of these reclassification efforts is provided in Fig. 1B.
Our analysis presented here demonstrated that a transition to the 2017 AMP/ASCO/CAP guidelines is likely to correlate well with current evidence-based classification systems. Although similar sources of evidence were used to guide clinical interpretations, the strength of the evidence affected the AMP/ASCO/CAP variant assignment to a greater degree. A standardized approach to evaluating the literature with discretely defined literature sources could reduce the observed variability. As such, we anticipate adoption of the AMP/ASCO/CAP guidelines in parallel with an ongoing somatic variant assessment quality assurance program. Ultimately, the development and utilization of a standardized approach to evaluating the literature with discretely defined literature sources could reduce the observed variability and effectively increase adoption of the AMP/ASCO/CAP guidelines. The standards should be harmonized with those for curation of variant-related data in emerging cancer knowledgebases. Alignment of clinical laboratory and oncology physicians about the utility of these consensus guidelines will also be critical for their ultimate adoption.
CRediT authorship contribution statement
Bijal A. Parikh: Conceptualization, Methodology, Formal analysis, Writing - original draft. Latisha Love-Gregory: Conceptualization, Methodology, Writing - original draft. Eric J. Duncavage: Writing - original draft, Supervision. Jonathan W. Heusel: Conceptualization, Writing - original draft, Visualization, Supervision.
Declaration of competing interest
No conflicts of interest are declared by the authors.
References
- 1.Li M.M., Datto M., Duncavage E.J. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the association for molecular Pathology, American society of clinical oncology, and college of American pathologists. J. Mol. Diagn. 2017;19(1):4–23. doi: 10.1016/j.jmoldx.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cottrell C.E., Al-Kateb H., Bredemeyer A.J. Validation of a next-generation sequencing assay for clinical molecular oncology. J. Mol. Diagn. 2014;16(1):89–105. doi: 10.1016/j.jmoldx.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ando Y., Iwasa S., Takahashi S. Phase I study of alpelisib (BYL719), an α-specific PI3K inhibitor, in Japanese patients with advanced solid tumors. Canc. Sci. 2019;110(3):1021–1031. doi: 10.1111/cas.13923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Welch J.S., Petti A.A., Miller C.A. TP53 and decitabine in acute myeloid leukemia and myelodysplastic syndromes. N. Engl. J. Med. 2016;375(21):2023–2036. doi: 10.1056/NEJMoa1605949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertheau P., Turpin E., Rickman D.S. Exquisite sensitivity of TP53 mutant and basal breast cancers to a dose-dense epirubicin-cyclophosphamide regimen. PLoS Med. 2007;4(3):e90. doi: 10.1371/journal.pmed.0040090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cortinovis D.L., Andriani F., Livio A. FHIT and p53 status and response to platinum-based treatment in advanced non-small cell lung cancer. Curr. Cancer Drug Targets. 2008;8(5):342–348. doi: 10.2174/156800908785133204. [DOI] [PubMed] [Google Scholar]
- 7.Oden-Gangloff A., Di Fiore F., Bibeau F. TP53 mutations predict disease control in metastatic colorectal cancer treated with cetuximab-based chemotherapy. Br. J. Canc. 2009;100(8):1330–1335. doi: 10.1038/sj.bjc.6605008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McNulty S.N., Parikh B.A., Duncavage E.J., Heusel J.W., Pfeifer J.D. Optimization of population frequency cutoffs for filtering common germline polymorphisms from tumor-only next-generation sequencing data. J. Mol. Diagn. 2019;21(5) doi: 10.1016/j.jmoldx.2019.05.005. 903-312. [DOI] [PubMed] [Google Scholar]