Abstract
Novel lanthanide (Ln) compounds [Ln(L)2]Cl.xH2O (Ln = La3+, Ce3+, Sm3+) containing aromatic N,O-chelate ligands [HL1 = 4-amino-2-(1H-benzimidazol-2-yl)phenol; HL2 = 5-amino-2-(1H-benzimidazol-2-yl)phenol] have been synthesized and structurally characterized by elemental analysis, NMR and IR spectroscopy, molar conductance measurements, and mass spectrometry (MS). The spectroscopic data suggested that the benzimidazolyl-phenol ligands act as N,O-chelate ligands through the iminic nitrogen and phenolic oxygen atoms. Elemental analysis indicated that lanthanide compounds were formed in a 1:2 stoichiometry (metal:ligand). In vitro biological evaluation was carried out using these complexes, exhibiting moderate cytotoxicity against six different human tumor cell lines (U251, human glioblastoma; HCT-15, colorectal carcinoma; MCF-7, breast epithelial adenocarcinoma; PC-3, prostate cancer; K562, myelogenous leukemia; SKLU-1, lung carcinoma) and lower toxicity against a non-cancerous cell line (COS-7, primate kidney). In addition, the antibacterial activity of the compounds was assessed against two gram-positive strains (Staphylococcus aureus ATCC 25923, Listeria monocytogenes ATCC 19115) and two gram-negative strains (Escherichia coli ATCC 25922, Pseudomonas aeruginosa ATCC 27583) using the microdilution method. The results obtained show that the metal complexes exhibit higher biological activity than the free ligands, confirming a synergistic effect. Further benzimidazolyl-phenol derivatives were explored for the detection of bacteria using fluorescence imaging studies. Interestingly, the fluorescent properties of these compounds make them potential candidates to monitor the morphology of bacteria at different compound concentrations. Hence, the interaction of the ligand and complexes with model membranes mimicking those of bacteria was studied by using differential scanning calorimetry (DSC) and molecular dynamics (MD), showing that both compounds decreased the enthalpy of transition in two model membranes as the concentration of the compounds increased. In addition, the main transition temperature was slightly reduced as a result of these interactions.
Keywords: Inorganic chemistry, Cytotoxicity, Antibacterial activity, Model membranes, Molecular dynamics, Fluorescence, Lanthanide complexes, Benzimidazole, Cancer
Inorganic chemistry; Cytotoxicity; Antibacterial activity; Model membranes; Molecular dynamics; Fluorescence; Lanthanide complexes; Benzimidazole; Cancer.
1. Introduction
Metal coordination complexes have become very important in medicinal chemistry, in great part due to their physicochemical properties and their multiple oxidation states and stereochemistry, making them suitable candidates for the development of new metal-based therapeutic drugs. Besides, the biological effects and reactivity of metal-based drugs can be easily tuned depending on the structure of the ligand. Thus, the interaction of metal ions with biologically active ligands, incorporates multifunctionality with a single metal coordination complex.
Benzimidazoles are a class of bioactive heterocyclic compounds that display a wide range of useful biological and clinical applications [1]. The benzimidazole backbone represents a privileged structure in medicinal chemistry due to its broad spectrum of pharmacological and therapeutic applications, which include, antimicrobial, antiviral, antiparasitic, anti-inflammatory, antidiabetic and anticancer activities, among others. However, the mechanism of action of these compounds is still not sufficiently understood; but, it has been shown that benzimidazole-based drugs are membrane-active, inhibitors of tubulin polymerization and cause oxidative damage to DNA for the production of reactive oxygen species (ROS) [2, 3].
The benzimidazole scaffold can be readily modified by adequate choice of substituents to modify their biological and pharmacological activities, for instance, to be used in membrane labeling studies in living organisms taking advantage of their fluorescent properties in some conjugated systems [4, 5]. In addition to their biological relevance, this motif is broadly used as ligand in organometallic and coordination chemistry. For example, 2-arylbenzimidazoles can form bonds through substituents on the benzene ring (non-core) and the nitrogen atom (with sp2 hybridization) of the imidazole, forming stable complexes with various metals, either as monodentate ligands [6, 7], N,O-chelate bidentate ligands [8, 9], or N,N,O-chelate tridentate ligands [10]. However, the studies of complexes of 2-arylbenzimidazoles with rare earth elements are scarce [11]. Lanthanide (Ln) coordination complexes are of interest due to their biological activities [12, 13, 14], specific physicochemical properties, unusual electronic configurations, and distinctive multielectron redox chemistry. These properties being ideal for the design and development of novel metal-based pharmaceutical drugs [15, 16, 17, 18].
Thus, considering the potential therapeutic application of lanthanide complexes with N,O-chelate ligands based on 2-arylbenzimidazoles, and in continuation of our work on synthesis and biological activity of metal coordination complexes [19, 20, 21], we herein report the synthesis of new lanthanide complexes with 4 and 5-amino-2-(1H-benzimidazol-2-yl)phenol ligands, as well as their cytotoxic activities against COS-7 non-cancerous cell line (primate kidney) and a number of human cancer cell lines i.e. U251 (human glioblastoma), PC-3 (prostate cancer), K562 (myelogenous leukemia), HCT-15 (colorectal carcinoma), MCF-7 (breast epithelial adenocarcinoma) and SKLU-1 (lung carcinoma). The study of the in vitro antibacterial activity was carried out against S. aureus (ATCC 25923), L. monocytogenes (ATCC 19115), E. coli (ATCC 25922), and P. aeruginosa (ATCC 27583). In addition, the fluorescent properties, and the interactions between model membranes and the synthesized compounds, applying DSC and molecular dynamics were also studied.
2. Experimental Section
2.1. Materials
All chemicals were purchased commercially and used directly without further purification. Microanalyses were performed using a Flash EA 1112 Series CHN Analyzer. Metal concentrations were determined by complexometric titration. Conductivity measurements of the lanthanide complexes were performed in an Orion™ 131S using 1 × 10−3 M solutions in ethanol. The infrared spectra were obtained using a Shimadzu Affinity 1 (FT-IR) spectrometer. 1H and 13C{1H} NMR spectra of the benzimidazolyl-phenol ligands and its lanthanide complexes were recorded on the Bruker Avance II 400 spectrometer using DMSO-d6 as a solvent. The thermal analysis was performed on the TGA 550 TA Instruments analyzer under nitrogen atmosphere between 50 and 500 °C, with a heating rate of 10 °C·min−1. The EPR measurements were recorded on a JEOL JES-TE300 spectrometer operating at X-Band fashions at 100 kHz modulation frequency. The spectrometer settings for all spectra were as follows: center field 336.5 mT; microwave power 8 mW; microwave frequency 9.15 GHz; sweep width ±7.5 mT; modulation width 0.1 mT; time constant 0.1 s; amplitude 200; sweep time 120 s; accumulation 3 scans. The g-factor values were calculated according to Weil [22]. For ligands, mass spectra were recorded on a Shimadzu-GCMS-QP2010 device with electronic impact ionization at 70 eV. The series of complexes were also analyzed by mass spectra in a JEOL AccuTOF JMS-T100LC with DART (Direct Analysis in Real Time) ionization system.
2.2. Synthesis of ligands
The syntheses of the ligands 4-amino-2-(1H-benzimidazol-2-yl)phenol (HL1) and 5-amino-2-(1H-benzimidazol-2-yl)phenol (HL2) were performed by condensation reactions of o-phenylenediamine with the corresponding carboxylic acid, by a slight modification of previously reported procedures [23, 24, 25].
2.2.1. Synthesis of HL1
4-aminosalicylic acid (1.53 g, 10 mmol) and o-phenylenediamine (1.08 g, 10 mmol) were mixed and stirred in polyphosphoric acid (10 mL) for 4 h at 150 °C. After cooling the reaction mixture was neutralized with potassium carbonate 20% and filtered. The gray precipitate was then washed with distilled water (2 × 20 mL) and then recrystallized in ethanol to give the final product HL1 as a beige powder. Yield: 1.58 g, 70%. C13H11N3O (225.2 g·mol−1): Calc. C, 69.32; H, 4.92; N, 18.66. Found: C, 69.07; H, 4.81; N, 18.72%. IR (ATR cm−1): 3340 m, 3222 m, 2373 w, 1615 s, 1578 s, 1509 s, 1472 s, 1402 w, 1349 w, 1240 m, 1208 m, 1155 m, 942 w, 901 w, 845 m, 810 w, 754 s, 694 w, 622 w. 1H NMR (DMSO-d6) δ (ppm) 13.00 (s, 1H), 12.14 (s, 1H), 7.69 (d, 1H, 3J = 7.2 Hz), 7.58 (d, 1H, 3J = 6.8 Hz), 7.26 (t, 2H, 3J = 7.3 Hz), 7.23 (d, 1H, 3J = 2.5 Hz), 6.79 (d, 1H, 3J = 8.5 Hz), 6.72 (dd, 1H, 3J = 8.7, 2.6 Hz), 4.69 (s, 2H). 13C{1H} NMR (DMSO-d6) δ (ppm) 152.3, 150.7, 139.4, 123.1, 120.5, 118.0, 112.9, 112.2. MS (EI, m/z (%)): 225/226 (M+/M+1+, 100/16), calc. 225/226 (M+/M+1+, 100/15); 196 (26), 169 (16), 149 (17), 129 (17), 97 (39), 81 (49), 69 (100), 43 (89).
2.2.2. Synthesis of HL2
The ligand HL2 was obtained as a white powder according to general procedure described for HL1 starting from 5-aminosalicylic acid (1.53 g, 10 mmol) and o-phenylenediamine (1.08 g, 10 mmol). Brown powder, yield: 1.69 g, 75%. C13H11N3O (225.2 g·mol−1): Calc. C, 69.24; H, 4.73; N, 18.79. Found: C, 69.07; H, 4.81; N, 18.72%. IR (ATR cm−1): 3403 m, 3309 m, 1638 w, 1590 w, 1505 s, 1454 m, 1404 m, 1322 m, 1275 m, 1253 m, 1225 m, 1146 w, 1077 w, 1013 w, 982 w, 944 w, 912 w, 849 m, 818 m, 799 m, 740 s, 699 m, 683 w, 636 w. 1H NMR (DMSO-d6) δ (ppm) 7.69 (d, 1H, 3J = 8.5 Hz), 7.57 (dd, 2H, 3J = 5.9, 3.2 Hz), 7.24 (dd, 2H, 3J = 6.0, 3.1 Hz), 6.25 (dd, 1H, 3J = 8.4, 2.1 Hz), and 6.19 (d, 1H, 3J = 2.1 Hz). 13C{1H} NMR (DMSO-d6) δ (ppm) 160.1, 153.6, 152.5, 128.2, 123.0, 114.2, 107.0, 100.4. MS (EI, m/z (%)): 225/226 (M+/M+1+, 100/16), calc. 225/226 (M+/M+1+, 100/15); 196 (30), 169 (16), 149 (14), 129 (15), 98 (29), 81 (50), 69 (100), 41 (65).
2.3. Synthesis of lanthanide compounds
The lanthanide complexes were prepared according to the literature [26] (Figure 1). The numbering of the protons in the complexes is the same used for the ligands.
Figure 1.
Synthesis of lanthanide complexes with ligands HL1 and HL2.
2.3.1. Synthesis of [La(L1)2(Cl)]·CH3CN (La-L1)
A solution of LaCl3⋅7H2O (83 mg, 0.22 mmol) in MeOH (1 mL) was added dropwise into 10 mL of acetonitrile solution of HL1 (2 eq: 100 mg, 0.44 mmol) giving place to the immediate formation of a precipitate. The reaction mixture was set to reflux under stirring for 2 h and cooled to room temperature. The separated brown solid was then filtered, washed with ethyl acetate (2 × 20 mL) and then dried under vacuum. Yield: 123 mg, 84%. C28H23ClLaN7O2 (663.9 g·mol−1): Calc. C, 50.66; H, 3.49; N, 14.77; La, 20.92. Found: C, 50.50; H, 3.59; N, 14.54; La, 20.34%. IR (ATR cm−1): 3382 w, 3306 w, 3164 w, 3048 w, 1618 m, 1558 s, 1492 s, 1464 m, 1322 m, 1262 s, 1238 m, 1143 m, 1029 m, 1007 w, 900 w, 812 m, 749 s, 623 w. 1H NMR (DMSO-d6) δ (ppm) 7.66 (dd, 4H, 3J = 6.0, 3.2 Hz), 7.53 (d, 2H, 3J = 2.6 Hz), 7.28 (dd, 4H, 3J = 6.0, 3.2 Hz), 6.98–6.89 (m, 4H), 4.14 (s, 4H). 13C{1H} NMR (DMSO-d6) δ (ppm) 152.5, 149.3, 141.2, 139.5, 134.5, 119.7, 118.0, 115.2, 100.0. TGA mass loss 6.23% (160–220 °C, 1 step, calc. 1 X CH3CN = 6.18%), 27.62% (250–380 °C, 1 step, calc. La2(CO3)3 formation = 26.49 %). MS (DART+) m/z: [M+1]+ calculated for C28H24ClLaN7O2+ = 664.0743, found = 664.2148. (Ethanol, 26 °C) (−1·cm2 ·mol−1): 0,007.
2.3.2. Synthesis of [La(L2)2(Cl)] (La-L2)
The synthesis of complex La-L2 was performed using the same procedure as that for compound La-L1. To afford a pink colored powder, yield: 121 mg, 88%. C26H20ClLaN6O2 (622.8 g·mol−1): Calc. C, 50.14; H, 3.24; N, 13.49; La, 22.30. Found: C, 50.25; H, 3.34; N, 13.87; La, 22.11%. IR (ATR cm−1): 3533 w, 3215 w, 3048 w, 2723 w, 2569 w, 1624 m, 1548 w, 1508 m, 1382 w, 1312 w, 1275 w, 1240 m, 1004 s, 824 m, 786 w, 742 s, 623 m. 1H NMR (DMSO-d6) δ (ppm) 7.86 (d, 2H, 3J = 8.7 Hz), 7.74 (dd, 4H, 3J = 6.0, 3.2 Hz), 7.44 (dd, 4H, 3J = 6.0, 3.2 Hz), 6.37 (s, 4H). 13C{1H} NMR (DMSO-d6) δ (ppm) 160.2, 153.3, 140.7, 137.0, 129.5, 128.1, 122.7, 106.8, 100.5, 100.0. TGA mass loss 66.12% (50–500 °C, 1 step, calc. LaClO formation = 69.44 %). MS (DART+) m/z: [M+1]+ calculated for C26H22ClLaN6O2+ = 624.0556, found = 624.2272. (Ethanol, 26 °C) (−1·cm2·mol−1): 0,030.
2.3.3. Synthesis of [Ce(L1)2(Cl)]·CH3CN (Ce-L1)
The synthesis of complex Ce-L1 was performed using the same procedure as that for compound La-L1. To afford an olive green colored powder, yield: 132 mg, 90%. C28H23CeClN7O2 (664.1 g·mol−1): Calc. C, 50.56; H, 3.49; N, 14.74; Ce, 21.07. Found: C, 50.68; H, 3.60; N, 14.57; Ce, 21.24%. IR (ATR cm−1): 3564 m, 3180 w, 3048 w, 2560 w, 2361 m, 2333 m, 1665 m, 1621 s, 1555 s, 1496 s, 1464 m, 1325 s, 1272 s, 1237 w, 1149 m, 1074 m, 1007 m, 884 m, 812 m, 752 s, 670 w, 623 w. TGA mass loss 6.34% (160–230 °C, 1 step, calc. 1 X CH3CN = 6.18%), 27.70% (230–500 °C, 1 step, calc. Ce2(CO3)3 formation = 26.25 %). MS (DART+) m/z: [M+1]+ calculated for C28H23CeClN7O2+ = 664.0656, found = 664.2174. (Ethanol, 26 °C) (−1·cm2·mol−1): 0,005.
2.3.4. Synthesis of [Ce(L2)2(Cl)]·(CH3CN) (CH3OH) (Ce-L2)
The synthesis of Ce-L2 was performed using the same procedure as that for compound La-L1. To afford a salmon colored powder, yield: 114 mg, 74%. C29H27CeClN7O3 (697.1 g·mol−1): Calc. C, 49.96; H, 3.90; N, 14.06; Ce, 20.10. Found: C, 50.10; H, 3.99; N, 14.18; Ce, 20.24%. IR (ATR cm−1): 3514 m, 3208 w, 3041 w, 2830 m, 2585 m, 2361 w, 1665 w, 1621 s, 1571 m, 1549 m, 1505 m, 1479 m, 1385 m, 1348 m, 1310 m, 1278 m, 1237 s, 1051 s, 1001 s, 881 m, 821 s, 793 m, 740 s, 623 m. TGA mass loss 5.02% (50–100 °C, 1 step, calc. 1 X CH3OH = 4.59%), 5.89% (180–240 °C, 1 step, calc. 1 X CH3CN = 6.18%). MS (DART+) m/z: [M+1]+ calculated for C29H29CeClN7O3+ = 698.1075, found = 698.2314. (Ethanol, 26 °C) (−1·cm2·mol−1): 0,010.
2.3.5. Synthesis of [Sm(L1)2(Cl)]·2H2O (Sm-L1)
The synthesis of Sm-L1 was performed using the same procedure as that for compound La-L1. To afford an olive green colored powder, yield: 135 mg, 97%. C26H20ClN6O2Sm (634.3 g·mol−1): Calc. C, 49.23; H, 3.18; N, 13.25; Sm, 23.70. Found: C, 49.40; H, 3.19; N, 13.34; Sm, 23.65%. IR (ATR cm−1): 3394 w, 3274 w, 3174 m, 3104 w, 2991 w, 2368 w, 2081 w, 1668 w, 1612 s, 1558 s, 1496 s, 1461 m, 1416 w, 1385 w, 1325 s, 1278 s, 1231 w, 1152 m, 1080 w, 1007 w, 963 w, 941 w, 884 w, 821 s, 784 m, 755 s, 623 m. TGA mass loss 5.46% (50–100 °C, 1 step, calc. 2 X H2O = 5.52%), 23.93% (230–450 °C, 1 step, calc. Sm2(CO3)3 formation = 24.21 %). MS (DART+) m/z: [M+1]+ calculated for C26H20ClN6O2Sm+ = 635.0534, found = 635.2128. (Ethanol, 26 °C) (−1·cm2·mol−1): 0,002.
2.3.6. Synthesis of [Sm(L2)2(Cl) (H2O)]·(CH3OH) (Sm-L2)
The synthesis of Sm-L2 was performed using the same procedure as that for compound La-L1. To afford a light pink colored powder, yield: 132 mg, 88%. C27H25ClN6O4Sm (683.3 g·mol−1): Calc. C, 47.39; H, 3.83; N, 12.28; Sm, 21.97. Found: C, 47.50; H, 3.91; N, 12.51; Sm, 22.14%. IR (ATR cm−1): 3419 m, 3218 w, 3041 w, 2834 m, 2582 w, 2361 m, 2336 m, 1665 w, 1624 s, 1568 m, 1508 m, 1483 w, 1385 m, 1350 m, 1316 m, 1278 m, 1240 s, 1162 w, 1067 s, 1001 s, 907 m, 821 m, 743 s, 689 w, 626 m. TGA mass loss 4.30% (100–160 °C, 1 step, calc. 1 X CH3OH = 4.68%), 3.06% (160–220 °C, 1 step, calc. 1 X H2O = 2.76%), 25.85% (250–500 °C, 1 step, calc. Sm2(CO3)3 formation = 24.21 %). MS (DART+) m/z: [M+1]+ calculated for C27H25ClN6O4Sm+ = 684.0823, found = 684.23106. (Ethanol, 26 °C) (−1·cm2·mol−1): 0,016.
2.4. Cytotoxic activity
2.4.1. Tumor cell culture
U251 (human glioblastoma), PC-3 (prostate cancer), K562 (myelogenous leukemia), HCT-15 (colorectal carcinoma), MCF-7 (breast epithelial adenocarcinoma) and SKLU-1 (lung carcinoma) cells were grown at 37 °C in RPMI-1640 medium (Gibco) enriched with 10% fetal bovine serum (FBS, Gibco), 1% penicillin:streptomycin:amphotericin B solution, 1% nonessential amino acids (Gibco), and 2 mM L-glutamine (Invitrogen) in a humidified atmosphere of 5% (v/v) CO2. Human tumor cytotoxicity was determined under protocols approved by the NCI 1 [27]. Trypan blue was used as indicator of cell viability.
2.4.2. Cytotoxicity assay
Cytotoxic activity of the compounds was determined using a fluoresecent dye sulforhodamine B (SRB) in a microculture assay [27]. 100 μL cell suspensions in fresh media were pipetted into 96-well microtiter plates (Costar), and the material was incubated for 24 h at 37 °C (5% CO2). Consequently, dilutions of the lanthanide complexes (100 μL in DMSO) were added to each well. The same concentration of vehicle was used as positive control. The cancer cell lines were exposed to the tested compounds at concentrations of 25 μM for 48 h. Cell culture medium without tumor cells and compounds was tested as a negative control, while cisplatin was used as reference drug.
2.5. Antibacterial activity
Four bacterial strains (S. aureus ATCC 25923, L. monocytogenes ATCC 19115, E. coli ATCC 25922, P. aeruginosa ATCC 27583) were used to determine the in vitro antibacterial activity of the benzimidazolyl-phenol ligands and their complexes by broth microdilution assays [28, 29]. The final concentration of the compounds was in the range 4–2000 μg·mL−1. Free medium (or with DMSO) was used as a negative control, while the positive control consisted of bacterial medium without an inhibiting agent. Furthermore, the standard antibiotics ciprofloxacin (CP) and silver nitrate were used as additional controls. After overnight incubation at 37 °C, the minimum inhibitory concentration (MIC) was determined. Assays were carried out in triplicate to verify that the inhibition observed in the wells corresponded to the same MIC in all cases.
2.5.1. Stability studies of the complexes in culture media
The stability of the complexes in culture solution was evaluated by electronic absorption experiments using an UV-visible Evolution 220 Thermo Scientific spectrophotometer equipped with Single Cell Peltier System for temperature control. Quartz cells 1 cm of thickness were used and measurements were carried out in a range of 300–700 nm. Mueller Hinton broth (MHB) was used as a cell culture media. Stock solutions of compounds (10−3 M in DMSO) were prepared on the day of experiments, then, 2, 4, 6, 8 and 10 μL of each solution were added to the MHB media, obtaining 2 mL of each with concentrations between 1–5 × 10−6 M; immediately, the UV-Vis spectra were taken at 37 °C. The solutions were then left under the same bacterial growth conditions for 24 h and the UV-Vis spectra were measured again. For blanks, the corresponding amount of DMSO added to the culture media was used.
2.6. Fluorescence studies
The ultraviolet-visible spectra were collected using a UV-Visible V-630BIO spectrophotometer (JASCO) equipped with peltier cell changer (JASCO, PAC-743). The fluorescence spectra were obtained using an FP-8500 spectrofluorometer (JASCO), using quartz cells 1 cm of thickness over the range from 200 to 700 nm. The fluorescence microscopy images were obtained using a Unico G500 Epi-Fluorescence microscope equipped with blue (B-1), green (G-1), and UV-1 filters.
2.6.1. Bacteria labeling
E. coli (ATCC 25922) labeling was performed, according to the literature [30]. The culture media (MHB) was inoculated with the bacterial strain and incubated overnight, at 37 °C. Then, the culture was diluted 1:200 in fresh culture media until reaching the exponential growth phase at 37 °C. To label the bacteria at a fixed concentration, 1 mL of the bacterial solution was added to media containing compound La-L1, at a final concentration of 125 μg·mL−1, and further incubated for 1 h at 37 °C. To study the effects of different compound concentrations on bacterial morphology, 100 μL of the bacterial solution was aliquoted each well of a 96-well plate, and media containing different concentrations of ligand or complex were added, followed by incubation for 12 h at 37 °C. After labeling, the samples were washed 3 times with culture media and subjected to chemical fixation (2.5% formaldehyde in 1 X PBS), for 1 h at room temperature. The slides were imaged using fluorescence microscope equipment. Fluorescence was excited at the wavelength of maximum absorption.
2.7. Interaction with model membranes
2.7.1. Membrane preparation
In a 2:1 (v/v) chloroform/methanol mixture, 1,2-dimyristoyl-sn-glycero-3-phosphocholine (PC) and 1,2-dimyristoyl-sn-glycero-3-phosphorylglycerol (PG) lipids were suspended in a 3:1 ratio (PC/PG), mimicking a model of synthetic bacterial membranes [31]. The PC/PG mixture was dried with nitrogen and under vacuum to obtain lipid films that were subsequently hydrated with HEPES buffer. After vortexing and incubating at 37 °C, multilamellar vesicles (MLVs) were obtained [32].
2.7.2. Differential scanning calorimetry
MLVs with ratios 1:50 and 1:10 (compound/lipids) were obtained from 1 mg of lipids. The samples and the reference solution (HEPES buffer) were placed in DSC Tzero hermetic pans and taken to a DSC Q25 TA instruments equipment to obtain the thermograms over a range of 10–40 °C at a heating rate of 1 °C·min−1. The phase transition temperature (Tm), as well as the enthalpy of transition (DH), were obtained using the Trios software (TA Instruments).
2.7.3. Construction of the ligand 3D model
The ligand molecule was modeled in 3D using the Molview tool [33]; later, its topology was obtained with Swissparam web service [34] which offers data for small organic molecules compatible with the CHARMM or GROMACS force field.
2.7.4. Construction of gram-negative and gram-positive bacterial membrane models
The CHARMM-GUI platform [35] was used to develop a gram-negative membrane model (E. coli) and another of normal human cells including neutral and negative phospholipids, following the methodology suggested by Epand and Wasan et al. [36, 37]. 160 molecules of 1-palmitoyl-2-oleoyl-sn-glycerol-3-phosphatidylethanolamine (POPE) and 40 molecules of 1-palmitoyl-2-oleyl-sn-glycerol-3-[phospho-rac-(1-glycerol)] (POPG) were distributed in internal and external monolayers for the construction of the gram-negative membrane. Moreover, the phospholipids used for the liposome model were organized in both outer and inner monolayers with 50 molecules of dimyristoylphosphatidylglycerol (DMPG) and 150 molecules of dimyristoylphosphatidylcholine (DMPC) in both outer and inner monolayers. The four molecules of 5-amino-2-(1H-benzimidazol-2-yl)phenol (HL2) were localized over the surface of each membrane in each one of the systems.
2.7.5. Molecular dynamic simulation
GROMACS software version 2019.3 was used for molecular dynamics simulation [38]. The ion placement method was used with 0.15 M KCl, with a water thickness of 22.5 Å and CHARMM36 as a force field, which is suitable for describing the distribution of molecules within large systems such as membranes [39]. To ensure there are no steric clashes between an HL2 molecule and the system, it was adjusted by heating to a temperature of 310 K at 1 fs (femtosecond)/step for 75 ps (picosecond), relaxing the structure in a process called energy minimization with 300 ps at 2 fs/step for the equilibration step. With a descent algorithm (tolerance value of 1000 kJ·mol−1 nm−1) in 5000 steps, the energy minimization of the system was obtained. To equilibrate the temperature and pressure, the Berendsen algorithm was used. After the system was equilibrated, we ran production MD for data collection for 10 ns, using the Nose-Hoover and Parrinello–Rahman algorithms to adjust the temperature and pressure. The particle mesh Ewald (PME) summation was applied to correct for long-range electrostatic interactions [40]. The SHAKE algorithm was used to minimize the energy values [41], which allowed a numerical integration time step of 2 fs to be used in the simulation [40].
The interaction analysis was performed between the molecule and the phospholipids to obtain relevant information that could suggest the possible mechanism of action and its correlation with the experiment performed. For this process, GROMACS [38] and VMD (visual molecular dynamics) [42] were used. In the VMD analysis, phospholipid interactions with the molecule were observed to determine how to penetrate the membrane model and whether the molecules remained insite or outside of the membrane.
The root-mean-square deviation (RMSD) and the hydrogen bonds of the molecule (HL2) were determined using the gmx rms and gmx hbond commands, respectively. The partial density distribution of the molecule and phospholipids in the z-axis was counted using the gmx density command.
3. Results and discussion
3.1. Synthesis and characterization
3.1.1. Synthesis and characterization of ligands
Benzimidazolyl-phenol ligands (HL1 and HL2), were obtained, in traditional manner by the condensation reaction of o-phenylenediamine with the corresponding carboxylic acid. The ligands were isolated as white crystalline solids that were soluble in methanol, ethanol, acetone, DMSO and DMF but slightly soluble in water and acetonitrile. The results obtained by NMR spectroscopy, MS and elemental analysis are in good agreement with previous reports [23, 24]. Due to tautomerism (ketoenolic equilibrium) among these ligands and because hydroxyl protons are interchangeable with the deuterated solvent, the peaks corresponding to OH and NH (imidazole) could decrease or disappear from the 1H NMR spectrum [24]. Hydrogen bonding elongates the O-H bond and decreases the electron density around the proton (deshielding effect), resulting in a higher frequency shift compared to NH, as suggested by other authors [25, 43].
3.1.2. Synthesis and characterization of lanthanide complexes
The reaction of LnCl3⋅nH2O (Ln = La(III), Ce(III), Sm(III)) with the corresponding benzimidazolyl-phenol ligand in acetonitrile affords complexes La-L1, La-L2, Ce-L1, Ce-L2, Sm-L1 and Sm-L2 in good yields. The Lanthanide complexes were obtained as microcrystalline powders and were soluble in DMSO and DMF and slightly soluble in methanol and acetone. The results obtained by elemental analysis (C, H and N), EDTA complexometry, TGA analysis and mass spectrometry agree well with the structures proposed (metal:ligand ratio 1:2) [23, 44]. The molar conductivity measurements of 1 × 10−3 M complexes in ethanol at 26 °C were very low, in the range of 0,002–0,030 Ω−1·cm2·mol−1, indicative of the nonelectrolyte nature of the complexes [45, 46].
FT-IR spectroscopy provides complementary information to elucidate the way that the ligands bind to the metal ions. Experimental FT-IR data of HL1 and HL2 and their complexes are presented in Table 1 (S1–S8. Supplementary Material). The characteristic absorption bands (O-H and N-H) of the ligands appear separately because of weak intramolecular H-bond between the OH and the N atom of the heterocyclic ring.
Table 1.
IR bands of HL1 and HL2 ligands and their lanthanide complexes (values in cm−1).
| Compound | υ (N-H) | δ(N-H) | υ(C=N) | υ(C=C) | υ(C-O) | δ(-OH)ip1 | δ(-OH)op2 | δ(C-H) |
|---|---|---|---|---|---|---|---|---|
| HL1 | 3500–3300 | 1615 | 1578 | 1509 | 1240 | 1402 | 942 | 754 |
| La-L1 | 3500–3300 | 1618 | 1558 | 1492 | 1262 | - | - | 749 |
| Ce-L1 | 3500–3300 | 1621 | 1555 | 1496 | 1272 | - | - | 752 |
| Sm-L1 | 3500–3300 | 1612 | 1558 | 1496 | 1278 | - | - | 755 |
| HL2 | 3500–3300 | 1638 | 1590 | 1505 | 1253 | 1404 | 944 | 740 |
| La-L2 | 3500–3300 | 1624 | 1548 | 1508 | 1275 | - | - | 742 |
| Ce-L2 | 3500–3300 | 1621 | 1549 | 1505 | 1278 | - | - | 740 |
| Sm-L2 | 3500–3300 | 1624 | 1568 | 1508 | 1278 | - | - | 743 |
ip = in plane.
op = out of plane.
In the infrared spectra of the metal complexes, the ν(C=N) vibration frequencies are shifted to lower wavenumbers (20−40 cm−1) indicating coordination of nitrogen to the metal atom (Ln-N). The lack of the ν(O–H) band in the FT-IR spectra of the complexes indicates coordination through the deprotonated phenolic oxygen to the Ln (III) ions. This conclusion is also supported by the phenolic ν(C–O) stretching vibrations that are shifted to higher wavenumbers upon complex formation. All these metal-based vibrations were similar to those of other metal coordination complexes previously published [47].
For all the compounds, the characteristic bands of different vibrations of the N-H bonds corresponding to primary and secondary amines (N-H of imidazole) were observed between 3550–3300 (N-H stretching) and 1640–1610 (N-H bending). The frequencies between 1600-1490 cm−1 are attributed to υ(C=C) and υ(C=N) stretching absorptions in the aromatic rings, while the strong bands ca. 750 cm−1 could be assigned to C-H out-of-plane bending vibrations of the aromatic hydrogen atoms [48].
The complexes (except those of La(III)) are paramagnetic. Complexes La-L1 and La-L2 were characterized by nuclear magnetic resonance (1H and 13C{1H}) (see Experimental Section). Thus, 1H NMR spectra of complexes La-L1 and La-L2 exhibit the expected signals in line with the proposed structures (Figure 2 and Figure S20).
Figure 2.
1H-NMR spectra: (a) superposition of HL1 (red) and La- L1 (blue); (b) comparative 1H-NMR spectrum expansion of (a).
The phenolic proton of the ligand (HL1 and HL2) are missing in the 1H NMR spectra of the lanthanum complexes upon deprotonation of the benzimidazolyl-phenol compounds. The absence of the signal corresponding to the N-H group of the imidazole could be explained by the rapid exchange between this proton and the solvent, and both the OH signal and the NH signal could disappear in the presence of metal [23]; however, in the 1H-NMR spectrum of La-L2 (Figure S20), a low-intensity signal was observed near 11.5 ppm, which confirms that the proton signal corresponding to NH is that with the lowest frequency between the two signals, suggesting that the NH signal can be found in the low field, whereas the signal corresponding to OH does not appear upon formation of the La-O bond. On the other hand, the chemical shifts due to H3, H5, H6 and H7' protons were also affected by deprotonation of phenolic group and nitrogen coordination (C=N) to the La(III) ion (for La-L1). Finally, the H3 signal in La-L1 is shifted downfield with respect to the L1 ligand, whereas NH2 protons resonances are shielded (Figure 2). 13C{1H} NMR spectra showed the expected carbon signals for the ligands. For La(III) complexes, the hydroxilic carbon signal was recorded at δ ≈ 141 ppm and 160 ppm for La-L1 and La-L2, respectively, while the carbon of the amine group was observed at 140 ppm and 153 ppm, respectively. Quaternary carbons of benzimidazole (C1′, C3′, C8′) were found in the characteristic chemical shift range, between 130-140 ppm, and were confirmed by dept-135 experiment.
The presence of the paramagnetic lanthanide ions in the complexes were confirmed via room temperature X-band EPR spectroscopy (Figures S22–S25). The EPR spectra showed broad resonance peaks, with g values (approx. 2.00) and line widths similar to those reported for other lanthanide complexes at room temperature [49, 50, 51]. A highly anisotropic complex spectrum was obtained in all cases; however, a hyperfine interaction generated by the Ln-N bond, could be observed if the experiments are brought at 5 K, as suggested by other authors [52]. For Ce-L2, the fine structure is lacking, due to Ce(III) zero-field splitting. The broad signals obtained by EPR showed that the Ce(III) complex is located in a disordered environment. Random H-bonds formed by solvent molecules (in this case methanol) and complexes induce small distortions, resulting in line broadenings. This phenomenon, called g-strain, explains the broad asymmetric EPR line shapes [53, 54]. In addition, mass spectrometric analysis (DART-MS) provides complementary information on the formation of the metal complexes, showing the molecular ion peak [M+1] with percentage error values of less than 0.03% respect to the calculated values reported in the Experimental Section. Figures S28–S33 show the similarity of isotopic patterns for molecular ion peaks compared to simulated mass spectra (isotopes).
3.2. Biological studies
In vitro biological properties of the series of compounds was explored through cytotoxicity and antibacterial susceptibility tests, while fluorescence microscopy and membrane model studies supported the results found.
3.2.1. Cytotoxic activity
The cytotoxic activity of all benzimidazolyl-phenol derivatives (ligands and complexes) was explored over six tumor cell lines and one noncancerous cell line (primate kidney). Table 2 presents the results of these experiments. From here one can easily see that the free ligands exhibited lower or null activities compared to their complexes. Thus, it is clear that the presence of the lanthanides produces an enhanced synergistic regardless of the metal. Hence, the derivatives of HL1 produced higher inhibition percentages compared to complexes including ligand HL2, being clear that the position of the -NH2 substituent group, plays a key role in the biological behavior of this complexes.
Table 2.
Inhibition of the growth (%) of human tumor cell lines at 25 μM.∗
| Compound | U251 | PC-3 | K562 | HCT-15 | MCF-7 | SKLU-1 | COS-7∗∗ |
|---|---|---|---|---|---|---|---|
| HL1 | NA | 19.6 ± 0.7 | 21.6 ± 1.1 | NA | NA | 29.3 ± 1.1 | NA |
| HL2 | NA | NA | NA | NA | NA | NA | NA |
| La-L1 | 1.1 ± 0.9 | 51.3 ± 1.8 | 70.9 ± 2.3 | 15.7 ± 1.0 | 23.3 ± 0.7 | 44.1 ± 0.9 | 13.7 ± 0.5 |
| Ce-L1 | 59.6 ± 2.1 | 53.3 ± 0.7 | 55.8 ± 0.6 | 23.2 ± 0.9 | 30.9 ± 1.2 | 37.0 ± 0.8 | 10.4 ± 0.4 |
| Sm-L1 | 30.3 ± 1.3 | 47.7 ± 1.6 | 56.3 ± 1.5 | 42.8 ± 1.2 | 18.7 ± 0.5 | 47.7 ± 1.1 | 10.4 ± 0.7 |
| La-L2 | NA | 31.5 ± 1.3 | 42.1 ± 2.2 | NA | NA | 27.3 ± 1.6 | NA |
| Ce-L2 | NA | 26.1 ± 0.7 | 34.9 ± 1.4 | NA | NA | 14.3 ± 1.0 | NA |
| Sm-L2 | NA | 17.5 ± 0.5 | 36.5 ± 1.0 | 7.9 ± 1.2 | NA | 13.4 ± 0.7 | NA |
| Cisplatin∗∗∗ | 90.3 ± 2.0 | 64.8 ± 1.3 | 55.3 ± 2.2 | 50.2 ± 1.4 | 79.9 ± 1.6 | 94.8 ± 2.3 | ND |
The results are the average of three runs. Values are expressed as mean ± SD. NA = Not active, ND = Not determined.
Non-cancerous cell line (primate kidney).
Reference drug.
For the series of complexes including HL1, some activity was presented against the examined cancerous cell lines. Having complex La-L1 exhibiting 70.9 ± 2.3 of % inhibition for K562, even more active than reference drug (55.3 ± 2.2), while for the rest of cancer cell lines i.e. HCT-15, MCF-7, PC-3, and SKLU-1, the activities observed were from low to moderate. A similar trend was observed for the Ce(III) and Sm(III) derivatives, with modest cytotoxic activities in all tumor cell lines. Not very much change was observed for the series of lanthanide complexes containing ligand HL2, producing low to modest activities for the PC-3, K562 and SKLU-1 cell lines. However, worth to note the fact that they did not show activity against the healthy cell line (COS-7), thus revealing the better specificity of the lanthanide derivatives of HL2. Based on the above, potential targets can be provided from benzimidazole compounds to understand this selectivity. For instance, in a previous study, triaryl-substituted imidazole served as a telomeric G-quadruplex ligand altering telomere maintenance, a crucial event for the unlimited proliferative potential of cancer cells. Additionally, the membrane interaction studies discussed below showed that hydrogen bonds can occur in the benzimidazole region of the ligand, which can be related to mechanisms involving inhibition of human DNA topoisomerase I, a process well known to occur with benzimidazole derivatives [55]. Another selective target can be the cell membrane of the tumor cells, since it contains phosphatidylserine, an anionic phospholipid, located only in the outer membrane leaflet of tumor cells. Thus, the cancer cell membrane surface could electrostatically attract selectively the ligand when is positively charged in a state of keto-enol equilibria.
3.2.2. Antibacterial activity
All the benzimidazolyl-phenol ligands and its complexes were tested for their antibacterial activities in vitro against gram-positive bacteria (S. aureus ATCC 25923, L. monocytogenes ATCC 19115) and gram-negative bacteria (E. coli ATCC 25922, P. aeruginosa ATCC 27583). The minimal inhibitory concentrations (MIC) of the ligands and its metal complexes were determined by the microdilution assays.
As Figure 3 shows, the synthesized ligands (HL1 and HL2) and its lanthanide complexes were found to be inactive against P. aeruginosa, suggesting a pronounced resistance towards the antibacterial action of the series of compounds tested, this being in agreement with previous reports where these bacteria had greater resistance to antibiotics [56, 57]. With regard to E. coli, S. aureus and L. monocytogenes, the MIC values obtained for the HL2 ligand and their lanthanide complexes were lower to those obtained from HL1 ligand and its complexes. The remarkable activity of the HL2 ligands and their lanthanide complexes may arise from the position of -NH2 substituent group in the phenolic ring, which may play a very important role in the biological behavior of the compounds.
Figure 3.
Minimal inhibitory concentrations (MICs) found for the series of compounds in antibacterial assays (μg·mL−1): (a) gram-negative strains; (b) gram-positive strains.
The MIC values obtained for the gram-positive and gram-negative bacteria are evidence of the broad antibacterial activity of the synthesized compounds [58]. Interestingly, for the analysis with gram-positive strains, the presence of the metal decreases the bacteriostatic effect compared to the ligand, while for the E. coli the same MIC values were observed; being complex Sm-L2 the only one exhibiting a lower value (125 μg·mL−1). Hence, based on these results above we can suggest that indeed coordination of the metal to the ligand hampers the antibacterial activity of the free ligand, since the -OH group of the aminophenol is not able to participate anymore in the interaction with the membranes via H-bonds (DMPG-aminophenol). And the decreasing in the MIC value for Sm-L2 could be rationalized in terms of the molecules present on the external coordination sphere of the complex, being the only compound of the series having hydration water molecules, thus enhancing its solubility in physiological media and thus its biodisponibility ultimately increasing also its activity.
3.2.3. Fluorescence studies
All compounds exhibited fluorescence when their ethanolic solutions were exposed to ultraviolet light (λ 365 nm). Further studies were performed to obtain the fluorescence spectra. First, the UV-Visible spectra used to determine the excitation wavelength, which indicated that the fluorescence experiments should be performed at a fixed excitation of 343 nm, as strong absorption was expected for all compounds at this wavelength. For the subsequent epifluorescence study, a fixed excitation wavelength was also used. The UV-Visible spectrum for HL2, and fluorescence spectra for the whole series of ligands and complexes are presented in Figure 4.
Figure 4.
Fluorescence studies: (a) the series of compounds dissolved in ethanol, in the absence of light (above), in the presence of normal light (medium) and under ultraviolet light (λ 365 nm) (below); (b) UV-Visible spectra for ligand HL2 and their complexes of La(III), Ce(III) and Sm(III); (c) emission spectra (at λ 343 nm) of ligand HL1 and their complexes; (d) emission spectra (at λ 343 nm) of ligand HL2 and their complexes.
Electronic absorption spectra of all compounds were collected using UV-Visible spectroscopy in the 200–800 nm wavelength range. The absorption spectra of HL1 and HL2 display a broad band at ca. 343 nm, which correspond to π→ π∗ transitions of the conjugated system in the ligand and it was shifted to higher wavelengths (red shift) upon complexation. The occurrence of red shifts for lanthanide complexes indicates a decrease in the corresponding energy gaps (HOMO-LUMO) in the electronic energy levels of the ligands as a result of chelation [59]. In addition, fluorescence spectra were taken at the same concentration (10−6 M) for each compound in ethanol at an excitation wavelength of 343 nm, finding maximum emission bands at λ 424 nm for the complexes including ligand HL1 and at λ 365 nm for the series of complexes having ligand HL2. Noteworthy the fact that the intensities of the bands increased with the presence of the lanthanide ions, confirming the expected enhancing effect after chelation.
From the fluorescence spectra, it is found that for the HL1 ligand and its complexes, the wavelength range where emission occurs reaches almost up to 600 nm for HL1 and their complexes, while for HL2 and their lanthanide derivatives, the emission occurred at almost λ 450 nm. Because of this, epifluorescence was studied in E. coli using Blue (B-1) and Green (G1) filters emitting at λ 450 and λ 535 nm respectively. Both filters were used to obtain images with the La-L1 complex at 125 μg·mL−1 (concentration below the MIC). Figure 5 (above) presents the comparative images using the two filters, where it is possible to observe the morphology of a bacillus at 100X.
Figure 5.
Images obtained by fluorescence microscopy on the E. coli: contrast with two microscope filters for E. coli using a solution of compound La-L1 at a concentration of 125 μg·mL−1 (above) and the effect of increasing the concentration of HL1 (below).
In order to improve the definition of the antibacterial effect of the series of compounds, 40X images were obtained with a blue filter for the bacteria media using different concentrations of ligand HL1. In Figure 5 (below) it can be observed that in the absence of the compound, it is not possible to observe any bacillus of E. coli, while even at concentrations as low as 125 μg·mL−1 marked bacillus units can be clearly observed. However, at a concentration of 250 μg·mL−1, agglomeration and elongation of the bacillus begins to be observed, a phenomenon that increases with concentration, to a maximum of concentration of 2000 μg·mL−1 where the morphology of the bacteria is highly affected, suggesting growth inhibition could occur due to genotoxic, epigenetic effects [60] or membrane disturbance [61]. These results motivated the further studies on the effect of the compounds on the integrity of the bacterial membrane.
The excited state intramolecular proton transfer (ESIPT) sites of the ligands explain the ability of the compounds to act as fluorochromes in imaging by epifluorescence microscopy, the use of similar ligands for the morphological analysis of fungi [62] and confluent monolayer of human stem-cells (tissue) has been reported with results comparable to DAPI [63]. Since, the fluorescence microscopy images allowed the observation of the morphology of the bacilli of E. coli, we can conclude that both the ligands and the series of complexes may serve as biomarkers for bacterial membrane. This statement it is supported by molecular dynamics studies (see below section 3.3.2), from which we concluded that there is a membrane insertion mechanism where the benzimidazole moiety of the ligand remains on the hydrophobic region of the membrane. In addition, it was found that, of the four ligands, after only 10 ns one manages to fully enter the membrane, while the other three remain stoked on the membrane surface.
3.2.4. Stability studies of the complexes in culture media
The stability studies for the complexes in MHB media were performed by monitoring changes in the electronic absorption spectra at different concentrations and interaction times. UV-Vis spectroscopy is one of the most widely used methods to determine binding constants and the stability of systems because it is a simple and easy-to-understand method for monitoring complex formation or decomposition [64, 65]. To compare the stability results of the complexes, with respect to time, stability constants were determined at the beginning of the test and 24 h after the compound-culture media interaction. The absorption band was followed at 340 nm and 330 nm, for the Ln-L1 and La-L2 series complexes, respectively, and a constant increase in intensity was observed as the concentrations of each complex increased (Figure 6 and Figures S40–S42).
Figure 6.
Absorption spectra of the Sm-1 complex at different concentrations (1.0–5.0 × 10−6 M) in MHB media and the double reciprocal plot of 1/(A−Ao) vs. 1/[Sm-1] (a) at the beginning of the experiment (b) after 24 h of interaction.
These studies were based on the methodology suggested by the literature for the determination of binding constants [66], assuming one type of interaction between the complexes and the media:
| (1) |
| (2) |
where Ln (III)L is either La(III), Ce(III) or Sm(III) complex, K is conditional stability constant for the complex in the culture media. Assuming :
| (3) |
where and are the analytical concentration of complexes and media, respectively. According to the Beer-Lambert law:
| (4) |
| (5) |
where, Ao = absorbance of MHB media in the absence of the lanthanide complexes, and A = absorbance of MHB media in the presence of the lanthanide complexes. and are the corresponding molar extinction coefficient, and is the light path of the cuvette (1 cm). By replacing Eqs. (4) and (5) in Eq. (3), Eq. (6) can be deduced [64]:
| (6) |
The stability constant (K) can be obtained from the double reciprocal linear graph, by the relationship between the slope and the intercept. Absorption values and stability constants are reported in Table 3. The values found at the beginning of the experiment were similar to those after 24 h of interaction with each complex in the culture medium, suggesting that these lanthanide complexes were stable in MHB medium and that biological activity was not dependent on the interaction between the compound and the medium but, instead, was reliant on the structure and properties of the molecule.
Table 3.
Absorption values (A) obtained from lanthanide complexes at different concentrations (λ = 330 or 340 nm) and stability constants in MHB media at the beginning of the experiment and after 24 h.
| Concentration x 10−6 [M] | La-L1 |
La-L2 |
Ce-L1 |
Ce-L2 |
Sm-L1 |
Sm-L2 |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 h | 24 h | 0 h | 24 h | 0 h | 24 h | 0 h | 24 h | 0 h | 24 h | 0 h | 24 h | |
| 1.0 | 0.1992 | 0,1980 | 0.4362 | 0.5542 | 0.1526 | 0.1838 | 0.1963 | 0.2141 | 0.1048 | 0.1199 | 0.2562 | 0.2897 |
| 2.0 | 0.3923 | 0,3829 | 0.8409 | 0.9483 | 0.3317 | 0.3634 | 0.3990 | 0.4028 | 0.2166 | 0.2247 | 0.4748 | 0.5517 |
| 3.0 | 0.5995 | 0,5580 | 1.2763 | 1.2763 | 0.4968 | 0.5188 | 0.6075 | 0.5980 | 0.3327 | 0.3390 | 0.7272 | 0.7882 |
| 4.0 | 0.8297 | 0,7394 | 1.5980 | 1.4708 | 0.6993 | 0.6963 | 0.7951 | 0.7930 | 0.4491 | 0.4371 | 0.9871 | 1.0198 |
| 5.0 | 1.0291 | 0,9252 | 1.7367 | 1.6156 | 0.8831 | 0.8487 | 0.9527 | 1.0410 | 0.5583 | 0.5441 | 1.2376 | 1.3005 |
| Constant, K x 104[M−1] | 8.37 | 8.91 | 9.24 | 9.90 | 4.51 | 4.38 | 2.95 | 3.53 | 3.75 | 3.29 | 3.55 | 3.59 |
3.3. Model membrane studies
3.3.1. Thermotropic behavior of model membranes
In order to shed further light on the antibacterial mechanism of the series of compounds, interactions with model membranes imitating bacterial membranes were performed applying DSC. Thermograms of MLVs composed of DMPC/DMPG (3:1) in the presence and absence of compounds are represented in Figure 7, where the pretransition and transition peaks showed an endothermic behavior. Ligand HL2 showed a modest effect on the model membrane at a 1:50 M ratio (Table 4), slightly increasing the Tm from 22.92 to 23.11 °C. Additionally, in all cases, the pretransition peak disappeared after the compounds were added (Figure 7), suggesting that small amounts of either HL2 or La-L2 may alter these MLVs since pretransition is very sensitive to the presence of other molecules in the polar region of the phospholipids [67]. On the other hand, by increasing the amount of either La-L2 or HL2 at a molar ratio of 1:10, the Tm and enthalpy were considerably reduced (Table 4). Based on MD simulations (see below), a lower gel-to-fluid phase transition is observed because the series of compounds can penetrate the phospholipid bilayer [61]. Thus, the hydrophobic moiety of aryl ring of the benzimidazole would be inserted into the hydrophobic core of the bilayer, hence decreasing the enthalpy by hampering the interactions between acyl lipid chains [68], such as inter and intra-molecular van der Waals interactions.
Figure 7.
DSC thermograms showing peaks of the phase transition of MLVs formed by DMPC/DMPG (3:1) before and after the addition of HL2 (a) and La-L2 (b) at different compound-lipids molar ratios: 0:1 ( ); 1:50 (
); 1:50 ( ); 1:10 (
); 1:10 ( ).
).
Table 4.
Tm and enthalpy transition (DH) values of MLVs constituted by DMPC/DMPG (3:1) before and after the addition of either HL2 and La-L2 at different compound-lipid molar ratios.
| MLV | Compound-lipid molar ratio | Pretransition temperature (°C) | Tm (°C) | DH (J.g−1) |
|---|---|---|---|---|
| DMPC/DMPG (3:1) | 0:1 | 12 | 22.92 | 0.39 |
| DMPC/DMPG (3:1)-HL2 | 1:50 | 23.11 | 0.39 | |
| 1:10 | 22.51 | 0.08 | ||
| DMPC/DMPG (3:1)-La-L2 | 1:50 | 22.85 | 0.22 | |
| 1:10 | 21.95 | 0.11 |
Interestingly, the complex showed a stronger effect on the membranes than the free ligand, in accordance with the antimicrobial activity observed, suggesting that the mechanism for bacterial death would be related to cell membrane disruption. The differences between HL2 and La-L2 could be due to the contributions of the La to the destabilization of the bilayer, specifically in the polar head of phospholipids, since this has the ability to make contact with the phosphate group of the phosphatidylcholine [69].
3.3.2. Analysis of molecular dynamics
Due to the effect of the molecule HL2 on both the E. coli membrane (POPE and POPG) and the membrane composed of DMPC and DMPG, interaction analysis was performed between this molecule and the phospholipids with the aim to shed further insight on the mechanism of biological action of these compounds and their correlation with the experiment results above. For this process, GROMACS [38] and VMD (visual molecular dynamics) [42] were used. In the VMD analysis, phospholipid interactions with the molecules were observed to determine how the molecule would penetrate them and whether they would remain in or out of the membrane.
Figure 8 shows the behavior of the ligand through the 10 ns of the simulation, for both membrane models DMPC-DMPG and POPG-POPE. It can be observed that the larger variation for both models occurs at the beginning of the simulation, this being due to the starting position of the molecules, because they were located above the membrane without being in contact with it. As time passes by, the variation decreases at 6 ns for the DMPC-DMPG model and at 8 ns for the POPE-POPG. This effect is caused by the interaction between the ligand molecules and the phospholipids, allowing the molecules to be anchored on the surface of the membrane, moderating its structural variation.
Figure 8.
RMSD analysis of the HL2 atoms in DMPC-DMPG (a) and E. coli (b) model membranes.
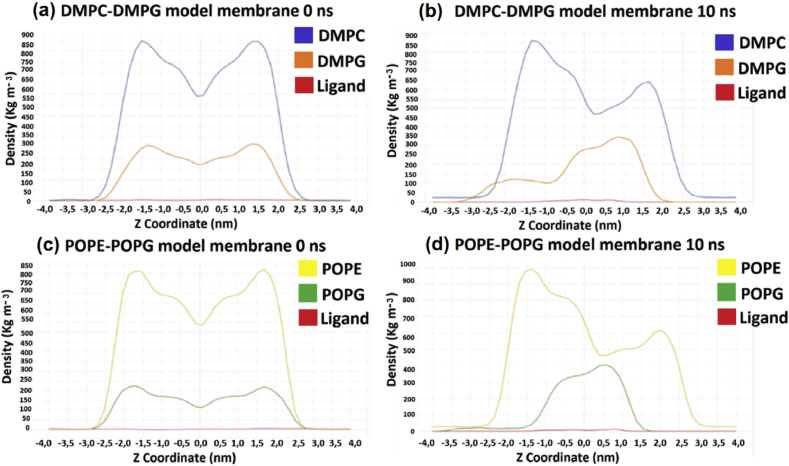
We obtained the density profile of the DMPC-DMPG and POPE-POPG bilayers in the presence of the ligand along the z-axis (Figure 9) in order to analyze the distribution of these components in the system at 0 ns and 10 ns. The peak ligand density is found in the lowest region of all phospholipids at both 0 ns and 10 ns. This indicating that the ligand is preferentially located near the phospholipids type DMPG and POPG. The decrease in density after 10 ns in both models is an indicator of the interaction and insertion of the molecule into the membrane.
Figure 9.
Partial density profile in the z direction in the DMPC-DMPG and POPE-POPG membrane systems: (a) represents the DMPC-DMPG model membrane at 0 ns; (b) represents the DMPC-DMPG model membrane at 10 ns; (c) represents the POPE-POPG model membrane at 0 ns; (d) represents the POPE-POPG model membrane at 10 ns.
Figure 10 shows the mechanism of insertion of the ligand HL2 in the DMPC-DMPG and POPE-POPG membrane systems through 10 ns. In both models, it is detailed how a molecule begins by interacting with phospholipid heads through the nitrogen atom located in its aminophenol moiety; however, this type of interaction is not strong enough to allow the molecule to be trapped, but the stronger interaction with the oxygen atom of the phenolic moiety of the ligand causes its anchorage. Subsequently, the molecule is inserted into the membrane and remains stable (slight changes in its position) with the aminophenol facing towards the membrane surface and the benzimidazole facing towards the hydrophobic region of the membrane.
Figure 10.
Mechanism of insertion of the ligand into the DMPC-DMPG (a) and POPE-POPG (b) membrane systems. The ligand shown is constituted by the red spheres representing oxygen, blue for nitrogen, white for hydrogen and green for carbon.
Figure 11 show the H-bonds between the aminophenol/benzimidazole regions of the ligand and the phospholipids DMPC-DMPG (Figure 11a) and POPE-POPG (Figure 11b). The null interaction between the aminophenol/benzimidazole region and DMPC is clear. However, with DMPG, H-bonds are formed from the middle and end of the simulation. This being due to the fact that the molecules were placed on the membrane models without being in contact with it. Additionally, it is clear how up to two H-bonds are formed with the DMPG and the benzimidazole region of the ligand.
Figure 11.
Number of H-bonds between the two parts of the ligand, namely, the aminophenol region and the benzimidazole region, with DMPC-DMPG (a) and POPE-POPG (b).
The POPE-POPG membrane model shows a null formation of H-bonds in the aminophenol region with the phospholipids. But, the benzimidazole region exhibits interactions with both the POPG and POPE, the latter being the one forming the largest number of H-bonds. The preference for H-bond formation of the membrane models with the benzimidazole region of the ligand is clear. This being caused by the increased presence of atoms prone to form H-bonds such as nitrogen and oxygen. This interaction is critical for the insertion and stabilization of the ligand in the membrane models shown.
Declarations
Author contribution statement
Alberto Aragón-Muriel: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Yamil Liscano-Martínez: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data.
Ernesto Rufino-Felipe: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
David Morales-Morales: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Jose Oñate-Garzón: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Dorian Polo-Cerón: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This work was supported by the Universidad del Valle, Universidad Nacional Autónoma de México (UNAM), Universidad Santiago de Cali, grant number DGI-COCEIN No 512-621116-A30 and the “Doctorados Nacionales Colciencias Convocatoria 727–2015” program.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The authors are grateful to Secretaría de Salud del Valle del Cauca - Complejo Integral de Servicios de Salud Pública Aníbal Patiño Rodríguez for providing the space and the bacterial strains for the antimicrobial assays.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Suman G.R., Bubbly S.G., Gudennavar S.B., Muthu S., Roopashree B., Gayatri V., Nanje Gowda N.M. Structural investigation, spectroscopic and energy level studies of Schiff base: 2-[(3′-N-salicylidenephenyl)benzimidazole] using experimental and DFT methods. J. Mol. Struct. 2017;1139:247–254. [Google Scholar]
- 2.Sahoo S.K., Crisponi G. Recent advances on iron(III) selective fluorescent probes with possible applications in bioimaging. Molecules. 2019;24 doi: 10.3390/molecules24183267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balamurugan G., Velmathi S. Coplanarity driven fluorescence turn-on sensor for chromium(III) and its application for bio-imaging. Photochem. Photobiol. Sci. 2018;17:239–244. doi: 10.1039/c7pp00425g. [DOI] [PubMed] [Google Scholar]
- 4.Liu H., Zhang B., Tan C., Liu F., Cao J., Tan Y., Jiang Y. Simultaneous bioimaging recognition of Al3+ and Cu2+ in living-cell, and further detection of F− and S2− by a simple fluorogenic benzimidazole-based chemosensor. Talanta. 2016;161:309–319. doi: 10.1016/j.talanta.2016.08.061. [DOI] [PubMed] [Google Scholar]
- 5.Wu Y.C., You J.Y., Jiang K., Wu H.Q., Xiong J.F., Wang Z.Y. Novel benzimidazole-based ratiometric fluorescent probes for acidic pH. Dyes Pigments. 2018;149:1–7. [Google Scholar]
- 6.Cheng Z., Yin Q., Wu H., He T., Luo L., Liu X. Regulating Cu(II)-benzimidazole coordination structure in rigid-rod aramid fiber and its composites enhancement effects. Compos. Sci. Technol. 2019;184:107837. [Google Scholar]
- 7.Rani V., Singh H.B., Butcher R.J. Synthesis and structure of the mercury chloride complex of 2,2′-(2-bromo-5-tert-butyl-1,3-phenyl-ene)bis-(1-methyl-1H-benzimidazole) Acta Crystallogr. Sect. E Crystallogr. Commun. 2017;73:341–344. doi: 10.1107/S2056989017001888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tong Y.P., Zheng S.L., Chen X.M. Syntheses, structures, photoluminescence and theoretical studies of two dimeric Zn(II) compounds with aromatic N,O-chelate phenolic ligands. J. Mol. Struct. 2007;826:104–112. [Google Scholar]
- 9.Tong Y.P., Zheng S.L., Chen X.M. Syntheses, structures, photoluminescence, and theoretical studies of a class of beryllium(II) compounds of aromatic N,O-chelate ligands. Inorg. Chem. 2005;44:4270–4275. doi: 10.1021/ic0501059. [DOI] [PubMed] [Google Scholar]
- 10.Hussain A., AlAjmi M.F., Rehman M.T., Amir S., Husain F.M., Alsalme A., Siddiqui M.A., AlKhedhairy A.A., Khan R.A. Copper(II) complexes as potential anticancer and Nonsteroidal anti-inflammatory agents: in vitro and in vivo studies. Sci. Rep. 2019;9:1–17. doi: 10.1038/s41598-019-41063-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olea-Román D., Solano-Peralta A., Pistolis G., Petrou A.L., Kaloudi-Chantzea A., Esturau-Escofet N., Durán-Hernández J., Sosa-Torres M.E., Castillo-Blum S.E. Lanthanide coordination compounds with benzimidazole-based ligands. luminescence and EPR. J. Mol. Struct. 2018;1163:252–261. [Google Scholar]
- 12.Sun H.J., Wang A.L., Bin Chu H., Zhao Y.L. Fluorescent studies on the interaction of DNA and ternary lanthanide complexes with cinnamic acid-phenanthroline and antibacterial activities testing. Luminescence. 2015;30:131–136. doi: 10.1002/bio.2701. [DOI] [PubMed] [Google Scholar]
- 13.Staszak K., Wieszczycka K., Marturano V., Tylkowski B. Lanthanides complexes – Chiral sensing of biomolecules. Coord. Chem. Rev. 2019;397:76–90. [Google Scholar]
- 14.Alptürk O., Rusin O., Fakayode S.O., Wang W., Escobedo J.O., Warner I.M., Crowe W.E., Král V., Pruet J.M., Strongin R.M. Lanthanide complexes as fluorescent indicators for neutral sugars and cancer biomarkers. Proc. Natl. Acad. Sci. U. S. A. 2006;103:9756–9760. doi: 10.1073/pnas.0603758103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cotruvo J.A. The Chemistry of Lanthanides in Biology: Recent Discoveries, Emerging Principles, and Technological Applications. ACS Cent. Sci. 2019;5:1496–1506. doi: 10.1021/acscentsci.9b00642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moksharagni B., Hussain Reddy K. A review on pharmaceutical applications of lanthanide complexes with nicotinoyl and isonicotinoyl hydrazones. Eur. J. Biomed. Pharmaceut. Sci. 2018;5:810–817. [Google Scholar]
- 17.Teo R.D., Termini J., Gray H.B. Lanthanides: applications in cancer diagnosis and therapy. J. Med. Chem. 2016;59:6012–6024. doi: 10.1021/acs.jmedchem.5b01975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Misra S.N., Gagnani M.A., I.D. M., Shukla R.S. Biological and clinical aspects of lanthanide coordination compounds. Bioinorgan. Chem. Appl. 2004;2:155–192. doi: 10.1155/S1565363304000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Polo-Cerón D. Cu(II) and Ni(II) complexes with new tridentate NNS thiosemicarbazones: synthesis, characterisation, DNA interaction, and antibacterial activity. Bioinorgan. Chem. Appl. 2019;2019 doi: 10.1155/2019/3520837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Londoño-Mosquera J.-D., Aragón-Muriel A., Polo Cerón D. Synthesis, antibacterial activity and DNA interactions of lanthanide(III) complexes of N(4)-substituted thiosemicarbazones. Univ. Sci. 2018;23:141–169. [Google Scholar]
- 21.Aragón-Muriel A., Camprubí-Robles M., González-Rey E., Salinas-Castillo A., Rodríguez-Diéguez A., Gómez-Ruiz S., Polo-Cerón D. Dual investigation of lanthanide complexes with cinnamate and phenylacetate ligands: study of the cytotoxic properties and the catalytic oxidation of styrene. Polyhedron. 2014;80:117–128. [Google Scholar]
- 22.Weil J.A., Bolton J.R., Wertz J.E., Buckmaster H.A. Electron paramagnetic resonance: Elementary theory and Practical applications. Phys. Today. 1995;48:71–72. [Google Scholar]
- 23.Tong Y.P., Zheng S.L., Chen X.M. Structures, photoluminescence and theoretical studies of two Zn II complexes with substituted 2-(2-hydroxyphenyl)benzimidazoles. Eur. J. Inorg. Chem. 2005:3734–3741. [Google Scholar]
- 24.Bansal Y., Kaur M., Silakari O. Benzimidazole-ibuprofen/mesalamine conjugates: potential candidates for multifactorial diseases. Eur. J. Med. Chem. 2015;89:671–682. doi: 10.1016/j.ejmech.2014.10.081. [DOI] [PubMed] [Google Scholar]
- 25.Holler M.G., Campo L.F., Brandelli A., Stefani V. Synthesis and spectroscopic characterisation of 2-(2′-hydroxyphenyl)benzazole isothiocyanates as new fluorescent probes for proteins. J. Photochem. Photobiol. Chem. 2002;149:217–225. [Google Scholar]
- 26.Ain Q., Pandey S.K., Pandey O.P., Sengupta S.K. Synthesis, spectroscopic, thermal and antimicrobial studies of neodymium(III) and samarium(III) complexes derived from tetradentate ligands containing N and S donor atoms. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2015;140:27–34. doi: 10.1016/j.saa.2014.12.040. [DOI] [PubMed] [Google Scholar]
- 27.Monks A., Scudiero D., Skehan P., Shoemaker R., Paull K., Vistica D., Hose C., Langley J., Cronise P., Vaigro-wolff A., Gray-goodrich M., Campbell H., Mayo J., Boyd M. Feasibility of a high-flux anticancer drug screen using a diverse panel of cultured human tumor cell lines. J. Natl. Cancer Inst. 1991;83:757–766. doi: 10.1093/jnci/83.11.757. [DOI] [PubMed] [Google Scholar]
- 28.Andrews J.M. Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 2001;48:5–16. doi: 10.1093/jac/48.suppl_1.5. [DOI] [PubMed] [Google Scholar]
- 29.Andrews J.M. Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 2002;49:1049. doi: 10.1093/jac/48.suppl_1.5. [DOI] [PubMed] [Google Scholar]
- 30.Lepore A., Taylor H., Landgraf D., Okumus B., Jaramillo-Riveri S., McLaren L., Bakshi S., Paulsson J., El Karoui M. Quantification of very low-abundant proteins in bacteria using the HaloTag and epi-fluorescence microscopy. Sci. Rep. 2019;9:1–9. doi: 10.1038/s41598-019-44278-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aragón-Muriel A., Ausili A., Sánchez K., Rojas A O.E., Londoño Mosquera J., Polo-Cerón D., Oñate-Garzón J. Studies on the interaction of alyteserin 1c peptide and its cationic analogue with model membranes imitating mammalian and bacterial membranes. Biomolecules. 2019;9:527. doi: 10.3390/biom9100527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oñate-Garzón J., Manrique-Moreno M., Trier S., Leidy C., Torres R., Patiño E. Antimicrobial activity and interactions of cationic peptides derived from Galleria mellonella cecropin D-like peptide with model membranes. J. Antibiot. (Tokyo) 2017;70:238–245. doi: 10.1038/ja.2016.134. [DOI] [PubMed] [Google Scholar]
- 33.Smith T.J. MOLView: a program for analyzing and displaying atomic structures on the Macintosh personal computer. J. Mol. Graph. 1995;13:122–125. doi: 10.1016/0263-7855(94)00019-o. [DOI] [PubMed] [Google Scholar]
- 34.Zoete V., Cuendet M.A., Grosdidier A., Michielin O. SwissParam: a fast force field generation tool for small organic molecules. J. Comput. Chem. 2011;32:2359–2368. doi: 10.1002/jcc.21816. [DOI] [PubMed] [Google Scholar]
- 35.Jo S., Kim T., Iyer V.G., Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 36.Epand R.F., Savage P.B., Epand R.M. Bacterial lipid composition and the antimicrobial efficacy of cationic steroid compounds (Ceragenins) Biochim. Biophys. Acta Biomembr. 2007;1768:2500–2509. doi: 10.1016/j.bbamem.2007.05.023. [DOI] [PubMed] [Google Scholar]
- 37.Wasan K.M., Brazeau G.A., Keyhani A., Hayman A.C., Lopez-Berestein G. Roles of liposome composition and temperature in distribution of amphotericin B in serum lipoproteins. Antimicrob. Agents Chemother. 1993;37:246–250. doi: 10.1128/aac.37.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berendsen H.J.C., van der Spoel D., van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995;91:43–56. [Google Scholar]
- 39.Huang J., Rauscher S., Nawrocki G., Ran T., Feig M., De Groot B.L., Grubmüller H., MacKerell A.D. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods. 2016;14:71–73. doi: 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Legge F.S., Treutlein H., Howlett G.J., Yarovsky I. Molecular dynamics simulations of a fibrillogenic peptide derived from apolipoprotein C-II. Biophys. Chem. 2007;130:102–113. doi: 10.1016/j.bpc.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 41.Ciccotti G., Ryckaert J.P. Molecular dynamics simulation of rigid molecules. Comput. Phys. Rep. 1986;4:346–392. [Google Scholar]
- 42.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 43.Padalkar V.S., Patil V.S., Gupta V.D., Phatangare K.R., Umape P.G., Sekar N. Synthesis, characterization, thermal properties, and antimicrobial activities of 5-(Diethylamino)-2-(5-nitro-1 H -benzimidazol-2-yl)phenol and its transition metal complexes. ISRN Org. Chem. 2011;2011:1–7. doi: 10.5402/2011/738361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xia Q., Cui Y., Yuan D., Wang Y., Yao Y. Synthesis and characterization of lanthanide complexes stabilized by N-aryl substituted β-ketoiminato ligands and their application in the polymerization of rac-lactide. J. Organomet. Chem. 2017;846:161–168. [Google Scholar]
- 45.Bošković P., Sokol V., Tomaš R., Prkić A. Conductometric study of potassium chloride in ethanol-water mixtures. Int. J. Electrochem. Sci. 2013;8:10961–10975. [Google Scholar]
- 46.Geary W.J. The use of conductivity measurements in organic solvents for the characterisation of coordination compounds. Coord. Chem. Rev. 1971;7:81–122. [Google Scholar]
- 47.Hernández-Morales A., Rivera J.M., López-Monteon A., Lagunes-Castro S., Castillo-Blum S., Cureño-Hernández K., Flores-Parra A., Villaseñor-Granados O., Colorado-Peralta R. Complexes containing benzimidazolyl-phenol ligands and Ln(III) ions: synthesis, spectroscopic studies and preliminary cytotoxicity evaluation. J. Inorg. Biochem. 2019;201:110842. doi: 10.1016/j.jinorgbio.2019.110842. [DOI] [PubMed] [Google Scholar]
- 48.Abood N.A., AL-Askari M., Saeed B.A. Structures and vibrational frequencies of Imidazole,benzimidazole and its 2-alkyl derivatives determined by DFT Calculations. Basrah J. Sci. 2012;30:119–131. [Google Scholar]
- 49.Ortu F., Liu J., Burton M., Fowler J.M., Formanuik A., Boulon M.-E., Chilton N.F., Mills D.P. Analysis of lanthanide-Radical magnetic interactions in Ce(III) 2,2′-Bipyridyl complexes. Inorg. Chem. 2017;56:2496–2505. doi: 10.1021/acs.inorgchem.6b02683. [DOI] [PubMed] [Google Scholar]
- 50.Gudasi K.B., Shenoy R.V., Vadavi R.S., Patil M.S., Patil S.A., Hanchinal R.R., Desai S.A., Lohithaswa H. Lanthanide(III) and yttrium(III) complexes of benzimidazole-2-acetic acid: synthesis, characterisation and effect of La(III) complex on germination of wheat. Bioinorgan. Chem. Appl. 2006;2006:1–8. doi: 10.1155/BCA/2006/75612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dunbar T.D., Warren W.L., Tuttle B.A., Randall C.A., Tsur Y. Electron paramagnetic resonance investigations of lanthanide-doped barium titanate: Dopant site occupancy. J. Phys. Chem. B. 2004;108:908–917. [Google Scholar]
- 52.Ortu F., Zhu H., Boulon M.E., Mills D.P. Synthesis and reactivity of a cerium(III) scorpionate complex containing a redox non-innocent 2,2’-bipyridine ligand. INORGA. 2015;3:534–553. [Google Scholar]
- 53.Bencini A., Benelli C., Caneschi A., Carlin R.L., Dei A., Gatteschi D. Crystal and molecular structure of and magnetic Coupling in two complexes containing Gadolinium(III) and Copper(II) ions. J. Am. Chem. Soc. 1985;107:8128–8136. [Google Scholar]
- 54.George G.N., Prince R.C., Bare R.E. Electron paramagnetic resonance spectroscopy of the iron-molybdenum Cofactor of Clostridium pasteurianum Nitrogenase. Inorg. Chem. 1996;35:434–438. doi: 10.1021/ic950740m. [DOI] [PubMed] [Google Scholar]
- 55.Blaszczak-Światkiewicz K., Mirowski M., Kaplinska K., Kruszynśki R., Trzesowska-Kruszyńska A., Mikiciuk-Olasik E. New benzimidazole derivatives with potential cytotoxic activity - study of their stability by RP-HPLC. Acta Biochim. Pol. 2012;59:279–288. [PubMed] [Google Scholar]
- 56.Pachori P., Gothalwal R., Gandhi P. Emergence of antibiotic resistance Pseudomonas aeruginosa in intensive care unit; a critical review. Genes Dis. 2019;6:109–119. doi: 10.1016/j.gendis.2019.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deredjian A., Colinon C., Brothier E., Favre-Bonté S., Cournoyer B., Nazaret S. Antibiotic and metal resistance among hospital and outdoor strains of Pseudomonas aeruginosa. Res. Microbiol. 2011;162:689–700. doi: 10.1016/j.resmic.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 58.Calvo J., Martínez-Martínez L. Mecanismos de acción de los antimicrobianos. Enferm. Infecc. Microbiol. Clín. 2009;27:44–52. doi: 10.1016/j.eimc.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 59.Aragón-Muriel A., Upegui Y., Muñoz J.A., Robledo S.M., Polo-Cerón D. Synthesis, characterization and biological evaluation of rare earth complexes against tropical diseases Leishmaniasis, Malaria and Trypanosomiasis. Av. En Quim. 2016;11:53–61. [Google Scholar]
- 60.Chindera K., Mahato M., Kumar Sharma A., Horsley H., Kloc-Muniak K., Kamaruzzaman N.F., Kumar S., McFarlane A., Stach J., Bentin T., Good L. The antimicrobial polymer PHMB enters cells and selectively condenses bacterial chromosomes. Sci. Rep. 2016;6:1–13. doi: 10.1038/srep23121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hyde A.J., Parisot J., McNichol A., Bonev B.B. Nisin-induced changes in Bacillus morphology suggest a paradigm of antibiotic action. Proc. Natl. Acad. Sci. U. S. A. 2006;103:19896–19901. doi: 10.1073/pnas.0608373104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scroferneker M.L., de Oliveira I.P., Carissimi M., Stefani V., Stopiglia C.D.O., Corbellini V.A. The use of 2-[2’-hydroxy-5’-aminophenyl]benzoxazole (HAMBO), a new fluorochrome for the morphological analysis of Fonsecaea pedrosoi ATCC 46428 using a microculture technique. Rev. Brasileira Biociências. 2010;8:406. http://ezproxy.uniandes.edu.co:8080/login?url=http://search.ebscohost.com/login.aspx?direct=true&db=edb&AN=65173522&lang=es&site=eds-live&scope=site [Google Scholar]
- 63.Oliveira F.F.D., Santos D.C.B.D., Lapis A.A.M., Corrêa J.R., Gomes A.F., Gozzo F.C., Moreira P.F., De Oliveira V.C., Quina F.H., Neto B.A.D. On the use of 2,1,3-benzothiadiazole derivatives as selective live cell fluorescence imaging probes. Bioorg. Med. Chem. Lett. 2010;20:6001–6007. doi: 10.1016/j.bmcl.2010.08.073. [DOI] [PubMed] [Google Scholar]
- 64.Abo El-Maali N., Wahman A.Y., Aly A.A.M., Nassar A.Y., Sayed D.M. Estimating of the binding constant of the anticancer 5-fluorouracil with Samarium and Europium. Inorg. Chem. Commun. 2019;107:107440. [Google Scholar]
- 65.Le M., Rathje O., Levina A., Lay P.A. High cytotoxicity of vanadium(IV) complexes with 1,10-phenanthroline and related ligands is due to decomposition in cell culture medium. J. Biol. Inorg. Chem. 2017;22:663–672. doi: 10.1007/s00775-017-1453-4. [DOI] [PubMed] [Google Scholar]
- 66.Ackermann T., Connors K.A. Binding constants - the measurement of molecular complex stability, John Wiley & Sons, New York, Chichester, Brisbane, Toronto, Singapore 1987. 411 Seiten, Preis: £ 64.15. Berichte Der Bunsengesellschaft Für Phys. Chemie. 1987;91:1398. [Google Scholar]
- 67.Ezer N., Sahin I., Kazanci N. Alliin interacts with DMPC model membranes to modify the membrane dynamics: FTIR and DSC Studies. Vib. Spectrosc. 2017;89:1–8. [Google Scholar]
- 68.Papahadjopoulos D., Moscarello M., Eylar E.H., Isac T. Effects of proteins on the thermotropic phase transitions of phospholipid membranes. BBA - Biomembr. 1975;401:317–335. doi: 10.1016/0005-2736(75)90233-3. [DOI] [PubMed] [Google Scholar]
- 69.Hauser H., Phillips M.C., Levine B.A., Williams R.J.P. Ion-binding to phospholipids: interaction of Calcium and lanthanide ions with phosphatidylcholine (Lecithin) Eur. J. Biochem. 1975;58:133–144. doi: 10.1111/j.1432-1033.1975.tb02357.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.