Abstract
Aging is associated with reduced liver function that may increase the risk for adverse drug reactions in older adults. We hypothesized that age-related changes to epigenetic regulation of genes involved in drug metabolism may contribute to this effect. We reviewed published epigenome-wide studies of human blood and identified the cytochrome P450 2E1 (CYP2E1) gene as a top locus exhibiting epigenetic changes with age. To investigate potential functional changes with age in the liver, the primary organ of drug metabolism, we obtained liver tissue from mice aged 4–32 months from the National Institute on Aging. We assayed global DNA methylation (5-methylcytosine, 5mC), hydroxymethylation (5-hydroxymethylcytosine, 5hmC), and locus-specific 5mC and histone acetylation changes around mouse Cyp2e1. The mouse livers exhibit significant global decreases in 5mC and 5hmC with age. Furthermore, 5mC significantly increased with age at two regulatory regions of Cyp2e1 in tandem with decreases in its gene and protein expressions. H3K9ac levels also changed with age at both regulatory regions of Cyp2e1 investigated, while H3K27ac did not. To test if these epigenetic changes are associated with varying rates of drug metabolism, we assayed clearance of the CYP2E1-specific probe drug chlorzoxazone in microsome extracts from the same livers. CYP2E1 intrinsic clearance is associated with DNA methylation and H3K9ac levels at the Cyp2e1 locus but not with chronological age. This suggests that age-related epigenetic changes may influence rates of hepatic drug metabolism. In the future, epigenetic biomarkers could prove useful to guide dosing regimens in older adults.
Electronic supplementary material
The online version of this article (10.1007/s11357-020-00181-5) contains supplementary material, which is available to authorized users.
Keywords: DNA methylation, Histone acetylation, Aging, Cyp2e1, Pharmacokinetics, Drug metabolism
Introduction
Adverse drug reactions (ADRs) are estimated to be between the fourth and sixth leading cause of death in the USA (Lazarou et al. 1998; Routledge et al. 2004). The impact and management of ADRs are complex and have been estimated to cost up to $30.1 billion annually (Sultana et al. 2013). Previous research suggests that rates of ADRs increase as people age, have more chronic health conditions, and take more medications (McLean and Le Couteur 2004; ElDesoky 2007; Budnitz et al. 2011). Human life expectancy has more than doubled in the last two centuries, and while mortality has been delayed, aging is still accompanied by a significantly elevated risks for many diseases (Issa 2002; Duron and Hanon 2008; Barzilai et al. 2012). Comorbid chronic conditions in individuals older than 65 years cause a high degree of polypharmacy in this population. According to a 2006 survey, 40% of persons 65 years of age or older were taking five to nine medications, while almost one-fifth (18%) were taking 10 or more (Slone Epidemiology Center 2006). Ultimately, older adults are almost seven times more likely than younger persons to have ADRs that require hospitalization (Budnitz et al. 2006). While ADRs are a serious problem in the aging population, up to 80% of ADRs in older patients are dose related and, therefore, are potentially avoidable (Routledge et al. 2004). This implies that effective methods for predicting the correct dose for the individual patient could make a significant impact in geriatric healthcare.
Age-associated changes to hepatic metabolism of drugs increase risk for ADRs in older adults (McLachlan et al. 2009; McLachlan and Pont 2012), but the determinants of these changes are not fully understood. One possible mechanism that may influence rates of drug metabolism in older adults is epigenetics (Seripa et al. 2015; Fisel et al. 2016; Kronfol et al. 2017). Aging is associated with substantial changes to the epigenome (Benayoun et al. 2015; Pal and Tyler 2016; Horvath and Raj 2018), and genes encoding drug metabolizing enzymes in the human liver are under epigenetic control (Bonder et al. 2014; Park et al. 2015). Furthermore, treatment with epigenetic drugs affects the metabolic capacity of cultured cells (Ruoß et al. 2019). These considerations led us to hypothesize that age-associated epigenetic changes at genes encoding drug metabolizing enzymes could affect rates of drug metabolism. Epigenome-wide association studies (EWAS) in human blood have found significant changes to DNA methylation at several genes encoding phase I (oxidative) drug metabolism enzymes with age (Heyn et al. 2012; Hannum et al. 2013; Horvath 2013; Steegenga et al. 2014; Reynolds et al. 2014; Marttila et al. 2015; Peters et al. 2015). However, the extent to which these age-related changes are present and affect enzymatic activity in the liver, the primary site of drug metabolism, is unclear. Therefore, the goal of this study is to identify age-related epigenetic changes at genes encoding phase I drug metabolizing enzymes in the liver and test if these epigenetic changes are associated with rates of drug metabolism. Due to the experimental control afforded and availability of the relevant tissue, we chose to conduct the experiments in mice.
To date, the number of published studies on epigenetics and drug metabolism in aging is limited. As a starting point to identify potential genes of interest, we reviewed published EWAS and genome-wide gene expression studies in human blood and chose the phase I drug metabolism genes showing the best empirical evidence of change with age. The rationale for using human blood studies to guide gene selection is because (1) the largest number of aging EWAS have been conducted in this tissue, (2) epigenetic aging effects are significantly correlated across tissues and species (Horvath 2013), and (3) consistent patterns of gene expression changes with age have been observed across several species (McCarroll et al. 2004). We identified two phase I drug metabolism genes, CYP2E1 and CYP1B1, showing strong evidence for age-associated epigenetic changes in human blood. We mapped the associated regions in the human genome to their homologous mouse regions and tested for epigenetic changes in the mouse liver. Only Cyp2e1 showed differential methylation with age. Based on these results, we focused on Cyp2e1 and conducted a detailed analysis of regulation at this locus including assays for DNA methylation and histone post-translational modifications (PTMs) with reported associations with age (McClay et al. 2014; Dozmorov 2015). Finally, we investigated if these effects were associated with CYP2E1 metabolic function by isolating liver microsomes and applying Michaelis–Menten kinetics to determine the intrinsic clearance (CLint) of the probe drug chlorzoxazone (CZ), which is predominantly metabolized by CYP2E1 (Lucas et al. 1999).
Methods
Extended methods are available in the Supplementary Material.
Mice
Liver tissue samples from 20 male CB6F1 mice (five subjects in each of four age groups: 4, 18, 24, and 32 months) were obtained from the National Institute on Aging (NIA) rodent tissue bank.
DNA and RNA extraction
Genomic DNA was extracted using the AllPrep DNA/RNA kit (Qiagen, Hilden, Germany) according to manufacturer’s protocol. DNA and RNA purity and quantity were measured using a Nanodrop spectrometer (Thermo Fisher, Waltham, MA).
Global 5mC and 5hmC
Global 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC) levels were measured using MethylFlash ELISA-based colorimetric assays (Epigentek, Farmingdale, NY). 5mC- or 5hmC-specific antibodies, provided in the kit, were incubated with 100 ng genomic DNA. Optical density at 450 nm was measured on a Synergy HT plate reader (BioTek Instruments, Winooski, VT). Known standards provided in the kit consisting of 0.1, 0.2, 0.5, 1, 2, and 5% for 5mC and 0.02, 0.04, 0.1, 0.2, 0.4, and 1% for 5hmC were also assayed. The optical density of liver samples was used to determine the percentage of 5mC and 5hmC of each sample by interpolation on respective standard curves. Each sample and standard was run in duplicate.
Selection of genomic regions of interest
Aging EWAS findings for human blood were obtained from published studies (see Dozmorov 2015), and a list of genes encoding drug metabolizing enzymes was obtained from the ADME (absorption, distribution, metabolism, and excretion) gene list from the pharmaADME consortium (pharmaADME.org), see Supplementary Table S1. Significant findings by ADME gene were summed across studies, and the top two phase I drug metabolism genes showing the most significant findings were selected for study.
Genomic location of human age-associated cytosine–phosphate–guanine (CpG) positions were used to extract the homologous regions in mouse using the “convert” function in the University of California Santa Cruz (UCSC) genome browser (Kent et al. 2002). A complete list of the genomic coordinates of all loci investigated in mouse is found in Supplementary Table S2. An additional regulatory region around human CYP2E1 was identified using the GeneHancer track (Fishilevich et al. 2017) on the UCSC genome browser (Fig. 1). Data from the mouse Encyclopedia of DNA Elements (ENCODE) (Stamatoyannopoulos et al. 2012) and Ludwig Institute for Cancer Research (LICR) (Barrera et al. 2008) chromatin immunoprecipitation (ChIP) sequencing runs on young (8 weeks) male mouse liver tissue were used to identify two regions with high levels of histone 3 lysine 9 acetylation (H3K9ac) and histone 3 lysine 27 acetylation (H3K27ac) PTMs (GSM1000153, GSM1000140).
Fig. 1.
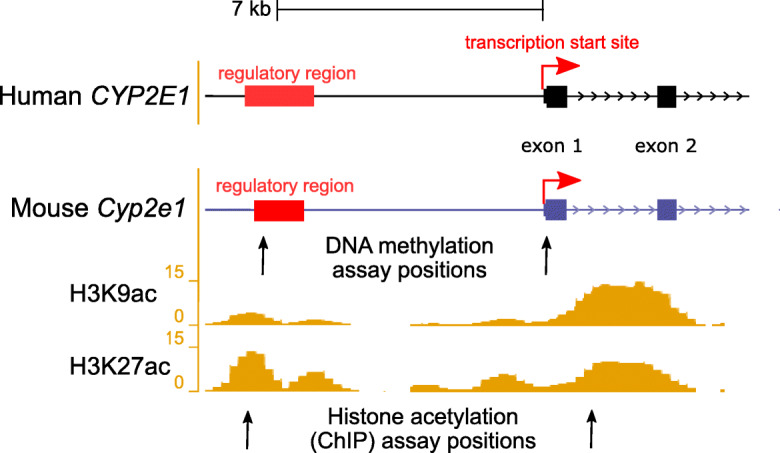
Illustration showing the transcription start site (TSS) and upstream regulatory regions of the human CYP2E1 and the homologous mouse Cyp2e1 gene. Upstream regulatory element positions were obtained from GeneHancer (Fishilevich et al. 2017) and ORegAnno (Lesurf et al. 2016). DNA methylation assays were conducted in the current study at both the TSS and upstream regulatory region in mouse at positions marked by the arrows. Reference histone 3 lysine 9 acetylation (H3K9ac) and histone 3 lysine 27 acetylation (H3K27ac) in 8-week-old mouse livers are from the ENCODE/LICR track in UCSC Genome Browser. Two loci with high liver histone acetylation levels were chosen for analysis using ChIP-qPCR in the current study, at positions marked by the arrows. For exact assay coordinates, see Supplementary Tables S2 and S3
Bisulfite conversion of genomic DNA and HRM analysis
Two hundred nanograms of liver genomic DNA was treated with sodium bisulfite according to the EZ DNA Methylation kit protocol (Zymo Research, Irvine, CA). Mouse genomic DNA standards of known percentage 5mC (EpigenDx, Hopkinton, MA) were used to create a standard curve. High-resolution melt (HRM) assays used MeltDoctor HRM MasterMix (Applied Biosystems, Foster City, CA) on a Quantstudio 3 instrument. HRM was used to measure 5mC levels at LINE1 elements using the method of Newman et al. (2012) with minor modifications (see Supplementary Methods) and at a 319-bp region on the 5′UTR of mouse Cyp2e1 (chr7: 147,949,616–147,949,934, mouse genome assembly mm9 NCBI37/ build 9, July 2007) encompassing 7 CpGs (Supplementary Table S2). This region at Cyp2e1 was amplified by qPCR as follows: 10 min hold at 95 °C followed by 40 cycles of 15 s at 95 °C, 30 s at 57 °C, and 30 s at 72 °C, followed by a final extension at 72 °C for 7 min and a melt curve stage with temperature range of 57 to 95 °C with fluorescence capture at 0.025° per s increment. Each reaction included 20 ng bisulfite-converted DNA and 0.2 μM each of forward and reverse primer (Supplementary Table S2) in 1× MeltDoctor HRM MasterMix. Liver samples and standards were run in triplicate. The net temperature shift (NTS) values (Newman et al. 2012) of the liver samples were interpolated on the standard curve to yield their 5mC percentage.
Gene expression analysis by RT-qPCR
For each sample, 1 μg of total liver mRNA was reverse transcribed using the iScript kit (Bio-Rad, Hercules, CA). Aliquots of cDNA were amplified in triplicate using TaqMan gene expression master mix and TaqMan Cyp2e1 Mouse Gene Expression Assay (Mm00491127_m1, Thermo Fisher) (Martinez et al. 2010; Koh et al. 2011). qPCR conditions were 40 cycles of 15 s at 95 °C and 60 s at 60 °C. Mouse Gapdh endogenous control was also assayed in triplicate (Mm99999915_g1, Thermo Fisher) (Scarzello et al. 2016; Wilhelm et al. 2016; Jia et al. 2018). A 1:2 serial dilution of control cDNA was used to generate a 5-point standard curve. Quantification cycles (Cq) were determined using the Relative Quantification application on the Thermo Fisher Cloud. Normalized quantification cycles (ΔCq) were obtained by subtracting the mean Gapdh Cq from the mean Cyp2e1 Cq. ΔΔCq was calculated for each age group by subtracting the ΔCq of the 4 months (youngest age group as reference) from each of the other age group’s ΔCq to calculate fold differences.
Western blots
Twenty milligrams of liver tissue was homogenized in lysis Pierce RIPA buffer with 1% v/v Halt Protease Inhibitor buffer (Thermo Fisher). All other reagents were from Bio-Rad (Hercules, CA) unless otherwise specified. Ten micrograms of total protein in β-mercaptoethanol (βME)-Laemmli buffer was separated on 10% Mini-PROTEAN precast SDS-PAGE gels and transferred onto PVDF membrane using a TransBlot Turbo at 1.3 A for 5 min. Membranes were blocked in 5% milk in 1× TBST (Tris-buffered saline, Tween 20). Blots were probed with anti-CYP2E1 antibody (1:2500, ab28146, Abcam, Cambridge, UK) overnight at 4 °C and with HRP-coupled rabbit IgG secondary antibody (1:10,000, Cell Signaling, Danvers, MA) for 1 h at room temperature, prior to treatment with ECL and imaging on a ChemiDoc instrument. Then, membranes were washed with 1× TBST and stripped using Restore stripping buffer (Thermo Fisher), followed by incubation with GAPDH primary antibody (1:5000, MA5-15738, Thermo Fisher) and then mouse IgG secondary antibody (1:20,000, 7076S, Cell Signaling, Danvers, MA). CYP2E1 bands were quantified using Image Lab v 6.0 (Bio-Rad) and normalized to GAPDH.
ChIP-qPCR
Eighty milligrams of mouse liver per sample was processed using the TruChIP tissue shearing kit (Covaris, Woburn, MA). Minced tissue was fixed in 1% formaldehyde for 2 min, washed, transferred to a tissueTUBE (Covaris), flash-frozen in liquid N2, and pulverized. After cell lysis, chromatin was sheared on a Covaris M220 for 8 min. Two percent of sheared chromatin per IP was set aside as input control. ChIP used 5 μl of anti-H3K9ac (39137, Active Motif, Carlsbad, CA), or 5 μg of anti-H3K27ac (39133, Active Motif), or 5 μg of Rabbit IgG isotype control (ab171870, Abcam) incubated with 2 μg sheared chromatin from each sample overnight (16 h) at 4 °C. This mixture was added to Dynabeads Protein G (Thermo Fisher) for 4 h at 4 °C before washing and elution of ChIP DNA by heating to 65 °C for 1 h. After elution, samples were treated with RNAse and Proteinase K and DNA was purified using QIAquick (Qiagen).
Each ChIP’ed DNA was amplified in triplicate using PowerUp SYBR Green (Applied Biosystems), with 0.2 μM of forward and reverse primers (Supplementary Table S2) as follows: 2 min at 50 °C, 2 min at 95 °C followed by 40 cycles of 15 s at 95 °C, 30 s at 59 °C, and 1 min at 72 °C, followed by melt curve stage. Cq values for each plate were obtained as above, and the mean threshold cycle (Cq) for each sample normalized to the dilution factor (2% = 1/50) corrected Cq value (Log2 (50) = 5.6438) of the input control to obtain ΔCq. The percentage of input was calculated by multiplying 100 by 2ΔCq.
Pyrosequencing
Quantitative methylation measures of individual CpGs were obtained via pyroseqeuncing of bisulfite converted DNA (Tost et al. 2003). Primers targeting the loci of interest were designed with PyroMark Assay Design version 2.0.1.15 (Qiagen) (Supplementary Table S3). Amplification was performed using the PyroMark PCR Kit (Qiagen) and the vendor specified program for bisulfite converted DNA. Next, the amplicons were bound to Streptavidin Sepharose beads (Fisher, Hampton, NH) and prepared for sequencing using the PyroMark Q96 vacuum station followed by sequencing on the PyroMark Q96MD platform with PyroMark Gold Q96 reagents (Qiagen). A standard consisting of 0, 5, 10, 25, 50, 75, and 100% methylated mouse genomic DNA was included on each plate (EpigenDx). Both the standard and the liver test samples were pyrosequenced in duplicate.
CYP2E1 intrinsic clearance
Mouse liver microsomes (MLM) were prepared from 500 mg of liver tissue per sample, as described previously (Knights et al. 2016), and details are provided in the Supplementary Material. Standards for CZ and 6-OH-CZ (18869, 10009029, Cayman Chemical, Ann Arbor, MI) were purchased. Chromatographic separation was achieved using an Agilent 1100 series HPLC (Agilent Technologies, Santa Clara, CA) system equipped with PerkinElmer series 200 degasser and auto injector (PerkinElmer, Waltham, MA) and C18 BDS hypersil 50 mm × 4.6 with 5 μm particle size (Thermo Fisher) column. Diode array detector was used to monitor the depletion of CZ and the formation of 6-OH-CZ at 280 and 299 nm for CZ and 6-OH-CZ respectively. Gradient elution was used with the initial mobile phase being 85% aqueous (2% acetic acid and 1% triethylamine, v/v) and 15% methanol (Supplementary Table S4). Total run time was 6.5 min with retention times of 1.86 and 5.17 min for 6-OH-CZ and CZ respectively. Linear standard curves with R2 ≥ 0.999 were obtained from 0.47 to 240 μM for 6-OH-CZ and 2.34–1200 μM for CZ (Supplementary Figs. S4 and S5).
Hydroxylation of chlorzoxazone (CZ) to 6-hydroxychlorzoxazone (6-OH-CZ) was measured to determine the catalytic activity of CYP2E1. Linearity of metabolite formation with time and MLM protein concentration was established (Supplementary Figs. S6 and S7). The final concentrations in the metabolic reactions were 50 mM potassium phosphate buffer pH 7.4, 10 mM MgCl2, 1 mg/ml MLM protein, 1 mM EDTA, and 1 mM NADPH with incubation time of 25 min at 37 °C. The final concentration of DMSO in each reaction was maintained at 0.5%. Each MLM sample was run in 8 MLM reactions that vary by the final concentration (10, 20, 40, 80, 160, 320, 640, 1000 μM) of the parent probe drug (CZ) to determine Michaelis–Menten kinetic constants. The reaction was stopped with methanol and centrifuged for 10 min at 20,000×g at room temperature. Reaction rates (pmol/min/mg protein) of 6-OH-CZ formation were plotted against CZ concentration (μM), and Km and Vmax values were estimated by GraphPad Prism using Eq. 1, where Y is the reaction rate (pmol/min/mg protein), X is CZ concentration (μM), Vmax is the maximal reaction rate of 6-OH-CZ formation (pmol/min/mg protein), and Km is CZ concentration at half maximal rate (μM).
| 1 |
Intrinsic clearance (CLint) is the enzyme-mediated activity toward a drug that would occur at concentration below Km without physiological limitations such as hepatic blood flow (Houston 1994). CLint by CYP2E1 for the hydroxylation reaction was calculated according to Eq. 2
| 2 |
Statistics
Linear regression and Pearson correlation tests were conducted in R version 3.6.1 (www.r-project.org) with α = 0.05. Beta values (β) and Pearson correlation coefficients (r) are reported. β indicates the degree of change in the outcome variable for every unit change in the predictor variable, while r indicates the degree of association found in the correlation test. Replicates beyond 2 standard deviations from the mean of any of the assays were discarded.
Results
Global epigenetic effects in aged mouse liver
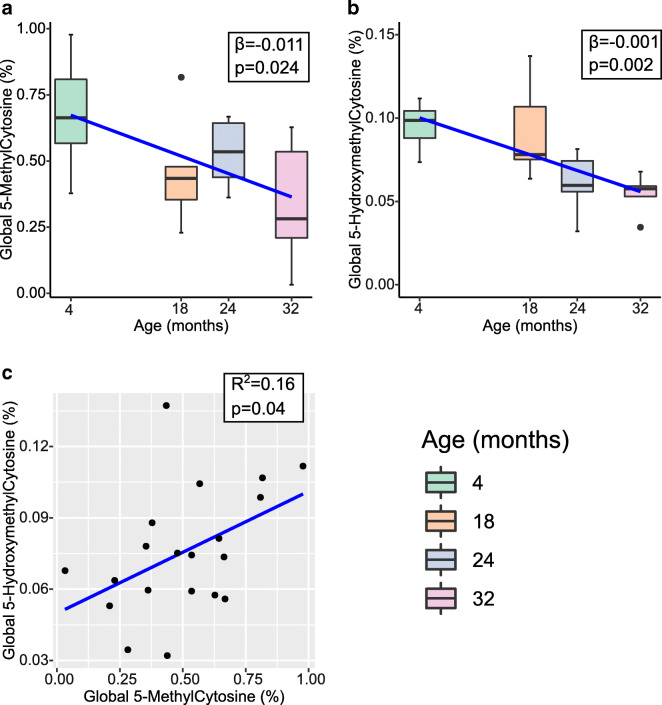
DNA and RNA extractions from the mouse liver samples were successful with an average 260/280 value of 1.94 [1.8–2.1] for DNA and 2.05 [1.94–2.1] for RNA. Prior to locus-specific analysis, we first assayed age-associated changes to the global abundance of epigenetic marks in the mouse liver. Using ELISA assays, we observed a significant decrease in global abundance of 5mC (β = − 0.011, SE = 0.004, p = 0.024) and 5hmC (β = − 0.001, SE = 0.0004, p = 0.002) (Fig. 2a, b). This translates to approximately 50% reduction in the 32 months age group compared to the 4 months age group for both modifications (0.6% vs 0.32% for 5mC and 0.1% vs 0.06% for 5hmC). We also measured 5mC at LINE1 elements using high-resolution melt (HRM) analysis of bisulfite converted DNA (Supplementary Figs. S1 and S2). We observed a significant decrease in abundance of 5mC at LINE1 elements with increasing age in our sample (β = − 2.062, p = 1 × 10−8), with an average reduction of 2% methylation per month of age. These results demonstrate that, in a broad sense, epigenetic changes were occurring with age in this study’s liver samples.
Fig. 2.
Box plots with regression line (blue) of age-associated changes to global a 5-methylcytosine (n = 20) and b 5-hydroxymethylcytosine (n = 20). Data represent median (middle hinge), 25% (lower hinge), and 75% (upper hinge) quantiles. Data points beyond upper or lower 1.5 × interquantile range are represented as individual black dots. c Scatter plot of 5-methylcytosine and 5-hydroxymethylcytosine with regression line (blue) (n = 20)
Age-associated changes to Cyp2e1 5′UTR methylation and gene and protein expressions
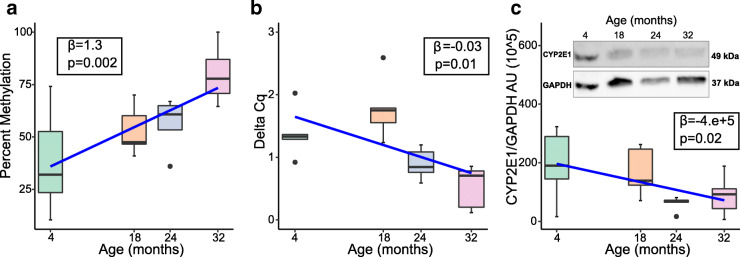
We reviewed published EWAS and genome-wide gene expression studies of aging in human blood (Supplementary Table S1) and identified CYP2E1 and CYP1B1 as the phase I drug metabolism genes showing most evidence for age-associated changes. We mapped the human age-associated differentially methylated regions from EWAS to their homologous mouse regions and tested for epigenetic changes at these loci in our aged mouse liver samples. The human CYP2E1 region mapped to the mouse Cyp2e1 5′UTR (Fig. 1) and HRM analysis revealed that methylation at this locus increased significantly with age in mouse liver DNA (β = 1.3, SE = 0.0038, p = 0.002) (Fig. 3a). However, no changes to Cyp1b1 methylation with age were detected in our sample (Supplementary Fig. S3) so we focused on Cyp2e1.
Fig. 3.
Box plots with regression line (blue) of age-associated changes to Cyp2e1a 5′UTR percent methylation (n = 19) and b gene expression (n = 20). c Box plot of age-associated changes to Cyp2e1 protein expression with representative western blot (n = 20). Data represent median (middle hinge), 25% (lower hinge), and 75% (upper hinge) quantiles. Data points beyond upper or lower 1.5 × interquantile range are represented as individual black dots
The observed Cyp2e1 5′UTR methylation increase with age corresponds to a 1.3% increase in methylation per month of increased age. In tandem, Cyp2e1 gene expression decreased significantly with age in the same samples (β = − 0.03, SE = 0.011, p = 0.01) (Fig. 3b). Using the 4 months group as reference, we observed a 2.15% reduction in expression of Cyp2e1 per month of increased age. Furthermore, we observed a significant decrease of CYP2E1 protein expression with age, as measured by change in chemiluminescence detected in Western blot (β = − 4.0e+5, SE = 0.01, p = 0.02) (Fig. 3c).
Base-resolution 5mC analysis of Cyp2e1 5′UTR and upstream regulatory region
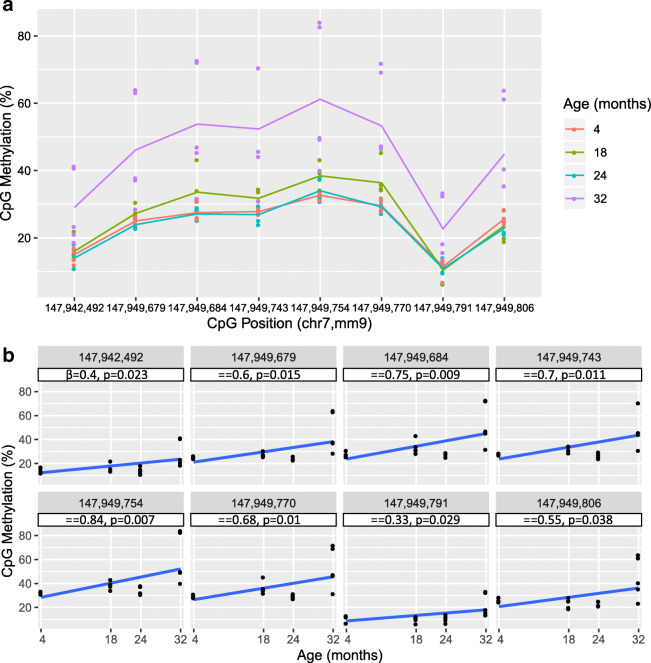
Having established that DNA methylation changes were occurring with age at the Cyp2e1 5′UTR, we aimed to obtain a fuller picture of Cyp2e1 epigenetic regulation. Our first priority was to assay 5mC levels at each of the seven CpGs in the 5′UTR region individually, in contrast to the aggregate measure obtained via HRM. We also identified an upstream regulatory region in human CYP2E1 (Fig. 1) that we used to obtain the homologous region in mouse that harbored a single CpG. Therefore, we subjected both regions to bisulfite pyrosequencing that allows highly quantitative methylation measurements at single-base resolution. This revealed that the single CpG in the upstream regulatory region (position 1, chr7: 147,942,492, mm9) was significantly hypermethylated with age (p = 0.023; Fig. 4 a and b, position 1). Pyrosequencing confirmed the methylation increase at the 5′UTR with age observed via HRM (Supplementary Table S6), with all individual CpGs showing significant increase in methylation with age (Fig. 4 a and b, positions 2 through 8). The CpG at position 5 (chr7: 147,949,754, mm9) was the most significantly hypermethylated (p = 0.007) with the largest beta value of 0.84% increase per month of age (Fig. 4b).
Fig. 4.
Pyrosequencing data for Cyp2e1. a Scatter plot of percent methylation of cytosine–phosphate–guanine (CpG) and all investigated CpG positions with superimposed line plot for each age group connecting the average methylation percentage at each CpG (n = 20 per CpG, N = 80 total). X-axis not drawn to scale. chr 7 chromosome 7, mm9 mouse genome assembly NCBI37/build 9, July 2007. b Scatter plot of CpG methylation and age of individual CpG positions with regression line and statistics under each location. A simple linear regression was performed on all 20 data points for each CpG (n = 20) against each age 4, 18, 24, and 32 months (N = 80). Positions 1–8: chr7: 147,942,492; 147,949,679; 147,949,684; 147,949,743; 147,949,754; 147,949,770; 147,949,791; 147,949,806, mm9
Histone acetylation analysis of Cyp2e1 5′UTR and upstream regulatory region
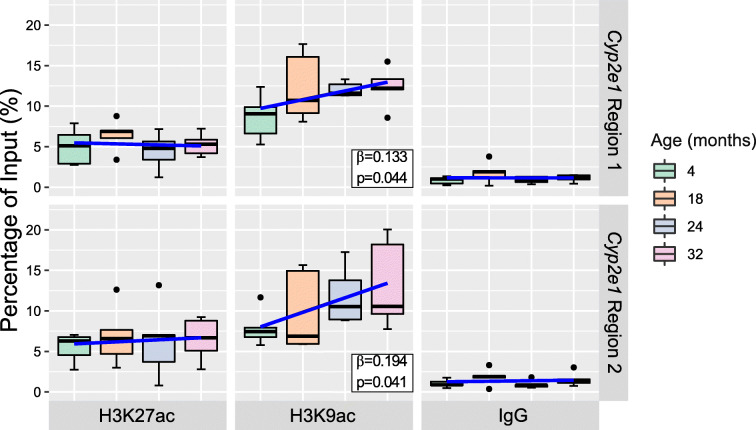
We viewed publicly available mouse ChIP-Seq data (GSM1000153, GSM1000140) in UCSC genome browser to identify two regions neighboring and/or overlapping the 5′UTR and the upstream regulatory region that showed high histone 3 lysine 9 acetylation (H3K9ac) and histone 3 lysine 27 acetylation (H3K27ac) occupancy rates in young mouse liver (Fig. 1). To assay histone acetylation levels in these regions, we used chromatin immunoprecipitation coupled to quantitative PCR (ChIP-qPCR). We observed a significant increase in H3K9ac at Cyp2e1 intron 1, adjacent to the 5′UTR (region 1, chr7: 147,950,223–147,950,367, mm9) (β = 0.133, SE = 0.06, p = 0.044) and at the upstream regulatory region (region 2, chr7: 147,942,350–147,942,468, mm9) (β = 0.194, SE = 0.08, p = 0.041) when regressing ChIP-qPCR percentage of input on age (Fig. 5). This corresponds to a 0.133% and 0.194% increase in H3K9ac per month of increased age at each site, respectively. However, H3K27ac levels were stable with age at both Cyp2e1 regions in this sample.
Fig. 5.
Chromatin immunoprecipitation quantitative polymerase chain reaction (ChIP-qPCR) data. Box plots with regression line (blue) of age-associated changes to percentage of input occupancy of histone 3 lysine 9 acetylation (H3K9ac) (n = 20 per region), histone 3 lysine 27 acetylation (H3K27ac) (n = 20 per region) at Cyp2e1 intron 1 (region 1, chr7: 147,950,223–147,950,367, mm9) and promoter (region 2, chr7: 147,942,350–147,942,468, mm9). IgG percentage of input shows a low background noise signal for each of the sample’s age groups (n = 20 per region). Data represent median (middle hinge), 25% (lower hinge), and 75% (upper hinge) quantiles. Data points beyond upper or lower 1.5 × interquantile range are represented as individual black dots
CYP2E1 pharmacokinetics, chronological age, and epigenetics
As shown above, 5mC and H3K9ac levels at Cyp2e1 changed with age in tandem with reduced gene and protein expressions in our sample. To determine if these effects impacted CYP2E1 metabolic activity, we assayed metabolism of the CYP2E1-specific probe drug chlorzoxazone (CZ) in microsome extracts from the same livers. The average microsome yield was 1.07% w/w [0.55–1.69], which was within expected range (Knights et al. 2016). Linearity of the HPLC assays of R2 ≥ 0.999 was established for 2.34–1200 μM CZ and 0.46–240 μM 6-OH-CZ (Supplementary Figs. S4 and S5). Linearity of 6-OH-CZ formation with reaction time and MLM final concentration was established, and the final reaction time and MLM concentration were 25 min and 1 mg/ml respectively (Supplementary Figs. S6 and S7). Vmax and Km values were estimated and used to calculate intrinsic clearance (CLint). Initial analyses established that chronological age was not significantly associated with CLint of CYP2E1 despite a relatively large correlation coefficient (r = 0.31, p = 0.8). Average Michaelis–Menten constants and CLint per age group are reported in Supplementary Table S7. Representative Michaelis–Menten curves of each age group are found in Supplementary Fig. S8. This result suggests that chronological age is not a robust independent predictor of CLint, corroborating prior research (Schmucker et al. 1990; Hunt et al. 1990; Mach et al. 2016).
We then tested Cyp2e1 epigenetic measures for association with the pharmacokinetic variables. All CpGs were negatively correlated with CLint (Fig. 6), and the most significant was with CpG at position 8 (chr7: 147,949,806, mm9) (r = − 0.29, p = 0.0008). Furthermore, H3K9ac levels at the upstream regulatory region, but not intron 1, of Cyp2e1 were positively correlated with CLint (r = 0.49, p = 0.005). All p values are provided in Supplementary Table S5.
Fig. 6.
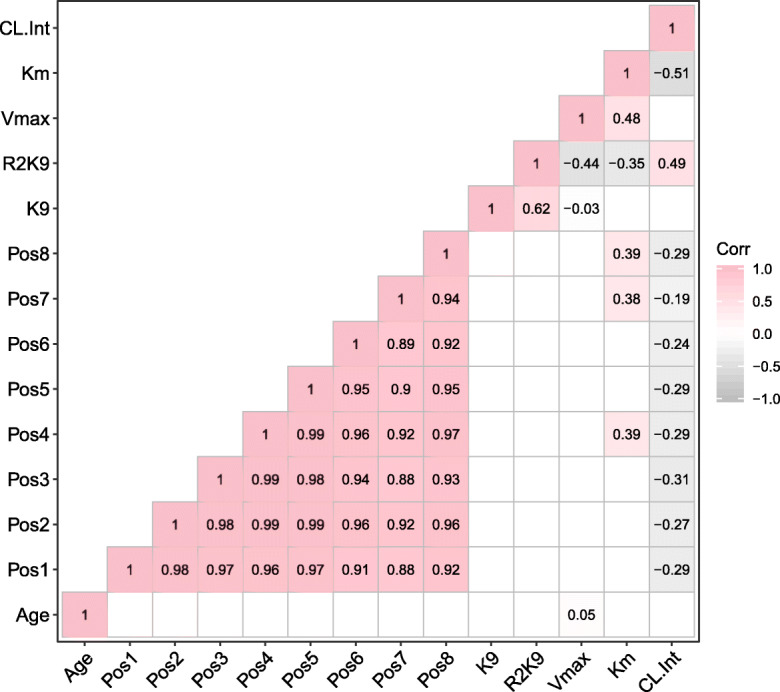
Correlation matrix reporting Pearson correlation statistical test result (r). White blank cells indicate non-significant association (p > 0.05). Refer to Supplementary Table S5 for individual p values of each Pearson correlation test of a given pair. Color gradient indicates the direction of effect of the association with dark pink representing the strongest positive association of 1 while dark gray representing the strongest negative association of − 1. Only lower half of the plot is shown to prevent redundancy in reporting the results. Age: chronological age, Pos 1–8: Positions 1–8 chr7: 147,942,492; 147,949,679; 147,949,684; 147,949,743; 147,949,754; 147,949,770; 147,949,791; 147,949,806, mm9. Vmax: maximal rate of hydroxylation reaction of chlorzoxazone by Cyp2e1, Km: chlorzoxazone concentration at half maximal rate, CL.Int: intrinsic clearance, K9: histone 3 lysine 9 acetylation in Cyp2e1 intron 1 (chr7: 147,950,223–147,950,367, mm9), R2K9 histone 3 lysine 9 acetylation in Cyp2e1 promoter (chr7: 147,942,350–147,942,468, mm9)
Discussion
In this study, we demonstrated that 5mC and H3K9ac levels change with age at Cyp2e1 in the mouse liver. Furthermore, we showed that these epigenetic changes were significantly associated with rates of CYP2E1-mediated drug metabolism in microsome extracts from the same livers, while chronological age was not. This finding suggests that epigenetic marks may be better predictors of drug metabolism in advanced age than chronological age itself.
Considering our findings in the context of published work, we found that global 5mC levels diminished with increasing age and this effect has been shown before in studies of different tissues (Booth and Brunet 2016), including the liver (Wilson et al. 1987). Previous reports have also shown reduced global 5hmC in the mouse liver with age, as we observed here (Tammen et al. 2014). However, recent genome-wide bisulfite sequencing studies in the mouse liver have shown either a modest excess of hypermethylated sites (Hahn et al. 2017) or no overall excess in either direction with age (Gravina et al. 2016). One possible reason for this discrepancy is that “genome-wide” approaches such as next-generation sequencing (NGS) are not truly representative of the whole genome. For example, NGS approaches typically underrepresent repetitive elements because reads in these regions may not align unambiguously to the reference genome and so are discarded. This may diminish the influence of repeats on the cumulative abundance of methylation in bisulfite sequencing studies. Several classes of repetitive elements reportedly lose methylation with age (Cardelli 2018). Therefore, underrepresentation of these elements could lead to underestimation of age-related hypomethylation in genome-wide NGS studies of aging. In our sample, we observed significant hypomethylation of LINE1 elements, with an average reduction of 2% per month of age. As LINE1 elements are the dominant repeat class in mouse and human, comprising almost 20% of the genome (Mouse Genome Sequencing Consortium et al. 2002), this result could partly explain the discrepancy between global 5mC studies and genome-wide bisulfite sequencing studies of aging in liver DNA.
Considering our locus-specific results, we found that DNA methylation of the Cyp2e1 5′UTR and upstream regulatory region increased significantly with age, while its gene expression declined with age. This supports and extends prior observations in human blood studies. For example, Peters et al. (2015) reported a hypermethylated CpG at human CYP2E1 and decreased gene expression with age. Reduced Cyp2e1 expression was also reported in the mouse liver tissue aged 28 months compared to 4 months, as detected via RNA sequencing (White et al. 2015). To date, prior studies of histone PTM changes in the context of aging are limited and we are not aware of available published data for the liver. Park et al. (2015) found that histone deacetylase inhibitors influenced transcription of cytochrome P450s in cultured hepatocytes, suggesting histone acetylation affects expression, but this study did not examine the effect of aging nor specific histone acetylation marks. Further work is needed in this area, in particular genome-wide analysis of H3K9ac with age, given our findings with respect to H3K9ac in this study.
We observed a significant negative correlation between CLint and Cyp2e1 methylation and a significant positive correlation between CLint and Cyp2e1 histone acetylation. The correlations are substantial, e.g., − 0.31 for CpG 5mC and 0.49 for H3K9ac, suggesting that epigenetics plays a significant role in regulating CYP2E1 hepatic activity. This finding may have clinical relevance because CYP2E1 is responsible for the metabolism of hepatotoxic substrates such as ethanol, acetaminophen, chlorzoxazone (CZ), pro-carcinogens (benzene, chloroform, and N-nitroso-nicotine), and endogenous compounds such as estrogen, acetone, and linoleic acid (Lieber 1997; Caro and Cederbaum 2004; Ingelman-Sundberg 2004; Porubsky et al. 2008). It comprises 5.5–16.5% of the hepatic P450 pool (Zanger and Schwab 2013), and its substrate profile makes it relevant to older adults due to the detrimental and unpredictable ADRs of these substrates. For example, CYP2E1 is inducible at heavy ethanol intake and is involved, along with alcohol dehydrogenases (ADH), in alcohol-mediated liver toxicity hospitalizations (Caro and Cederbaum 2004). In addition, acetaminophen-related liver injury is the primary over-the-counter drug-related hospitalization (Lee 2017). Acetaminophen hepatotoxicity is mediated by the excessive formation of the toxic byproduct N-acetyl-p-benzoquinone imine (NAPQI) and subsequent thiol residue depletion upon acetaminophen overdose. CYP2E1 is responsible for NAPQI formation at high acetaminophen doses, which highlights its importance in acetaminophen-induced liver damage (French 2013). Finally, CZ is a high-risk drug listed on the 2019 American Geriatrics Society Beers criteria for potentially inappropriate use in older adults (American Geriatrics Society 2019). CZ is a centrally acting skeletal muscle relaxant that is strongly recommended to avoid in the elderly due to moderate evidence of poor toleration by this population according to these criteria. Taken together, these examples provide evidence for clinical significance of the metabolism of drugs that are CYP2E1 substrates.
A limitation of the study as presented is that we could not manipulate epigenetic levels in our post-mortem samples, so the association between epigenetics and drug metabolism is correlational, rather than causal. Nevertheless, the use of the probe drug CZ renders the association highly specific to the action of CYP2E1. In addition, sex differences in drug metabolism have been described; however, our sample was comprised of only male mice. Hence, female-specific effects should be investigated in future studies. Age-related changes to pharmacokinetic attributes such as plasma protein binding and hepatic blood flow should be the subject of future in vivo investigations that pertain to drugs that are highly protein bound or have high hepatic extraction ratio due to the possible changes to these attributes that affect the overall disposition of drugs with advanced age (McLachlan and Pont 2012). Looking to the future, this work may have clinical applications if extended to human populations. Age-related epigenetic changes at human CYP2E1 and other drug metabolizing genes should be studied, and if these changes are linked to altered clearance clinically, epigenetic biomarkers of altered drug metabolism could be potentially used to guide dosing decisions in older adults. One potential issue is that epigenetic modifications vary by cell and tissue type. Therefore, age-related effects mapped in the liver will need to be tested for equivalency in peripheral cells or tissues (e.g., blood) that are more readily accessible for the purposes of biomarker testing. However, prior work by Horvath (2013) and others (Spiers et al. 2016; Zhu et al. 2018) suggests that many age-related epigenetic changes are consistent across tissues and indeed in this study, we demonstrated continuity across mouse and human. Future work should investigate genome-wide epigenetic changes with age in the liver and blood concurrently, for the same organism.
Electronic supplementary material
(PDF 748 kb)
Acknowledgments
MMK completed this study in partial fulfillment of the doctoral requirements in Pharmaceutical Sciences at VCU. We are grateful to the staff at the National Institute on Aging Rodent Tissue Bank for providing us with the sample to carry out this study.
Funding information
MMK was funded through a graduate studentship from Virginia Commonwealth University (VCU) School of Pharmacy and through R15AG061649 from the US National Institute on Aging. Research reported in this publication was supported by the National Institute on Aging of the National Institutes of Health under Award Number R15AG061649 to JLM.
Compliance with ethical standards
Disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest
The authors declare that they have no conflicts of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- American Geriatrics Society American Geriatrics Society 2019 updated AGS Beers Criteria® for potentially inappropriate medication use in older adults: 2019 AGS BEERS CRITERIA® UPDATE EXPERT PANEL. J Am Geriatr Soc. 2019;67:674–694. doi: 10.1111/jgs.15767. [DOI] [PubMed] [Google Scholar]
- Barrera LO, Li Z, Smith AD, Arden KC, Cavenee WK, Zhang MQ, Green RD, Ren B. Genome-wide mapping and analysis of active promoters in mouse embryonic stem cells and adult organs. Genome Res. 2008;18:46–59. doi: 10.1101/gr.6654808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzilai N, Huffman DM, Muzumdar RH, Bartke A. The critical role of metabolic pathways in aging. Diabetes. 2012;61:1315–1322. doi: 10.2337/db11-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benayoun BA, Pollina EA, Brunet A. Epigenetic regulation of ageing: linking environmental inputs to genomic stability. Nat Rev Mol Cell Biol. 2015;16:593–610. doi: 10.1038/nrm4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonder MJ, Kasela S, Kals M, Tamm R, Lokk K, Barragan I, Buurman WA, Deelen P, Greve JW, Ivanov M, Rensen SS, van Vliet-Ostaptchouk J, Wolfs MG, Fu J, Hofker MH, Wijmenga C, Zhernakova A, Ingelman-Sundberg M, Franke L, Milani L. Genetic and epigenetic regulation of gene expression in fetal and adult human livers. BMC Genomics. 2014;15:860. doi: 10.1186/1471-2164-15-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth LN, Brunet A. The aging epigenome. Mol Cell. 2016;62:728–744. doi: 10.1016/j.molcel.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budnitz DS, Pollock DA, Weidenbach KN, Mendelsohn AB, Schroeder TJ, Annest JL. National surveillance of emergency department visits for outpatient adverse drug events. JAMA. 2006;296:1858–1866. doi: 10.1001/jama.296.15.1858. [DOI] [PubMed] [Google Scholar]
- Budnitz DS, Lovegrove MC, Shehab N, Richards CL. Emergency hospitalizations for adverse drug events in older Americans. N Engl J Med. 2011;365:2002–2012. doi: 10.1056/NEJMsa1103053. [DOI] [PubMed] [Google Scholar]
- Cardelli M. The epigenetic alterations of endogenous retroelements in aging. Mech Ageing Dev. 2018;174:30–46. doi: 10.1016/j.mad.2018.02.002. [DOI] [PubMed] [Google Scholar]
- Caro AA, Cederbaum AI. Oxidative stress, toxicology, and pharmacology of CYP2E1. Annu Rev Pharmacol Toxicol. 2004;44:27–42. doi: 10.1146/annurev.pharmtox.44.101802.121704. [DOI] [PubMed] [Google Scholar]
- Dozmorov MG. Polycomb repressive complex 2 epigenomic signature defines age-associated hypermethylation and gene expression changes. Epigenetics. 2015;10:484–495. doi: 10.1080/15592294.2015.1040619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duron E, Hanon O. Hypertension, cognitive decline and dementia. Arch Cardiovasc Dis. 2008;101:181–189. doi: 10.1016/s1875-2136(08)71801-1. [DOI] [PubMed] [Google Scholar]
- ElDesoky ES. Pharmacokinetic-pharmacodynamic crisis in the elderly. Am J Ther. 2007;14:488–498. doi: 10.1097/01.mjt.0000183719.84390.4d. [DOI] [PubMed] [Google Scholar]
- Fisel P, Schaeffeler E, Schwab M. DNA methylation of ADME genes. Clin Pharmacol Ther. 2016;99:512–527. doi: 10.1002/cpt.343. [DOI] [PubMed] [Google Scholar]
- Fishilevich S, Nudel R, Rappaport N, et al. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database (Oxford). 2017. 10.1093/database/bax028. [DOI] [PMC free article] [PubMed]
- French SW. The importance of CYP2E1 in the pathogenesis of alcoholic liver disease and drug toxicity and the role of the proteasome. Subcell Biochem. 2013;67:145–164. doi: 10.1007/978-94-007-5881-0_4. [DOI] [PubMed] [Google Scholar]
- Gravina S, Dong X, Yu B, Vijg J. Single-cell genome-wide bisulfite sequencing uncovers extensive heterogeneity in the mouse liver methylome. Genome Biol. 2016;17:150. doi: 10.1186/s13059-016-1011-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn O, Grönke S, Stubbs TM, Ficz G, Hendrich O, Krueger F, Andrews S, Zhang Q, Wakelam MJ, Beyer A, Reik W, Partridge L. Dietary restriction protects from age-associated DNA methylation and induces epigenetic reprogramming of lipid metabolism. Genome Biol. 2017;18:56. doi: 10.1186/s13059-017-1187-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, Klotzle B, Bibikova M, Fan JB, Gao Y, Deconde R, Chen M, Rajapakse I, Friend S, Ideker T, Zhang K. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49:359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyn H, Li N, Ferreira HJ, Moran S, Pisano DG, Gomez A, Diez J, Sanchez-Mut JV, Setien F, Carmona FJ, Puca AA, Sayols S, Pujana MA, Serra-Musach J, Iglesias-Platas I, Formiga F, Fernandez AF, Fraga MF, Heath SC, Valencia A, Gut IG, Wang J, Esteller M. Distinct DNA methylomes of newborns and centenarians. Proc Natl Acad Sci U S A. 2012;109:10522–10527. doi: 10.1073/pnas.1120658109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:3156. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet. 2018;19:371–384. doi: 10.1038/s41576-018-0004-3. [DOI] [PubMed] [Google Scholar]
- Houston JB (1994) Utility of in vitro drug metabolism data in predicting in vivo metabolic clearance. Biochemical Pharmacology 47(9):1469–1479. 10.1016/0006-2952(94)90520-7 [DOI] [PubMed]
- Hunt CM, Strater S, Stave GM. Effect of normal aging on the activity of human hepatic cytochrome P450IIE1. Biochem Pharmacol. 1990;40:1666–1669. doi: 10.1016/0006-2952(90)90470-6. [DOI] [PubMed] [Google Scholar]
- Ingelman-Sundberg M. Human drug metabolising cytochrome P450 enzymes: properties and polymorphisms. Naunyn Schmiedeberg's Arch Pharmacol. 2004;369:89–104. doi: 10.1007/s00210-003-0819-z. [DOI] [PubMed] [Google Scholar]
- Issa J-P. Epigenetic variation and human disease. J Nutr. 2002;132:2388S–2392S. doi: 10.1093/jn/132.8.2388S. [DOI] [PubMed] [Google Scholar]
- Jia L, Chang X, Qian S, Liu C, Lord CC, Ahmed N, Lee CE, Lee S, Gautron L, Mitchell MC, Horton JD, Scherer PE, Elmquist JK. Hepatocyte toll-like receptor 4 deficiency protects against alcohol-induced fatty liver disease. Mol Metab. 2018;14:121–129. doi: 10.1016/j.molmet.2018.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knights KM, Stresser DM, Miners JO, Crespi CL. In vitro drug metabolism using liver microsomes. Curr Protoc Pharmacol. 2016;74:7.8.1–7.8.24. doi: 10.1002/cpph.9. [DOI] [PubMed] [Google Scholar]
- Koh KH, Xie H, Yu A-M, Jeong H. Altered cytochrome P450 expression in mice during pregnancy. Drug Metab Dispos. 2011;39:165–169. doi: 10.1124/dmd.110.035790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronfol MM, Dozmorov MG, Huang R, Slattum PW, McClay J. The role of epigenomics in personalized medicine. Expert Rev Precis Med Drug Dev. 2017;2:33–45. doi: 10.1080/23808993.2017.1284557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarou J, Pomeranz BH, Corey PN. Incidence of adverse drug reactions in hospitalized patients: a meta-analysis of prospective studies. JAMA. 1998;279:1200–1205. doi: 10.1001/jama.279.15.1200. [DOI] [PubMed] [Google Scholar]
- Lee WM. Public health: acetaminophen (APAP) hepatotoxicity—isn’t it time for APAP to go away? J Hepatol. 2017;67:1324–1331. doi: 10.1016/j.jhep.2017.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesurf R, Cotto KC, Wang G, Griffith M, Kasaian K, Jones SJ, Montgomery SB, Griffith OL, Open Regulatory Annotation Consortium ORegAnno 3.0: a community-driven resource for curated regulatory annotation. Nucleic Acids Res. 2016;44:D126–D132. doi: 10.1093/nar/gkv1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber CS. Cytochrome P-4502E1: its physiological and pathological role. Physiol Rev. 1997;77:517–544. doi: 10.1152/physrev.1997.77.2.517. [DOI] [PubMed] [Google Scholar]
- Lucas D, Ferrara R, Gonzalez E, Bodenez P, Albores A, Manno M, Berthou F. Chlorzoxazone, a selective probe for phenotyping CYP2E1 in humans. Pharmacogenetics. 1999;9:377–388. doi: 10.1097/00008571-199906000-00013. [DOI] [PubMed] [Google Scholar]
- Mach J, Huizer-Pajkos A, Mitchell SJ, McKenzie C, Phillips L, Kane A, Jones B, de Cabo R, Cogger V, le Couteur DG, Hilmer SN. The effect of ageing on isoniazid pharmacokinetics and hepatotoxicity in Fischer 344 rats. Fundam Clin Pharmacol. 2016;30:23–34. doi: 10.1111/fcp.12157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez SM, Bradford BU, Soldatow VY, Kosyk O, Sandot A, Witek R, Kaiser R, Stewart T, Amaral K, Freeman K, Black C, LeCluyse E, Ferguson SS, Rusyn I. Evaluation of an in vitro toxicogenetic mouse model for hepatotoxicity. Toxicol Appl Pharmacol. 2010;249:208–216. doi: 10.1016/j.taap.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marttila S, Kananen L, Häyrynen S, Jylhävä J, Nevalainen T, Hervonen A, Jylhä M, Nykter M, Hurme M. Ageing-associated changes in the human DNA methylome: genomic locations and effects on gene expression. BMC Genomics. 2015;16:179. doi: 10.1186/s12864-015-1381-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll SA, Murphy CT, Zou S, Pletcher SD, Chin CS, Jan YN, Kenyon C, Bargmann CI, Li H. Comparing genomic expression patterns across species identifies shared transcriptional profile in aging. Nat Genet. 2004;36:197–204. doi: 10.1038/ng1291. [DOI] [PubMed] [Google Scholar]
- McClay JL, Aberg KA, Clark SL, Nerella S, Kumar G, Xie LY, Hudson AD, Harada A, Hultman CM, Magnusson PK, Sullivan PF, van den Oord E. A methylome-wide study of aging using massively parallel sequencing of the methyl-CpG-enriched genomic fraction from blood in over 700 subjects. Hum Mol Genet. 2014;23:1175–1185. doi: 10.1093/hmg/ddt511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLachlan AJ, Pont LG. Drug metabolism in older people—a key consideration in achieving optimal outcomes with medicines. J Gerontol A Biol Sci Med Sci. 2012;67A:175–180. doi: 10.1093/gerona/glr118. [DOI] [PubMed] [Google Scholar]
- McLachlan A, Hilmer S, Le Couteur D. Variability in response to medicines in older people: phenotypic and genotypic factors. Clin Pharmacol Ther. 2009;85:431–433. doi: 10.1038/clpt.2009.1. [DOI] [PubMed] [Google Scholar]
- McLean AJ, Le Couteur DG. Aging biology and geriatric clinical pharmacology. Pharmacol Rev. 2004;56:163–184. doi: 10.1124/pr.56.2.4. [DOI] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium. Waterston RH, Lindblad-Toh K, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Newman M, Blyth BJ, Hussey DJ, Jardine D, Sykes PJ, Ormsby RJ. Sensitive quantitative analysis of murine LINE1 DNA methylation using high resolution melt analysis. Epigenetics. 2012;7:92–105. doi: 10.4161/epi.7.1.18815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S, Tyler JK. Epigenetics and aging. Sci Adv. 2016;2:e1600584. doi: 10.1126/sciadv.1600584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H-J, Choi Y-J, Kim JW, Chun HS, Im I, Yoon S, Han YM, Song CW, Kim H. Differences in the epigenetic regulation of cytochrome P450 genes between human embryonic stem cell-derived hepatocytes and primary hepatocytes. PLoS One. 2015;10:e0132992. doi: 10.1371/journal.pone.0132992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters MJ, Joehanes R, Pilling LC, et al. The transcriptional landscape of age in human peripheral blood. Nat Commun. 2015;6. 10.1038/ncomms9570. [DOI] [PMC free article] [PubMed]
- Porubsky PR, Meneely KM, Scott EE. Structures of human cytochrome P-450 2E1. J Biol Chem. 2008;283:33698–33707. doi: 10.1074/jbc.M805999200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds LM, Taylor JR, Ding J, Lohman K, Johnson C, Siscovick D, Burke G, Post W, Shea S, Jacobs DR Jr, Stunnenberg H, Kritchevsky SB, Hoeschele I, McCall C, Herrington D, Tracy RP, Liu Y. Age-related variations in the methylome associated with gene expression in human monocytes and T cells. Nat Commun. 2014;5:5366. doi: 10.1038/ncomms6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routledge PA, O’Mahony MS, Woodhouse KW. Adverse drug reactions in elderly patients. Br J Clin Pharmacol. 2004;57:121–126. doi: 10.1046/j.1365-2125.2003.01875.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruoß M, Damm G, Vosough M, et al. Epigenetic modifications of the liver tumor cell line HepG2 increase their drug metabolic capacity. Int J Mol Sci. 2019;20:347. doi: 10.3390/ijms20020347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarzello AJ, Jiang Q, Back T, Dang H, Hodge D, Hanson C, Subleski J, Weiss JM, Stauffer JK, Chaisaingmongkol J, Rabibhadana S, Ruchirawat M, Ortaldo J, Wang XW, Norris PS, Ware CF, Wiltrout RH. LTβR signalling preferentially accelerates oncogenic AKT-initiated liver tumours. Gut. 2016;65:1765–1775. doi: 10.1136/gutjnl-2014-308810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmucker DL, Woodhouse KW, Wang RK, Wynne H, James OF, McManus M, Kremers P. Effects of age and gender on in vitro properties of human liver microsomal monooxygenases. Clin Pharmacol Ther. 1990;48:365–374. doi: 10.1038/clpt.1990.164. [DOI] [PubMed] [Google Scholar]
- Seripa D, Panza F, Daragjati J, Paroni G, Pilotto A. Measuring pharmacogenetics in special groups: geriatrics. Expert Opin Drug Metab Toxicol. 2015;11:1073–1088. doi: 10.1517/17425255.2015.1041919. [DOI] [PubMed] [Google Scholar]
- Slone Epidemiology Center. Patterns of medication use in the United States - a report from the Slone Survey. Boston University; 2006.
- Spiers H, Hannon E, Wells S, Williams B, Fernandes C, Mill J. Age-associated changes in DNA methylation across multiple tissues in an inbred mouse model. Mech Ageing Dev. 2016;154:20–23. doi: 10.1016/j.mad.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatoyannopoulos JA, Snyder M, Hardison R, et al. An encyclopedia of mouse DNA elements (Mouse ENCODE) Genome Biol. 2012;13:418. doi: 10.1186/gb-2012-13-8-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steegenga WT, Boekschoten MV, Lute C, Hooiveld GJ, de Groot PJ, Morris TJ, Teschendorff AE, Butcher LM, Beck S, Müller M. Genome-wide age-related changes in DNA methylation and gene expression in human PBMCs. AGE. 2014;36:9648. doi: 10.1007/s11357-014-9648-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultana J, Cutroneo P, Trifirò G. Clinical and economic burden of adverse drug reactions. J Pharmacol Pharmacother. 2013;4:S73–S77. doi: 10.4103/0976-500X.120957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tammen SA, Dolnikowski GG, Ausman LM, Liu Z, Sauer J, Friso S, Choi SW. Aging and alcohol interact to alter hepatic DNA hydroxymethylation. Alcohol Clin Exp Res. 2014;38:2178–2185. doi: 10.1111/acer.12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tost J, Dunker J, Gut IG. Analysis and quantification of multiple methylation variable positions in CpG islands by pyrosequencing. BioTechniques. 2003;35:152–156. doi: 10.2144/03351md02. [DOI] [PubMed] [Google Scholar]
- White RR, Milholland B, MacRae SL, Lin M, Zheng D, Vijg J. Comprehensive transcriptional landscape of aging mouse liver. BMC Genomics. 2015;16:899. doi: 10.1186/s12864-015-2061-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm A, Aldridge V, Haldar D, Naylor AJ, Weston CJ, Hedegaard D, Garg A, Fear J, Reynolds GM, Croft AP, Henderson NC, Buckley CD, Newsome PN. CD248/endosialin critically regulates hepatic stellate cell proliferation during chronic liver injury via a PDGF-regulated mechanism. Gut. 2016;65:1175–1185. doi: 10.1136/gutjnl-2014-308325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson VL, Smith RA, Ma S, Cutler RG. Genomic 5-methyldeoxycytidine decreases with age. J Biol Chem. 1987;262:9948–9951. [PubMed] [Google Scholar]
- Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 2013;138:103–141. doi: 10.1016/j.pharmthera.2012.12.007. [DOI] [PubMed] [Google Scholar]
- Zhu T, Zheng SC, Paul DS, et al. Cell and tissue type independent age-associated DNA methylation changes are not rare but common. Aging (Albany NY) 2018;10:3541–3557. doi: 10.18632/aging.101666. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 748 kb)