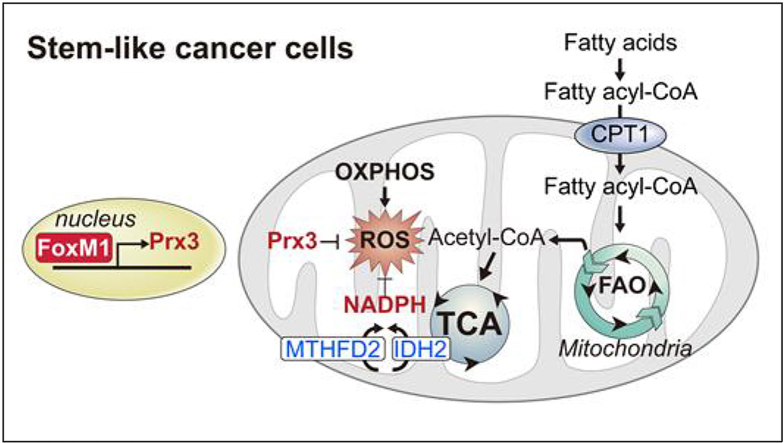
Abstract
Increased oxidative phosphorylation (OXPHOS) and reactive oxygen species (ROS) levels are inherently linked. ROS are essential signaling molecules, with detrimental effects when produced in excess during chemotherapy, leading to cell death. Cancer stem-like cells (CSCs) are a subpopulation of tumor cells resistant to chemotherapy, highly invasive and metastagenic, driving malignant cancer behavior. In this study, we demonstrated that CSCs exhibit increased OXPHOS but paradoxically low ROS levels. Considering the detrimental effects of large amounts of ROS, CSCs have developed potential mechanisms for quenching excess ROS to maintain redox homeostasis. We aimed to investigate the distinct metabolic features and mechanisms of ROS regulation in gastric CSCs and explore potential therapeutic strategies targeting CSCs. Human gastric cancer cell lines, AGS and MKN1, were subjected to liquid chromatography/mass spectrometry-based metabolomic and microarray analyses. Mitochondrial properties such as mitochondrial mass, membrane potential, and ROS were assessed by flow cytometric analysis. CSCs with increased OXPHOS levels maintained low ROS levels by coupling FoxM1-dependent Prx3 expression and fatty acid oxidation-mediated NADPH regeneration. Thus, interventions targeting ROS homeostasis in CSCs may be a useful strategy for targeting this drug-resistant tumor cell subpopulation.
Keywords: Oxidative phosphorylation, Reactive oxygen species, Cancer stem-like cell, FoxM1, Fatty acid oxidation, Prx3, NADPH
Abbreviations: OXPHOS, oxidative phosphorylation; ROS, reactive oxygen species; CSCs, cancer stem-like cells; TCA, citric acid cycle
Graphical abstract
Highlights
-
•
Metabolic features and ROS regulation mechanisms were investigated in gastric CSC.
-
•
CSCs with high OXPHOS levels maintained paradoxically low ROS levels.
-
•
FoxM1-dependent Prx3 expression was associated with low ROS levels and 5-FU resistance.
-
•
FAO fueled NADPH regeneration in mitochondria was coupled with FoxM1-Prx3 in CSC survival.
-
•
Mitochondrial ROS homeostasis-targeted approach may effectively overcome drug resistance in clinically intractable cancer.
1. Introduction
Central bioenergetic metabolic pathways include glycolysis and the citric acid (TCA) cycle. Cancer cells are traditionally known to have an altered canonical central metabolic pathway and largely depend on glycolysis uncoupling with mitochondrial energy transduction, which is known as the Warburg effect. Intermediary metabolites from glycolysis can be shunted to the pentose phosphate pathway or other branching metabolic submodules, resulting in sufficient biomass production to satisfy the high cell proliferation requirement of cancer cells. However, more quiescent cells, such as tumor cells in an invasive or metastatic state, or those in a dormant state that can evade anticancer therapy do not show this characteristic glycolytic phenotype [1,2]. Recent studies have suggested that highly malignant tumor cells exhibit a metabolic shift toward mitochondrial oxidative phosphorylation (OXPHOS) [[3], [4], [32]]. These subpopulations of cells with enhanced OXPHOS are drug-resistant, hence highlighting their clinical significance.
Concurrent with these previous findings, we recently reported that highly metastagenic and drug-resistant subpopulations of tumor cells exhibit a distinct form of energy metabolism that does not rely on glycolysis but rather depends on mitochondria-centric metabolism [[5], [6], [7]]. Since increased OXPHOS and reactive oxygen species (ROS) levels are inherently linked, a highly malignant subpopulation of cancer cells are expected to have elevated ROS levels [8]. However, ROS production beyond the physiological range can be detrimental, even to cancer cells.
The efficacy of many anticancer therapeutics is based on their ability to increase ROS levels in tumor cells. However, it is currently unclear how most malignant cancer cells can depend on the OXPHOS-prone metabolic phenotype while maintaining disproportionately low levels of ROS. We hypothesized that maintenance of low ROS levels is one of the mechanisms of drug resistance in cancer stem-like cells (CSCs). To test this hypothesis, we investigated the distinct metabolic features and mechanisms of ROS regulation in CSCs, selected from gastric cancer cell lines, to explore potential therapeutic strategies targeting CSCs.
2. Materials and methods
2.1. Cell culture conditions
The human gastric cancer cell lines, AGS and MKN1, were obtained from the Korean Cell line Bank (Seoul, Korea) and grown in RPMI-1640 medium with 10% fetal bovine serum in a humidified incubator containing 5% CO2 at 37 °C. Stem-like cancer cells (S-cells) were generated by in-vitro chronic metabolic stress culture, as described previously [5].
2.2. Intracellular metabolite extraction
Parental cells (P-cells) and S-cells were plated in the presence of 5.5 mM [13C6] glucose and 100 μM [13C16] palmitate (Cambridge Isotope Labs, Tewksbury, MA, USA) for 48 h. The cells were washed twice with ice-cold PBS, and intracellular metabolites were extracted with a cold solution of methanol, acetonitrile, and water (5:3:2). The cell extracts were centrifuged at 16,000×g for 10 min at 4 °C, and the supernatants were assessed via liquid chromatography-mass spectrometry (LC-MS) analysis.
2.3. LC-MS-based metabolomics
LC-MS analysis was performed as described previously [10].
2.4. Microarray analysis
The NuRNA™ Human Central Metabolism PCR Array (Arraystar, Inc., Rockville, MD, USA) was used to identify mRNA transcripts with differential expression between P-cells and S-cells. The array covers 373 transcripts encoding enzymes or proteins involved in cell metabolism. Samples were used for array analysis in accordance with the manufacturer's protocol and each analysis was performed in triplicate.
2.5. Fluorescence-activated cell sorting (FACS) and flow cytometry
Human gastric cancer cells (AGS and MKN1) were dissociated into single cells, washed with PBS, and stained with fluorescent antibodies for CD133-PE (BD Biosciences, Franklin Lakes, New Jersey) and CD44-FITC (BD Biosciences, Franklin Lakes, New Jersey). To determine the effect of ROS levels on M-and E-BCSCs in breast cancer cell lines, MCF7 cells were incubated with antibodies against CD24-PE (BD Biosciences, Franklin Lakes, New Jersey) and CD44-FITC. Content of ALDH+E-BCSCs was determined by Aldefluor assay (StemCell Technologies) per manufacturer's instructions. The cells were sorted using a BD FACSAria flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA).
2.6. Western blot analysis
Cells were lysed in lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, and 1% Triton-X100) containing 1 × protease inhibitor cocktail (Sigma, St. Louis, MO, USA) and 1 × phenylmethylsulfonyl fluoride (Sigma). Protein concentration was quantified using a BCA protein concentration assay kit (Thermo Fisher Scientific, Waltham, MA, USA). Equal amounts of protein were electrophoresed on sodium dodecyl sulfate-polyacrylamide gels and transferred onto polyvinylidene difluoride membranes. The membranes were incubated with primary antibodies in 2% skim milk containing 0.05% Tween-20 overnight at 4 °C. The membranes were incubated with horseradish peroxidase-conjugated secondary antibody for 1 h at room temperature and visualized by electrochemiluminescence (ThermoFisher Scientific).
2.7. Reverse transcription-quantitative PCR
Total RNA was isolated with TRIzol (Invitrogen, Carlsbad, CA, USA), and 1 μg of total RNA was used for cDNA synthesis using M-MLV reverse transcriptase (Mbiotech, Hanam-si, Korea). Quantitative PCR was carried out using SYBR Green PCR Master Mix (PhileKorea, Seoul, Korea). Experimental cycle threshold values were normalized to those of GAPDH, and relative mRNA expression was calculated versus GAPDH expression.
2.8. Lactate production
A lactate assay kit (Biovision Research Products, Milpitas, CA, USA) was used to measure extracellular lactate following the manufacturer's instructions. Briefly, equal numbers of cells were seeded into 6-well plates and cultured in serum-free media for 24 h. The culture medium was then mixed with the reaction solution. Lactate levels were measured at 570 nm using a microplate reader. The cells were trypsinized, and cell number was counted using trypan blue. Absorbance values were normalized to the cell number.
2.9. Membrane potential assay
Mitochondrial membrane potential was measured using JC-1 dye (Invitrogen) according to the manufacturer's instructions. Briefly, equal numbers of cells were seeded into 6-well plates; after 72 h, 2 μM JC-1 was added and the cells were incubated at 37 °C for 15 min. Carbonyl cyanide chlorophenylhydrazone (CCCP; Sigma) was used as a control to confirm that the JC-1 response was sensitive to changes in membrane potential. The cells were then trypsinized and washed twice with PBS, after which fluorescence was analyzed using a BD FACS LSRII flow cytometer.
2.10. Intracellular ROS
To measure intracellular ROS levels, 10 μM DCF-DA (Sigma) was used as a fluorescent dye. The cells were stained with DCF-DA for 30 min at 37 °C, trypsinized, washed thrice with PBS, and immediately analyzed with a BD FACS LSRII flow cytometer.
Mitochondrial ROS levels were assessed using a MitoSOX Red mitochondrial superoxide indicator (Thermo Fisher Scientific) in accordance with the manufacturer's protocol. The MitoSOX Red reagent is a live-cell permeant that is rapidly and selectively targeted to the mitochondria. The parental or CSCs were incubated with MitoSOX reagent for 10 min at 37 °C, while protecting them from light. After the cells were washed thrice with PBS, they were collected, and MitoSOX Red signal was detected by flow cytometry.
2.11. Imaging of ROS production
ROS production was visualized in cells loaded with 10 μM DCF-DA (Sigma) for 20 min at room temperature. ROS-induced green fluorescence of DCF was imaged using 488-nm laser excitation. The laser power was set to 1–3%.
2.12. Oxygen consumption rate
The oxygen consumption rate was measured using an optical oxygen sensor in a Seahorse Bioscience XF96 Extracellular Flux Analyzer (North Billerica, MA, USA). Briefly, the cells were seeded in quadruplicate at equal densities in XF96 culture plates. Cell media were changed, after 24 h of cell seeding, to unbuffered Dulbecco's modified Eagle's medium, in accordance with the manufacturer's protocol. Oxygen consumption was measured with sequential injection of oligomycin, FCCP, and rotenone/antimycin A.
2.13. Glucose uptake and mitochondrial mass
Cells were incubated for 30 min at 37 °C with 100 μM 2-N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)-2-deoxyglucose (2-NBDG) and 100 nM Mitotracker Deep Red FM (both from Life Technologies, Carlsbad, CA, USA) prior to FACS analysis.
2.14. NADP+/NADPH ratio determination
Colorimetric analysis of the cell lysates was performed using the NADP/NADPH Quantitation Kit (Biovision Research Products) in accordance with the manufacturer's instructions.
2.15. Quantification and statistical analysis
The overall survival data obtained from patients with gastric, breast, lung, and ovarian cancer were analyzed by the Kaplan-Meier Plotter (http://kmplot.com/analysis). Statistics was determined by paired Student's t-test and ANOVA using GraphPad Prism 7 software (GraphPad, Inc., La Jolla, CA, USA). Results are presented as the mean ± standard deviation for representative experiments with 3–4 independent biological repeats. A P-value < 0.05 was considered statistically significant.
3. Results
3.1. Gastric CSCs exhibited metabolic reprogramming and increased mitochondrial OXPHOS rather than glycolysis
We had previously reported a cell subpopulation, adapted to chronic metabolic stress, exhibiting cancer stem-like characteristics [[5], [6], [7]]. Accordingly, we used S-cells derived from the human gastric cancer cell lines MKN1 and AGS as a model system for gastric CSCs.
We first compared the expression of central metabolism genes between P- and S-cells, using a PCR array. Compared to P-cells, S-cells exhibited general repression of metabolic pathway genes. However, genes of the fatty acid oxidation (FAO) and one-carbon pool related purine nucleobase synthesis pathways were coordinately upregulated in S-cells (SMKN1 and SAGS, respectively) (Fig. S1A). These data confirmed that S-cells display reprogrammed metabolic activities. We further validated the protein expression of corresponding mRNAs between P- and S-cells under various metabolic stress conditions. Of note, the specific genes and modules that were differentially expressed varied between the two subtypes of CSCs (SMKN1 and SAGS; Fig. S1B). Among others, antioxidant enzymes such as SOD2, GPX4 and catalase were most prominently differently expressed between the two CSCs, while Prx3 expression was increased in both CSCs. Similarly, G6PDH involved in the oxidative PPP, and PHGDH, SHMT2 and MTHFD2 involved in one-carbon metabolism were increased only in SAGS, while IDH2, a mitochondrial NADPH-generating enzyme, was increased in SMKN1 alone, indicating heterogeneous metabolic properties in CSCs (Fig. S1B).
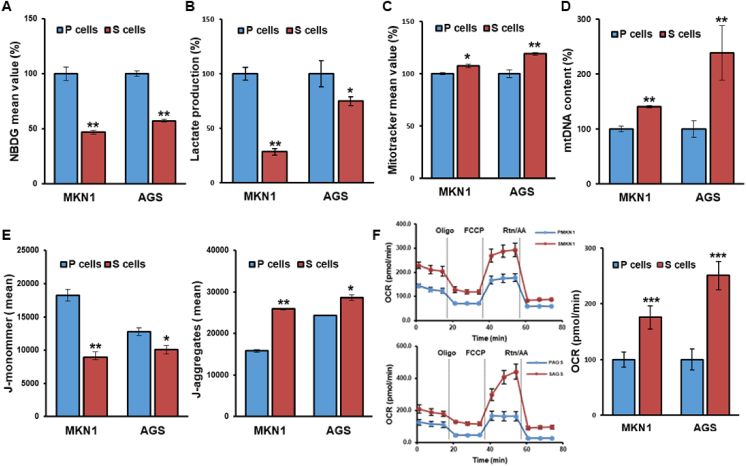
Accumulating evidence suggests that mitochondrial biogenesis is crucial for CSC maintenance and chemotherapy resistance in several tumor types [[2], [3], [4],9,10]. To assess mitochondria-prone metabolic reprogramming in CSCs, we examined the metabolic features related to mitochondrial function. Indeed, S-cells displayed reduced glucose uptake and lactate production in comparison to P-cells (Fig. 1A-B). Further, S-cells exhibited an increased mitochondrial mass and DNA content (Fig. 1C-D), indicating that mitochondrial biogenesis is activated. Consistently, S-cells demonstrated increased mitochondrial membrane potential, evident from an increased amount of aggregated JC-1 dye and mitochondrial oxygen consumption (Fig. 1E-F). Together, these results indicated that CSCs display distinct metabolic features that are more compatible with mitochondrial oxidative energy metabolism compared with P-cells.
Fig. 1.
Gastric cancer stem-like cells display metabolic features compatible with oxidative metabolism. (A–F) Glucose uptake by 2-NBDG incorporation measured by flow cytometry (A), lactate production (B), mitochondrial mass as determined by flow cytometry using MitoTracker Green-FM (C), mitochondrial DNA (mtDNA) content (D), membrane potential (E), and oxygen consumption rate (F) in parental and selected stem-like cells from the MKN1 and AGS gastric cancer cell lines. Error Bars represent standard deviation. Statistical significance was determined by student's T-test. All experiments were performed at least in triplicate not otherwise indicated. * P-value <0.05, ** P-value <0.01, *** P-value < 0.001. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2. CSCs have reduced ROS levels
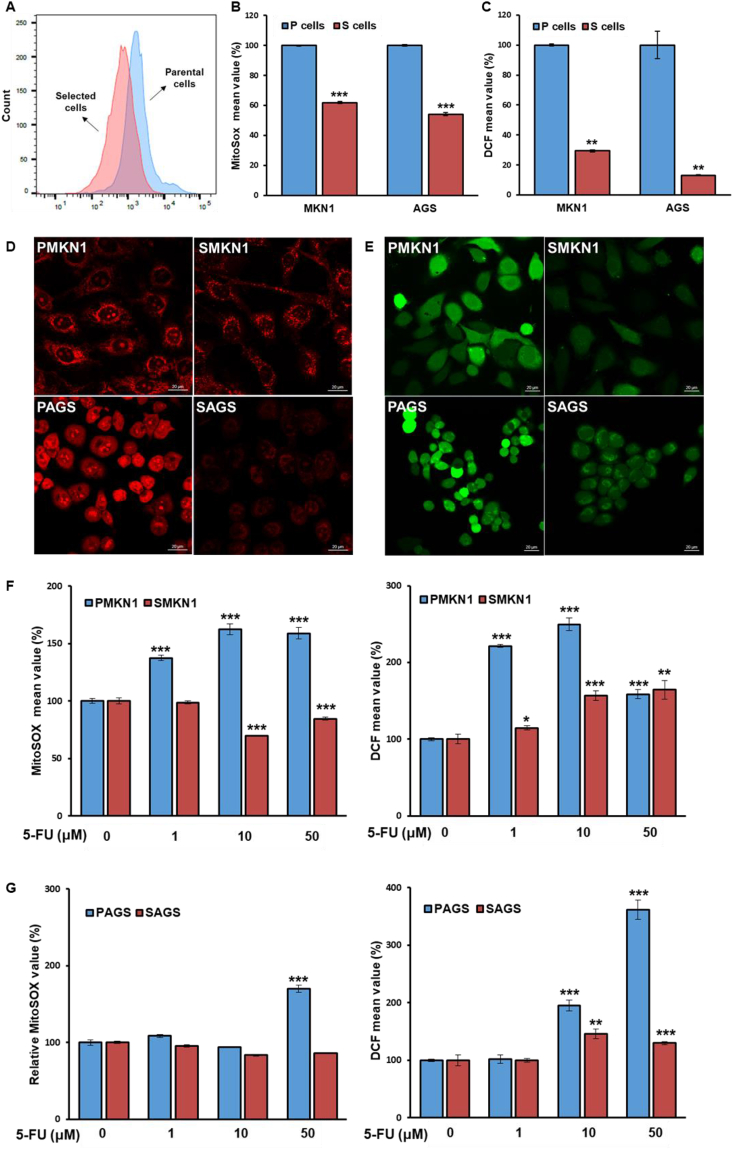
Increased OXPHOS and ROS levels are inherently linked. To investigate ROS levels in CSCs, we measured the mitochondrial superoxide levels using MitoSOX Red. Lower levels of superoxide (based on MitoSOX Red) staining were found in S-cells of both cell lines compared with those of their corresponding P-cells (Fig. 2A-B). Similarly, dichloro-dihydro-fluorescein diacetate (DCFH-DA) fluorescence detection (as a marker of intracellular hydrogen peroxide) revealed decreased ROS levels in S-cells (Fig. 2C). Furthermore, immunofluorescence analysis revealed that S-cells generally had reduced ROS levels both in the mitochondria and cytosol compared with P-cells (Fig. 2D and E).
Fig. 2.
Cancer stem-like cells (CSCs) maintain low reactive oxygen species (ROS) levels compared with non-CSCs. (A-C) Levels of mitochondrial superoxide (A,B) and intracellular hydrogen peroxide (C) were measured by fluorescence-activated cell sorting after staining with MitoSOX Red and DCFH-DA dye. (D–E) Representative confocal micrographs of immunofluorescence staining in parental cells and CSCs using MitoSOX Red (D) and DCFH-DA dye (E). (F–G) CSCs and non-CSCs were treated with the indicated doses of 5-FU for 72 h. Levels of mitochondrial ROS were measured by MitoSOX Red staining (F). Levels of intracellular ROS were measured by DCFH-DA fluorescence (G). Error Bars represent standard deviation. Statistical significance was determined by student's T-test. All experiments were performed at least in triplicate not otherwise indicated. * P-value <0.05, ** P-value <0.01, *** P-value < 0.001. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
To exclude the possibility of an idiosyncratic reduction of ROS in S-cells, we confirmed the ROS levels in a CSC subpopulation from P-cells, sorted by CD44 and CD133. CD44+CD133+ high-expressing cells (CSCs) displayed decreased ROS levels in comparison with CD44−CD133- cells (non-CSCs; Fig. S2A). To further confirm these findings in gastric CSCs, breast CSCs were subject to ROS measurement [11]. Indeed, SMCF7 displayed lower ROS levels than the parental cells (PMCF7) (Fig. S2B). Likewise, breast CSCs were sorted by flow cytometry to determine ROS levels. The two BCSC subtypes, CD24−CD44+ M-BCSCs and ALDH+ E-BCSCs, showed a low ROS phenotype compared with the non-CSC population (Figs. S2C–D). Together, these results indicated that maintenance of low ROS levels might be a universal metabolic feature of CSCs.
A CSC phenotype is a well-known feature among numerous chemotherapy-resistant tumors [12], and most chemotherapeutics induce ROS generation in cancer cells [13]. Concurrent with this general phenomenon, S-cells displayed reduced sensitivity compared with P-cells to the anticancer drug 5-fluorouracil (5-FU), which is widely used for gastric cancer treatment (Figs. S3A–B). Moreover, ROS levels increased after 5-FU treatment in P-cells in a dose-dependent manner, whereas relatively low levels of ROS were maintained in CSCs (Fig. 2F-G). These findings suggested a link between ROS levels and 5-FU sensitivity in CSCs, indicating that CSCs may have a cell-intrinsic drug resistance mechanism related to redox homeostasis.
3.3. FoxM1 transcriptionally activated Prx3 in gastric CSCs
Cancer cells have developed antioxidant mechanisms to protect themselves from oxidative stress by expressing various detoxifying enzymes such as superoxide dismutase, catalase, glutathione peroxidase, and peroxiredoxin [14].
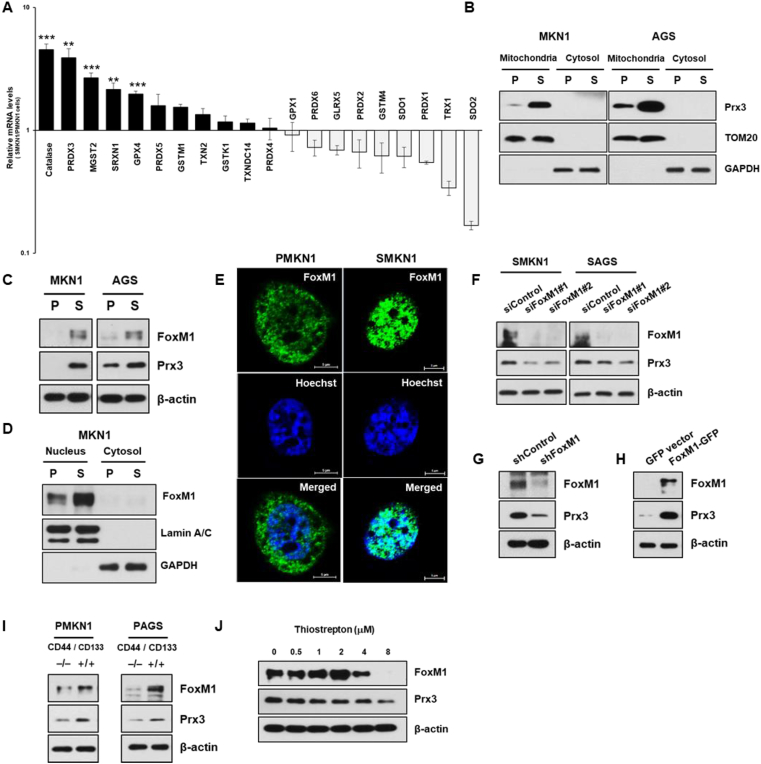
To determine whether ROS detoxification genes increase in S-cells and contribute to selective advantages, we detected the mRNA expression of key ROS detoxification genes through reverse transcription-quantitative PCR. Catalase and peroxiredoxin 3 (Prx3) mRNAs were highly upregulated in SMKN1 cells (Fig. 3A). Consistently, mitochondrial Prx3 protein was upregulated in both S-cells compared with that in P-cells (Fig. 3B).
Fig. 3.
FoxM1 increases Prx3 expression in CSCs. (A) mRNA expression levels of ROS detoxification genes in parental cells (PMKN1) and stem-like cancer cells (SMKN1). (B) Cytoplasmic and mitochondrial of Prx3 levels in parental cells and CSCs. (C) Immunoblot analysis of FoxM1 and Prx3 levels in parental cells and CSCs. (D–E) Cytoplasmic and nuclear levels of FoxM1 in parental cells and CSCs. (F) Prx3 expression, in cells transfected with siRNA against FoxM1 and control siRNA. (G–H) FoxM1-depleted cells showed downregulation of Prx3 expression (G), and FoxM1-overexpression cells showed upregulation of Prx3 expression (H) based on immunoblot analysis with the indicated antibodies. (I) FoxM1 and Prx3 expression confirmed by immunoblot analysis of CD44−CD133- and CD44+CD133+ non-CSC subpopulations. (J) SMKN1 cells were treated with thiostrepton, a potent FoxM1 inhibitor, and expression of Prx3 protein levels were determined by western blot. Error Bars represent standard deviation. Statistical significance was determined by student's T-test. All experiments were performed at least in triplicate not otherwise indicated. * P-value <0.05, ** P-value <0.01, *** P-value < 0.001.
Prx3 is reported to be transcriptionally upregulated by the transcription factor FoxM1 [14,15]. We also observed a positive correlation between basal levels of endogenous FoxM1 and Prx3 in our model systems (Fig. 3C). To investigate the regulation of Prx3 expression in S-cells, we analyzed the expression and nuclear localization of FoxM1. Indeed, the expression levels and nuclear localization of FoxM1 were increased in S-cells compared with those in P-cells (Fig. 3D and E). In contrast, FoxM1 siRNA-transfected cells displayed Prx3 downregulation in comparison with control siRNA-transfected cells (Fig. 3F). We also showed that stable knock down of FoxM1 led to a decrease in Prx3 expression and overexpression of exogenous FoxM1 led to an increase in Prx3 expression in S-cells and in P-cells, respectively (Fig. 3G-H). Furthermore, FoxM1 and Prx3 were upregulated in CD44+CD133+ isolated gastric cancer cells compared with their levels in CD44−CD133- cells (Fig. 3I). We also utilized thiostrepton, a selective FoxM1 inhibitor, and it reduced FoxM1 and Prx3 expression (Fig. 3J). Together, these results confirmed the mechanistic association between FoxM1 and Prx3 expression in gastric CSCs.
3.4. The transcription factor FoxM1 is required for redox homeostasis and survival of gastric CSCs against chemotherapeutics
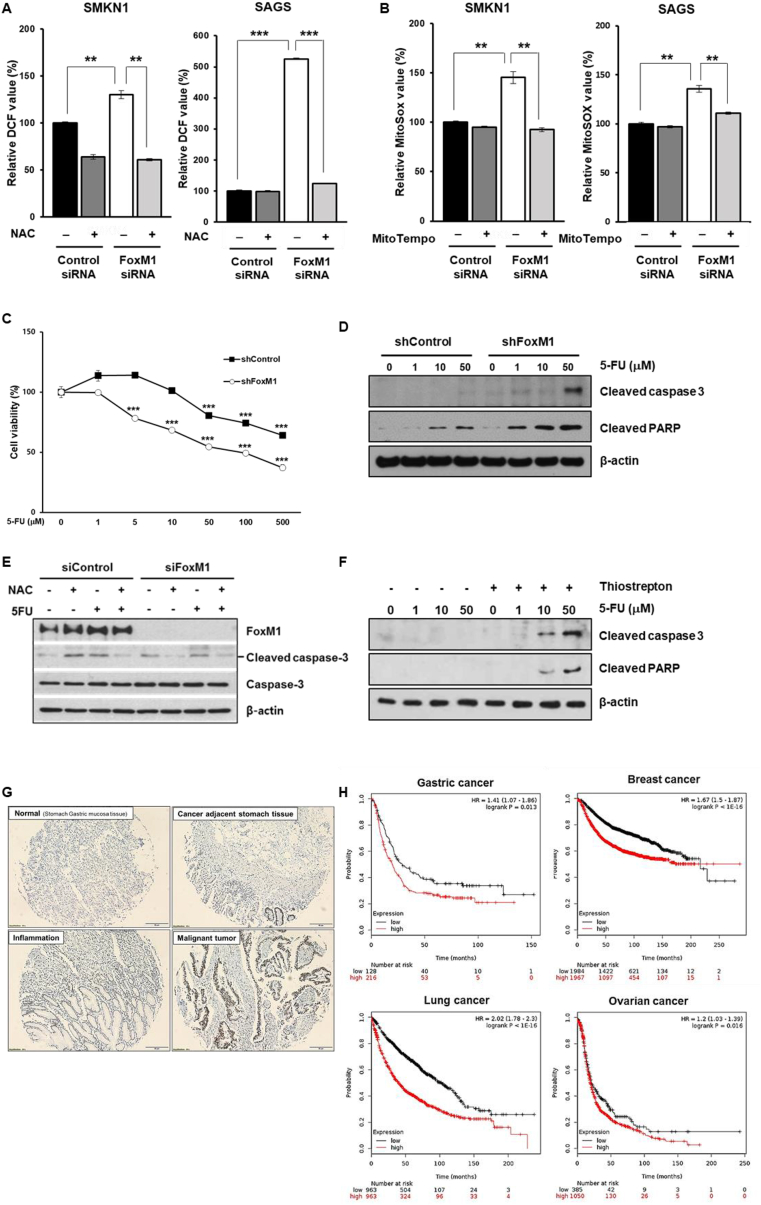
FoxM1 is suggested to regulate intracellular ROS levels, thus protecting cancer cells from oxidative stress [14,16]. Based on this link between FoxM1 and the mitochondria-specific antioxidant enzyme Prx3 in S-cells, we next explored the role of FoxM1 in regulating ROS homeostasis in relation to cell fitness. As expected, intracellular ROS concentrations were significantly increased in FoxM1-depleted cells, whereas treatment of FoxM1-depleted cells with the antioxidant N-acetylcysteine (NAC) substantially restored the ROS levels (Fig. 4A). Similarly, mitochondrial ROS levels were significantly higher in FoxM1-depleted cells, and MitoTEMPO, a specific scavenger of mitochondrial superoxide, efficiently suppressed the ROS accumulation in FoxM1-depleted S-cells (Fig. 4B).
Fig. 4.
FoxM1 mediates drug resistance through reducing mitochondrial ROS in CSCs. CSCs were transfected with control or FoxM1 siRNA for 72 h. Intracellular ROS levels were measured using DCF-DA at baseline and upon supplementation with NAC (A), and mitochondrial ROS levels were measured using MitoSOX at baseline and upon supplementation with MitoTempo (B). (C) Cell viability of FoxM1 knockdown and control cells treated with indicated concentrations of 5-FU (n = 3). (D) Immunoblot of cleaved caspase-3 and cleaved PARP in control and FoxM1 knockdown cells treated with indicated concentrations of 5-FU. (E) S-MKN1 cells were pretreated with NAC and then 5-FU was added to the mixture and incubated. Total and cleaved caspase-3 and FoxM1 were detected by western blot analysis using specific antibodies. (F) Immunoblot of cleaved caspase-3 and cleaved PARP in SMKN1 cells upon 5-FU and thiostrepton treatment at indicated concentrations. (G) Expression of FoxM1 in gastric cancer tissues and adjacent normal tissues from patients determined by immunohistochemistry. (H) FoxM1 levels correlated with poor patient survival in gastric cancer, breast cancer, lung cancer and ovarian cancer. Error Bars represent standard deviation. Statistical significance was determined by student's T-test. All experiments were performed at least in triplicate not otherwise indicated. * P-value <0.05, ** P-value <0.01, *** P-value < 0.001.
FoxM1 reportedly plays important roles in drug resistance [17,18]. To investigate the potential association between drug resistance and the capability of FoxM1 to mitigate oxidative stress induced by anticancer drugs, we analyzed the effect of chemotherapy in CSCs. We observed that FoxM1-knockdown cells became significantly more sensitive toward 5-FU than control cells (Fig. 4C). Consistently, western blot analysis showed increased cleaved caspase-3 and cleaved PARP in 5-FU-treated FoxM1-knockdown cells (Fig. 4D). Moreover, treatment with NAC substantially rescued 5-FU-induced cell death (Fig. 4E). Consistently, thiostrepton induced increased apoptosis in 5-FU-resistant cells in dose-dependent manner (Fig. 4F).
Together, these data showed that elevated ROS levels are associated with increased cell death after exposure to chemotherapeutics in CSCs, implying FoxM1-mediated ROS down-modulation as a mechanism underlying drug resistance.
Clinically, FoxM1 was upregulated in patients with malignant gastric cancer compared to that in the adjacent normal or benign inflamed tissues (Fig. 4G). Further, FoxM1 upregulation predicted poor survival in patients with various types of cancers, including gastric, breast, lung, and ovarian cancers (Fig. 4H).
3.5. CSCs have increased mitochondrial NADPH regeneration
ROS-quenching enzymes, including Prx3, requires cellular reducing power to function. NADPH, the primary source of cellular reducing power, serves as an electron carrier to maintain redox homeostasis and reductive biosynthesis, with compartmentalized cytosolic and mitochondrial pools to facilitate reduction reactions at corresponding sites [19].
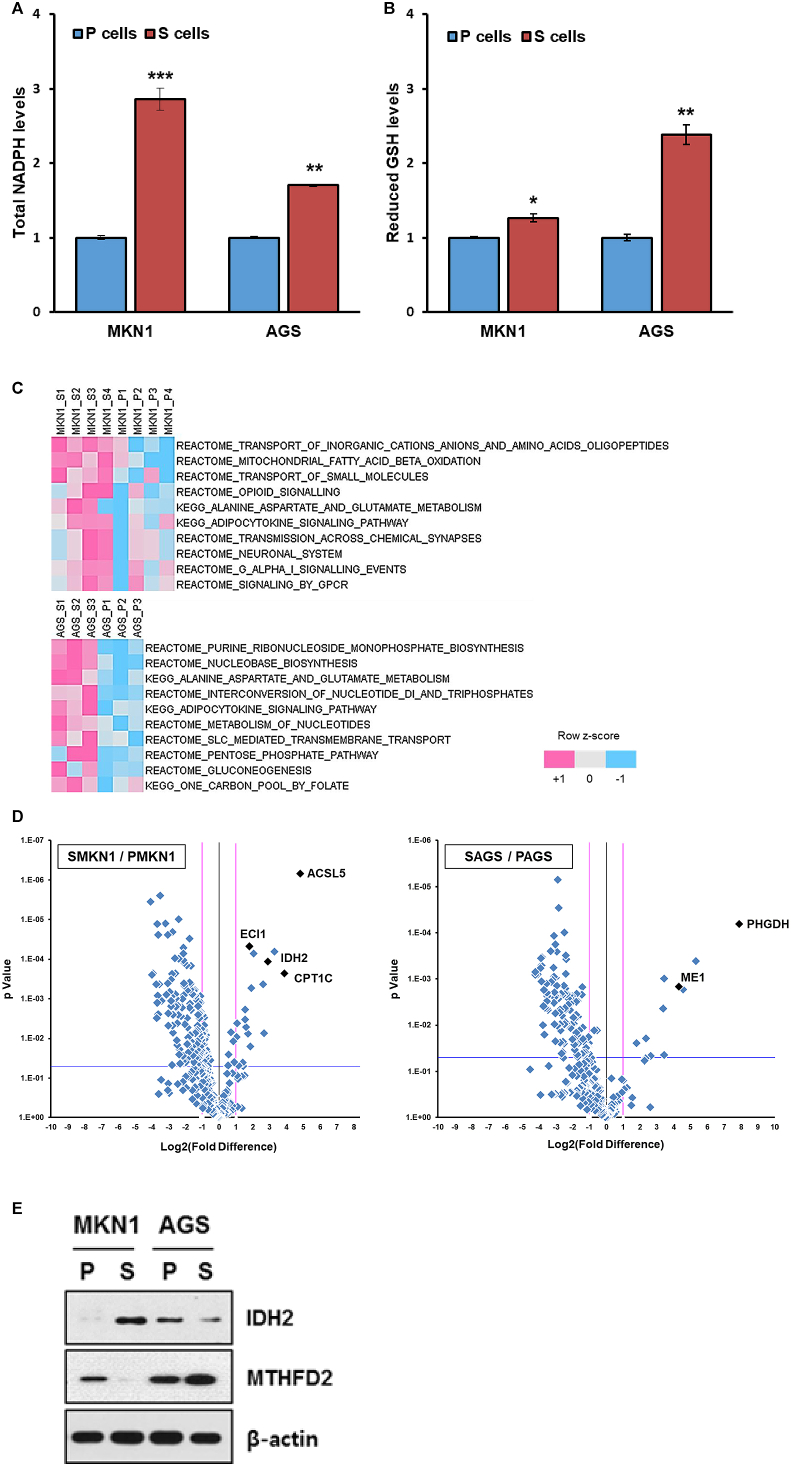
Indeed, intracellular NADPH levels were higher in CSCs, and were associated with an increase in reduced glutathione levels (Fig. 5A and B). PCR array analysis (Fig. S1A) revealed that genes encoding some NADPH-generating enzymes (e.g., IDH2, ME3, MTFHD2L, MTHFD1, ME1, and G6PD) were upregulated in both CSC subtypes in comparison with their respective P-cells. Single-sample gene enrichment analysis (ssGSEA) of PCR array result revealed that mitochondrial fatty acid beta-oxidation pathways were significantly enriched in SMKN1 cells, while one-carbon pool by folate and related purine nucleotide biosynthesis pathways as well as PPP were enriched in SAGS cells (Fig. 5C). Indeed, assessment of differential gene expression by volcano plots showed that essential genes involved in FAO (e.g., ACSL5, ECI1, and CPT1C) were coordinately upregulated in S-MKN1 cells. The most differentially expressed genes in SAGS over PAGS were PHGDH, ME1, and G6PDH, confirming the ssGSEA results (Fig. 5D). To assess the protein expression of corresponding genes, we performed immunoblotting, revealing that IDH2 and MTHFD2 were increased in SMKN1 and SAGS, respectively (Fig. 5E).
Fig. 5.
Mitochondrial NADPH regeneration increases in CSCs. (A) Total intracellular NADPH levels in non-CSCs and CSCs (n = 3). (B) Reduced glutathione levels in non-CSCs and CSCs (n = 3). (C) Single-sample gene set enrichment analysis (ssGSEA) with rank normalization method was performed on the PCR array result. It determines whether a predefined gene set is enriched at the top or bottom of the pre-ranked gene list for each sample. Molecular Signatures Database (MSigDB) gene sets curated from the KEGG and Reactome pathway database were used. The canonical pathways of which average enrichment score is greater at the S cells are sorted by p-value from student's t-test. (D) A total of 373 transcripts involved in cell metabolism are depicted in a volcano plot. The pink lines indicate a fold-change threshold of 2, and the blue lines indicate the P-value cut-off (P = 0.05). (E) Immunoblot analysis of IDH2 and MTHFD2 levels in parental cells and CSCs. Error Bars represent standard deviation. Statistical significance was determined by student's T-test. All experiments were performed at least in triplicate not otherwise indicated. * P-value <0.05, ** P-value <0.01, *** P-value < 0.001. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Therefore, both SMKN1 and SAGS cells displayed increased levels of mitochondrial enzymes compared with their parental counterparts, playing major roles in regenerating NAPDH from NADP+, mitochondrial isocitrate dehydrogenase (IDH2), and mitochondrial methylene tetrahydrofolate dehydrogenase (MTHFD2), respectively.
3.6. CSCs have enhanced fatty acid oxidation mediated NADPH regeneration responsible for drug resistance
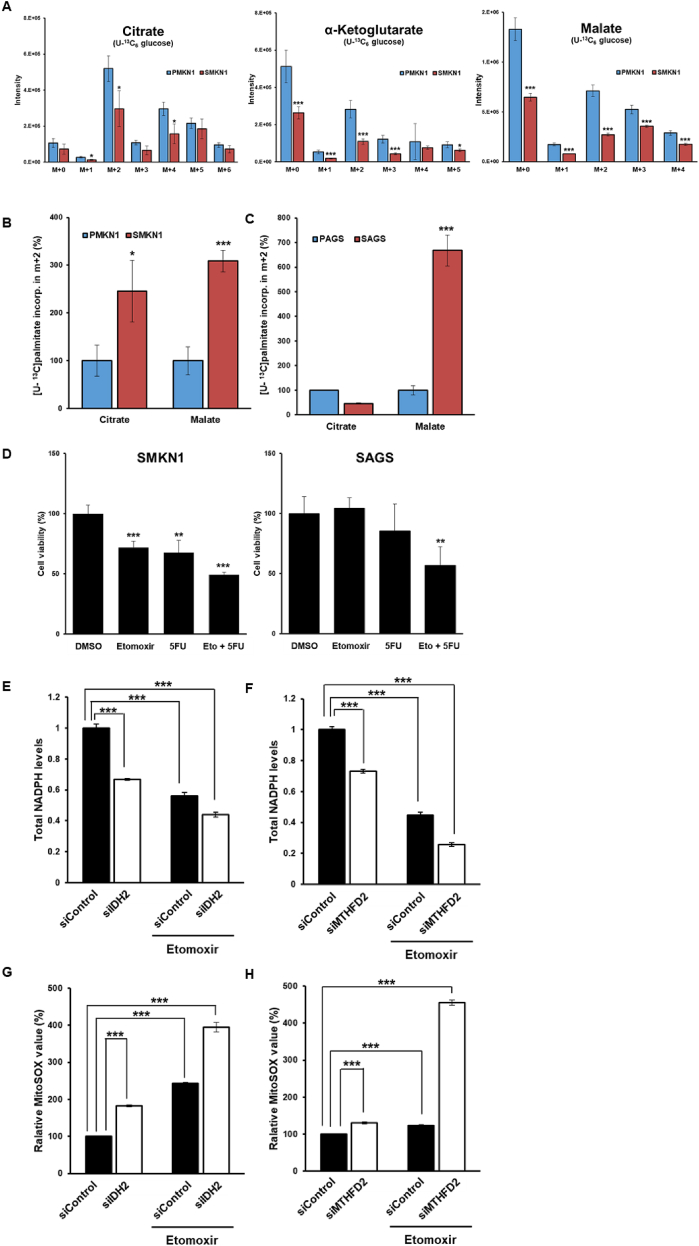
In cancer cells, FAO is an important source of mitochondrial NADPH by generating acetyl-CoA to sustain the TCA cycle [20]. To analyze how CSCs utilize nutrients, we performed uniformly-labeled [U–13C]glucose and [U–13C]palmitate flux analyses by LC-MS using cell extracts. Interestingly, the glucose-derived 13C signal was significantly reduced among TCA cycle intermediates, including citrate, α-ketoglutarate (α-KG), and malate in SMKN1 cells, compared to those in PMKN1 cells (Fig. 6A). Furthermore, S-AGS cells had reduced citrate, malate, and succinate levels, although α-KG was not detected (Fig. S4A). However, mass distribution analysis revealed an increase in m+2 malate in both SMKN1 and SAGS cells, suggesting that FA-derived carbons sustained the oxidative TCA cycle in CSCs (Fig. 6B and C).
Fig. 6.
Enhanced fatty acid oxidation mediated NADPH regeneration is a reversible cause of drug resistance in CSCs. (A–C) CSCs and non-CSCs cultured in RPMI media containing no glucose plus 10% dialyzed fetal bovine serum was respectively traced with 5.5 mM U–13C glucose and 100 μM U–13C palmitate for 16 h. Relative isotopolog distribution of the indicated metabolites measured by LC-MS. n = 3. FC, fold-change in glucose/palmitate-derived (13C ≥ 2) metabolite abundance relative to in parental cells. (D) Cell viability assay showing etomoxir sensitizes CSCs to 5-FU. (E–H) Total intracellular NADPH levels (E–F, n = 3) in control and deletion of IDH2 or MTHFD2 in CSCs and mitochondrial ROS levels (G-H, n = 3) in control and deletion of IDH2 or MTHFD2 in CSCs, in baseline and upon supplementation with etomoxir. Error Bars represent standard deviation. Statistical significance was determined by student's T-test. All experiments were performed at least in triplicate not otherwise indicated. * P-value <0.05, ** P-value <0.01, *** P-value < 0.001.
Furthermore, the mRNA expression levels of key genes associated with FAO were upregulated in CSCs (Fig. S4B), whereas the protein levels of ACC1, which encodes malonyl-CoA and directly inhibits CPT1, were decreased in CSCs (Fig. S4C).
Since NADPH, which is required to quench ROS, is generated through FAO, we hypothesized that FAO is associated with the mechanism underlying drug resistance in CSCs. To determine the contribution of FAO to 5-FU-acquired resistance, CSCs were treated with 5-FU in combination with the FAO inhibitor etomoxir. Etomoxir and 5-FU exhibited synergistic effects to significantly decrease CSC viability (Fig. 6D).
FAO is reportedly important for cells that have become detached from the extracellular matrix and those cultured in nutrient-deprived conditions [21,22]. To confirm the role of the FAO-NADPH-ROS pathway herein, we examined the effects of matrix detachment on ROS production. Extracellular matrix detachment significantly increased ROS levels in both CSCs and non-CSCs; however, the magnitudes of these increases were relatively lower in CSCs (Fig. S4D). Similarly, ROS levels were lower in CSCs under glucose-deprived conditions (Fig. S4E) and higher in those treated with etomoxir (Fig. S4F). These results also implied that FAO-mediated ROS homeostasis is associated with key features including CSC invasion and metastasis.
To investigate the role of mitochondrial NADPH-producing enzymes in CSCs, we depleted mitochondrial NAPDH-producing enzymes with siRNAs, which significantly decreased NADPH levels and was associated with increased mitochondrial ROS levels in CSCs (Fig. 6E–H). Furthermore, FAO inhibition with etomoxir, an irreversible CPT1 inhibitor, more significantly reduced NADPH levels and increased mitochondrial ROS levels in CSCs. Thus, FAO, via IDH2 or MTHFD2, substantially contributes to physiologically significant mitochondrial NADPH production.
4. Discussion
Tumor cells reprogram their metabolic pathways to adapt to the fluctuating metabolic environment. CSCs have evolved a flexible energy metabolic strategy to increase fitness in the ever-changing tumor microenvironment. In particular, to cope with the uncertainty in securing nutrients and appropriate energy substrates, CSCs harness OXPHOS as the dominant metabolic strategy, which is far more efficient than glycolysis but comes at a cost of overproduced ROS, thereby requiring large amounts of NADPH as a counteractive measure [23].
Although physiologically important, beyond a certain level, ROS can damage the cellular functions of cancer cells by subjecting them to oxidative stress. Thus, the maintenance of ROS at appropriate levels is imperative for regulating the stemness-associated properties of cancer cells [24]. This was confirmed in our CSC model, wherein both CSC lines displayed activated mitochondrial OXPHOS and maintained relatively low ROS levels to sustain their functions.
Scavenging of ROS is mediated by a set of antioxidant enzymes expressed in various subcellular compartments [25]. During adaptation to metabolic stress, our selected CSCs (S-cells) displayed metabolic remodeling of numerous antioxidant enzymes including catalase, superoxide dismutase 2, Gpxs, and Prxs. In particular, Prx3 levels were substantially increased as a common factor in the two CSC subtypes. Recent studies have reported that mitochondrial Prx3, transcriptionally activated by FoxM1, maintains mitochondrial function by eliminating ROS produced upon ATP production in the OXPHOS system in colonic CSCs [15]. Similarly, our S-cells also primarily eliminated ROS via the FoxM1-Prx3 axis through ROS-quenching mechanisms. Moreover, gastric cancer organoid models, highly expressing FoxM1 and Prx3, exhibited low levels of mitochondrial ROS (Figs. S5A–C).
In general and in glycolytic tumors, including proliferating tumor cells, most of the cellular reducing power in the form of NADPH is generated through the pentose phosphate pathway or hexose monophosphate shunt. However, tumor cells, including the S-cells established herein, do not rely on glycolysis but rather maintain sufficiently high levels of NADPH to effectively neutralize ROS. To explore the potential sources of this NAPDH, we performed metabolomic flux analysis and found that S-cells utilize fatty acid-derived metabolites to generate NADPH. Furthermore, in the absence of glucose, the S-cells were still viable and maintained ROS in the homeostatic range, suggesting that these S-cells use a different energy/nutrient substrate other than glucose. S-cells used fatty acid to feed the TCA cycle and generate NADPH, as is evident from metabolic flux analysis using stable isotope labeling for MS analysis.
Several recent studies have reported that mitochondrial IDH2 is essential for supplying the NADPH needed to defend cancer cells against mitochondrial oxidative damage and promote cell survival [[26], [27], [28]].
MTHFD2, which is frequently overexpressed in cancers [29], also contributes to NADPH homeostasis [30]. Moreover, MTHFD2-mediated mitochondrial one-carbon metabolism appears to be critical for cancer stem-like properties and drug resistance [31]. Notably, S-cells also appear to use different enzyme modules to generate NADPH. Differences have been observed between the two model cell lines, with one line displaying IDH2-dependent NADPH regeneration and the other depending on MTHFD2.
A range of anticancer therapeutics, such as chemotherapy or ionizing radiation, work by increasing cellular ROS levels, which are effective against cancer cells [13]. Thus, tumor cells with intrinsic mechanisms to mitigate ROS production induced by chemotherapy or ionizing radiation can survive and become resistant to the anticancer therapy. Indeed, S-cells exhibited higher cell viability than P-cells upon 5-FU treatment, which was associated with lower ROS levels, indicating the crucial role of ROS in chemotherapy-induced cytotoxicity. When we knocked down FoxM1 in S-cells or supplemented the cells with NAC, a potent ROS detoxifying agent, reversion of cell viability was observed, further confirming the mechanistic link between ROS and the tumoricidal effect of 5-FU. Therefore, anticancer strategies are required to harness the mechanisms underlying intracellular ROS regulation, which can help overcome drug resistance and consequently enhance the clinical benefits of anticancer therapeutics.
Although the two S-cell lines differ in terms of the specific enzyme modules used for NADPH regeneration, they both utilize the TCA cycle or are mitochondria-dependent, suggesting that CSCs have a mitochondria-centric energy metabo-phenotype. However, further studies would be required to investigate the regulatory mechanisms of these enzymes in CSCs.
5. Conclusions
This study showed that CSCs exhibit increased OXPHOS and paradoxically maintain a low level of ROS, through coupling of FoxM1-dependent Prx3 expression and FAO-mediated NADPH regeneration. Thus, a mitochondrial ROS homeostasis-targeted approach may be an effective strategy for controlling cancers that are refractory to conventional chemotherapy.
Author contributions
Hae-Ji Choi and Jae-Ho Cheong conceived and designed the study and interpreted the results. Hae-Ji Choi performed all the experiments and analyses. Yoo-Lim Jhe, Jungmin Kim, Ju Yeon Lim, Jae Eun Lee assisted and performed the experiments. Min-Kyue Shin analyzed the microarray data. Jae-Ho Cheong supervised all experiments and approved the submission.
Declaration of competing interest
None.
Acknowledgments
This work was supported by a grant from the Korean Health Technology R&D Project through the Korean Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea [grant number HI14C1324], and by the National Research Foundation of Korea MRC grant funded by the Korean government [MSIT; grant number 2018R1A5A2025079 to J.-H. Cheong].
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101589.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Lebleu V.S., O'Connell J.T., Gonzalez Herrera K.N., Wikman H., Pantel K., Haigis M.C., De Carvalho F.M., Damascena A., Domingos Chinen L.T., Rocha R.M., Asara J.M., Kalluri R. PGC-1alpha mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat. Cell Biol. 2014;16:992–1003. doi: 10.1038/ncb3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Viale A., Pettazzoni P., Lyssiotis C.A., Ying H., Sanchez N., Marchesini M., Carugo A., Green T., Seth S., Giuliani V., Kost-Alimova M., Muller F., Colla S., Nezi L., Genovese G., Deem A.K., Kapoor A., Yao W., Brunetto E., Kang Y., Yuan M., Asara J.M., Wang Y.A., Heffernan T.P., Kimmelman A.C., Wang H., Fleming J.B., Cantley L.C., DePinho R.A., Draetta G.F. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;514:628–632. doi: 10.1038/nature13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee K.M., Giltnane J.M., Balko J.M., Schwarz L.J., Guerrero-Zotano A.L., Hutchinson K.E., Nixon M.J., Estrada M.V., Sanchez V., Sanders M.E., Lee T., Gómez H., Lluch A., Pérez-Fidalgo J.A., Wolf M.M., Andrejeva G., Rathmell J.C., Fesik S.W., Arteaga C.L. MYC and MCL1 cooperatively promote chemotherapy-resistant breast cancer stem cells via regulation of mitochondrial oxidative phosphorylation. Cell Metabol. 2017;26:633–647. doi: 10.1016/j.cmet.2017.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roesch A., Vultur A., Bogeski I., Wang H., Zimmermann K.M., Speicher D., Korbel C., Laschke M.W., Gimotty P.A., Philipp S.E., Krause E., Pätzold S., Villanueva J., Krepler C., Fukunaga-Kalabis M., Hoth M., Bastian B.C., Vogt T., Herlyn M. Overcoming intrinsic multidrug resistance in melanoma by blocking the mitochondrial respiratory chain of slow-cycling JARID1B(high) cells. Canc. Cell. 2013;23:811–825. doi: 10.1016/j.ccr.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee E., Yang J., Ku M., Kim N.H., Park Y., Park C.B., Suh J.S., Park E.S., Yook J.I., Mills G.B., Huh Y.-M., Cheong J.-H. Metabolic stress induces a Wnt-dependent cancer stem cell-like state transition. Cell Death Dis. 2015;6 doi: 10.1038/cddis.2015.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee J., Kee H.J., Min S., Park K.C., Park S., Hwang T.H., Ryu D.H., Hwang G.S., Cheong J.H. Integrated omics-analysis reveals Wnt-mediated NAD+ metabolic reprogramming in cancer stem-like cells. Oncotarget. 2016;7:48562–48576. doi: 10.18632/oncotarget.10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park K.C., Kim S.W., Jeon J.Y., Jo A.R., Choi H.J., Kim J., Lee H.G., Kim Y., Mills G.B., Noh S.H., Lee M.G., Park E.S., Cheong J.H. Survival of cancer stem-like cells under metabolic stress via CaMK2α-mediated upregulation of sarco/endoplasmic reticulum calcium ATPase expression. Clin. Canc. Res. 2018;24:1677–1690. doi: 10.1158/1078-0432.CCR-17-2219. [DOI] [PubMed] [Google Scholar]
- 8.Mailloux R.J., Mcbride S.L., Harper M.E. Unearthing the secrets of mitochondrial ROS and glutathione in bioenergetics. Trends Biochem. Sci. 2013;38:592–602. doi: 10.1016/j.tibs.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 9.Sancho P., Burgos-Ramos E., Tavera A., Bou Kheir T., Jagust P., Schoenhals M., Barneda D., Sellers K., Campos-Olivas R., Grana O., Viera C.R., Yuneva M., Sainz B., Jr., Heeschen C. MYC/PGC-1alpha balance determines the metabolic phenotype and plasticity of pancreatic cancer stem cells. Cell Metabol. 2015;22:590–605. doi: 10.1016/j.cmet.2015.08.015. [DOI] [PubMed] [Google Scholar]
- 10.Kuntz E.M., Baquero P., Michie A.M., Dunn K., Tardito S., Holyoake T.L., Helgason G.V., Gottlieb E. Targeting mitochondrial oxidative phosphorylation eradicates therapy-resistant chronic myeloid leukemia stem cells. Nat. Med. 2017;23:1234–1240. doi: 10.1038/nm.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo M., Shang L., Brooks M.D., Jiagge E., Zhu Y., Buschhaus J.M., Conley S., Fath M.A., Davis A., Gheordunscu E., Wang Y., Harouaka R., Lozier A., Triner D., McDermott S., Merajver S.D., Luker G.D., Spitz D.R., Wicha M.S. Targeting breast cancer stem cell state equilibrium through modulation of redox signaling. Cell Metabol. 2018;28:69–86. doi: 10.1016/j.cmet.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beck B., Blanpain C. Unravelling cancer stem cell potential. Nat. Rev. Canc. 2013;13:727–738. doi: 10.1038/nrc3597. [DOI] [PubMed] [Google Scholar]
- 13.Yang H., Villani R.M., Wang H., Simpson M.J., Roberts M.S., Tang M., Liang X. The role of cellular reactive oxygen species in cancer chemotherapy. J. Exp. Clin. Canc. Res. 2018;37:266. doi: 10.1186/s13046-018-0909-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park H.J., Carr J.R., Wang Z., Nogueira V., Hay N., Tyner A.L., Lau L.F., Costa R.H., Raychaudhuri P. FoxM1, a critical regulator of oxidative stress during oncogenesis. EMBO J. 2009;28:2908–2918. doi: 10.1038/emboj.2009.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song I.S., Jeong Y.J., Jeong S.H., Heo H.J., Kim H.K., Bae K.B., Park Y.H., Kim S.U., Kim J.M., Kim N., Ko K.S., Rhee B.D., Han J. FOXM1-induced PRX3 regulates stemness and survival of colon cancer cells via maintenance of mitochondrial function. Gastroenterology. 2015;149:1006–1016. doi: 10.1053/j.gastro.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 16.Halasi M., Pandit B., Wang M., Nogueira V., Hay N., Gartel A.L. Combination of oxidative stress and FOXM1 inhibitors induces apoptosis in cancer cells and inhibits xenograft tumor growth. Am. J. Pathol. 2013;183:257–265. doi: 10.1016/j.ajpath.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carr J.R., Park H.J., Wang Z., Kiefer M.M., Raychaudhuri P. FoxM1 mediates resistance to herceptin and paclitaxel. Canc. Res. 2010;70:5054–5063. doi: 10.1158/0008-5472.CAN-10-0545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khongkow P., Karunarathna U., Khongkow M., Gong C., Gomes A.R., Yague E., Monteiro L.J., Kongsema M., Zona S., Man E.P., Tsang J.W., Coombes R.C., Wu K.J., Khoo U.S., Medema R.H., Freire R., Lam E.W. FOXM1 targets NBS1 to regulate DNA damage-induced senescence and epirubicin resistance. Oncogene. 2014;33:4144–4155. doi: 10.1038/onc.2013.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blacker T.S., Mann Z.F., Gale J.E., Ziegler M., Bain A.J., Szabadkai G., Duchen M.R. Separating NADH and NADPH fluorescence in live cells and tissues using FLIM. Nat. Commun. 2014;5:3936. doi: 10.1038/ncomms4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pike L.S., Smift A.L., Croteau N.J., Ferrick D.A., Wu M. Inhibition of fatty acid oxidation by etomoxir impairs NADPH production and increases reactive oxygen species resulting in ATP depletion and cell death in human glioblastoma cells. Biochim. Biophys. Acta. 2011;1807:726–734. doi: 10.1016/j.bbabio.2010.10.022. [DOI] [PubMed] [Google Scholar]
- 21.Schafer Z.T., Grassian A.R., Song L., Jiang Z., Gerhart-Hines Z., Irie H.Y. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature. 2009;461:109–113. doi: 10.1038/nature08268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeon S.M., Chandel N.S., Hay N. AMPK regulates NADPH homeostasis to promote tumour cell survival during energy stress. Nature. 2012;485:661–665. doi: 10.1038/nature11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pavlova N.N., Thompson C.B. The emerging hallmarks of cancer metabolism. Cell Metabol. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chandimali N., Jeong D.K., Kwon T. Peroxiredoxin II regulates cancer stem cells and stemness-associated properties of cancers. Cancers. 2018;10:305. doi: 10.3390/cancers10090305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sabharwal S.S., Schumacker P.T. Mitochondrial ROS in cancer: initiators, amplifiers or an Achilles' heel? Nat. Rev. Canc. 2014;14:709–721. doi: 10.1038/nrc3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jo S.H., Son M.K., Koh H.J., Lee S.M., Song I.H., Kim Y.O., Lee Y.S., Jeong K.S., Kim W.B., Park J.W., Song B.J., Huh T.L. Control of mitochondrial redox balance and cellular defense against oxidative damage by mitochondrial NADP+-dependent isocitrate dehydrogenase. J. Biol. Chem. 2001;276:16168–16176. doi: 10.1074/jbc.M010120200. [DOI] [PubMed] [Google Scholar]
- 27.Kil I.S., Kim S.Y., Lee S.J., Park J.W. Small interfering RNA-mediated silencing of mitochondrial NADP+-dependent isocitrate dehydrogenase enhances the sensitivity of HeLa cells toward tumor necrosis factor-alpha and anticancer drugs. Free Radic. Biol. Med. 2007;43:1197–1207. doi: 10.1016/j.freeradbiomed.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 28.Kil I.S., Chung K.H., Park J.W. Silencing of mitochondrial NADP+-dependent isocitrate dehydrogenase gene enhances selenite-induced apoptosis. Free Radic. Res. 2010;44:332–339. doi: 10.3109/10715760903494184. [DOI] [PubMed] [Google Scholar]
- 29.Nilsson R., Jain M., Madhusudhan N., Sheppard N.G., Strittmatter L., Kampf C., Huang J., Asplund A., Mootha V.K. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat. Commun. 2014;5:3128. doi: 10.1038/ncomms4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fan J., Ye J., Kamphorst J.J., Shlomi T., Thompson C.B., Rabinowitz J.D. Quantitative flux analysis reveals folate-dependent NADPH production. Nature. 2014;510:298–302. doi: 10.1038/nature13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishimura T., Nakata A., Chen X., Nishi K., Meguro-Horike M., Sasaki S., Kita K., Horike S.I., Saitoh K., Kato K., Igarashi K., Murayama T., Kohno S., Takahashi C., Mukaida N., Yano S., Soga T., Tojo A., Gotoh N. Cancer stem-like properties and gefitinib resistance are dependent on purine synthetic metabolism mediated by the mitochondrial enzyme MTHFD2. Oncogene. 2019;38:2464–2481. doi: 10.1038/s41388-018-0589-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shin Min-Kyue, Cheong Jae-Ho. Mitochondria-Centric Bioenergetic Characteristics in Cancer Stem-like Cells. 2019 doi: 10.1007/s12272-019-01127-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.