Abstract
During aging, organs such as skeletal muscle and heart require sufficient NAD+ both as a coenzyme for oxidative-reductive electron transfer and as a substrate for multiple signaling pathways. Sirtuins (SIRTs), a family of NAD+-dependent deacetylase, play an important role in regulating mitochondrial homeostasis and antioxidant defense by deacetylating transcription factors and enzymes such as PGC-1α, p65, GCN5, and SOD2. However, age-related DNA damage and increased SASP activate PARP-1 and CD38, the enzymes competing with SIRTs for NAD+. Thus, it is important to know how aging alters intracellular NAD+ status and NAD+-depending enzyme expression in muscles. In this study, we report that the acetylation level of muscle protein pool, as well as major SIRTs target proteins (PGC-1α, GCN5, p65, and SOD2), was significantly increased in hindlimb and cardiac muscles of 24-month old mice compared with their 6-month old counterparts, despite the fact that most members of the SIRT family were upregulated with aging. Aging increased the protein content of PARP-1 and CD38, whereas decreased NAD+ levels in both skeletal and heart muscles. Aged muscles demonstrated clear signs of mitochondrial dysfunction, oxidative stress, and inflammation. Taken together, our data suggest that despite the upregulation of SIRTs, aged muscles suffered from NAD+ deficit partly due to the competition of elevated CD38 and PARP-1. The enhanced acetylation of several key proteins involved in broad cellular functions may contribute to the age-related muscle deterioration.
Electronic supplementary material
The online version of this article (10.1007/s11357-020-00171-7) contains supplementary material, which is available to authorized users.
Keywords: Aging, CD38, Deacetylation, NAD+, PARP, SIRT, Skeletal muscle
Introduction
It is well-documented that deterioration of mitochondrial function plays a critical role in the process of muscle aging (Hepple 2014; Picca et al. 2018). A growing number of studies suggest that dysregulation of protein acetylation/deacetylation in the mitochondria is an important cause of mitochondrial dysfunction (Wagner and Payne 2011; Sack and Finkel 2012). Acetylation of mitochondrial proteins and enzymes involved in the tricarboxylic acid (TCA) cycle, electron transport chain (ETC), and antioxidant defense, as well as key transcription factors (TF) in cell signaling, can lead to their inactivation and, in turn, the defects in the respective biological functions under their control (Guan and Xiong 2011; Gómez and Hagen 2012). These include but are not limited to the peroxisome proliferator-activated receptor gamma, coactivator 1 alpha (PGC-1α), superoxide dismutase (SOD) 2, nuclear factor kappa B (NFκB), and forkhead box O family TF (FoxO) (Kang et al. 2013; Konopka et al. 2014; Kawahara et al. 2009; Palikaras and Tavernarakis 2014; Sakellariou et al. 2016; Sanchez et al. 2014). Aging has been shown to increase the acetylation of these enzymes and TFs, which is considered a major mechanism for age-associated decline of mitochondrial homeostasis and deteriorative diseases (Wagner and Payne 2011; Sack and Finkel 2012).
In the eukaryotic cells, deacetylation is executed by sirtuins (SIRTs), the nicotinamide adenine dinucleotide (NAD)+–dependent protein deacetylases (as well as demalonylases and desuccinylases), that control mitochondrial function, oxidative-reductive regulation, and antioxidant defense that are highly relevant to aging (Michan and Sinclair 2007). In the mammals, SIRTs are comprised of seven members, SIRT1–7, depending on their cellular location, substrate, and function (Kabiljo et al. 2019). SIRT1, 6, and 7 are in the nucleus; SIRT2 primarily localizes in the cytoplasm; and SIRT3, 4, and 5 are located in the mitochondrion (Michan and Sinclair 2007). All seven isoforms of SIRTs are strongly expressed in the skeletal muscle. Over the past decades, SIRT1, 3, and 6 have been extensively studied, and it is well-established that mitochondrial biogenesis is regulated by the SIRT1-PGC-1α axis, primarily because Sirt1 deacetylates and increases PGC-1α activity (Nemoto et al. 2005; Fernandez-Marcos and Auwerx 2011). In addition, Sirt1 regulates the activity of NFκB and FoxO, the key controllers for pro-inflammatory cytokine expression, ubiquitin proteolysis, and mitophagy (Webb and Brunet 2014). Importantly, SIRT3 deacetylates enzymes and proteins in the TCA cycle and ETC thereby promoting mitochondrial oxidative phosphorylation (Lombard et al. 2007). Another key role of SIRT3 is to deacetylate SOD2 at multiple lysine sites, the acetylation of which renders the enzyme inactive (Tao et al. 2010). The primary function of SIRT6 is promoting DNA repair and genome stability (Kugel and Mostoslavsky 2014). In addition, SIRT6 also inhibits NFkB activation at histone level thus inhibiting inflammation (Kawahara et al. 2009).
The enzymatic activity of SIRT is dependent on adequate levels of its substrate NAD+, which accepts the acetyl- (or malonyl/succinyl-) moiety to form 0-acetyl-ADP-ribose. Research evidence suggests that deacetylation capacity of SIRTs decreases with aging due in part to diminished cellular NAD+ pool (Gómez and Hagen 2012; Chini et al. 2017). The decline could be caused either by a decrease of NAD+ biosynthesis or by an increase in NAD+ degradation or a combination of both (Camacho-Pereira et al. 2016). NAD+ biosynthesis can be accomplished by a de novo pathway from tryptophan or by a salvage pathway controlled by nicotinamide phosphate transferase (NAMPT). NAD+ degradation is controlled by several NAD+-consuming enzymes such as the cluster of differentiation 38 (CD38), poly(ADP-Ribose) polymerase (PARP)-1, and SIRTs (Kolthur-Seetharam et al. 2006; Bai et al. 2011; Tarragó et al. 2018). CD38, also known as cyclic ADP ribose hydrolase, cleaves NAD+ to produce nicotinamide (NAM) and ADP-ribose (ADPR), as well as a small fraction of ADP-ribosyl cyclases (cADPR), which serves as a second messenger for calcium release from the endoplasmic reticulum (Chini 2009). A series of recent reports show that CD38 activity increases with aging in various tissues including skeletal muscle of mice, which lead to NAD+ decline and thus SIRTs activity (Camacho-Pereira et al. 2016; Tarragó et al. 2018). PARP-1 consumes NAD+ by using its ADP-ribose polymer backbone to repair damaged DNA base pair, thus competing with SIRTs for a shared NAD+ pool (Imai and Guarente 2014). Recent research suggests that PARP-1 is chronically activated in aging skeletal muscle, which also decreases cellular NAD+ level and negatively affects SIRTs activity (Bai et al. 2011; Verdin 2015). Furthermore, cleavage of PARP-1 by specific proteases such as caspases, calpains, and cathepsins has been recognized as indications of cell stress and death (Chaitanya et al. 2010). Because of the important relationship between SIRT and NAD+, several experimental approaches have been employed to maintain cellular NAD+ levels and boost SIRTs deacetylation activity in order to ameliorate mitochondrial homeostasis, including pharmacological inhibition of CD38, caloric restriction (CR), fasting, exercise training, and resveratrol treatment (Chini 2009; Chini et al. 2017, 2018; Badreh et al. 2019). However, few studies up to date have made comprehensive examination of the impact of aging on NAD+, SIRTs, and their target proteins’ acetylation in skeletal and cardiac muscles.
Thus, the primary purpose of this study was to investigate how aging affects SIRT expression, the acetylation status of muscle proteins and TFs, and their relationship with cellular NAD+ level in skeletal and cardiac muscle in a mouse model. We also intended to explore the cellular mechanisms behind and the implication of these changes in mitochondrial metabolic and antioxidant functions in aging muscles. Our primary hypothesis was that aging decreases SIRTs expression and increases muscle protein acetylation due to decreased cellular NAD+ level, which would result in mitochondrial dysfunction and redox disturbance in aged muscles.
Results
Protein acetylation was increased with aging
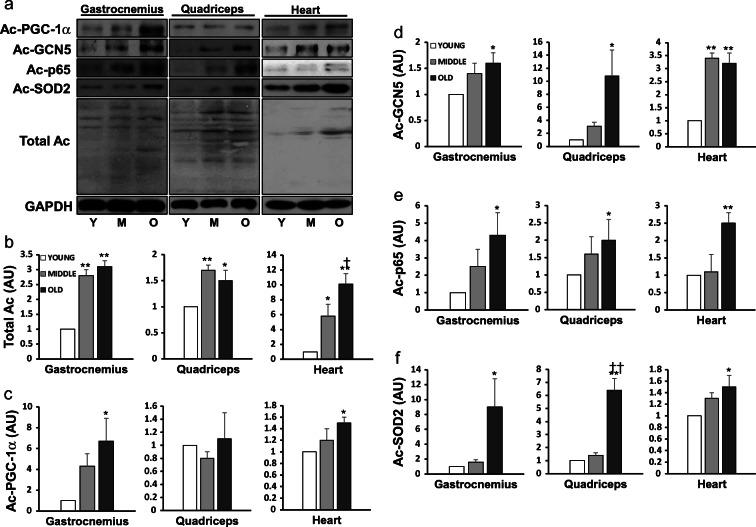
We first examined whether aging would enhance the acetylation of the total muscle protein pool and select enzymes and TFs involved in various important cellular functions in two major muscle groups in mouse hindlimb, the gastrocnemius (G) and quadriceps (Q), and in heart (H). The extent of acetylation in total muscle proteins showed a prominent increase in both mid-aged (M) and old (O) G, Q, and H, compared with their young (Y) counterparts (P < 0.05 or 0.01) (Fig. 1a, b). Acetylation of PGC-1α was increased in G and H muscle of O (6.7- and 1.5-fold, P < 0.05, respectively) vs. Y (Fig. 1c). Acetylated general control nonrepressed (GCN) 5 level was elevated by 1.6- (P < 0.05), 10.8- (P < 0.05), and 3.2-fold (P < 0.01), respectively, in the three muscles comparing O vs. Y mice (Fig. 1d). Aging also increased acetylated-p65 levels in G (P < 0.05) and H (P < 0.01) of O vs. Y mice (Fig. 1e). SOD2 showed a dramatic 9-fold higher acetylation level in G (P < 0.05) and Q (P < 0.01), as well as in H (P < 0.05), when comparing O vs. Y mice (Fig. 1f).
Fig. 1.
Protein acetylation was increased with aging. Representative Western blots (a) of total acetylated protein pool (b), acetylated PGC-1α (c), GCN5 (d), p65 (e), and SOD2 (f) in two skeletal muscles and cardiac muscle of young (N = 8), mid-aged (N = 8), and old mouse (N = 8), respectively. Data are mean ± SEM. *P < 0.05, **P < 0.01, Mid-aged or Old vs. Young; †P < 0.05, ††P < 0.01, old vs. mid-aged
Aging increased SIRT expression in skeletal muscle
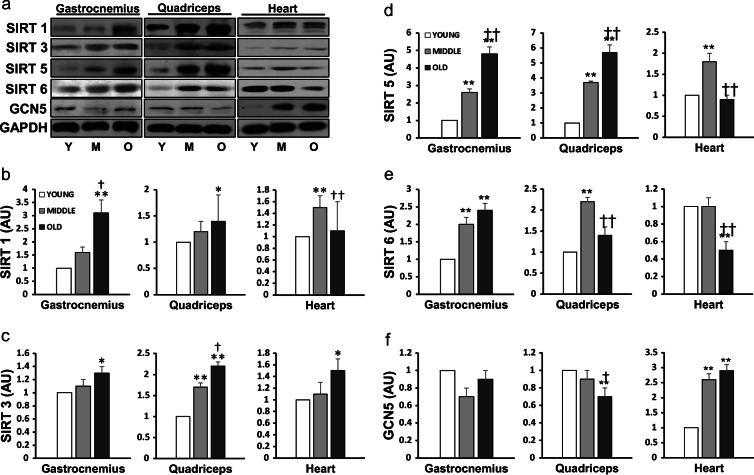
Since increased protein acetylation could have been caused by decreased SIRT levels in the cell, we next measured the protein content of major members of the SIRT family (Fig. 2). In skeletal muscle, SIRT1 protein level was unaltered at mid-age but significantly increased at old age (P < 0.05) (Fig. 2b). SIRT3 level was elevated in Q (P < 0.01) in M and in both G (P < 0.05) and Q (P < 0.01) in O (Fig. 2c). Increments of SIRT 5 and 6 in both G and Q were significant in M and O vs. Y mice (P < 0.01; Fig. 2d, e). In the cardiac muscle, age effect was diverse; SIRT 1 and 5 showed an increase (P < 0.01) in M, but no change in O mice. SIRT 3 level was increased, but SIRT 6 level decreased in O vs. Y group (P < 0.05). In order to determine whether increased protein acetylation in aged muscles was related to increased cellular acetylation potential, we assessed GCN5 expression as a biomarker. GCN5 protein content was unaffected in G but was significantly decreased in Q (P < 0.01) of O vs. Y mice (Fig. 2f). Thus, increased acetylation of protein in skeletal muscle during aging was not caused by elevated GCN5 expression. In contrast to skeletal muscle, GCN5 level in H was elevated by 2.6- and 2.9-fold (P < 0.01), respectively, comparing M and O vs. Y mice.
Fig. 2.
Aging increased SIRT expression in skeletal muscle. Representative Western blots (a) of the protein levels of SIRT1 (b), SIRT3 (c), SIRT5 (d), SIRT6 (e), and GCN5 (f) in two skeletal muscles and cardiac muscle of young (N = 8), mid-aged (N = 8), and old mouse (N = 8), respectively. Data are mean ± SEM. *P < 0.05, **P < 0.01, Mid-aged or Old vs. Young; †P < 0.05, ††P < 0.01, old vs. mid-aged
NAD+ content decreased with aging
Because NAD+ is required as a substrate for SIRT and is controlled by CD38 and PARP activities, we examined concentration of NAD+ in various muscles during aging. NAD+ levels in all three muscles were decreases by more than 40% in both M and O mice (P < 0.05; Fig. 3b). To gain some insight into the distribution of NAD+ across cellular compartments, we measured NAD+ levels in the nuclear extraction and soluble fraction of the hindlimb muscles, realizing that nuclear pores are large enough for NAD+ to exit making the cytosolic NAD+ pool a mixture of nuclear and perhaps also mitochondrial NAD+. The data are shown in supplementation figures. Both NAD+ (Fig. 1Sb) and NAD+/NADH ratio (Fig. 1Sc) were decreased by more than half (P < 0.05) in G and Q muscles, comparing M and O with Y mice. Cytoplasmic NAD+ levels were affected less, showing a significant reduction only in G of M mice (P < 0.05) and in Q of O mice (P < 0.05). NAD+/NADH ratio was lowered with aging in both G and Q muscles (P < 0.05).
Fig. 3.
NAD+ content was decreased with aging. The level of NAD+, expressed as pmol/mg protein, in two skeletal muscles and cardiac muscle of young (N = 8), mid-aged (N = 8), and old mouse (N = 8), respectively. Data are mean ± SEM. *P < 0.05 Mid-aged or Old vs. Young
NAD+-consuming enzymes were upregulated with aging
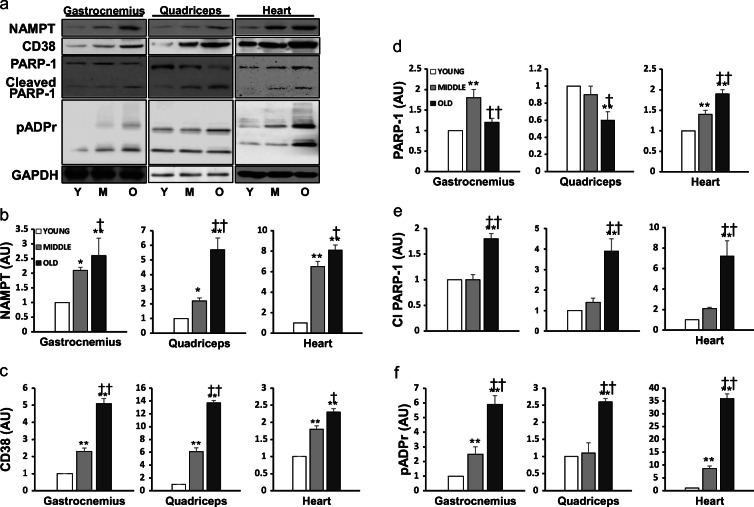
Since NAD+ turnover is controlled by both its synthesis (mainly by the salvage pathway) and degradation, we next measured the expression of NAD+ salvaging enzyme nicotinamide phosphoribosyltransferase (NAMPT) in the various muscles. NAMPT content was elevated by 2.5- and 5.5-fold in G and Q of aged mice (P < 0.01), and the increases were significant in the mid-aged mice as well (Fig. 4b). Interestingly, NAMPT levels in H showed a dramatic 8-fold increment in both M and O vs. Y (P < 0.05).
Fig. 4.
NAD+-consuming enzymes were upregulated with aging. Representative Western blots (a) of the protein content of NAMPT (b), CD38 (c), PARP-1 (d), cleaved form of PARP-1 (e), and pADPr (f) in two skeletal muscles and cardiac muscle of young (N = 8), mid-aged (N = 8), and old mouse (N = 8), respectively. Data are mean ± SEM. *P < 0.05, **P < 0.01, Mid-aged or Old vs. Young; †P < 0.05, ††P < 0.01, old vs. mid-aged
We then measured the protein content of CD38, a major NAD+-consuming enzyme in the cell. CD38 level was elevated by 2- and 6-fold (P < 0.01) in M vs. Y in G and Q muscles, respectively (Fig. 4c). Strikingly, old G and Q muscle showed more than 5- and 13-fold increases in CD38 levels compared with their Y counterparts (P < 0.01). In addition, CD38 expression was significantly increased in H muscle in M and O vs. Y (P < 0.01).
Yet another NAD+-degrading enzyme is PARP-1, which cleaves NAD+ to provide ADP-ribose backbone for DNA repair. PARP-1 level was increased in G of M (P < 0.01) but not O mice (Fig. 4d). PARP-1 level in Q was unchanged in M but decreased in O vs. Y (P < 0.01). H muscle showed a significant increase in PARP-1 level in both M and O (P < 0.01). Furthermore, the cleaved form of PARP-1 was dramatically elevated by ~2-, 4-, and 7-fold (P < 0.01) in O vs. Y mice (Fig. 4e). As an additional marker of age-associated changes of PARP-1, the extent of poly(ADP-ribosylation) (pADPr) increased strikingly at old age in all three muscles (P < 0.01, Fig. 4f).
Aging resulted in decreased mitochondrial function
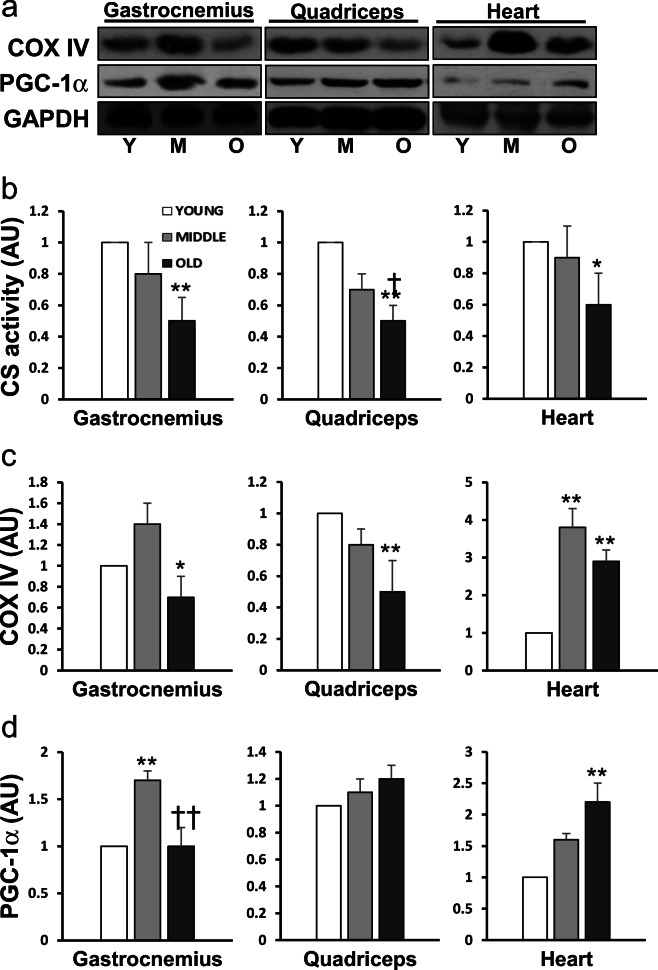
Aging caused a significant alteration of mitochondria-related protein contents and enzyme activities in the muscles (Fig. 5). As the rate-limiting enzyme of TCA cycle, citrate synthase (CS) activity showed an age-dependent reduction by ~50% in both G and Q muscles (P < 0.01) and in H (P < 0.05; Fig. 5b). The protein content of cytochrome c oxidase subunit 4 (COX IV), a classic enzyme marker of ETC, also decreased significantly in G (P < 0.05) and Q muscles (P < 0.01) of O vs. Y mice. Intriguingly, COXIV level in H was elevated by 3.8- and 2.9-fold in M and O vs. Y, respectively (P < 0.01; Fig. 5c). PGC-1α protein content was unaltered by old age in G and Q muscles, except an increase (P < 0.01) in G muscle of M mice. In contrast, there was a 2-fold increase in PGC-1α content in H of O mice (P < 0.01; Fig. 5d). It seems that the aging effect on PGC-1α is not the downregulation of protein expression but decreased activity due to acetylation.
Fig. 5.
Representative Western blots (a) of the protein content of mitochondrial COX IV (c) and PGC-1α (d), and citrate synthase (CS) enzyme activity (b) in two skeletal muscles and cardiac muscle of young (N = 8), mid-aged (N = 8), and old mouse (N = 8), respectively. Data are mean ± SEM. *P < 0.05, **P < 0.01, Mid-aged or Old vs. Young; †P < 0.05, ††P < 0.01, old vs. mid-aged
Aging altered muscle antioxidant defense and increased oxidative stress
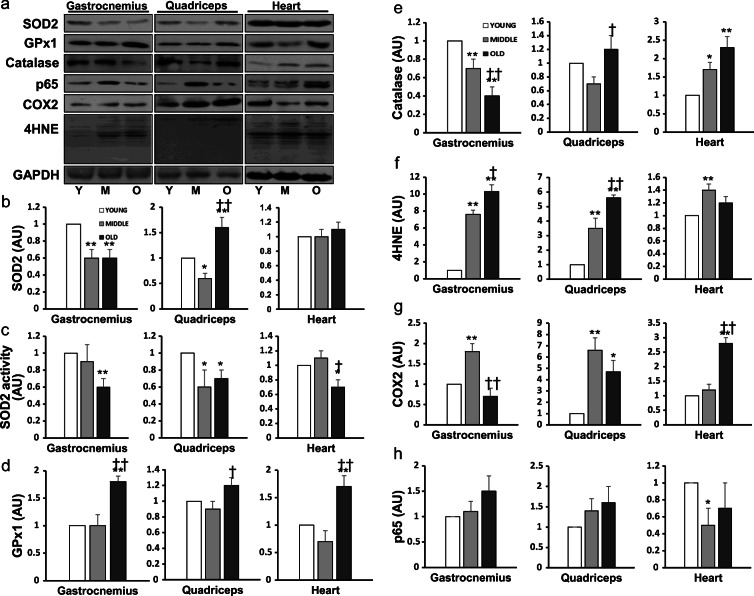
SOD2 protein content was downregulated by 40% (P < 0.01) in G of both M and O vs. Y (Fig. 6b). In Q muscle, SOD2 level showed a reduction in M (P < 0.05) but an increase in O (P < 0.01). SOD2 level in H was not affected by aging. As shown in Fig. 6c, SOD2 activity was decreased by 40% (P < 0.01) in G, and 30% (P < 0.05) in Q and H, comparing O vs. Y mice. GPx1 protein level was significantly increased in G, Q, and H in O vs. Y mice (P < 0.05 or 0.01; Fig. 6d). Catalase content showed differential effects with aging, with a reduction in G muscle of M and O vs. Y mice (P < 0.01) but an increase in Q (P < 0.05) and H (P < 0.01) of O mice (Fig. 6e).
Fig. 6.
Representative Western blots (a) of the protein content of SOD2 (b), GPx1 (d), catalase (e), 4-HNE (f), COX2 (g) and p65 (h), and SOD2 enzyme activity (c) in two skeletal muscles and cardiac muscle of young (N = 8), mid-aged (N = 8), and old mouse (N = 8), respectively. Data are mean ± SEM. *P < 0.05, **P < 0.01, Mid-aged or Old vs. Young; †P < 0.05, ††P < 0.01, old vs. mid-aged
Aged skeletal and cardiac muscles demonstrated clear signs of oxidative stress, reflected by an increase in lipid peroxidation marker 4-hydroxynonenal (4-HNE) levels in both G and Q muscles of M and O vs. Y mice (P < 0.01). H muscle showed an increase in 4-HNE only in M (P < 0.01; Fig. 6f). Cyclooxygenase (COX) 2, a well-studied inflammatory marker, was increased by 80% (P < 0.01) in G of M vs. Y mice (Fig. 6g). COX2 content in Q muscle was elevated by 6.6-fold (P < 0.01) and 4.7-fold (P < 0.05), respectively, in M and O vs. Y. There was a 2.8-fold increase in COX2 level in H of O vs. Y mice (P < 0.01). P65 content, measured as a marker of NFκB activation, was not altered in skeletal muscle but showed a decrease in H of M mice (P < 0.05; Fig. 6h).
Discussion
A major finding in the present study is that global protein acetylation levels were significantly increased at old age in skeletal and cardiac muscles. To our knowledge, this has never been reported before. Proteomic analysis of the acetylated peptides has identified over 100 lysine acetylation sites in mitochondrial proteins and established lysine acetylation as one of the common post-translational modification in the mitochondrion (Kim et al. 2006). Acetylation can influence multiple mitochondria-related protein functions, including mitochondrial turnover, TCA cycle, ETC, and antioxidant defense (Guan and Xiong 2011; Gómez and Hagen 2012). Aging is a prominent inducer of mitochondrial protein hyperacetylation, which may be one of the major causes for mitochondrial enzyme dysfunction in the TCA cycle and ETC, loss of redox homeostasis, and increased protein and organelle oxidative damage (Wagner and Payne 2011; Sack and Finkel 2012). We identified several important enzymes and proteins that showed elevated acetylation in the aged muscle. For example, acetylation is known to render PGC-1α incapable of coactivating mitochondrial biogenesis, and this could be a reason for decreased mitochondrial oxidative capacity, demonstrated by declines of CS activity and COXIV content (Fig. 5; Yeo et al. 2019). Acetylation also has significant implication in cellular antioxidant defense and redox balance, as increased acetylation of SOD2 has long been recognized as a major factor for its decreased catalytic activity as shown in muscle immobilization (Kang et al. 2015) and aging (Yeo et al. 2019). The 8- to 10-fold increase in SOD2 acetylation level (Fig. 1f), coupled with the downregulation of SOD2 protein, resulted in a ~40% reduction of total SOD2 activity in the hindlimb muscles of old mice (Fig. 6c). This could represent a devastating effect to aged muscle because elevated superoxide radical (O2●-) can reactive with hydrogen peroxide (H2O2) or nitric oxide (NO) to form hydroxyl radical (OH●) or peroxynitrite (ONOO●), respectively. Thus, the increased GPx1 protein content found in the older muscles could represent a compensatory response to elevated H2O2 level. Conceivably, the increased oxidative stress could be the main explanation for increased lipid peroxidation in aged animals (Fig. 6f).
Another serious threat to aged organelle is inflammation termed “inflammaging” (Chini et al. 2017). Increased ROS generation and decreased antioxidant defense could lead to NFκB activation and pro-inflammatory cytokine production thus escalating cellular oxidative damage and dysfunction. We observed a large increase in acetylation level of p65, the subunit of NFκB that binds with target gene DNA (Fig. 1e), although the trend of increase in p65 protein content did not reach significant level (Fig. 6h). Acetylation has been shown to increases the binding activity of p65, a crucial step in the transactivation of pro-inflammatory cytokine expression (Yeung et al. 2004; Kawahara et al. 2009). Indeed, aged muscles showed higher levels of COX2, the enzyme that is known to be activated by TNFα (Fig. 6g).
Because dysregulation of SIRTs has been considered a major explanation for age-related diseases such as cardiovascular disease, neurodegenerative disease, and cancer (Haigis and Guarente 2006), we hypothesized that increased acetylation of mitochondrial protein pool could be caused by decreased SIRT protein expression due to aging. However, this hypothesis was not supported by our data in the present study, which provided clear evidence that, except for SIRT6 in the heart, protein levels of all SIRTs increased significantly in an age-dependent manner in mouse skeletal and cardiac muscles (Fig. 2). These data are in partial agreement with several other studies reporting a positive correlation between SIRT1 protein and age in muscle (Koltai et al. 2010; Kilic et al. 2015). However, some studies suggested that aging could downregulate SIRT1 protein content (Michishita 2005; Sasaki et al. 2006; Khanh et al. 2018; Badreh et al. 2019; Kabiljo et al. 2019). Since most of these studies mostly investigated other organs and tissues than skeletal muscle, the discrepancies likely reflected differences in species, organs, and employed methodology. With regard to age-related changes in other SIRT isoforms, there have been only a few recent studies reporting SIRT3 and 5 level decrease with aging in human adipose tissue-derived mesenchymal stem cells and in Nothobranchius furzeri (Khanh et al. 2018; Badreh et al. 2019). Thus, it is plausible that skeletal muscle represents a unique organ that expresses SIRTs differently from non-muscle organs during the course of aging (Kabiljo et al. 2019).
The question arises as to what cellular signals and mechanisms could lead to increased SIRT protein expression at old age. More critically, why did aged muscles display higher levels of protein acetylation despite dramatically elevated SIRT content? In order to explore potential explanations, we focused on cellular NAD+ pool, a mandatory substrate for SIRT’s deacetylation activity. The role of NAD+ has been highlighted in the recent literature (Cantó et al. 2015; Camacho-Pereira et al. 2016; Tarragó et al. 2018). In addition to serving as a coenzyme to accept hydrogen ions as reducing power for a variety of metabolic enzymes, NAD+ is the required acceptor of acetyl group during the SIRT-catalyzed deacetylation reaction (Chini et al. 2017). Furthermore, a decrease in cellular NAD+ availability due to aging has been identified as a main reason for organ’s and tissue’s inability to deal with increased protein acetylation tendency despite high SIRT levels (Imai and Guarente 2014; Johnson and Imai 2018; Verdin 2015). Indeed, we confirmed in the present study that NAD+ levels in the whole muscle lysate and nucleus are severely decreased in mid-aged and old mouse hindlimb muscles (Fig. 3 and Fig. 1S). This finding agrees with several previous studies reporting age-related deficits of NAD+ in skeletal muscle (Koltai et al. 2010; Camacho-Pereira et al. 2016; Frederick et al. 2016; Schultz and Sinclair 2017; Tarragó et al. 2018). Furthermore, we demonstrated that the expression of NAMPT, the rate-limiting enzyme for the NAD+ salvage pathway, was not decreased, as reported previously (Koltai et al. 2010; Frederick et al. 2016; Schultz and Sinclair 2017), but dramatically upregulation with aging (Fig. 4b). Although NAMPT is considered the dominate enzyme in mammals, decreased de novo synthesis should not be ruled out in contributing to age-related NAD+ deficit (Mills et al. 2016; Mitchell et al. 2018). Moreover, decline of nicotinamide riboside (NR) with aging could be another cause for NAD+ depletion. A natural NAD+ precursor, NR can be directly converted to NMN, thereby bypassing NAMPT in the salvage pathway (Trammell et al. 2016; Schöndorf et al. 2018). Nevertheless, without experimental data, the role of NAD+ de novo synthesis in age-related decline of NAD+ remains speculative.
Recent literature indicates that besides SIRTs, the two main pathways that consume cellular NAD+ pool are CD38 and PARP-1 (Kolthur-Seetharam et al. 2006; Chini 2009; Bai et al. 2011; Tarragó et al. 2018). CD38 is a NADase and has been postulated as the main regulator of cellular NAD+ levels (Chini 2009). Cleavage of NAD+ generates cyclic ADPribose and nicotinamide (NAM), which is an endogenous inhibitor Sirt1. CD38 has been implicated in age-related pathogenic conditions such as obesity, diabetes, and chronic inflammation (Chini et al. 2018). Recent studies reported that aging gradually increases CD38 protein level and its NADase activity, possibly due to elevation of senescence-associated secretory phenotype (SASP) (Camacho-Pereira et al. 2016; Tarragó et al. 2018; Chini et al. 2019). In fact, 78c, a specific CD38 inhibitor, as well as CD38 gene knockout, has been shown to rescue intracellular NAD+ and preserved SIRTs activity (Camacho-Pereira et al. 2016; Tarragó et al. 2018). Our finding that CD38 expression was dramaticaly increased by 2–6-fold in the skeletal muscle of mid-aged mice and by 5–13-fold in the old mice strongly suggest that upregulation of this enzyme could be a main reason for muscle NAD+ deficit in aged muscles.
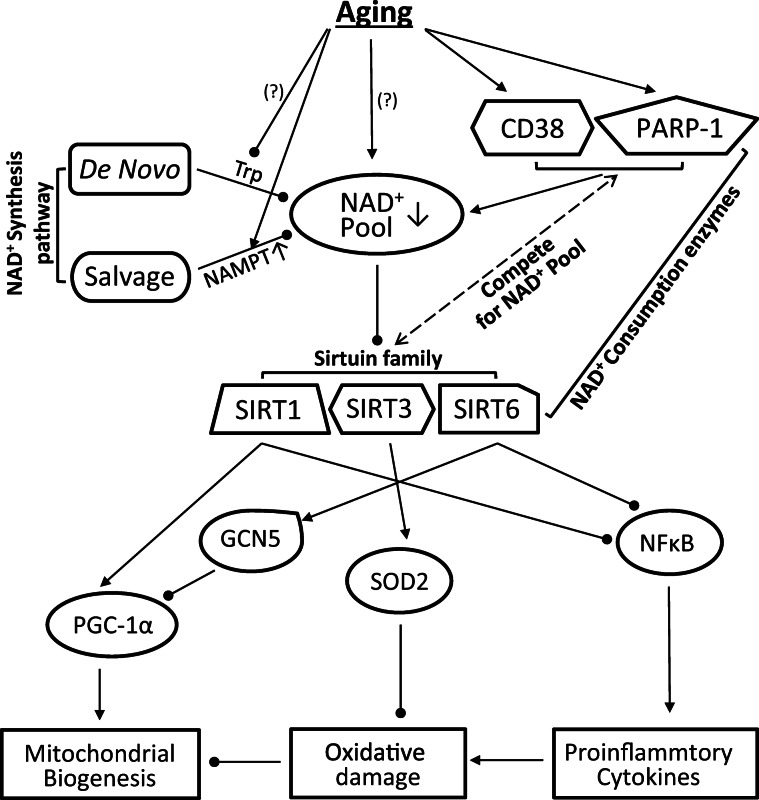
An alternative explanation of age-associated decrease in muscle NAD+ could be due to elevated PARP-1 expression. PARP-1 uses NAD + as a substrate to catalyze the covalent transfer of ADP-ribose to a variety of nuclear protein acceptors, such as during DNA base pairs (Imai and Guarente 2014). It has been suggested that increased PARP-1 level could be an inevitable process of aging due to accumulation of DNA damage (Fang et al. 2014). Indeed, recent studies using PARP-1 pharmacological inhibitor revealed an increase in NAD+ pool and enhanced SIRTs activity (Mouchiroud et al. 2013; Fang et al. 2014, 2016). Our data that skeletal muscles of old mice accumulated higher levels of cleaved PARP-1 and pADPr suggest that PARP-1 activity was increased during aging, although its protein level was not uniformly upregulated. Taken together, we demonstrate that aged muscles had suffered from a NAD+ deficit, which might be attributed to the upregulation of two enzymes that consumes NAD+, namely CD38, and PARP-1. Despite the upregulation of SIRT protein levels during aging, the declination of NAD+ as the acetyl acceptor, together with the inhibition of SIRT enzyme activity by NAM resulting from NAD+ degradation, could greatly hinder its deacetylation activity and cause hyperacetylation of key proteins, enzymes, and TFs at old age (Fig. 7).
Fig. 7.
Schematic overview of the aging impacts on various cellular pathways that control NAD+ homeostasis and protein acetylation status. Aging decreases NAD+ pool partly due to the upregulation of CD38 and PARP-1, which compete with SIRTs for NAD+ as substrate. Deficit of NAD+ pool reduces the deacetylation capacity of SIRTs, resulting in hyperacetylation of the enzymes and transcription factor/cofactor under its control. This enhanced acetylation of the key proteins may contribute to the age-related mitochondrial dysfunction and oxidative damage in muscle
Mitochondrial dysfunction resulting in oxidative stress and chronic inflammation are hallmarks of aging (Sack and Finkel 2012). As expected, aged muscles suffered from decreased mitochondrial functions marked by lowered CS activity and COXIV content (Fig. 5b, c). Because older muscles displayed increased acetylation of PGC-1α in the present study (Fig. 1c), the inactivation of PGC-1α leading to decreased mitochondrial biogenesis could be a possible explanation for mitochondrial deterioration in the aged muscle (Handschin 2009). Acetylation, as well as malonylation and succinylation, is known to cause inactivation of other key mitochondrial enzymes such as isocitrate dehydrogenase, succinate dehydrogenase, uncoupling proteins (UCP), SOD2, and enzymes involved in fatty acid metabolism, whereas SIRT3, the mitochondrial sirtuin, plays a critical role in deacetylating and activating the mentioned enzymes and PGC-1α (Camacho-Pereira et al. 2016). Besides the SIRT-PGC-1α axis, other pathways such as SIRT6-GCN5 and AMPK may also contribute to ameliorating PGC-1α activity in aged muscle. GCN5 can directly acetylate and inhibit PGC-1α, whereas SIRT6 attenuates the inhibition (Dominy et al. 2012). However, SIRT6 has relatively low deacetylation capability compared with SIRT1 and SIRT3, and was unlikely a major factor in regulating PGC-1α acetylation through GCN5 (Kugel and Mostoslavsky 2014). Another promoter of mitochondrial dysfunction in aging muscle is inflammation driven primarily by NFκB activation (Salminen and Kaarniranta 2009). Increased acetylated form of p65 and upregulation of COX2 reported in this study suggest that NFκB in aged muscle was activated, which could be yet another important consequence of declined NAD+ availability and subsequent hyperacetylation in aged muscles (Zhu et al. 2011). In fact, pro-inflammatory cytokines, especially TNFα and IL-6, are potent activators of CD38, which further reduces NAD+ pool and constitutes a vicious cycle (Chini et al. 2017).
Although our study was focused on skeletal muscle, several characteristics of aging cardiac muscle are noteworthy. In general, myocardium shares a prominent hyperacetylation profile with skeletal muscles, but excepted for SIRT3 in the mitochondria, other SIRT isoforms were either unchanged or downregulated with aging, whereas GCN5 was upregulated (Fig. 2). Thus, increased acetylation driven by GCN5 could have exacerbated observed hyperacetylation in the heart. Noticeably, PGC-1α level was increased during aging in myocardia, which might help resist age-related decline of mitochondrial COXIV (Fig. 5c) and SOD2 (Fig. 6b) expression. However, as in skeletal muscle, aging myocardia also suffered from decreased NAD+ level due to increased CD38 and PARP-1 levels.
In summary, our data indicate that aging can enhance protein acetylation in skeletal and cardiac muscles despite elevated SIRT expression. The imbalance between acetylation and deacetylation may be caused by diminished NAD+ pool, which is consumed by age-related upregulation of CD38 and PARP-1 in competition for NAD+. Hyperacetylation of key enzymes and transcription factors may play an important role in muscle aging.
Materials and methods
Animals and experimental design
Female C57BL/6 J mice at the age of 6 months (young, Y; N = 8), 12 months (middle, M; N = 8), and 24 months (old, O; N = 8) were housed individually in the University of Minnesota Twin Cities (UMTC) animal facilities in 22 °C rooms on a reverse 12-h light/dark cycle and fed a chow diet and tap water ad libitum. The animal user protocols were approved by the UMTC Research Animal Resource Center.
Muscle harvest and sample preparation
Whole G, Q, and H muscles were collected immediately after the mice were euthanized by pentobarbital (80 mg/kg) injection. The isolated muscles were immediately frozen in liquid nitrogen and the frozen muscles were homogenized with a bullet blender (Next Advance, Troy, NY, USA) for 3 min in ice-cold IP-Lysis or RIPA buffer (Thermo Scientific, Waltham, MA, USA) or mitochondrial isolation reagents (mitochondrial isolation kit for tissue, Thermo Scientific) or NE-PER™ nuclear and cytoplasmic extraction reagents (Thermo Scientific) in the presence of protease and phosphatase inhibitor cocktails (Thermo Scientific). All homogenized muscle samples were then stored at − 80 °C until analysis.
Immunoprecipitation and Western blot analysis
Protein lysates were pre-cleared with protein A beads for 30 min before immunoprecipitation with lysine-acetylation antibody (Santa Cruz Biotechnology, Dallas, TX, USA) for 4 h. After the incubation, the beads were pelleted by centrifugation at 8000×g for 30 s and washed with ice-cold buffer 3 times then the pellets were eluted to 2× sample buffer, vortexed, and boiled at 100 °C for 5 min. Thereafter, the mixtures were centrifuged at 8000×g for 30 s, and supernatant was stored at − 80 °C until western blotting.
Both whole lysate protein samples and lysine-acetylated precipitation samples were separated by SDS-PAGE, transferred to a PVDF membrane (GE Healthcare, Little Chalfont, UK), and blocked with 5% of skim milk prior to primary antibody incubation. The following antibodies were used for primary antibody incubation: Anti-SIRT1 (ab110304), SIRT6 (ab62739), catalase (ab16731), GPx1 (ab22604), COX2 (ab15191), and 4-HNE (ab46546) purchased from Abcam (Cambridge, UK); anti-SIRT3 (sc-99143), SIRT5 (sc-271635), GCN5 (sc-365321), SOD2 (sc-30080), Ac-lysine (sc-32268), NAMPT (sc-166946), CD38 (sc-374650), and pADPr (sc-56198) purchased from Santa Cruz Biotechnology; anti-p65 (#8242), PARP-1 (#9532), and GAPDH (#5174) purchased from Cell Signaling Technology (Danvers, MA, USA); anti-PGC-1a (ST1201) purchased from Calbiochem (Billerica, MA, USA); anti COX IV (A21348) purchased from Invitrogen (Carsbad, CA, USA). Following secondary antibody incubation, the membranes were developed by using Pierce™ ECL Western Blotting Substrate (Thermo Scientific) to expose Pierce CL-Xposure™ Film (Thermo Scientific) and quantified using ImageJ software (version 1.50i, National Institutes of Health, USA).
NAD+/NADH, ELISA, and enzyme activity assay
Total NAD+/NADH and NAD+ levels were measured according to manufacturer’s instruction (ab176723, NAD+/NADH Assay kit, Abcam). Briefly, 25 μl of NAD/NADH control buffer or NADH extraction buffer was added to whole, nuclear and cytoplasmic lysate sample, respectively, and then incubated at 37 °C for 15 min. 75 μl of NAD/NADH reaction mixture was added into each samples and incubated at RT for 1 h in the darkroom. Fluorescence signals were measured on a Synergy H1 microplate reader (BioTek, Winooski, VT, USA) at Ex/Em 540/590. ELISA assay was performed to quantify TNFα protein levels, following the manufacturer’s description (BD Bioscience, Franklin Lakes, NJ, USA). CS and SOD2 activity were measured in mitochondria using a kit according to the manufacturer’s protocol (Sigma, St. Louis, MO and Cayman, Ann Arbor MI, USA, respectively).
Statistical analysis
Experimental data were expressed as means ± SE, and group comparisons were made by one-way ANOVA. The Tukey-Kramer method was used as a post hoc test when ANOVA reached significance (P < 0.05).
Electronic supplementary material
(DOCX 203 kb)
Funding information
This research was supported by a grant from the Office of Vice President for Research at the University of Minnesota, Grant-in-Aid, 146825.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Badreh F, Joukar S, Badavi M, Rashno M, Dehesh T. The effects of age and fasting models on blood pressure, insulin/glucose profile, and expression of longevity proteins in male rats. Rejuvenation Res. 2019. 10.1089/rej.2019.2205. [DOI] [PubMed]
- Bai P, Cantó C, Oudart H, et al. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metab. 2011. 10.1016/j.cmet.2011.03.004. [DOI] [PMC free article] [PubMed]
- Camacho-Pereira J, Tarragó MG, Chini CS, et al. CD38 dictates age-related NAD decline and mitochondrial dysfunction through and SIRT3-dependent mechanism. Cell Metab. 2016. 10.1016/j.cmet.2016.05.006. [DOI] [PMC free article] [PubMed]
- Cantó C, Menzies KJ, Auwerx J. NAD+ metabolism and the control of energy homeostasis: a balancing act between mitochondria and the nucleus. Cell Metab. 2015. 10.1016/j.cmet.2015.05.023. [DOI] [PMC free article] [PubMed]
- Chaitanya GV, Alexander JS, Babu PP. PARP-1 cleavage fragments: signatures of cell-death proteases in neurodegeneration. Cell Commun Signal. 2010. 10.1186/1478-811X-8-31. [DOI] [PMC free article] [PubMed]
- Chini EN. CD38 as a regulator of cellular NAD: a novel potential pharmacological target for metabolic conditions. Curr Pharm Des. 2009. 10.2174/138161209787185788. [DOI] [PMC free article] [PubMed]
- Chini EN, Chini CS, Netto JME, et al. The pharmacology of CD38/NADase: an emerging target for cancer and aging diseases. Trends Pharmacol Sci. 2018. 10.1016/j.tips.2018.02.001. [DOI] [PMC free article] [PubMed]
- Chini CS, Hogan KA, Warner GM, et al. The NADase CD38 is induced by factors secreted from senescent cells providing a potential link between senescence and age-related cellular NAD (+) decline. Biochem Biophys Res Commun. 2019. 10.1016/j.bbrc.2019.03.199. [DOI] [PMC free article] [PubMed]
- Chini CS, Tarragó MG, Chini EN. NAD and the aging process: Role in life, death and everything in between. Mol Cell Endocrinol. 2017. 10.1016/j.mce.2016.11.003. [DOI] [PMC free article] [PubMed]
- Dominy JE, Lee Y, Jedrychowski MP, et al. The deacetylase Sirt6 activates the acetyltransferase GCN5 and suppresses hepatic gluconeogenesis. Mol Cell. 2012. 10.1016/j.molcel.2012.09.030. [DOI] [PMC free article] [PubMed]
- Fang EF, Scheibye-Knudsen M, Brace LE, et al. Defective mitophagy in XPA via PARP-1 hyperactivation and NAD+/SIRT1 reduction. Cell. 2014. 10.1016/j.cell.2014.03.026. [DOI] [PMC free article] [PubMed]
- Fang EF, Kassahun H, Croteau DL, et al. NAD+ replenishment improves lifespan and healthspan in ataxia telangiectasia models via mitophagy and DNA repair. Cell Metab. 2016. 10.1016/j.cmet.2016.09.004. [DOI] [PMC free article] [PubMed]
- Fernandez-Marcos PJ, Auwerx J. Regulation of PGC-1α, a nodal regulator of mitochondrial biogenesis. Am J Clin Nutr. 2011. 10.3945/ajcn.110.001917. [DOI] [PMC free article] [PubMed]
- Frederick DW, Loro E, Liu L, et al. Loss of NAD homeostasis leads to progressive and reversible degeneration of skeletal muscle. Cell Metab. 2016. 10.1016/j.cmet.2016.07.005. [DOI] [PMC free article] [PubMed]
- Gómez LA, Hagen TM. Age-related decline in mitochondrial bioenergetics: does supercomplex destabilization determine lower oxidative capacity and higher superoxide production? Semin Cell Dev Biol doi. 2012. 10.1016/j.semcdb.2012.04.002. [DOI] [PMC free article] [PubMed]
- Guan KL, Xiong Y. Regulation of intermediary metabolism by protein acetylation. Trends Biochem Sci. 2011. 10.1016/j.tibs.2010.09.003. [DOI] [PMC free article] [PubMed]
- Haigis MC, Guarente LP. Mammalian sirtuins - emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006. 10.1101/gad.1467506. [DOI] [PubMed]
- Handschin C. The biology of PGC-1α and its therapeutic potential. Trends Pharmacol Sci. 2009. 10.1016/j.tips.2009.03.006. [DOI] [PubMed]
- Hepple RT. Mitochondrial involvement and impact in aging skeletal muscle. Front Aging Neurosci. 2014. 10.3389/fnagi.2014.00211. [DOI] [PMC free article] [PubMed]
- Imai SI, Guarente L. NAD+ and sirtuins in aging and disease. Trends Cell Biol. 2014. 10.1016/j.tcb.2014.04.002. [DOI] [PMC free article] [PubMed]
- Johnson S, Imai S. NAD+ biosynthesis, aging, and disease. F1000Research. 2018. 10.12688/f1000research.12120.1. [DOI] [PMC free article] [PubMed]
- Kabiljo J, Murko C, Pusch O, Zupkovitz G. Spatio-temporal expression profile of sirtuins during aging of the annual fish Nothobranchius furzeri. Gene Expr Patterns. 2019. 10.1016/j.gep.2019.05.001. [DOI] [PubMed]
- Kang C, Chung E, Diffee G, Ji LL. Exercise training attenuates aging-associated mitochondrial dysfunction in rat skeletal muscle: role of PGC-1α. Exp Gerontol. 2013. 10.1016/j.exger.2013.08.004. [DOI] [PubMed]
- Kang C, Goodman CA, Hornberger TA, Ji LL. PGC‐1α overexpression by transfection attenuates mitochondrial deterioration of skeletal muscle caused by immobilization. FASEB J. 2015. 10.1096/fj.14-266619. [DOI] [PMC free article] [PubMed]
- Kawahara TLA, Michishita E, Adler AS, et al. SIRT6 links histone H3 lysine 9 deacetylation to NF-κB-dependent gene expression and organismal life span. Cell. 2009. 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed]
- Khanh VC, Zulkifli AF, Tokunaga C, et al. Aging impairs beige adipocyte differentiation of mesenchymal stem cells via the reduced expression of Sirtuin 1. Biochem Biophys Res Commun. 2018. 10.1016/j.bbrc.2018.04.136. [DOI] [PubMed]
- Kilic U, Gok O, Erenberk U, et al. A remarkable age-related increase in SIRT1 protein expression against oxidative stress in elderly: SIRT1 gene variants and longevity in human. PLoS One. 2015. 10.1371/journal.pone.0117954. [DOI] [PMC free article] [PubMed]
- Kim SC, Sprung R, Chen Y, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006. 10.1016/j.molcel.2006.06.026. [DOI] [PubMed]
- Koltai E, Szabo Z, Atalay M, et al. Exercise alters SIRT1, SIRT6 NAD and NAMPT levels in skeletal muscle of aged rats. Mech Ageing Dev. 2010. 10.1016/j.mad.2009.11.002. [DOI] [PMC free article] [PubMed]
- Kolthur-Seetharam U, Dantzer F, McBurney MW, et al. Control of AIF-mediated cell death by the functional interplay of SIRT1 and PARP-1 in response to DNA damage. Cell Cycle. 2006. 10.4161/cc.5.8.2690. [DOI] [PubMed]
- Konopka AR, Suer MK, Wolff CA, Harber MP. Markers of human skeletal muscle mitochondrial biogenesis and quality control: effects of age and aerobic exercise training. J Gerontol A Biol Sci Med Sci. 2014. 10.1093/gerona/glt107. [DOI] [PMC free article] [PubMed]
- Kugel S, Mostoslavsky R. Chromatin and beyond: the multitasking roles for SIRT6. Trends Biochem Sci. 2014. 10.1016/j.tibs.2013.12.002. [DOI] [PMC free article] [PubMed]
- Lombard DB, Alt FW, Cheng H-L, et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007. 10.1128/mcb.01636-07. [DOI] [PMC free article] [PubMed]
- Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J. 2007. 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed]
- Michishita E. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell. 2005. 10.1091/mbc.e05-01-0033. [DOI] [PMC free article] [PubMed]
- Mills KF, Yoshida S, Stein LR, et al. Long-term administration of nicotinamide mononucleotide mitigates age-associated physiological decline in mice. Cell Metab. 2016. 10.1016/j.cmet.2016.09.013. [DOI] [PMC free article] [PubMed]
- Mitchell SJ, Bernier M, Aon MA, et al. Nicotinamide improves aspects of healthspan, but not lifespan, in mice. Cell Metab. 2018. 10.1016/j.cmet.2018.02.001. [DOI] [PMC free article] [PubMed]
- Mouchiroud L, Houtkooper RH, Moullan N, et al. The NAD+/sirtuin pathway modulates longevity through activation of mitochondrial UPR and FOXO signaling. Cell. 2013. 10.1016/j.cell.2013.06.016. [DOI] [PMC free article] [PubMed]
- Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1α. J Biol Chem. 2005. 10.1074/jbc.M501485200. [DOI] [PubMed]
- Palikaras K, Tavernarakis N. Mitochondrial homeostasis: the interplay between mitophagy and mitochondrial biogenesis. Exp Gerontol doi. 2014. 10.1016/j.exger.2014.01.021. [DOI] [PubMed]
- Picca A, Calvani R, Bossola M, et al. Update on mitochondria and muscle aging: all wrong roads lead to sarcopenia. Biol Chem. 2018. 10.1515/hsz-2017-0331. [DOI] [PubMed]
- Sack MN, Finkel T. Mitochondrial metabolism, sirtuins, and aging. Cold Spring Harb Perspect Biol. 2012. 10.1101/cshperspect.a013102. [DOI] [PMC free article] [PubMed]
- Sakellariou GK, Pearson T, Lightfoot AP, Nye GA, Wells N, Giakoumaki II, Vasilaki A, Griffiths RD, Jackson MJ, McArdle A. Mitochondrial ROS regulate oxidative damage and mitophagy but not age-related muscle fiber atrophy. Sci Rep. 2016;6:1–15. doi: 10.1038/srep33944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen A, Kaarniranta K. NF-κB signaling in the aging process. J Clin Immunol. 2009. 10.1007/s10875-009-9296-6. [DOI] [PubMed]
- Sanchez AMJ, Candau RB, Bernardi H. FoxO transcription factors: their roles in the maintenance of skeletal muscle homeostasis. Cell Mol Life Sci. 2014. 10.1007/s00018-013-1513-z. [DOI] [PMC free article] [PubMed]
- Sasaki T, Maier B, Bartke A, Scrable H. Progressive loss of SIRT1 with cell cycle withdrawal. Aging Cell. 2006. 10.1111/j.1474-9726.2006.00235.x. [DOI] [PubMed]
- Schöndorf DC, Ivanyuk D, Baden P, et al. The NAD+ precursor nicotinamide riboside rescues mitochondrial defects and neuronal loss in iPSC and fly models of Parkinson’s disease. Cell Rep. 2018. 10.1016/j.celrep.2018.05.009. [DOI] [PubMed]
- Schultz MB, Sinclair DA. Why NAD+ declines during aging: it’s destroyed. Cell Metab. 2017. 10.1016/j.cmet.2016.05.022. [DOI] [PMC free article] [PubMed]
- Tao R, Coleman MC, Pennington JD, et al. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol Cell. 2010. 10.1016/j.molcel.2010.12.013. [DOI] [PMC free article] [PubMed]
- Tarragó MG, Chini CS, Kanamori KS, et al. A potent and specific CD38 inhibitor ameliorates age-related metabolic dysfunction by reversing tissue NAD+ decline. Cell Metab. 2018. 10.1016/j.cmet.2018.03.016. [DOI] [PMC free article] [PubMed]
- Trammell SAJ, Weidemann BJ, Chadda A, et al. Nicotinamide riboside opposes type 2 diabetes and neuropathy in mice. Sci Rep. 2016. 10.1038/srep26933. [DOI] [PMC free article] [PubMed]
- Verdin E. NAD + in aging, metabolism, and neurodegeneration. Science. 2015. 10.1126/science.aac4854. [DOI] [PubMed]
- Wagner GR, Payne RM. Mitochondrial acetylation and diseases of aging. J Aging Res. 2011. 10.4061/2011/234875. [DOI] [PMC free article] [PubMed]
- Webb AE, Brunet A. FOXO transcription factors: key regulators of cellular quality control. Trends Biochem Sci. 2014. 10.1016/j.tibs.2014.02.003. [DOI] [PMC free article] [PubMed]
- Yeo D, Kang C, Gomez-Cabrera MC, Vina J, Ji LL. Intensified mitophagy in skeletal muscle with aging is downregulated by PGC-1alpha overexpression in vivo. Free Radic Biol Med. 2019. 10.1016/j.freeradbiomed.2018.10.456. [DOI] [PubMed]
- Yeung F, Hoberg JE, Ramsey CS, et al. Modulation of NF-κB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004. 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed]
- Zhu X, Liu Q, Wang M, et al. Activation of Sirt1 by resveratrol inhibits TNF-α induced inflammation in fibroblasts. PLoS One. 2011. 10.1371/journal.pone.0027081. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 203 kb)