Abstract
Cellular senescence is a fundamental process that may play positive or detrimental roles for the organism. It is involved in tissue development and in tumor prevention although during aging is becoming a detrimental process contributing to the decline of tissue functions. In previous investigations, we have uncovered a better capacity to detect DNA damage in cells from long-lived mammals. Here, we report that cultured cells derived from long-lived species have a higher propensity to undergo senescence when challenged with DNA damage than cells derived from short-lived species. Using a panel of cells derived from six mammals, which range in lifespan from 3-4 years up to 120 years, we examined cell cycle response, induction of apoptosis and of cellular senescence. All species exhibited a cell cycle arrest while induction of apoptosis was variable. However, a significant positive correlation was found between the relative percent of cells, within a population which entered senescence following damage, and the lifespan of the species. We suggest that cellular senescence may have a positive role during development allowing it to contribute to the evolution of longevity.
Electronic supplementary material
The online version of this article (10.1007/s11357-019-00066-2) contains supplementary material, which is available to authorized users.
Keywords: Cellular senescence, Apoptosis, Longevity, Body mass, SA β-gal
Introduction
Evolutionary pressure on reproductive fitness is thought to indirectly dictate the lifespan of the species through the selection of traits that may be beneficial to survival but detrimental to longevity (Kirkwood and Rose 1991). Many cellular mechanisms have been proposed to contribute to the evolution of longevity; some of these mechanisms can be species or taxa specific, while others are common among species (Crawford et al. 2012; Croco et al. b; Tian et al. 2017). One fundamental cellular mechanism that has been implicated in the evolution of lifespan is DNA damage recognition and repair. We have previously shown that an enhanced capacity to recognize DNA double-strand breaks is positively correlated with longevity, which appears to be related to the recognition of DNA-ends by the Ku80-Ku70 heterodimer (Lorenzini et al. 2009), as well as a greater capacity to form DNA damage foci of the phosphorylated histone H2AX (γH2AX) and of 53BP1 (Fink et al. 2011; Croco et al. 2017a).
An improved capacity to detect DNA damage would be expected to facilitate repair. Indeed, we have observed lower levels of genomic damage in cultured cells derived from longer-lived species compared with short-lived species when the cultures were treated with similar amounts of genotoxic substances (Croco et al. 2017a). Additional mechanisms related to DNA damage may also contribute to enhanced longevity. For example, resistance to DNA damage could also be coupled with an enhanced capacity for the induction of apoptosis or cellular senescence. In fact, these cell responses bear functional similarities with DNA repair, facilitating the removal of damaged/dysfunctional cells to preserve the integrity of the cell population within a tissue. Apoptosis acts through the elimination of irreparably damaged cells, while cellular senescence blocks proliferation of damaged cells, permitting clearance by the immune system.
The aim of the present work is to examine the correlation between apoptosis, cellular senescence and species longevity, using primary fibroblast cultures of six mammalian species in which body mass is not correlated with lifespan, when challenged with DNA damage.
Adult body mass is a known confounding variable in any correlation with lifespan, as clearly explained by Speakman (2005), and correlation between damage responses and body mass was also considered.
Materials and methods
Cell cultures
Fibroblast strains from rat, mouse, little brown bat (LBB), dog, cow and human were established as previously described (Lorenzini et al. 2005). Cells were maintained in minimum essential medium (MEM) with Earle’s salts and L-glutamine containing 10% fetal bovine serum (FBS), minimum essential medium vitamins, amino acids, and penicillin-streptomycin (all from Sigma Aldrich, St. Louis, Mo, USA). Rat cells were from Sprague-Dawley rats. Mouse cells were from a first filial generation hybrid mice offspring of a cross between C57BL/6J females (B6) and 129S1/SvImJ males (129S). All strains displayed replicative senescence with the exception of LBB and were used for all experiments before two third of their expected replicative life span (see Lorenzini et al. 2005). LBB strains reached more than 160 population doublings (PD), indicating probable immortalization. Consequently, for this species, cultures were used for experiments between 6 and 11 PD, to exclude the possibility of using immortalized cultures. Fibroblasts were passaged weekly. For the experiments, cells from confluent monolayers were seeded at 10,000 cells/cm2.
Genotoxic treatments
To challenge the cultures, cells were treated with a combination of concentrations and exposure times of genotoxic drugs so to cause damage only to a fraction of the cell population. After the challenge, the cells were allowed to recover for different amounts of time in fresh media. This choice was dictated by the desire of studying cellular responses to mild and chronic insults as the one caused by the wear and tear of the aging process instead of studying the response to acute stressors. We used etoposide (ETO, an antineoplastic agent and well-known apoptosis inducer) at 100 μM for 30 min and neocarzinostatin (NCS, a chemotherapeutic antibiotic) at 0.13 μM for 2 h, both in serum-free media to avoid possible interaction with serum albumin. ETO and NCS were both from Sigma-Aldrich. After ETO treatment, the highest level of cell death (measured as percentage of necrotic plus apoptotic cells, see next paragraph) reached 38% at 48 h from treatment (Supplemental Fig. 1a). With NCS treatment, the highest percentage of cell death (measured by FCA Guava ViaCount assay) was around 30% at 120 h from the beginning of treatment (Supplemental method and Fig. 1b). See also the vitality results in our previously published work where we also used higher concentrations of ETO and NCS (Croco et al. 2017a).
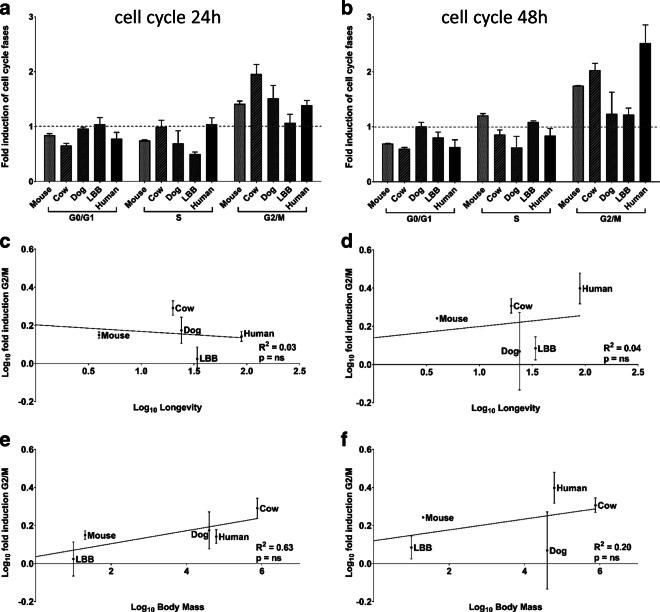
Fig. 1.
Cell cycle arrest following DNA damage: relationship with longevity and body mass. Fibroblast cultures were treated with 100 μM ETO for 30 min; data are expressed as fold induction in each cell cycle phase 24 h (a) and 48 h (b) following treatment relative to control cultures; species are ordered by increasing longevity. Fold induction specifically in the G2/M phase are plotted against maximum longevity (c for 24 h data; d for 48 h data) and against body mass (e for 24 h data; f for 48 h data). Linear regression trend lines, coefficient of determination, and significance of the regression are shown. The experiment was performed twice in duplicate
Cell cycle analysis by flow cytometry
Cell-cycle analysis was performed by flow cytometry (FCM, Guava easyCyte 5HT equipped with a class IIIb laser operating at 488 nm, Merck Millipore, Darmstadt, Germany) and analyzed with Guava Cell-Cycle software, 24 and 48 h after treatment with 100 μM ETO for 30 min. Cells were fixed and permeabilized with ice-cold 70% ethanol and washed with PBS (Sigma Aldrich). Fixed cells were then resuspended in Guava Cell-Cycle Reagent (Merck Millipore) containing the dye propidium iodide (PI) and each sample was incubated in dark at room temperature for 30 min. The PI is able to penetrate the membrane of permeabilized cells, bind covalently to DNA and emit red fluorescence. Cells in G2/M phase show double fluorescence compared to those in G0/G1 phase, while cells in S phase have intermediate fluorescence. The obtained results were calculated as the percentage of cells in different stages of the cell cycle in treated cultures relative to that present in the control cultures and expressed as fold induction.
Apoptosis quantitation by FCM
The percentage of viable, apoptotic and necrotic cells was assessed by FCM and analyzed with Guava Nexin software (Merck Millipore). After 24 and 48 h from treatment with 100 μM ETO for 30 min, Guava Nexin Reagent (Merck Millipore) was added to the cells, the reagent contains two dyes, 7-aminoactinomycin D (7-AAD) and Annexin-V-PE. 7-AAD allows the discrimination between live and dead cells, while Annexin-V-PE allows identification of apoptotic cells by binding to phosphatidylserine and emitting yellow fluorescence. In particular, live cells are 7-AAD and Annexin-V-PE negative, apoptotic cells are 7-AAD negative and Annexin-V-PE positive and necrotic cells are 7-AAD and Annexin-V-PE positive. Briefly, cells were incubated for 20 min at room temperature in the dark and samples analyzed. The obtained results were calculated as the percentage of cells in apoptosis in treated cultures relative to that present in the untreated cultures and expressed as fold induction.
Histochemical staining of senescence cells
After 24 h and 120 h from treatment with NCS 0.13 μM for 2 h, cells were fixed and stained for senescence-associated β-galactosidase (SA β-gal). Twenty-four-hour experiments were performed in AL laboratory at the University of Bologna, 120-h experiments were performed in CS laboratory at Drexel University College of Medicine with different cell strains. Cells staining was performed using the Senescence Cells Histochemical Staining Kit (Sigma Aldrich). The Kit contains X-gal (5-bromo-4-chloro-3-indolyl-b-D-galactopyranoside) which in presence of β-galactoside produces galactile and an insoluble blue compound. Briefly, cells were fixed with fixation buffer for 6–7 min at room temperature, washed with PBS, and incubated with staining mixture at 37 °C without CO2 for 24 h. The percentage of senescent cells was evaluated by microscopy at 200× magnification using Olympus IX50 fluorescence microscope (Tokyo, Japan). A total number of 400 cells were counted for each sample. The obtained results were calculated as the percentage of stained cells in treated cultures relative to that present in the control cultures and expressed as fold induction.
p21 quantitation by FCM
The mean fluorescence intensity value of p21 protein was analyzed in mouse and human strains, after 24 h and 48 h from treatment with NCS 0.13 μM for 2 h, by FCM with Guava Incyte software (Merck Millipore). Cells were fixed in PBS plus formaldehyde 4% and permeabilized in 90% cold methanol. Cells were then incubated with Anti-p21 antibody (#AHZ0422, Thermo Fisher, Waltham, MA, USA), washed, incubated with AlexaFluor488-conjugated goat anti-mouse secondary antibody (#4408S; Cell Signaling, Danvers, MA, USA), and then analyzed. The obtained results were expressed as fold induction: mean fluorescence intensity values of cells in treated cultures relative to control culture values.
p21 mRNA quantitation by qRT-PCR
Total RNA extraction was performed 72 h after NCS 0.13 μM/2-h treatment using TRIzol reagent (Thermo Fischer) and according to the manufacturer’s instructions. RNA samples were purified by RNeasy Mini kit (Qiagen, Hilden, Germany). Two hundred nanograms of total RNA was used for qRT-PCR amplification of p21. p21-specific primers for all the species were designed by using NCBI Primer-BLAST software. The primer sequences are provided as supplemental information (S2). The expression of housekeeping gene GAPDH was used as an internal control to normalize the variability in the expression levels. qRT-PCR was performed by using Verso SYBR Green 1-Step qRT-PCR Low ROX Mix kit (Thermo Fisher). The run was set according to the manufacturer’s instructions: cDNA synthesis (50 °C for 15 min, 1 cycle), Thermo-Start activation (95 °C for 15 min, 1 cycle), denaturation (95 °C for 15 s), annealing (60 °C for 30 s), and extension (72 °C for 30 s) (40 cycles). Reactions were run in triplicate. Expression data were normalized to the geometric mean of housekeeping gene GAPDH to control the variability in expression levels.
p21 visualization by immunofluorescence
Mouse and human fibroblasts were seeded onto glass coverslips and treated with NCS 0.13 μM for 2 h. Cells were fixed at 24 and 48 h after damage in 4% paraformaldehyde for 10 min and permeabilized in 0.2% Triton X-100 in phosphate-buffered saline (PBS) for 5 min. Cells were washed once in PBS and blocked for 30 min in 4% bovine serum albumin (BSA) in PBS containing Tween-20 (PBST), after which they were incubated with the p21 primary antibody (#AHZ0422, Thermo Fisher) in 1% BSA–PBST buffer for 2 h at room temperature in a humidified chamber. Slides were washed three times in PBST and incubated with secondary antibody (#4408S; Cell Signaling) in 0.1% BSA–PBST for 1 h. Cells were washed three times, stained with 4′,6-diamidine-2′-phenylindole dihydrochloride (DAPI, Sigma-Aldrich), and mounted with Vectashield mounting medium (Vector Laboratories, Burlingame, CA, USA) before analysis. Images were captured using Olympus IX50 fluorescence microscope equipped with a Canon camera G16 (Tokyo, Japan).
Statistical analysis
All results are expressed as mean ± standard error of the mean (SEM).
For the analysis of the relation between cell cycle or apoptosis with longevity and body mass, we used linear regression. For the analysis of the relations between senescence with longevity, body mass, sexual maturity, and post-natal growth rate, we used linear regression, correlation and partial correlation. To compare apoptosis induction among species, we used the one-way analysis of variance (ANOVA) for unpaired data, followed by Tukey test for multiple post comparison. For p21 analysis between mouse and human, we used unpaired t test.
All the statistical analyses were performed using Prism Software 7, with the exception of partial correlations that were performed using the VassarStats website.
Species information
Species data are all from AnAge database (Tacutu et al. 2018). Maximum longevity in the text is referred simply as longevity. Body mass is taken from weight value in AnAge. For sexual maturity, we used the average between male and female data. Growth rate in AnAge is calculated by fitting empirical data taken from published growth curves to sigmoidal growth functions (Table 1).
Table 1.
Traits of analyzed species
| Species | Common name | Sample size* | Maximum longevity (years) | Body mass (g) | Sexual maturity (days) | Postnatal growth rate (days−1) |
|---|---|---|---|---|---|---|
| Rattus norvegicus | Rat | Large | 3.8 | 300 | 80 | 0.0207** |
| Mus musculus | House Mouse | Large | 4.0 | 20.5 | 42 | 0.0298 |
| Bos taurus | Domestic cattle; cow | Large | 20 | 750,000 | 548 | 0.0031 |
| Canis familiaris | Dog (Beagle) | Large | 24 | 40,000 | 510 | 0.0244 |
| Myotis lucifugus | Little brown bat (LBB) | Medium | 34 | 10 | 210 | 0.116 |
| Homo sapiens | Human | Huge | 90*** | 62,000 | 4927 | 0.0005 |
Values are means (except for maximum longevity) obtained from the AnAge database, available at http://genomics.senescence.info/species/ (Tacutu et al. 2018).
*Medium, 100–1000; large, over 1000; huge, to indicate the special status of the human data.
**For the growth rate of rat (rattus norvegicus), value absent in AnAge was used, instead, the value from rattus rattus.
***Maximum human longevity is 122.5 years. In our analysis, we consider a maximum longevity of 90 years to account for the fact that, for the others species, only small cohorts were used to determine maximum life span, and 90 years seem a more realistic estimate for a random sample of humans
Table 1 Legend: Values are means (except for maximum longevity) obtained from the AnAge database, available at: http://genomics.senescence.info/species/ (Tacutu et al. 2018). * vMedium, 100–1000; large, over 1000; huge, to indicate the special status of the human data. ** For the growth rate of rat (rattus norvegicus), value absent in AnAge was used, instead, the value fromRattus rattus. *** Maximum human longevity is 122.5 years. In our analysis, we consider a maximum longevity of 90 years to account for the fact that, for the others species, only small cohorts were used to determine maximum life span, and 90 years seem a more realistic estimate for a random sample of humans.
Results
Cell cycle analysis
We initially sought to determine if cells, from species which vary in their lifespan, have a similar capacity to control and arrest the cell cycle when faced with DNA damage. In order to ascertain this, cell cycle profiling was performed following damage with etoposide (ETO), a topoisomerase II inhibitor known to induce an S phase arrest. A short pulse of ETO was used to induce damage specific to S phase, allowing visualization of G2/M arrest as a readout of cell cycle arrest.
The percent of cells in G2/M was compared at different time points (24 h and 48 h) following damage in untreated cultures and treated cultures, Fig. 1a, b. Cultured cells derived from mouse, dog, cow, little brown bat and human were examined. In these comparisons, there was an increase in G2/M phase in all species, demonstrating that all are capable of cell cycle arrest in response to DNA damage.
An analysis of the relationship of the magnitude of the cell cycle arrest, as determined by the percent of the population in G2/M following this response, with longevity gives a negative result (Fig. 1c, r2 = 0.03, p = n.s.; Fig. 1d, r2 = 0.04, p = n.s.). Although not significant, a relationship was observed between cell cycle arrest and adult body mass (Fig. 1er2 = 0.63, p = n.s.; Fig. 1fr2 = 0.20, p = n.s.).
Induction of apoptosis
Next, we sought to determine if longevity correlates with the intrinsic capacity to induce apoptosis. Using ETO as a damaging agent, the fold induction of apoptosis was determined for each species. A subset of species showed little response to the concentrations of ETO used (mouse and dog) while among others (rat, cow, LBB, and human), the response was evident. The fold induction levels for apoptosis in cultures after 24 h and 48 h following DNA damage are reported in Fig. 2a, b. Regression analysis of the results revealed no meaningful relationship between fold induction of apoptosis and either longevity or body mass (Fig. 2c–f). We also investigated the possibility that a relationship may exist between longevity or body mass and the total number of dying cells (apoptotic + necrotic cells). Even when these parameters were combined, no relationship was detected between these data and either longevity or body mass (data not shown).
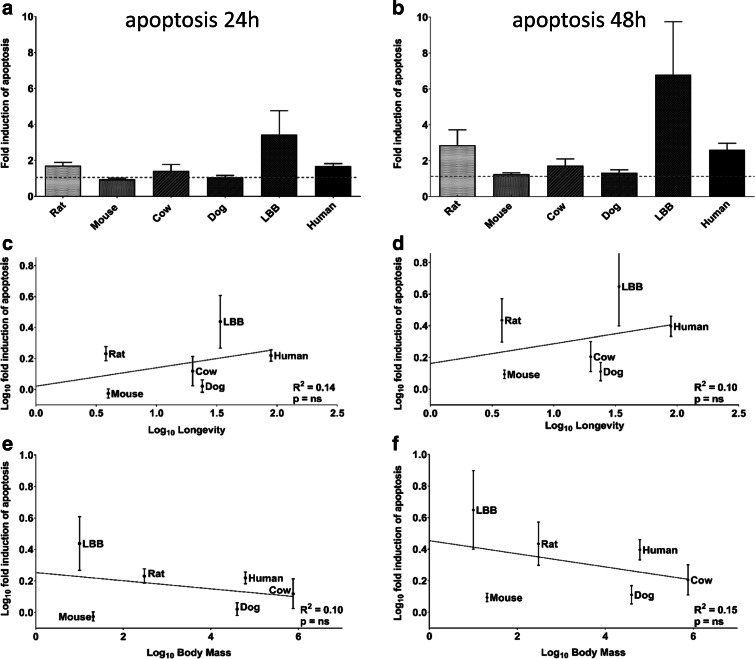
Fig. 2.
Induction of apoptosis relative to longevity and body mass. Fibroblast cultures were treated with 100 μM ETO for 30 min; the percentage of cells in apoptosis after 24 h and 48 h following treatment is reported relative to the percentage in untreated cultures (a, b, respectively); species are ordered by increasing longevity. The fold induction of apoptosis is plotted against longevity (c for 24 h and d for 48 h) and against body mass (e for 24 h and f for 48 h). Linear regression trend lines, coefficient of determination and significance of the regression are shown. Experiments were performed three times in duplicate
Apoptosis is a fundamental cellular process and the results may suggest that apoptosis represents a species-specific or taxa-specific longevity-assurance strategy. For example, Zhang et al. have reported positive selection for p53, a key mediator of apoptosis, in a species of vesper bat, Myotis davidii (Zhang et al. 2013). Our data is consistent with the concept that p53-mediated apoptosis is a longevity-assurance mechanism for the LBB (an extremely long-lived species for its size) since only this species shows a strong response. This interpretation, however, is weakened by the high variation of this species response, and an ANOVA analysis failed to show a significant difference among the species in terms of apoptotic response. We also analyzed LBB cells at high PD (between 80 and 90), and although these data were not used in our comparative analyses, the response was intense and similar to the response of LBB at low PD.
Induction of stress-induced senescence
In order to assess the relative capacity to induce senescence following genotoxic damage in each species, we used an antibiotic that has effects on DNA which are biologically similar to the effects of ionizing radiation: neocarzinostatin (NCS) (Goldberg 1987) followed by an evaluation of the percentage of cells positive for SA-β-gal.
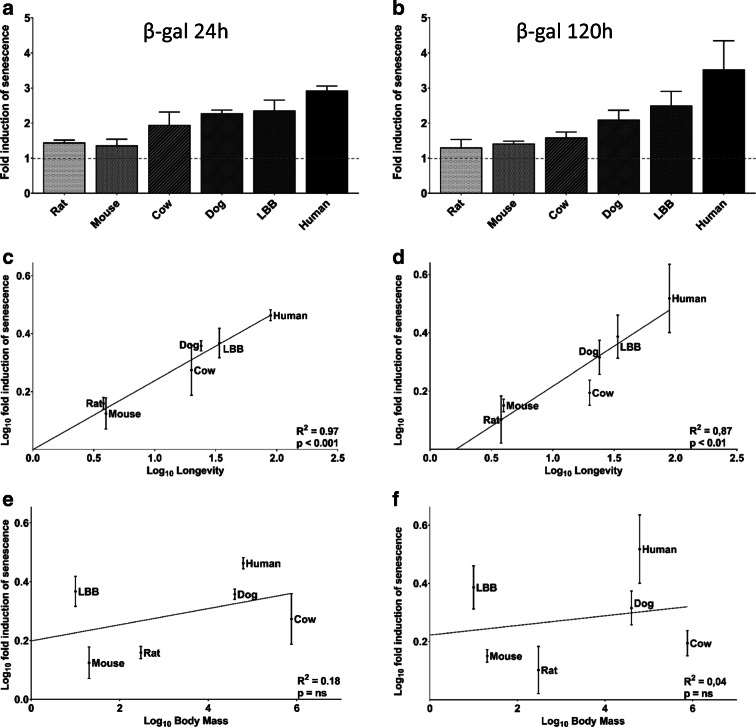
All species responded to the genotoxic stressor with an increase in the percentage of SA β-gal-stained cells. The increase varied from a minimum of 1.31-fold for mouse cultures at 120 h to a maximum of 3.52-fold for human cultures also at 120 h. Regression analysis demonstrated a positive, strong and highly significant relationship with longevity both at 24 h and 120 h (Fig. 3c, r2 = 0.97, p < 0.001; Fig. 3d, r2 = 0.87, p < 0.01). No significant relationship was found with body mass at either time point (Fig. 3e, r2 = 0.18, p = n.s.; Fig. 3f, r2 = 0.04, p = n.s.). The lack of relationship with body mass is important as it is well known that body mass and longevity are related. For example, De Magalhães and colleagues, analyzed data from 856 mammals and reported that body mass explained 66% of the longevity variation (De Magalhães et al. 2007). However, body mass is not significantly related to lifespan among the six species we used in this analysis (Fig. 3d). To further explore the possible role of body mass in the relationship between senescence and longevity, we performed correlation and partial correlation analysis (Table 2). This exploration confirms that longevity is the parameter primarily related to the ability to induce cellular senescence.
Fig. 3.
Induction of senescence and its relationship with longevity and body mass. The percentage of SA β-gal stained cells relative to the percentage in untreated cultures, 24 h and 120 h following treatment with NCS (0.13 μM for 2 h) is reported in a and b, respectively; species are ordered for increasing longevity. The fold induction of SA β-gal stained cells was plotted against longevity (c for 24 h and d for 120 h) and against body mass (e for 24 h and f for 120 h). Linear regression trend lines, coefficient of determination, and significance of the regression are shown. Twenty-four-hour experiments were performed at least three times in duplicates, 120-h experiments were performed twice in duplicates
Table 2.
Correlations and partial correlations
| Correlation 24 h | r of correlation | Controlling for | r of partial correlation |
| Senescence vs longevity | 0,983 | Body mass | 0,98 |
| Senescence vs body mass | 0,425 | Longevity | − 0.20 |
| Longevity vs body mass | 0,466 | Senescence | 0.29 |
| Correlation 120 h | r of correlation | controlling for | r of partial correlation |
| Senescence vs longevity | 0,938 | Body mass | 0,97 |
| Senescence vs body mass | 0,212 | Longevity | − 0,73 |
| Longevity vs body mass | 0,466 | Senescence | 0,79 |
Table 2 legend: Senescence is presented as fold induction of DNA damage stimulated senescence (data used are plotted in Fig. 3).
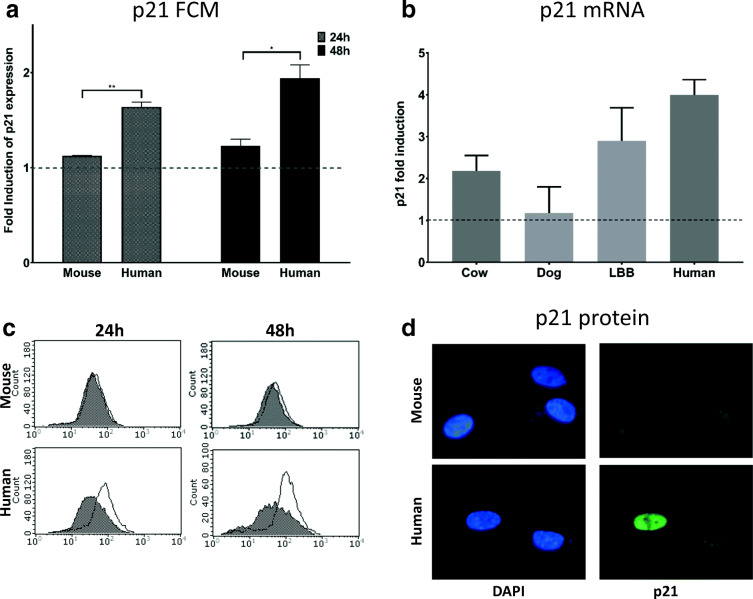
As an additional proof for our finding, we used p21 as a further marker for cellular senescence. In the presence of unresolved DNA damage, the ATM kinase activates p53 and its transcriptional target p21, which arrests proliferation by inhibiting cell-cycle-dependent kinases [reviewed in (Bitto et al. 2014)]. Stimuli that elicit a DNA damage response induce senescence primarily through the p53-p21 pathway (Campisi and d’Adda di Fagagna 2007). p21, additionally, seems essential for developmentally programmed senescence (Storer et al. 2013). We assayed p21 protein expression in human and mouse cells using an antibody that recognized an epitope conserved among these species. Direct comparisons with other species were not possible since few antibodies recognize an epitope conserved across multiple species. The results of this analysis are presented in Fig. 4c and quantified in Fig. 4a. Human cells increased p21 expression to a significantly greater extent than mouse cells at both 24 h and 48 h following DNA damage. Additionally, to compensate for the lack of a single antibody able to recognize p21 in all species, we performed quantitative real-time PCR (qRT-PCR) to measure the relative abundance of p21 mRNA. Data presented in Fig. 4b show the relative abundance of p21 mRNA in species for which we were able to design primers that gave reliable results. We observed the same trend for the steady-state p21 mRNA levels as we did with SA β-gal, i.e., greater induction of senescence in long-lived species.
Fig. 4.
Expression of p21 following DNA damage. FCM determination of p21 expression in mouse and human cells 24 h and 48 h from treatment with NCS (0.13 μM for 2 h) or untreated is shown as fold induction relative to untreated cells. In c representative histogram plots are presented. In b qRT-PCR values for p21 expression among different species normalized to GADPH are presented. In d, pictures of treated human and mouse cells after immunofluorescence staining with the p21 antibody are presented. FCM and immunofluorescence experiments were performed twice in duplicates. qRT-PCR experiment was performed once in triplicates
A positive relationship between longevity and slow development is well supported by large comparative analyses (De Magalhães et al. 2007; Ricklefs 2010). Consequently, we decided to use available data on two developmental characteristic of species to see if a regression analysis will show a relationship between development and the capacity to induce senescence. Supplemental Fig. 2a, b shows that we found a significant positive relationship between the capacity to induce cellular senescence and time to sexual maturity. For growth rate, we observed a negative trend although the relationship is not significant (Supplemental Fig. 2c, d).
Discussion
The evolution of large body mass requires the adaptation of cellular mechanisms to allow the high number of cell divisions required to maintain the larger body mass (reviewed in Croco et al. 2017a, b). Accordingly, species with higher body mass tend to have a more accurate erythropoiesis (Croco et al. 2016). Similarly, the evolution of greater longevity should also be accompanied by an adaptation of cellular functions that support the longer period of functionality associated with enhanced longevity. For example, it was observed that cells from long-lived species retain higher viability when challenged with different stressors (Kapahi et al. 1999). In this study, we sought to determine if the evolution of longevity was accompanied by an enhanced regulation of cell cycle progression, apoptosis, or senescence using equivalent levels of damage as inducers.
Longevity vs inhibition of growth and induction of apoptosis
Our analysis reveals no evident relationship between the capacity to arrest the cell cycle or to induce apoptosis with either species longevity or body mass.
There are several potential explanations for these negative results. Cell cycle regulation and apoptosis are highly conserved cellular mechanisms present in all metazoan, important for maintaining a homeostatic balance within tissues during development and in adulthood (Crawford et al. 2012; Meier et al. 2000). Consequently, all mammalian species are equipped with effective cell cycle regulation and apoptotic machineries and it may be that these processes are too fundamental to allow significant variation beyond that required for evolutionary fitness.
An alternative hypothesis is that enhanced cell cycle control and/or apoptosis may be associated with longevity in a subset of long-lived species having been associated with selective pressure for reproduction in a few instances. This hypothesis is supported by reports of a remarkably efficient apoptotic response in elephants (Abegglen et al. 2015; Vazquez et al. 2018).
There are several caveats to this study. The small number of species involved, and the fact that for cow, dog and the laboratory rodent strains used, we cannot ignore the potential confounding impact of human-based artificial selection to which they were exposed. Consequently, the possibility exists that our study is not sufficiently powered to detect a weak correlation with longevity. In this regard, it is noteworthy that there is a tendency for higher capacity to induce apoptosis in species with higher longevity (Fig. 2c, d).
Longevity is positively associated with stress induced senescence
Using the SA β-gal staining method, we detected a highly significant relationship between longevity and the capacity to induce senescence following DNA damage. The expression of the senescence marker p21 is also higher in longer lived species, reinforcing this observation. Although correlation does not imply causation, the relevance of this finding is underscored by the fact that SA β-gal staining is an established marker for cellular senescence (Dimri et al. 1995) and represents a reliable early sign of replicative senescence (Maier et al. 2007), while multiple studies have demonstrated that this marker is capable of detecting stress induced senescence (Debacq-Chainiaux et al. 2005; Dumont et al. 2000; Frippiat et al. 2001; Zhao et al. 2018). In addition, stress-induced senescence appears to be a more universal response than replicative senescence, which can be absent in both short-lived and long-lived species (Zhao et al. 2018). Furthermore, cells of the long lived rodent, the naked mole rat, favor senescence over apoptosis when compared to mouse cells (Zhao et al. 2018).
A possible role for stress induced senescence during development
Because cellular senescence is considered one of the hallmarks of the aging process (López-Otín et al. 2013) and data are available demonstrating an increase in senescent markers in individuals of increasing age (Lawrence et al. 2018; Waaijer et al. 2016), the positive relationship between the capacity to induce senescence and longevity that we have observed may appear counter intuitive.
Our hypothesis is that spontaneous DNA damage may be induced at any time including early development, the moment in life when tissues require exponential growth and when, consequently, DNA replication errors would be expected to be more frequent. The appearance of a damaged but surviving cell during development poses a great challenge if the cell is not repaired or eliminated by apoptosis. However, the rate of apoptosis may be limited by other evolutionary constraints. In this case, a compromised cell could retain the capacity to proliferate with the consequence that all descendants will carry the same inherent genomic damage. For this reason, efficiently inducing cellular senescence to avoid the accumulation of a sub-population of damaged cells, may represent a pro longevity assurance mechanism. Consistent with this notion, a physiological role for senescence during early development has been demonstrated in both humans and in mice (Chuprin et al. 2013; Storer et al. 2013).
Our previous studies have led us to consider developmental time as an important factor in the theory of aging (Lorenzini et al. 2011). It is possible that cellular mechanisms supporting functional integrity during development may be more significant than those of adult life in explaining the functional declines of aging. Ameur and colleagues, for example, have observed that most somatic mtDNA mutations do not result from DNA damage buildup in adult life, but occur during development (Ameur et al. 2011). With a reduced rate of development, cells damaged by errors such as replication fork collapse or transcription/replication fork collisions may be able to undergo a more prolonged arrest, to either repair the damage or undergo apoptosis or senescence.
Argüelles and colleagues propose that efficient apoptosis could be considered a pleiotropic aging factor, positive during development but negative during adulthood (Argüelles et al. 2019). We suggest that the same logic applies to senescence: enhanced senescence may be a positive feature during development although accumulation of senescent cells in adulthood remains pro aging.
Finally, Peto’s paradox suggests that species with higher body mass and great longevities should succumb to cancer more often than small short-lived species due to the requirement for increased cell division (representing more potential targets for oncogenic transformation) and greater time available to accumulate mutations. The paradox is that the opposite is observed (Peto 2015). Our findings represent an additional explanation to this puzzle.
Electronic supplementary material
(DOCX 356 kb)
Abbreviations
- ETO
Etoposide
- NCS
Neocarzinostatin
- LBB
Little brown bat
- FCM
Flow cytometry
- SA β-gal
Senescence-associated β-galactosidase
- qRT-PCR
quantitative real time PCR
Funding information
This work was supported by RFO funding of University of Bologna to A. Lorenzini and National Institutes of Health/National Institute on Aging (Grant AG39799) to C. Sell.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Amany Attaallah, Email: sayedahmed.amany@yahoo.com.
Monia Lenzi, Email: m.lenzi@unibo.it.
Silvia Marchionni, Email: silvia.marchionni@unibo.it.
Giacomo Bincoletto, Email: binco77@gmail.com.
Veronica Cocchi, Email: veronica.cocchi4@unibo.it.
Eleonora Croco, Email: eleonora.croco@gmx.de.
Patrizia Hrelia, Email: patrizia.hrelia@unibo.it.
Silvana Hrelia, Email: silvana.hrelia@unibo.it.
Christian Sell, Email: christian.sell@drexelmed.edu.
Antonello Lorenzini, Email: antonello.lorenzini@unibo.it.
References
- Abegglen LM, Caulin AF, Chan A, Lee K, Robinson R, Campbell MS, Kiso WK, Schmitt DL, Waddell PJ, Bhaskara S, Jensen ST, Maley CC, Schiffman JD. Potential mechanisms for Cancer resistance in elephants and comparative cellular response to DNA damage in humans. JAMA. 2015;314(17):1850–1860. doi: 10.1001/jama.2015.13134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameur A, Stewart JB, Freyer C, Hagström E, Ingman M, Larsson N-G, Gyllensten U. Ultra-deep sequencing of mouse mitochondrial DNA: mutational patterns and their origins. PLoS Genet. 2011;7(3):e1002028. doi: 10.1371/journal.pgen.1002028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argüelles S, Guerrero-Castilla A, Cano M, Muñoz MF, Ayala A (2019) Advantages and disadvantages of apoptosis in the aging process. Ann N Y Acad Sci. Mar 610.1111/nyas.14020 [DOI] [PubMed]
- Bitto A, Crowe EP, Lerner C, Torres C, Sell C (2014) The senescence arrest program and the cell cycle. Methods Mol Biol, 1170:145-54. 10.1007/978-1-4939-0888-2_8 [DOI] [PubMed]
- Campisi J, d'Adda di Fagagna F (2007) Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol Sep, 8(9):729-40. https://doi.org/10.1038/nrm2233 [DOI] [PubMed]
- Chuprin A, Gal H, Biron-Shental T, Biran A, Amiel A, Rozenblatt S, Krizhanovsky V. Cell fusion induced by ERVWE1 or measles virus causes cellular senescence. Genes Dev. 2013;27(21):2356–2366. doi: 10.1101/gad.227512.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford ED, Seaman JE, Barber AE, David DC, Babbitt PC, Burlingame AL, Wells JA. Conservation of caspase substrates across metazoans suggests hierarchical importance of signaling pathways over specific targets and cleavage site motifs in apoptosis. Cell Death Differ. 2012;19(12):2040–2048. doi: 10.1038/cdd.2012.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croco E, Marchionni S, Lorenzini A. Genetic instability and aging under the scrutiny of comparative biology: a meta-analysis of spontaneous micronuclei frequency. Mech Ageing Dev. 2016;156:34–41. doi: 10.1016/j.mad.2016.04.004. [DOI] [PubMed] [Google Scholar]
- Croco E, Marchionni S, Bocchini M, Angeloni C, Stamato T, Stefanelli C, … Lorenzini A (2017a) DNA damage detection by 53BP1: relationship to species longevity. J Gerontol A Biol Sci Med Sci, 72(6), 763–770. doi:10.1093/gerona/glw170 [DOI] [PubMed]
- Croco E, Marchionni S, Storci G, Bonafè M, Franceschi C, Stamato TD, Sell C, Lorenzini A. Convergent adaptation of cellular machineries in the evolution of large body masses and long life spans. Biogerontology. 2017;18(4):485–497. doi: 10.1007/s10522-017-9713-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Magalhães JP, Costa J, Church GM. An analysis of the relationship between metabolism, developmental schedules, and longevity using phylogenetic independent contrasts. J Gerontol A Biol Sci Med Sci. 2007;62(February):149–160. doi: 10.1093/gerona/62.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debacq-Chainiaux F, Borlon C, Pascal T, Royer V, Eliaers F, Ninane N, … Toussaint O (2005) Repeated exposure of human skin fibroblasts to UVB at subcytotoxic level triggers premature senescence through the TGF-beta1 signaling pathway. J Cell Sci, 118(Pt 4), 743–58. doi:10.1242/jcs.01651 [DOI] [PubMed]
- Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C., … Pereira-Smith O (1995) A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A, 92(20), 9363–9367 10.1073/pnas.92.20.9363 [DOI] [PMC free article] [PubMed]
- Dumont P, Burton M, Chen QM, Gonos ES, Frippiat C, Mazarati JB., … Toussaint O (2000) Induction of replicative senescence biomarkers by sublethal oxidative stresses in normal human fibroblast. Free Radic Biol Med, 28(3), 361–73. 10.1016/S0891-5849(99)00249-X [DOI] [PubMed]
- Fink LS, Roell M, Caiazza E, Lerner C, Stamato T, Hrelia S, Lorenzini A, Sell C (2011) 53BP1 contributes to a robust genomic stability in human fibroblasts. Aging 3(9):836–845 10.18632/aging.100381 [DOI] [PMC free article] [PubMed]
- Frippiat C, Chen QM, Zdanov S, Magalhaes JP, Remacle J, Toussaint O. Subcytotoxic H2O2 stress triggers a release of transforming growth factor-beta 1, which induces biomarkers of cellular senescence of human diploid fibroblasts. J Biol Chem. 2001;276(4):2531–2537. doi: 10.1074/jbc.M006809200. [DOI] [PubMed] [Google Scholar]
- Goldberg IH. Free radical mechanisms in neocarzinostatin-induced DNA damage. Free Radic Biol Med. 1987;3(1):41–54. doi: 10.1016/0891-5849(87)90038-4. [DOI] [PubMed] [Google Scholar]
- Kapahi P, Boulton ME, Kirkwood TB (1999) Positive correlation between mammalian life span and cellular resistance to stress. Free Radic Biol Med 26(5–6):495–500 10.1016/S0891-5849(98)00323-2 [DOI] [PubMed]
- Kirkwood TB, Rose MR. Evolution of senescence: late survival sacrificed for reproduction. Philos Trans R Soc Lond Ser B Biol Sci. 1991;332(1262):15–24. doi: 10.1098/rstb.1991.0028. [DOI] [PubMed] [Google Scholar]
- Lawrence I, Bene M, Nacarelli T, Azar A, Cohen JZ, Torres C, Johannes G, Sell C. Correlations between age, functional status, and the senescence-associated proteins HMGB2 and p16INK4a. GeroScience. 2018;40(2):193–199. doi: 10.1007/s11357-018-0015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Otín C, Blasco M a, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzini A, Tresini M, Austad SN, Cristofalo VJ. Cellular replicative capacity correlates primarily with species body mass not longevity. Mech Ageing Dev. 2005;126(10):1130–1133. doi: 10.1016/j.mad.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Lorenzini A, Johnson FB, Oliver A, Tresini M, Smith JS, Hdeib M, Sell C, Cristofalo VJ, Stamato TD. Significant correlation of species longevity with DNA double strand break recognition but not with telomere length. Mech Ageing Dev. 2009;130(11–12):784–792. doi: 10.1016/j.mad.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzini A, Stamato T, Sell C (2011) The disposable soma theory revisited: time as a resource in the theories of aging. Cell Cycle 10:3853–3856 10.4161/cc.10.22.18302 [DOI] [PubMed]
- Maier AB, Westendorp RGJ, Van Heemst D. beta-Galactosidase Activity as a Biomarker of Replicative Senescence during the Course of Human Fibroblast Cultures. Ann N Y Acad Sci. 2007;1100(1):323–332. doi: 10.1196/annals.1395.035. [DOI] [PubMed] [Google Scholar]
- Meier P, Finch A, Evan G. Apoptosis in development. Nature. 2000;407:796–801. doi: 10.1038/35037734. [DOI] [PubMed] [Google Scholar]
- Peto R (2015) Quantitative implications of the approximate irrelevance of mammalian body size and lifespan to lifelong cancer risk. Philos Trans R Soc Lond B 370(1673):20150198 10.1098/rstb.2015.0198 [DOI] [PMC free article] [PubMed]
- Ricklefs RE. Life-history connections to rates of aging in terrestrial vertebrates. Proc Natl Acad Sci. 2010;107(22):10314–10319. doi: 10.1073/pnas.1005862107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speakman JR. Correlations between physiology and lifespan--two widely ignored problems with comparative studies. Aging Cell. 2005;4(4):167–175. doi: 10.1111/j.1474-9726.2005.00162.x. [DOI] [PubMed] [Google Scholar]
- Storer M, Mas A, Robert-Moreno A, Pecoraro M, Ortells MC, Di Giacomo V, … Keyes WM (2013) Senescence is a developmental mechanism that contributes to embryonic growth and patterning. Cell, 155(5), 1119–1130. doi:10.1016/J.CELL.2013.10.041 [DOI] [PubMed]
- Tacutu R, Thornton D, Johnson E, Budovsky A, Barardo D, Craig T, Diana E, Lehmann G, Toren D, Wang J, Fraifeld VE, de Magalhães JP. Human ageing genomic resources: new and updated databases. Nucleic Acids Res. 2018;46(D1):D1083–D1090. doi: 10.1093/nar/gkx1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, Seluanov A, Gorbunova V. Molecular mechanisms determining lifespan in short- and long-lived species. Trends Endocrinol Metab. 2017;28(10):722–734. doi: 10.1016/j.tem.2017.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez JM, Sulak M, Chigurupati S, Lynch VJ. A zombie LIF gene in elephants is upregulated by TP53 to induce apoptosis in response to DNA damage. Cell Rep. 2018;24(7):1765–1776. doi: 10.1016/j.celrep.2018.07.042. [DOI] [PubMed] [Google Scholar]
- Waaijer MEC, Croco E, Westendorp RGJ, Slagboom PE, Sedivy JM, Lorenzini A, Maier AB (2016) DNA damage markers in dermal fibroblasts in vitro reflect chronological donor age. Aging 8(1):147–157 10.18632/aging.100890 [DOI] [PMC free article] [PubMed]
- Zhang G, Cowled C, Shi Z, Huang Z, Bishop-Lilly KA, Fang X, Wynne JW, Xiong Z, Baker ML, Zhao W, Tachedjian M, Zhu Y, Zhou P, Jiang X, Ng J, Yang L, Wu L, Xiao J, Feng Y, Chen Y, Sun X, Zhang Y, Marsh GA, Crameri G, Broder CC, Frey KG, Wang LF, Wang J. Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science. 2013;339(6118):456–460. doi: 10.1126/SCIENCE.1230835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Tyshkovskiy A, Muñoz-Espín D, Tian X, Serrano M, de Magalhaes JP, Nevo E, Gladyshev VN, Seluanov A, Gorbunova V. Naked mole rats can undergo developmental, oncogene-induced and DNA damage-induced cellular senescence. Proc Natl Acad Sci. 2018;115(8):1801–1806. doi: 10.1073/pnas.1721160115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 356 kb)