Abstract
The immune regulation of cancer growth and regression has been underscored by the recent success of immunotherapy. The possibility that immune microenvironmental factors may impact on clinical outcome and treatment response still requires intense investigations. Hereby, supporting data of the research article “Integrated CT Imaging and Tissue Immune Features Disclose a Radio-Immune Signature with High Prognostic Impact on Surgically Resected NSCLC” [1], are presented. With the ultimate aim to provide non-invasive prognostic scores, we report on our approach to correlate different Tumor Immune Microenvironment (TIME) profiles with CT imaging-derived qualitative (semantic, CT-SFs) and quantitative (radiomic, CT-RFs) features in a cohort of 60 surgically resected NSCLC. The renowned characterization of TIME, essentially based on the score evaluation of Programme Death Ligand-1 (PD-L1) and Tumor Infiltrating Lymphocytes (TILs), was implemented here by the assessment of effector and suppressor phenotypes including the analysis of Programme Death receptor 1 (PD-1). Thus, we defined two main TIME categories: hot inflamed (PD-L1high, CD8/CD3high and PD-1/CD8low) as opposed to cold inactive (PD-L1low, CD8/CD3lowand PD-1/CD8high). Importantly, as reported in the extended publication [1], these distinctive immune contextures identified different prognostic classes and were decoded by radiomics. To corroborate our radiomic approach, a comparative estimation of CT-RFs extracted from 60 NSCLC and 13 non neoplastic tissues was undertaken, documenting high discrimination ability. Moreover, we tested the potential association of qualitative radiologic features with clinico-pathological and TIME parameters. Taken together, our findings suggest that CT-SFs and CT-RFs may underlay specific patterns of lung cancer.
Keywords: Lung cancer, CT imaging, Immune contexture, Radiomics, Prognostic signature
Specifications Table
| Subject | Oncology |
| Specific subject area | Prognostic Biomarkers in Non Small Cell Lung Cancer |
| Type of data | Table Image Graph Fig. Raw data |
| How data were acquired | Data were retrospectively collected. Radiology. - Digital Imaging and Communications in Medicine (DICOM) datasets retrieved from PACS; - CT scanners: 6-slice and second generation dual source 128-slice; - Open-source software for quantitative analyses: 3dSlicer 4.9.0, www.slicer.org[9]. Scanned Assisted Microscopy. - Optical and fluorescence. - Immunohistochemistry. Calculations and graphics. - IBM SPSS version 25 (IBM Corporation, Armonk, NY, USA). - Morpheus software (Broad, Institute, Cambridge, MA, USA). - GraphPad PRISM, version 5 (GraphPad Software, Inc., La Jolla, CA, USA). |
| Data format | Table 1, Table 2, Table 3, Table 4, Table 5 .pptx, Fig. 1, Fig. 2 .tiff and raw data to Tables and Fig.s in excel format |
| Parameters for data collection | Clinico-pathological data. CT scan images. Immunohistochemical parameters. |
| Description of data collection | Clinico-pathological data were collected retrospectively from the local electronic hospital information system or the electronic patient record. Immunohistochemical (IHC) analysis was performed on tissue samples archived at the Pathology Unit of our Institution. IHC data were retrospectively investigated on surgical specimens of NSCLC patients who underwent surgical resection at the Thoracic Surgery Unit Entered data was double checked to reduce the possibility of potential errors. Raw data are provided in Excel file format. |
| Data source location | Department of Medicine and Surgery and University Hospital of Parma, Medical Oncology, Radiology, Pathology Units Via Gramsci, 14 43126 Parma ITALY |
| Data accessibility | Raw data to Table 1, Table 2, Table 3, Table 4, Table 5 and Fig. 1, Fig. 2 are provided as supplementary files in excel format within the article |
| Related research article | Giulia Mazzaschi, Gianluca Milanese, Paolo Pagano, Denise Madeddu, Letizia Gnetti, Francesca Trentini, Angela Falco, Caterina Frati, Bruno Lorusso, Costanza Lagrasta, Roberta Minari, Luca Ampollini, Mario Silva, Nicola Sverzellati, Federico Quaini, Giovanni Roti and Marcello Tiseo Integrated CT Imaging and Tissue Immune Features Disclose a Radio-Immune Signature with High Prognostic Impact on Surgically Resected NSCLC Lung Cancer ID-19-01653 |
Value of the data
-
•
These data may help to achieve the goal to non-invasively decipher the tumor immune microenvironment by advanced imaging analysis.
-
•
These data would benefit the translational research to generate new clinically applicable algorithms.
-
•
These data can serve as potential benchmark for other researchers involved in the analysis of tumor immune contexture and its integration in multiparametric models.
-
•
These data may represent the basis for the development of predictive scores of the response to immunotherapy in NSCLC.
-
•
These data may prospectively contribute to the selection of early stage NSCLC patients candidate to receive neoadjuvant or adjuvant treatment
1. Data Description
1.1. Tumor Immune Signature
The morphometric analysis of NSCLC samples documented a large variability in tissue composition within patients and between Squamous Cell Carcinoma (SCC) and Adenocarcinoma (ADC) (Table 1). Compared to ADC, SCC showed higher amount of fibrosis and necrosis with a relative reduced fractional volume occupied by neoplastic cells (Table 1). In addition, CD3pos TILs were more abundant in ADC while SCC were richer in PD-1pos lymphocytes (Table 1). No differences among histotypes were observed in PD-L1 expression, as measured by both H-score and QIF, nor in CD8pos TILs number.
Table 1.
Morphometric Analysis.
| Tissue Composition | ||||||
|---|---|---|---|---|---|---|
| SCC = 39 |
ADC = 30 |
NSCLC = 69 |
||||
| Range | Mean ± St. Err | Range | Mean ± St. Err | Range | Mean ± St. Err | |
| Tumour Volume, cm3 | 0.27-717 | 53.12±23.80 | 0.6-297 | 44.32±12.25 | 0.27-717 | 48.87±13.55 |
| Neoplastic cells, % | 18.82-61.57 | 41.26±1.93 | 14.93-85.10 | 57.26± 2.85 (*) | 14.93-85.10 | 49.01±6.74 |
| TILs, % | 1.11-35.09 | 11.09±1.52 | 0.08-43.29 | 11.09±1.82 | 0.08-43.29 | 11.09±4.17 |
| Fibrosis, % | 7.58-33.62 | 18.95±1.21 | 0.69-29.17 | 14.61±1.22 (*) | 0.69-33.62 | 16.84±1.98 |
| Necrosis, % | 0.01-55.35 | 9.79±1.76 | 0.01-21.38 | 4.35±0.98 (*) | 0.01-55.35 | 7.15±1.59 |
| Tumour Immune Microenvironment | ||||||
| SCC = 39 |
ADC = 30 |
NSCLC = 69 |
||||
| Range | Mean ± St. Err | Range | Mean ± St. Err | Range | Mean ± St. Err | |
| CD3+, n/mm2 | 55.99-2372.05 | 691.05±77.37 | 92.05-6016.75 | 1197.27±201.82 (*) | 55.99-6016.75 | 893.76±107.11 |
| CD8+, n/mm2 | 13.00-382.34 | 118.11±12.54 | 8.73-397.72 | 95.86±15.06 | 8.73-397.72 | 104.71±9.75 |
| CD4+, n/mm2 | 13.07-287.53 | 74.66±12.97 | 0.00-76.80 | 39.59±9.32 (*) | 0.001-287.53 | 61.17±9.47 |
| PD-1+, n/mm2 | 0.001-158.20 | 47.25±6.36 | 2.67-135.96 | 31.83±5.26 (*) | 0.001-158.20 | 39.34±4.25 |
| CD57+, n/mm2 | 0.001-14.07 | 1.66±0.72 | 0.001-1.32 | 0.59±0.17 | 0.001-14.07 | 1.27±0.56 |
| CD25+, n/mm2 | 4.78-16.05 | 8.96±1.47 | 1.59-14.08 | 7.12±1.55 | 1.59-16.05 | 8.10±1.01 |
| FOXP3+, n/mm2 | 1.94-4.89 | 3.42±0.99 | 0.12-1.33 | 0.73±0.61 | 0.12-4.89 | 2.07±1.01 |
| GZB +, n/mm2 | 0.001-12.46 | 5.58±1.46 | 0.001-8.20 | 2.96±1.23 (*) | 0.001-12.46 | 4.27±1.14 |
| PD-L1 | ||||||
| H-Score | 68.00-300.00 | 178.18±20.76 | 0.001-295.00 | 104.35±15.22 | 0.001-300.00 | 122.07±13.52 |
| QIF (x106) | 29.14 -288.13 | 135.01±18.41 | 0.049-130.94 | 48.54±8.46 | 0.049-288.13 | 80.30±11.39 |
TILs: Tumour Infiltrating Lymphocytes; GZB: Granzyme B; QIF: quantitative immunofluorescence; (*) P<0.05 vs SCC
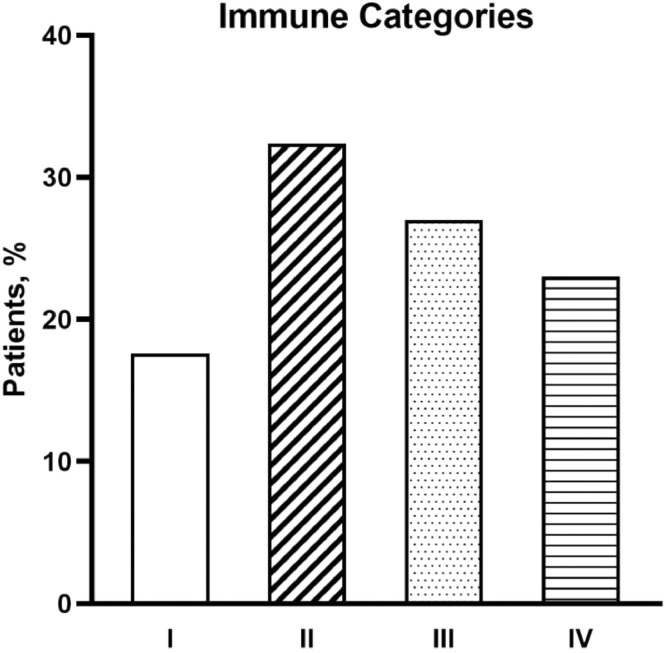
To define a tumor immune signature, first we immunohistochemically assessed PD-L1 levels and the incidence of TILs as major determinants of the widely reported classification of TIME into four immune classes [2]. In our cohort of NSCLC, more than 1/3 of cases belonged to class II implying a rather desert immune contexture (Fig. 1).
Fig. 1.
Distribution of TIME categories among the overall patient population of NSCLC.
The quantitative values and the relative contribution of PD-L1 and TILs phenotypes to the composition of these classes of TIME are reported in Table 2.
Table 2.
TILs incidence and PD-L1 Expression according to TIME categories.
| Parameters | Type I |
Type II |
Type III |
Type IV |
||||
|---|---|---|---|---|---|---|---|---|
| Range | Mean ± St. Err | Range | Mean ± St. Err | Range | Mean ± St. Err | Range | Mean ± St. Err | |
| CD3+, n/mm2 | 420.75-6016.72 | 1441.9±429.8 | 92.05-1144.87 | 626.7±76.01 | 148.78-3140.19 | 546.7±167.6 | 55.9-2719.9 | 1247.5±147.4 |
| CD8+, n/mm2 | 39.41-397.71 | 131.11±29.26 | 8.73-174.42 | 68.45±10.43 | 13.92-254.68 | 83.84±11.75 | 11.04-435.93 | 159.08±30.06 |
| CD4+, n/mm2 | 34.00-104.66 | 73.60±14.69 | 0.00-51.40 | 30.44±15.57 | 0.00-94.05 | 54.09±8.36 | 5.01-287.53 | 79.80±31.42 |
| PD-1+, n/mm2 | 2.66-104.52 | 39.90±7.38 | 0.00-135.65 | 41.89±6.67 | 9.89-79.99 | 34.43±6.28 | 2.00-143.66 | 51.45±11.39 |
| CD8-to-CD3 | 0.03-0.24 | 0.11±0.02 | 0.008-0.25 | 0.08±0.016 | 0.04-0.57 | 0.21±0.03 | 0.008-0.41 | 0.15±0.04 |
| PD-1-to-CD8 | 0.03-1.45 | 0.91±0.14 | 0.001-3.92 | 1.01±0.23 | 0.08-1.56 | 0.48±0.12 | 0.034-3.65 | 0.65±0.24 |
| CD57+, n/mm2 | 0.25-1.01 | 0.56±0.16 | 0.01-0.77 | 0.27±0.24 | 0.01-14.06 | 2.31±1.68 | 0.11-1.96 | 0.72±0.16 |
| CD25+, n/mm2 | 3.69-11.24 | 7.44±2.17 | 0.00-3.75 | 3.62±1.56 | 1.59-11.03 | 6.98±1.39 | 4.80-16.05 | 9.54±1.86 |
| FOXP3+, n/mm2 | 0.11-2.24 | 0.99±0.19 | 0.29-3.13 | 2.07±1.01 | 0.12-1.94 | 1.03±0.91 | 1.33-4.88 | 3.11±1.77 |
| GZB+, n/mm2 | 2.29-8.20 | 4.84±1.75 | 0.00-3.75 | 1.68±1.10 | 5.89-12.46 | 9.17±3.28 | 0.00-9.06 | 3.31±2.05 |
| PD-L1 | ||||||||
| H-Score | 62-295 | 181.56±20.74 | 0-300 | 98.79±19.92 | 0-280 | 162.83±17.88 | 14-170 | 80-88±13.83 |
| QIF (x106) | 72.72-210.52 | 137.8±20.8 | 2.13-102.25 | 43.74±8.53 | 61.32-288.12 | 148.15±23.72 | 0.488-116.68 | 44.8±11.4 |
TILs: Tumour Infiltrating Lymphocytes; GZB: Granzyme B; QIF: quantitative immunofluorescence
Nonetheless, in our patient population the clinical relevance of the above-mentioned classification was faint [1]. Thus, to further distinguish active from dormant TIME profiles, we introduced the combined evaluation of PD-L1 with both the relative proportion of CD8pos cells over the total population of T CD3pos lymphocytes (CD8-to-CD3 ratio) and the incidence of PD-1 inhibitory receptor on CD8pos cytotoxic cells (PD-1-to-CD8 ratio). By this approach, high PD-L1 levels, high CD8-to-CD3 ratio and low expression of PD-1 on CD8pos cytotoxic lymphocytes depicted hot TIME (PD-L1high-CD8/CD3high-PD-1/CD8low) while PD-L1low/neg-CD8/CD3low-PD-1/CD8high characterized cold immune background (raw data see Appendix A).
In support to our categorization in hot and cold TIME, effector phenotypes (CD57+ NK and GrzB+) were prominent in hot while suppressor cells (CD25+ Treg) prevailed in cold TIME (Table 3).
Table 3.
Incidence of specific TILs phenotypes in Hot and Cold TIME.
| Hot n=15 | Cold n=15 | |||
|---|---|---|---|---|
| Parameters | Range | Mean ± St. Err | Range | Mean ± St. Err |
| CD57+, n/mm2 | 0.00-7.08 | 2.26±1.02 | 0.00-1.16 | 0.20±0.08 (*) |
| CD25+, n/mm2 | 0.00-1.05 | 0.12±0.06 | 1.09-5.28 | 3.12±1.78 (*) |
| FOXP3+, n/mm2 | 3.94-10.89 | 7.40±2.59 | 5.09-11.33 | 8.50±0.77 |
| GZB +, n/mm2 | 4.01-12.46 | 8.44±1.73 | 0.001-8.20 | 1.05±0.80 (*) |
GZB: Granzyme B; (*) P<0.05 vs Hot TIME
The differential impact of hot and cold TIME on NSCLC survival outcome is clearly documented in the main manuscript [1]. All raw datasets referring to Table 1-3 and Fig. 1 are included in the supplementary data (see raw data in Appendix A).
1.2. CT-derived Semantic Features (CT-SFs)
To initially assess the impact of qualitative CT descriptors on patients’ outcome, we analyzed shape, margins, texture, structure and effect on parenchyma, whose clinical relevance in NSCLC had been previously demonstrated [3]. The distribution of semantic CT features in our cohort of patients is reported in Table 4. We observed that association of clinical parameters with CT-SFs could identify specific patterns of NSCLC. For example, a trend toward a correlation of active smoking with CT non-homogeneous structure was documented (P=0.09). Moreover, an association between pTNM staging and CT structure (P=0.074) and margins (P=0.032) was present (Table 5). All raw datasets referring to Table 4-5 are included in the supplementary data (see raw data Appendix A).
Table 4.
CT-derived Semantic Features (CT-SFs).
| SCC = 39 | ADC = 30 | NSCLC = 69 | ||
|---|---|---|---|---|
| n (%) | n (%) | n (%) | ||
| Shape | ||||
| Spherical | 22 (56) | 15 (50) | 37 (54) | |
| Non spherical | 17 (44) | 15 (50) | 32 (46) | |
| Margins | ||||
| Well defined | 5 (13) | 4 (13) | 8 (12) | |
| Undefined | 9 (23) | 8 (27) | 18 (26) | |
| Lobulated | 10 (26) | 8 (27) | 19 (27) | |
| Spiculated | 15 (38) | 10 (33) | 24 (35) | |
| Texture | ||||
| Solid | 36 (92) | 26 (87) | 63 (91) | |
| Non solid | 3 (8) | 4 (13) | 6 (9) | |
| Structure | ||||
| Homogeneous | 13 (33) | 20 (67) | 33 (48) | |
| Non homogeneous | 26 (67) | 10 (33) | 36 (52) | |
| Effect on parenchyma | ||||
| No effect | 17 (44) | 13 (43) | 31 (45) | |
| Pleural retraction | 13 (33) | 9 (30) | 23 (33) | |
| Scissural displacement | 8 (21) | 5 (17) | 12 (18) | |
| Overinflation | 1 (2) | 3 (10) | 3 (4) |
Table 5.
Clinico-pathological and CT-SFs Correlations.
| Effect |
Margins |
Texture |
Shape |
Structure |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Presence | Absence |
Well defined | Spiculated |
Solid | Non solid |
Spherical | Non spherical |
Homogeneous | Non homogeneous |
||||||
| n (%) | n (%) | P | n (%) | n (%) | P | n (%) | n (%) | P | n (%) | n (%) | P | n (%) | n (%) | P | |
| Sex | |||||||||||||||
| Male | 30 (60) | 20 (40) | 0.057 | 18 (36) | 32 (64) | 0.786 | 48 (96) | 2 (4) | 0.185 | 24 (48) | 26 (52) | 0.165 | 22 (44) | 28 (56) | 0.384 |
| Female | 7 (37) | 12 (63) | 7 (37) | 12 (63) | 14 (74) | 5 (26) | 13 (32) | 6 (32) | 11 (58) | 8 (42) | |||||
| Smoking status | |||||||||||||||
| Never | 4 (67) | 2 (33) | 0.883 | 3 (50) | 3 (50) | 0.667 | 5 (83) | 1 (17) | 0.425 | 3 (50) | 3 (50) | 0.680 | 5 (83) | 1 (17) | 0.043 (*) |
| Current | 13 (56) | 10 (44) | 9 (39) | 14 (61) | 20 (83) | 4 (17) | 15 (65) | 8 (35) | 7 (30) | 16 (70) | |||||
| Ex | 22 (55) | 18 (45) | 16 (33) | 24 (67) | 35 (87) | 5 (13) | 19 (48) | 21 (52) | 20 (50) | 20 (50) | |||||
| Histotype | |||||||||||||||
| SCC | 22 (56) | 17 (44) | 0.589 | 14 (36) | 25 (64) | 0.455 | 37 (95) | 2 (5) | 0.287 | 22 (56) | 17 (44) | 0.643 | 13 (33) | 26 (67) | 0.008 (#) |
| ADC | 17 (57) | 13 (43) | 12 (40) | 18 (20) | 26 (87) | 4 (13) | 15 (50) | 15 (50) | 20 (67) | 10 (33) | |||||
| Stage | |||||||||||||||
| I | 16 (55) | 13 (45) | 0.872 | 20 (69) | 9 (31) | 0.028 (*) | 27 (94) | 2 (6) | 0.222 | 15 (52) | 14 (48) | 0.956 | 19 (65) | 10 (35) | < 0.001 |
| II | 13 (57) | 10 (43) | 13 (57) | 10 (43) | 20 (87) | 3 (13) | 11 (48) | 12 (52) | 3 (13) | 20 (87) | |||||
| III | 7 (41) | 10 (59) | 5 (29) | 12 (71) | 15 (88) | 2 (12) | 10 (59) | 7 (41) | 7 (41) | 10 (59) | |||||
| EGFR Mutation | |||||||||||||||
| WT | 10 (52) | 11 (48) | 0.578 | 8 (38) | 13 (62) | 0.657 | 16 (76) | 5 (34) | 0.343 | 9 (43) | 12 (57) | 0.635 | 11 (52) | 10 (48) | 0.127 |
| Mut | 2 ()50 | 2 (50) | 2 (50) | 2 (50) | 4 (100) | 0 (0) | 2 (50) | 2 (50) | 4 (100) | 0 (0) | |||||
| KRAS Mutation | |||||||||||||||
| WT | 12 (66) | 6 (34) | 0.255 | 5 (28) | 13 (72) | 0.101 | 16 (89) | 2 (11) | 0.368 | 11 (61) | 7 (39) | 0.535 | 11 (61) | 7 (39) | 0.535 |
| Mut | 1 (25) | 3 (75) | 3 (75) | 1 (25) | 3 (75) | 1 (25) | 2 (50) | 2 (50) | 2 (50) | 2 (50) | |||||
1.3. CT-derived Radiomic Features (CT-RFs)
Specificity and reliability of our proposed radiomic approach to tumor lesions (NSCLC) were validated using control tissues (muscles and lymph nodes).
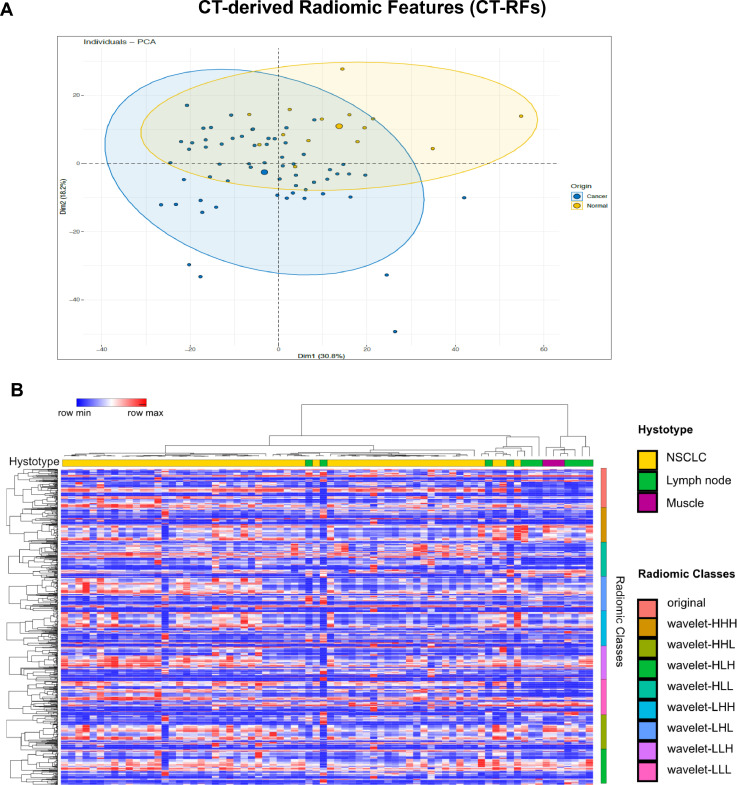
Considering the 841 CT-RFs as correlated variables, we employed specific algorithms for dimensionality reduction. Thus, principal component analysis (PCA) was applied to CT-RFs extracted from 60 lung cancers and 13 normal tissues (10 uninvolved lymph nodes, 3 skeletal muscles). As shown in Fig. 2A, two vectors contain nearly the 50% of data variance, sufficient to preliminary distinguish tumor versus normal samples.
Fig. 2.
A: Principal Component Analysis (PCA) score plots representing the congruence of CT-RFs extracted from tumor (blue dots) and normal tissues (yellow dots). B: Heatmap illustrating the unsupervised cluster analysis of RFs extracted from CT slices of 60 NSCLC (yellow), 10 uninvolved lymph nodes (green) and 3 skeletal muscles (purple). While strings from two lymph nodes are intercalated to NSCLC samples, the sharp right-sided CT-RFs clustering of most normal tissues is apparent (see histotype bar code).
To further confirm our PCA results, the unsupervised hierarchical clustering analysis demonstrated that normal and tumor samples cluster in different branches, indicating that tissue heterogeneity may be intercepted by radiomic features (Fig. 2B). All raw datasets referring to Fig. 2 are included in the supplementary data (see raw data Appendix B).
2. Experimental Design, Materials, and Methods
2.1. Patient Population
The patient population included in our analysis consisted of surgically resected stage I-IIIa NSCLC according to the 8th TNM edition [1]. Tissue specimens and CT scans were collected at the time of diagnosis from 60 consecutive patients undergoing radical major lung resection for primary NSCLC.
2.2. Morphometric Analysis
The quantification of the fractional volume occupied by neoplastic cells, inflammatory infiltrate, fibrosis, vascular-stromal compartment and necrosis was performed at 200X magnification applying a morphometric grid defining a tissue area of 0.23 mm2 and containing 42 sampling points each covering an area of 0.0052 mm2. The number of points overlying each tissue components was counted and expressed as the percentage of the total number of points explored. The morphometric quantification of tumor necrosis was assessed by morphology, the amount of fibrosis on Masson's Trichrome while the fractional volume of neoplastic cells and TILs was determined in Cytokeratin (CK) and CD3 immunostained sections, respectively.
2.3. Immunohistochemical Analysis
Most parameters were acquired based on a methodology widely employed by our laboratory and already reported in several publications [4,5].
2.3.1. Programmed Death Ligand-1 (PD-L1)
The quantification of PD-L1 expression was assessed after a comparative evaluation between immunoperoxidase and immunofluorescence on control tissues (placenta) and on serial sections from the same cases. After careful evaluation and consultation with expert pathologists (L.G.), reliable and consistent results were obtained using clone 28-8 (Abcam #ab205921, o/n at 1:100 concentration) that was employed for quantitative analysis.
-
-
Immunoperoxidase: tissue sections (5 μm thick) were cut from formalin-fixed, paraffin-embedded blocks containing representative tumor and processed for IHC. For immunoperoxidase the reaction was revealed by DAB staining and hematoxylin counterstaining. The analysis of PD-L1 expression in tumor cells involved the counting of a minimum of 32,000 to a maximum of 800,000 tumor cells. For the quantification, an algorithm was used to obtain the PD-L1 score (H-score; 0–300), which is computed on the basis of both extent and intensity of PD-L1 staining [6].
-
-
Confocal Immunofluorescence: lung sections were double labelled by anti-PD-L1 antibody followed by FITC conjugated secondary antibody (Sigma Aldrich) and by Pan-Cytokeratin antibody (Mouse monoclonal, clone AE1/AE3, Dako M3515, 1:50 o/n 4°C) followed by TRITC conjugated secondary antibody. Images were digitally captured at 200X final magnification with “LAS Advanced Fluorescence” software (Leica) connected to a motorized epifluorescent microscope (Leica DMI6000B) provided by a digital camera (LeicaDFC350FX) and a Z-stack automation system. For each sample a tissue surface from a minimum of 1.23 mm2 to a maximum of 46.95 mm2 was analyzed.
The emission signals of FITC, TRITC and DAPI were excited at 495nm, 552nm and 359nm respectively and their fluorescence intensities recorded. The specificity of the reaction and autofluorescence were carried out by omitting the primary antibody.
For each region of interest, a graph plotting mean pixel intensity and the emission wavelength was generated. The spectrum obtained from PD-L1 positive cells exhibited a major peak at ∼519 nm. In contrast, the spectrum of autofluorescence was more uniformly spread across the range of wavelengths and did not show a clearly defined peak of emission. To compare the shape of each curve obtained from PD-L1 positive and negative structures, the values of emission spectra were normalized by dividing the intensity of each wavelength by the peak signal.
Scores were normalized to the exposure time and bit depth at which the images were captured, allowing scores collected at different exposure times to be comparable. The fractional area occupied by the fluorescent signal and its intensity, expressed as Integrated Optical Density (IOD) per unit area, was then evaluated using a software for image analysis (Image Pro Plus 4.0).
2.3.2. Tumor Infiltrating Lymphocytes (TILs)
TILs were analyzed by the immunohistochemical detection of CD3, CD8, PD-1, CD25, CD57 and Granzyme B. Immunoperoxidase was performed using an automated staining system (OptiView DAB IHC Detection Kit - Ventana Medical Systems) with antibodies against CD3 (rabbit monoclonal, clone 2GV6, ready-to-use o/n a 4°C, Ventana), CD8 (rabbit monoclonal, clone SP16, 1:50, o/n a 4°C, Neomarkers), PD-1 (mouse monoclonal, clone NAT105, 1:100, 30’, 37°C, Roche), CD25 (mouse monoclonal, clone 4C9,ready to use, 30’, 37°, Roche), FOXP3 (rabbit monoclonal, clone 5H10L18, 1:20 o/n, ThermoFisher Scientific), Granzyme B (rabbit polyclonal, ready to use, 30’, 37°, Roche) and CD57 (mouse monoclonal, clone NK1, ready to use, 30’, 37°, Roche).
T lymphocyte subpopulations and PD-L1 were also studied by confocal immunofluorescence in order to simultaneously evaluate multiple epitopes (Zeiss LSM 700 system with Axio Observer). To this purpose specific secondary antibodies (anti-mouse or anti-rabbit –FITC/-TRITC/-Cy5, 1:20, 60’ 37°C, Sigma-Aldrich) were employed. Nuclei were counterstained with DAPI.
The density (n/mm2) of CD3, CD8, PD-1, CD57, FOXP3 and CD25 labelled cells was computed analyzing a tissue area of a minimum of 2.97 mm2 to a maximum of 18.77 mm2 for SCC samples and a minimum of 2.51 mm2 to a maximum of 17.17 mm2 for ADC samples. In addition, based on their relative incidence or double immunofluorescence confocal analysis, the ratio of CD8pos cells among the whole population of T (CD3pos) lymphocytes (CD8-to-CD3) and the fraction of PD-1 expressing cells over the cytotoxic (CD8pos) lymphocytes (PD-1-to-CD8) were computed.
2.4. TIME
2.4.1. Immune Categories
Cut off values, established according to regression tree (CART) analysis, were applied to PD-L1 levels and TILs density to define PD-L1 high/low and TILs rich/poor cases. On this basis Type I-IV classification according to Teng M et al [2] was defined as follow:
-
-
Type I adaptive immune resistance (PD-L1 positive and high TILs with effector phenotype)
-
-
Type II immune ignorance (PD-L1 negative and low TILs with immunosuppressive phenotype)
-
-
Type III intrinsic induction (PD-L1 positive and low TILs with residual effector phenotype)
-
-
Type IV immune tolerance (PD-L1 negative and high TILs with immunosuppressive phenotype).
To characterize effector and suppressor phenotypes, the number of CD57+ NK and granzyme B, and CD25+ FOXP3+ Treg phenotypes, respectively, was computed.
2.4.2. Hot and Cold TIME
To assess these two immune contextures, an approach was followed based on our [4] and other findings [7, 8] indicating the relevance of the presence of PD-1 receptor at PD-L1 mediated checkpoint and the extent of T effector lymphocytes in the immune response to cancer. Specifically, the incidence of CD8+ lymphocytes over the whole CD3+ T cell compartment and the magnitude of the available pool of PD-1 negative cytotoxic CD8+ cells were combined with PD-L1 levels. Thus, hot TIME included tumor samples displaying PD-L1high, CD8-to-CD3high and PD-1-to-CD8low while cold were defined as PD-L1low, CD8-to-CD3low and PD-1-to-CD8high.
2.5. CT Examination
2.5.1. Qualitative analysis
CT datasets reconstructed with sharp and soft tissue convolution kernel (B70 and B30) with lung (window width, 1600 HU; window level, −600 HU) and mediastinal (window width, 400 HU; window level, 60 HU) window settings were made available for qualitative analysis performed by a trained radiologist (PP, 5 years of experience in chest imaging). The reader was instructed to carefully review all CT images and was allowed to modify both window width and window level to identify qualitatively assessable morphological findings (hereinafter termed semantic features, SF).
The five categories of Semantic Features evaluated in the present work according to reports from the Radiology Society of North America [3] were:
-
-
Shape: the reader was instructed to differentiate between spherical and non-spherical lesions. Lesions were deemed spherical when displaying a round morphology on axial, coronal and sagittal CT reformatted images;
-
-
Margins: margins were stratified into four categories: well-defined, poorly-defined, lobulated and spiculated);
-
-
Texture: lesions were divided into solid and sub-solid lesions (i.e. lesions displaying a ground-glass component). A sub-solid lesion consists of both non-solid and solid soft-tissue attenuation components;
-
-
Structure: homogeneous lesions were differentiated from heterogeneous. Heterogeneous lesions were those displaying hypoattenuating areas because of cavitation (air within the lesion) or necrosis (hypodense areas within the lesion);
-
-
Effect on parenchyma: an effect on surrounding structures was identified for lesions displaying indentations, scissural and/or pleural retraction.
2.5.2. Quantitative analysis
Pre-surgical CT scans were retrospectively retrieved and loaded into a dedicated open-source software (3dSlicer, version 4.9.0, www.slicer.org, #ref.9), which allowed the extraction of radiomic features (RF). The vast majority of CT images were obtained without intravenous administration of contrast media, however, in a number of cases (15 CT datasets, 21.7%) only enhanced CT images were available and were included in the analysis. For the purpose of quantitative analysis, the same reader who performed the qualitative evaluation (PP) was instructed to semi-automatically delineate regions of interest (ROIs). Non-tumoral structures (e.g. healthy parenchyma, vessels, airways) were excluded. Volume of interest (VOI) outlining pulmonary lesions acted as source of information for quantitative analyses, allowing the extraction of 841 CT-RFs belonging to 8 main categories. Highly-correlated features were excluded to prevent overfitting.
As a result of the retrospective fashion of our analysis, technical parameters could vary between scanning protocols. Therefore, image compensation approach was applied to realign feature distributions. Radiomic models obtained from either original or image-compensated features were compared to evaluate the optimal reproducibility of radiomic signatures under different scan protocols. Additionally, to overcome, at least in part, the potential inter-software and inter-reader variability, we performed a comparison between different radiomic software platforms and tested the inter-reader reproducibility in extracting radiomic features.
To reinforce the specificity and reliability of our radiomic investigation, the same high-throughput approach was applied to CT images of mediastinal lymph nodes (n=10) and latissimus dorsi, trapezius and deltoid muscles (n=3) from subjects unaffected by cancer (controls). CT-RFs extracted from these 13 control tissues together with 60 NSCLC were simultaneously subjected to cluster analysis.
2.6. Statistical analysis
The Fisher's exact test was used to examine the differences in categorical variables, and the Mann-Whitney U test to detect differences in continuous variables between groups of patients, given that the distribution of data was not normal (Kolmogorov-Smirnov test).
P value of 0.05 was set as a threshold of statistical significance. IBM SPSS Statistics v 25.0 (IBM) and Stata 13 with Cart module (Statacorp) were used to perform all computational analyses. Heatmaps and matrix analyses were performed by freely available modules in “Morpheus” software (Broad, Institute, Cambridge, MA, USA).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships which have, or could be perceived to have, influenced the work reported in this article.
Acknowledgments
Acknowledgments
We thank Emilia Corradini, Gabriella Becchi and Nicoletta Campanini for their technical assistance.
Ethic Statement
The study was performed following the approval of the Internal Review Board and of the Ethical Committee. Patients were enrolled after informed consent and in accordance with Helsinki principles.
Contributor Information
Giulia Mazzaschi, Email: giulia.mazzaschi@studenti.unipr.it.
Federico Quaini, Email: federico.quaini@unipr.it.
References
- 1.G. Mazzaschi, G. Milanese, P. Pagano et al. manuscript co-submitted to Lung Cancer, ID-19-01653
- 2.Teng M.W.L., Ngiow S.F., Ribas A. Classifying Cancers Based on T-cell Infiltration and PD-L1. Cancer Res. 2015;75:2139–2145. doi: 10.1158/0008-5472.CAN-15-0255. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hansell D.M., Bankier A.A., MacMahon H. Fleischner Society: Glossary of Terms for Thoracic Imaging. Radiology. 2008;246:697–722. doi: 10.1148/radiol.2462070712. https://doi.org/ [DOI] [PubMed] [Google Scholar]
- 4.Mazzaschi G., Madeddu D., Falco A. Low PD-1 expression in cytotoxic CD8 þ tumor-Infiltrating lymphocytes confers an immune-privileged tissue microenvironment in NSCLC with a prognostic and predictive value. Clin. Cancer Res. 2018;24:407–419. doi: 10.1158/1078-0432.CCR-17-2156. https://doi.org/ [DOI] [PubMed] [Google Scholar]
- 5.Mazzaschi G, Facchinetti F, Missale G. The circulating pool of functionally competent NK and CD8+ cells predicts the outcome of anti-PD1 treatment in advanced NSCLC. Lung Cancer. 2019 Jan;127:153–163. doi: 10.1016/j.lungcan.2018.11.038. [DOI] [PubMed] [Google Scholar]
- 6.Igarashi T., Teramoto K., Ishida M. Scoring of PD-L1 expression intensity on pulmonary adenocarcinomas and the correlations with clinicopathological factors. ESMO Open. 2016;1(4) doi: 10.1136/esmoopen-2016-000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Binnewies M., Roberts E.W., Kersten K. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018;24:541–550. doi: 10.1038/s41591-018-0014-x. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahmadzadeh M., Johnson LA., Heemskerk B. Tumor antigen- specific CD8 T cells infiltrating the tumor express high levels of PD-1 and are functionally impaired. Blood. 2009;114:1537–1544. doi: 10.1182/blood-2008-12-195792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fedorov A., Beichel R., Kalpathy-Cramer J. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn. Reson. Imaging. 2012;30:1323–1341. doi: 10.1016/j.mri.2012.05.001. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]