Abstract
The NIN-LIKE PROTEIN (NLP) family of transcription factors were identified as nitrate-responsive cis-element (NRE)-binding proteins, which function as transcriptional activators in the nitrate-regulated expression of downstream genes. This study was aimed at genome-wide analysis of NLP gene family in rice and the expression profiling of NLPs in response to nitrogen (N) supply and deficiency in rice genotypes with contrasting N use efficiency (NUE). Based on in silico analysis, 6 NLP genes (including alternative splice forms 11 NLPs) were identified from rice. Expression of NLPs was promoted by nitrate supply as well as N deficiency (NLP1, NLP3, NLP4 and NLP5). Four rice genotypes APO (high NUE under sufficient N), IR83929-B-B-291-3-1-1 (IR-3-1-1), Nerica-L-42 (NL-42) (High NUE at low N), and Pusa Basmati 1 (PB1, low NUE) to correlate traits governing NUE and expression of NLPs. Analysis of rate of nitrate uptake and expression of N assimilatory and uptake genes established that IR-3-1-1 has high uptake and assimilation efficiency, translating into high NUE, whereas PB1 is efficient in uptake only when N availability is high. Along with the transcriptional upregulation of NLPs, genotype IR-3-1-1, displayed highest expression of OsNRT1.1B gene, the closest rice homologue of nitrate transceptor AtNRT1.1 and plays major role in nitrate uptake, translocation and signaling in rice. The results showed that high NUE rice genotypes has both high Nitrogen uptake efficiency (NUpE) and Nitrogen utilization efficiency (NUtE), resulting from the effective and coordinated signal transduction network involving the rice homologue of nitrate transceptor OsNRT1.1B, the probable primary nitrate response (PNR) regulator OsNLP1 and the master response regulator OsNLP3, a homologue of AtNLP6/7.
Subject terms: Plant sciences, Plant physiology
Introduction
Nitrogen (N) is an essential nutrient and major component of proteins, chlorophyll, nucleotides and plant hormones, and therefore has immense role in determining plant growth and economic yield1,2. In order to meet the food demand of ever-growing human population, enormous amounts of N fertilizers are applied inorder to tap the maximum crop yield potential worldwide3. The global demand for N fertilizers in 2014 was 1.13 M tonnes and is projected to grow at approximately 1.4% per year, reaching 1.22 M tonnes by 20204. On the other hand, around 50% of the applied N fertilizer is lost to the environment depending on the cropping conditions and plant species. The loss of fertilizer N results in contamination of soil water and water bodies and production of nitrogenous greenhouse gases like nitrous oxide (N2O) which has high global warming potential5. Nitrogen use efficiency (NUE) of rice is particularly low (around 40%), though genetic variation for the trait has been reported6. Consequently, there is an impending requirement to improve the NUE of rice to maintain the steadiness of high crop yields vis-a vis low N fertilizer inputs7.
Transgenic manipulation is one of the potent way to achieve the current demand for high NUE, which necessitates comprehensive understanding of mechanisms regulating N uptake, transport, assimilation and signaling8. Amongst various N forms that plants are able to take up, nitrate is the most abundant form of N in aerobic agricultural soils and is prone to leaching due to its chemical nature9. Recent finding indicates the role of nitrate as a signalling molecule along with its nutritional role in plants10. Components mediating nitrate signaling mechanism were recognized in recent past11. In particular; the regulator of nitrate assimilation, Nitrilase (NIT2), containing DNA-binding RWP-RK domain, was a found to be a key regulator of nitrate signalling in Chlamydomonas12. NIT2 protein is structurally similar to NODULE INCEPTION (NIN) proteins of leguminous plants12. In higher plants the first NIN gene was identified in Lotus japonicus and imperative for symbiotic nodule formation13. NIN protein also contains Phox and Bem1 (PB1) domain with the conserved RWP-RK domain, and it is considered the founding member of the NIN-like proteins (NLPs) in plants14. Phylogenetic analyses identified homologues of NIN proteins in legumes, NLPs in both legumes and other non- legumes such as rice, Arabidopsis, wheat, and maize14,15.
Recently, several studies enhanced the comprehensive understanding on nitrate signalling and its role in the regulation of N uptake and assimilation16,17. Konishi and Yanagisawa18 deciphered that, AtNLPs play important role in nitrate signaling by bimding to nitrate-responsive cis-elements (NREs) and coordinates nitrate-regulated expression of genes19. The discovery of nitrate-CPK-NLP network threw more light on the role of NLPs in nitrate signalling and the importance of phosphorylation of NLPs in nutrient-growth coordination20. Liu et al. (2017) also proposed CPK10, CPK30, and CPK32, as major calcium mediated regulators in primary nitrate responses (PNR), and the downstream signaling20. Ca2+chelator EGTA (ethylene glycol-bis (β-aminoethyl ether)-N, N,N′,N′-tetra acetic acid) could arrest phosphorylation of NLP7 by CPK10. Approximately 50 transcription factor encoding genes including NLP7 were targeted by nitrate-CPK module21. These findings clearly points out that manoeuvring Ca2+- CPK–NLP signalling cascade is indeed an efficient strategy for improving the NUE22. Further, the roles of NLPs in the N starvation adaptation, nodulation, N and phosphate (P) interactions, and root growth have been clarified in recent years23. Loss of major part of applied N as gaseous N in the submerged paddies is one of the reason for low NUE in rice24. However, aerobic rice soils have a ratio of 6.5:1 of nitrate and ammonical N25, indicating a significant role of understanding nitrate signaling for improving rice NUE in drought prone agriculture situations. Current study encompasses the genome wide analysis of NLPs from rice, it’s in silico expression profiling and expression characterisation in response to N treatments in rice genotypes having contrasting NUE. We hypothesize that differential expression of NLPs may have regulatory role in NUE of contrasting rice genotypes.
Materials and Methods
Genome wide identification and Chromosomal localization of NLP homologues
Genome sequence of Rice Genome Annotation Project Release 7 (RGAP)26 was Basic local alignment search tool (BLAST) searched using Maize /Arabidopsis candidate NLP genes. The nucleotide (gene and cDNA) sequences and protein sequences were downloaded for further use. Protein sequences were used for protein BLAST to verify conserved RWP-RK and PB1 domains (RWP-RK. hmm, PF02042; PB1.hmm, PF00564). Information about the physical locations of all identified NLP genes on chromosomes was mapped using CIRCOS tool (www.circos.ca/).
Gene structure prediction and sub cellular protein localisation of NLP homologues
Gene structure of NLPs, showing their exon-intron boundaries and UTR regions, was generated using GSDS server (http://gsds.cbi.pku.edu.cn/). Sub-cellular localisation of NLP proteins was predicted using Target P1.1 (www.cbs.dtu.dk/services/TargetP/).
Identification of NLP homologues and their putative promoter elements
NLP sequences were searched in Plant Ensembl Database (www.plants.ensembl.org/) in each rice species and sub-species and then by name/function search. The procedure for identification of homologues and promoter elements were followed as described in Verma et al.27. The protein sequences of the identified NLP members were then used as queries in multiple databases to ensure that no additional related genes were missed from the database. Gene and cDNA sequences of 11 rice species, Oryza barthii, Oryza brachyantha, Oryza glaberrima, Oryza glumipatula, Oryza longistaminata, Oryza meridionalis, Oryza nivara, Oryza punctata, Oryza rufipogon, Oryza sativa Indica Group, Oryza sativa Japonica Group, were retrieved for further use. Promoter sequence (1000 bp upstream of 5’ UTR) of 11 rice species were retrieved from Plant Ensembl Database (www.plants.ensembl.org/) or Plant PAN (http://plantpan2.itps.ncku.edu.tw) and analysed for the presence of important cis-regulatory elements using PLANTCARE (http://bioinformatics.psb. ugent.be/webtools/plantcare/html/).
Phylogenetic analysis of NLPS from Oryza spp
The protein sequences of putative NLPS downloaded from RGAP and EnsemblPlants (http://plants.ensembl.org/index.html) were used for phylogenetic analysis using Molecular Evolutionary Genetics Analysis software version 7.0 (MEGA7)28. For tree construction, the amino acid sequences were aligned using the Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo) program. Neighbour joining analysis was performed with the pair-wise deletion option with Poisson correction, bootstrap analysis was conducted with 1000 replicates and the Poisson correction method was used for computing evolutionary distances.
In-silico expression analysis of NLPS from rice using Genvestigator and NCBI-GEO
Bioinformatics analysis of publically available transcriptome data was used to decipher expression response of NLPs in response to various stress treatments and developmental stages. The in silico gene expression analysis was done by Genevestigator software29 (https://genevestigator.com/gv/index.jsp). The locus IDs of all the genes were given as input in the development and perturbations tool and searched against experiments involving anatomy, development, hormones, nutrients, drought, salt, cold stresses, germination and others. Expression data of relevant experiments (GSE61370 and GSE66807) were downloaded from NCBI-GEO (https://www.ncbi.nlm.nih.gov/gds) and analysed to study NLPs expression.
Analysis of protein-protein interaction networks
To study protein-protein interaction network, NLP protein sequences were analyzed in SMART (http://smart.embl-heidelberg.de/) followed by prediction of interaction partners and networks using STRING tool (http://string-db.org/).
Plant material
Based on our previous lab and field experiments (unpublished) four rice genotypes APO (high NUE under sufficient N), IR83929-B-B-291-3-1-1 and Nerica-L-42 (High NUE at low N), and Pusa Basmati 1 (PB1) (N responsive and low NUE) were selected for studying the gene expression and N responses. Genotypes were collected from Division of Genetics, ICAR-IARI and Division of Crop Research, ICAR Research Complex for Eastern Region, Patna, Bihar.
Field evaluation of rice genotypes and determination of NUE
The plants were raised during Kharif season (June-Oct 2018) under field conditions with recommended N, 120 Kg ha−1 (N+) and without N fertilizer application (N-) at Division of Plant Physiology, ICAR-IARI, New Delhi. Seeds of the rice varieties were washed with double distilled water and then surface sterilized with 0.1% mercuric chloride HgCl2 for 5 min. To remove the traces of HgCl2, seeds were thoroughly washed for 5-6 times with double distilled water. The surface sterilized seeds were planted in nursery by broadcasting method and 25-30 days old seedlings were transplanted into the field with identification tags for each genotype. The average soil N content before transplanting of crop was 198 Kg ha−1 in N + plot and 148 Kg ha−1 in N− plot. Full dosage of P2O5 (as Single Super Phosphate) and K (as Muriate of Potash) was applied as a basal dose. 50% N was applied during field preparation before planting in N + field, remaining 50% was applied as 2 split doses during early- and late- vegetative stages of the crop. At physiological maturity, plants were harvested, oven dried and biomass and grain yield per plant (g) were recorded. Plant material was then powdered and N content of different tissues (leaf, stem and grain) were estimated following Kjeldahl’s method30. Nitrogen utilization efficiency (NUtE) was calculated as (g grain/g total N uptake).
Hydroponic culture rice seedlings for determination of root traits
Plants were raised in hydroponics (Supplementary Fig. 1) in controlled environment glass house (National Phytotron Facility, ICAR-IARI). Plants were supplied with three different nitrogen treatments, High nitrate: Low ammonia (T1: 6.5 mM Nitrate: 1 mM Ammonium), Low nitrate: High ammonia (T2: 6.5 mM Ammonium: 1 mM Nitrate), Low N (T3: 0.24 mM Ammonium Nitrate). The N treatments were selected based on the previous reports that aerobic rice soils have a ratio of 6.5:1 of nitrate and ammonical nitrogen31 and for screening of rice genotypes for low N tolerance, 0.24 mM of N is optimum32. Seeds of the rice genotypes were sterilized with 0.1% (HgCl2) and wrapped in moistened germination paper. Five days post germination; seedlings having similar growth were used for experimental treatments. The plants were held with cotton plugs in Styrofoam sheets kept on plastic trays containing 10 liters of nutrient solution. Two seedlings were maintained per hill and each treatment comprised of at least three trays. After every 4 days, nutrient solution was changed, pH of the nutrient solution was maintained using a pocket pH meter at 5.0 throughout the study. The growing media was prepared in de-ionized water with modified IRRI nutrient solution (Yoshida et al.,1972) and the composition of the nutrient solution is as follows, 0.035 mM K2SO4, 0.1 mM magnesium sulphate hepta hydrate (MgSO4 7H2O), 0.1 mM calcium chloride (CaCl2), 0.1 mM potassium bi sulphate (KH2PO4), 0.01 mM micronutrient solution (H3BO3, MnCl2 4H2O, ZnSO4 7H2O, CuSO4 5H2O, Na2MoO4.2H2O). Concentration of nitrate and ammonium ions was adjusted depending on the treatments using 1 M potassium nitrate (KNO3), 1 M ammonium sulphate ((NH4)2SO4) and 1 M ammonium nitrate (NH4 NO3). Nitrification inhibitor Dicyandiamide32 was also included in the media to prevent conversion of ammonium to nitrate.
After thirty days of transfer to treatment trays, plants were harvested to record different observations (Supplementary Fig. 2). For recording root traits, roots were scanned in a root scanner (Epson, Expression 11000XL, Graphic Art Model) representative plants were taken for each replication and scanning was done in triplicates for each treatment. Root scanning data were analyzed by Win-RHIZO, Regent Instruments to calculate total root length, total root volume, total root surface area, average diameter, no of root tips.
Rate of nitrate uptake in rice genotypes
Seeds of rice genotypes were germinated and transplanted to trays containing nutrient solution as described earlier. Plants were provided with T1 treatment for 2 weeks followed by nitrogen deprivation (0 mM N) for one week. The seedlings were shifted to growth chambers (Model PGW 36, Conviron, Winnipeg, Canada) maintained at: day/night temperature 27 °C/18 °C, photoperiod-10 h, photon flux density of 400 μmol m−2 s−1 (PAR) and relative humidity (RH) - 80 to 90%. After another week of N deprivation in the growth chambers, seedlings were incubated in 100 µM, and 1.0 mM nitrate concentration for two hours to study kinetics of nitrate uptake. The photon flux density (PAR) was raised to 500 μmol m−2 s−1 during incubation. Rate of nitrate uptake (µmol NO3− g−1 FW h−1)33 was computed based on the nitrate remaining in the incubation solution. Estimation of nitrate in the medium and dried tissue powder was based on hydrazine sulphate reduction method34,35. Solution nitrate content was also confirmed using nitrate selective electrode (Go Direct Nitrate Ion-Selective Electrode, Vernier).
Expression of nitrogen metabolism and NLP genes in rice
Rice genotypes were germinated as described earlier; seedlings were then shifted to glass tubes holding 50 ml nutrient solution and were held with cotton plugs. Two seedlings were maintained per tube and each treatment comprised of at least 25 tubes. The whole experiment was laid out at National Phytotron Facility, Indian Agricultural Research Institute (IARI), New Delhi in growth chambers (Model PGW 36, Conviron, Winnipeg, Canada) maintained at following growth conditions: day/night temperature 27 °C/18 °C, photoperiod-10 h, photon flux density of 400 μmol m−2 s−1 (PAR) and relative humidity (RH) - 80 to 90%. Plants were provided with T1 treatment for 2 weeks followed by nitrogen deprivation (0 mM N) for two weeks. Plants were then supplied with three different nitrogen treatments, High nitrate: Low ammonia ratio (T1: 6.5 mM Nitrate: 1 mM Ammonium), Low nitrate: High ammonia (T2: 6.5 mM Ammonium: 1 mM Nitrate), Low N (T3: 0.24 mM Ammonium Nitrate) for 24 hours, leaf and root tissues were sampled includes three biological replication in liquid nitrogen for RNA extraction. Since amplicons of NLP1 was not detected in samples after 24-hour exposure to treatments, the time course of NLP1 expression after 30 min, 2 hr, 24 hr, 48 hr and 72 hr after exposure to 7.5 mM nitrate was analyzed in high NUE genotype IR-3-1-1. Total RNA was extracted using QIAGEN RNeasy plant mini kit followed by DNase I treatment to obtain DNA free RNA. RNA quantification and purity check were done using Thermo nanodrop 2000c spectrophotometer. First strand cDNA was synthesized from 1 µg of total RNA using Superscript–III reverse transcriptase (Invitrogen, USA). To study the expression level of candidate genes, qRT-PCR was carried using Power SYBR Green Master Mix (Applied Biosystems, USA) on real time PCR detection system (Applied Biosystems). qRT-PCR was done using gene specific primers (Supplementary Table 1). Melt curve data collection and analysis was enabled. qRT-PCR products were also visualised by agarose gel electrophoresis to confirm the single specific amplicon. Normalization of the data for each transcript was carried out using OsUBQ1 as an internal control and level of expression were analysed using ∆CT and 2−∆∆CT methods36.
Statistical analysis
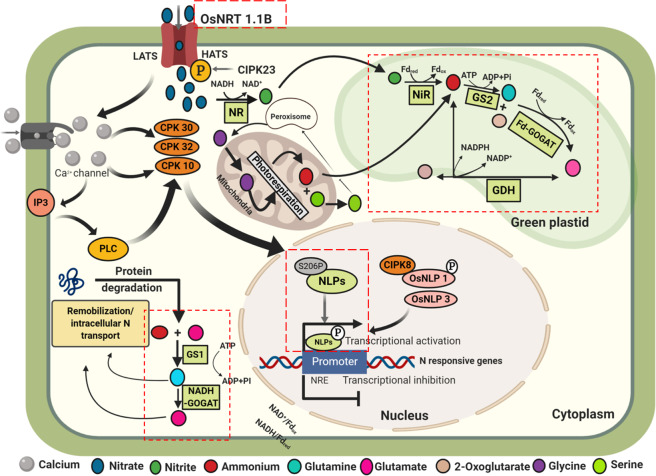
Two-way analysis of variance (ANOVA) was carried out in GraphPad Prism version 8 (La Jolla, California, USA) with variety, N treatments as treatment effects to compute adjusted P values and level of significance. Mean separation was done using Sidak’s multiple comparisons test following one-way ANOVA37 (Supplementary Tables 2–5). Graphs and heatmaps were prepared using GraphPad Prism version 8 (La Jolla, California, USA) and illustrations in Fig. 9 was made with Biorender (https://biorender.com/).
Figure 9.
Schematic model summarizing gene regulation by nitrogen in plants. Illustrations was made with Biorender (https://biorender.com/) based on the information available in the public domain. The transcriptional changes of the steps analyzed in the study are enclosed in dashed outlines.
Results
Rice genotypes showed variation in different traits governing NUE
To correlate the role of NLPs in governing NUE of rice, we used four rice genotypes identified based on previous lab studies (unpublished). The variability in traits governing NUE such as N content (N%), rate of nitrate uptake, root system architechture and NUE are presented.
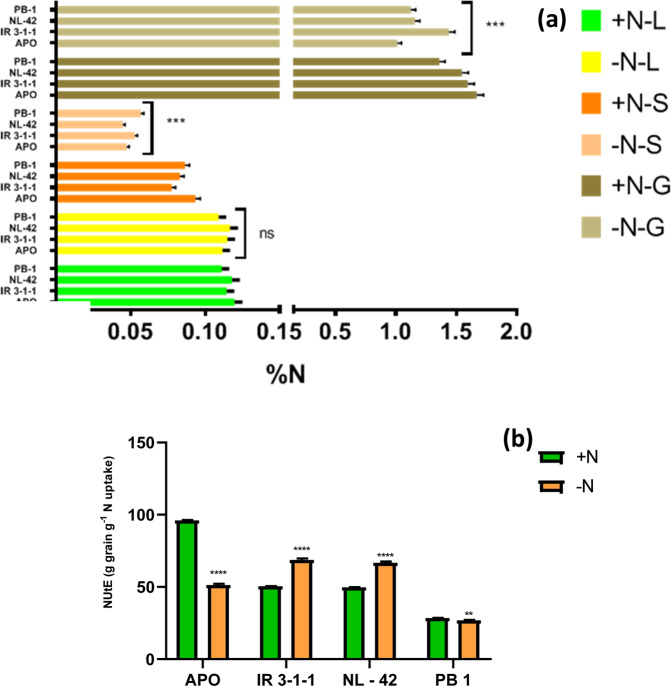
Plants grown under N- showed overall decrease in N % of leaves, culm and grain in all the genotypes. However, differences in treatment means were significant only in culm and grains. In culms there was 40% reduction in N content in N- w.r.to N + treatment, whereas in grains the reduction was 23%. There was significant treatment effect on % reduction in varietal means of culm and grain N%: APO (49% and 39%), IR-3-1-1 (32% and 9%), NL-42 (46% and 25%) and PB1 (34% and 17%) as shown in Fig. 1a. Under low N, maximum culm N was shown by PB1 (indicating impaired N remobilization). Highest Grain N% was displayed by IR-3-1-1 followed by NL-42. Varietal means of NUtE (Fig. 1b) were significantly different as indicated by Sidak’s multiple comparisons test: with respect to N + treatment N- treatment means decreased by 46% and 5.6% in APO and PB1 whereas increased by 36% and 34% in IR-3-1-1 and NL-42 respectively.
Figure 1.
Effect of nitrogen deficient (no applied N: N−) and nitrogen sufficient (120 kg ha−1 applied N: N+) field conditions on (a) Tissue N% (L-leaf, S-culm, G-grain) (b) nitrogen utilization efficiency (NUtE) of rice genotypes Apo, IR-83929-B-B-291-3-1-1 (IR-3-1-1), and Nerica L-42 (NL-42), and Pusa Basmati 1 (PB1). Values are means (±SE) of 3 biological replicates. Sidak’s multiple comparisons test for influence of +N and –N is indicated with astericks. (P values less than 0.001 are summarized with three asterisks, and P values less than 0.0001 are summarized with four asterisks).
The rate of nitrate uptake was studied at 2 h after incubating rice seedlings in either at high (1 mM) or low (100 µM) nitrate concentrations (Fig. 2). Significant difference in rate of uptake was observed among genotypes, IR-3-1-1 and APO displayed higher rate of uptake in response to low nitrate supply. Whereas IR-3-1-1 and PB1 were more efficient in taking up nitrate as the nitrate availability was higher. PB1 being a N responsive genotype is efficient in uptake when N availability is high, IR-3-1-1 has high uptake efficiency, at both high and low external nitrate concentrations.
Figure 2.
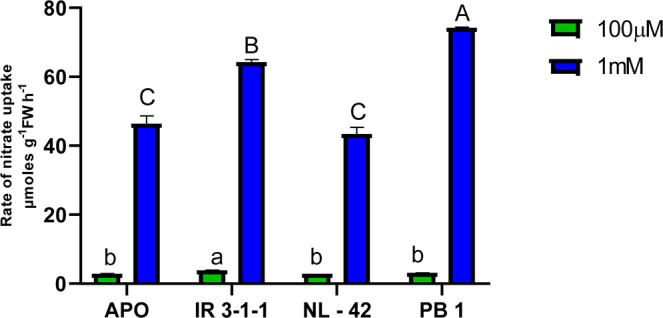
Comparison of rate of nitrate uptake (µmol g−1 FW h−1) of rice genotypes Apo, IR-83929-B-B-291-3-1-1 (IR-3-1-1), and Nerica L-42 (NL-42), and Pusa Basmati 1 (PB1) in hydroponics and receiving 0.1 and 1.0 mM nitrate treatments. Values are means (±SE) of 3 biological replicates. Duncan’s multiple comparisons test for varietal differences indicated with different letters for each nitrate level.
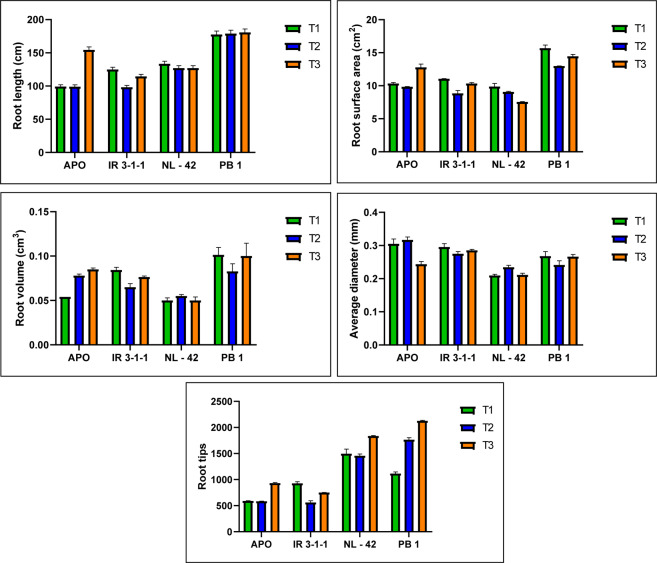
Rate of nitrate uptake is often determined by root system responses. Significant difference was observed in root traits viz., total root length, total root volume and total root surface area with N treatments and varieties (Fig. 3a–e). Nitrate nutrition (T1) and n deficiency (T3) triggered root growth and root surface area in rice. N deficiency also significantly promoted root proliferation. Genotypic variation in root traits was also significant. Root proliferation also has significant correlation with nitrate uptake kinetics. Genotypes PB1 and IR-3-1-1 were having statistically significant robust root system. The treatment effects on root diameter were not significant; however, the genotypic effects were significant.
Figure 3.
Comparison of total root parameters of rice genotypes Apo, IR-83929-B-B-291-3-1-1 (IR-3-1-1), and Nerica L-42 (NL-42), and Pusa Basmati 1 (PB1) in hydroponics and receiving different nitrogen treatments; T1: 6.5 mM Nitrate: 1 mM Ammonium, T2: 6.5 mM Ammonium: 1 mM Nitrate, T3: 0.24 mM Ammonium Nitrate. Values are means (±SE) of 3 biological replicates. Values are means (±SE) of 3 biological replicates. Sidak’s multiple comparisons test for varietal differences indicated with astericks. (P values less than 0.001 are summarized with three asterisks, and P values less than 0.0001 are summarized with four asterisks).
Genome wide identification and analyses of rice NLP homologues
Genome sequence of Rice Genome Annotation Project Release 7 (RGAP26) was BLAST searched for Arabidopsis candidate NLP genes. Protein sequences were used for protein BLAST to verify conserved RWP-RK and PB1 domains (RWP-RK. hmm, PF02042; PB1.hmm, PF00564). Six NLPs were identified from Oryza sativa Japonica. These genes were named as NLP1 to NLP 6 based on similarity to Arabidopsis NLPs and previous reports in rice14. Including alternative splice forms 11 NLPs were identified in rice genome (Table 1) Gene structure of NLP genes, showing their exon-intron boundaries and UTR regions, was generated using GSDS server. Number of exons ranged from 1 in NLP5.5 to 7 in NLP6. There were four exons in NLP1, five exons each in NLP2 and 3, four in NLP4 (Supplementary Fig. 3a). Information about the physical locations of all identified NLP genes on chromosomes (Chr.) was mapped using CIRCOS tool (www.circos.ca/) (Supplementary Fig. 3b). NLPs were located in 6 out of 12 chromosomes in rice: NLP3 in Chr.1, NLP6 in Chr.2, NLP2 in Chr.4, NLP4 in Chr.9, NLP5 in Chr.11 and NLP1 in Chr.3. Subcellular localisation of NLP proteins was studied by using Target P (Table 2). Location with the highest score is the most likely according to Target P. Except NLP5.3, NLP5.5 (predicted as a secretory peptide) and NLP5.4 (predicted as mitochondria localised), NLPs are preferably having either cytosolic or nuclear localisation in agreement with At NLPs.
Table 1.
Details of NLP homologues in Oryza sativa Japonica.
| Gene name | Locus ID | Locus ID Of alternative splice forms |
|---|---|---|
| OsNLP1 | LOC_Os03g03900.1 | — |
| OsNLP2 | LOC_Os04g41850.1 | — |
| OsNLP3 | LOC_Os01g13540.1 | — |
| OsNLP4 | LOC_Os09g37710 |
LOC_Os09g37710.1(NLP4.1) LOC_Os09g37710.2 (NLP4.2) |
| OsNLP5 | LOC_Os11g16290 |
LOC_Os11g16290.1 (NLP5.1) LOC_Os11g16290.2 (NLP5.2) LOC_Os11g16290.3 (NLP5.3) LOC_Os11g16290.4 (NLP5.4) LOC_Os11g16290.5 (NLP5.6) |
| OsNLP6 | LOC_Os02g04340.1 | — |
Table 2.
Protein localization of NLP homologues in Oryza sativa Japonica predicted by TargetP 1.1 Server. Reliability Class (RC) is a measure of the size of the difference (‘diff’) between the highest (winning) and the second highest output scores.
| Name | Len | Chloroplast Transit Peptide (cTP) | Mitochondral targeting Peptide mTP | Signal Peptide (SP) | Others | Loc | Reliability Class (RC) |
|---|---|---|---|---|---|---|---|
| OsNLP1 | 942 | 0.187 | 0.084 | 0.051 | 0.897 | — | 2 |
| OsNLP2 | 936 | 0.087 | 0.114 | 0.046 | 0.920 | — | 1 |
| OsNLP3 | 938 | 0.281 | 0.037 | 0.012 | 0.884 | — | 2 |
| OsNLP4.1 | 842 | 0.066 | 0.121 | 0.040 | 0.847 | — | 2 |
| OsNLP4.2 | 877 | 0.144 | 0.055 | 0.031 | 0.902 | _ | 2 |
| OsNLP5.1 | 886 | 0.187 | 0.034 | 0.086 | 0.566 | _ | 4 |
| OsNLP5.2 | 858 | 0.187 | 0.034 | 0.086 | 0.566 | _ | 4 |
| OsNLP5.3 | 329 | 0.009 | 0.063 | 0.801 | 0.105 | S | 2 |
| OsNLP5.4 | 299 | 0.081 | 0.677 | 0.011 | 0.392 | M | 4 |
| OsNLP5.5 | 274 | 0.012 | 0.165 | 0.854 | 0.171 | S | 2 |
| OsNLP6 | 669 | 0.022 | 0.284 | 0.048 | 0.898 | — | 2 |
| Cut-off | 0.730 | 0.860 | 0.430 | 0.840 | |||
There are 5 reliability classes, defined as 1: diff > 0.800, 2: 0.800 > diff > 0.600, 3: 0.600 > diff > 0.400, 4: 0.400 > diff > 0.200, 5: 0.200 > diff.
The protein sequences of putative OsNLPS were downloaded from RGAP, and used for BLAST search of Arabidopsis thaliana genome (TAIR10) at Ensembl Plants (http://plants.ensembl.org/index.html) to identify the homologs of OsNLPs from Arabidopsis and other eleven rice spp. The protein sequences of rice NLPs and their Arabidopsis homologs were used for phylogenetic analysis using Molecular Evolutionary Genetics Analysis software version 7.0 (MEGA7)28. Based on phylogenetic analysis rice NLPs were classified in to 3 groups. Group I is the smallest group consists of four rice (NLP6, NLP6.1, NLP5, NLP5.1) and three Arabidopsis NLPs (NLP1, NLP2 and NLP3). Group II consists of twenty one rice (NLP1, NLP3, NLP4) and two Arabidopsis NLPs (NLP4 and NLP5). Group III consists of forty-two rice (NLP2, NLP3, NLP4, NLP5, NLP6) and four Arabidopsis (NLP6, NLP7, NLP8 and NLP9) NLPs (Supplementary Fig. 4). Based on the bootstrap values and tree branching, rice NLP3 and NLP4 and Arabidopsis NLP6 and NLP7 can be considered as paralogs.
The protein sequences of the identified NLP members were then used as queries in Plant Ensembl Database to collect homologues in each rice species and sub-species. Gene, cDNA and putative promoter sequences of eleven rice species, Oryza barthii, Oryza brachyantha, Oryza glaberrima, Oryza glumipatula, Oryza longistaminata, Oryza meridionalis, Oryza punctata, Oryza rufipogon, Oryza sativa Indica Group, Oryza sativa Japonica Group, Oryza nivara were retrieved for further use. The information on NLP homologues, the chromosome localization, protein Length (aa) and molecular weight (kDa) are compiled as (Supplementary Table 6). Promoter sequence (1000 bp upstream of 5’ UTR) were analysed for the presence of cis-regulatory elements using PLANTCARE (http://bioinformatics.psb. ugent.be/web tools/plantcare/html/). Total 95 putative promoter elements were found (including few unnamed elements) in eleven Rice species. Putative promoter elements (RE) including Abscisic acid responsive element (ABA RE), Auxin RE, Light RE, Anaerobic RE, Gibberellin RE, drought RE, salicylic acid RE, methyl jasmonate (MeJA) RE were identified, REs related to defence and stress responsiveness, circadian control, cell cycle regulation, in seed-specific regulation were also found. Light responsive elements were most represented followed by ABA RE. NLPs bind to NREs and regulate expression of N- metabolism and signaling genes in response to external cues (Supplementary Table 7).
In silico Expression analysis of NLPS
To infer the regulatory role of NLPs in rice development and stress response the expression potential was retrieved from Genevestigator. Expression potential is the normalized expression value for a gene across all experiments available in the database, and the darkest red color represents the “maximum” level of expression for the given probe.
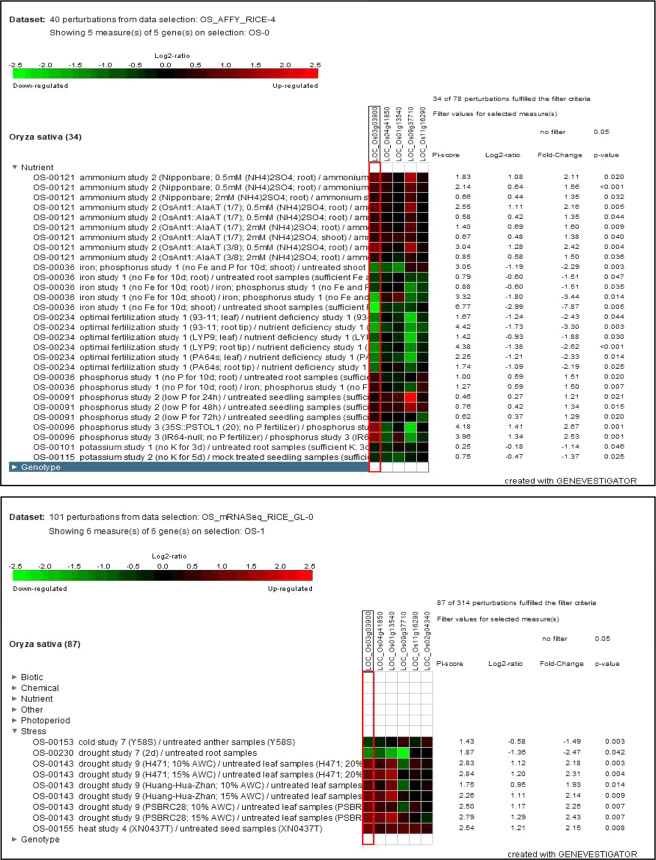
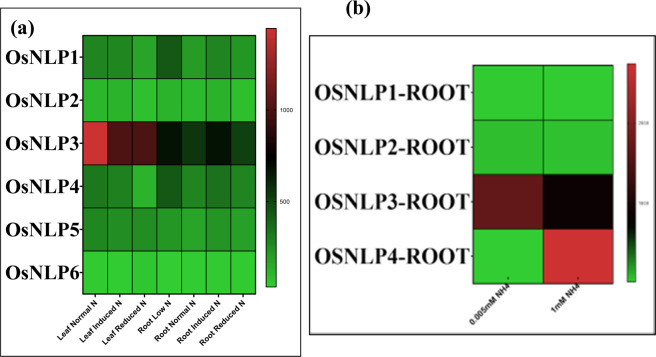
Expression of five NLPs (except NLP6) in 22 anatomical parts and six NLPs in 20 parts were analysed in silico in microarray and mRNA seq data (Supplementary Fig. 5). High expression in Flag leaf, shoot and callus. Tissue specificity of expression implies role in development and coordination of developmental triggers. Most of the NLPs showed moderate to high levels of expression in different developmental stages of rice. High expression of NLPs coincides with early vegetative stage, early and late reproductive phases. Stage specificity of expression implies role in crop establishment and reproductive success in response to nutritional and environmental cues. Stress responsive expression pattern of NLPs were analysed using transcriptome data of nutrient level, hormones drought, cold and salt stresses (Supplementary Figs. 5–7). Expression of five NLPs (except NLP6) in were differentially regulated in response to ABA, salicylic acid and Trans-zeatin treatments (P ≤ 0.05). The presence of the respective putative promoter elements in NLP promoter region reinstates the expression data. The hormone responsive expression also indicates the role of NLPs in sensing and transducing hormone signals to modulate downstream metabolism. Low (0.5 mM) and high (2 mM) ammonium and low P up- regulate NLP expression. Expression levels of six NLPs were differentially regulated in response to cold, heat and drought treatments (P ≤ 0.05) (Fig. 4). The presence of the respective putative promoter elements in NLP promoter region reinstates the expression data. The stress responsive expression also indicates the regulation of N metabolism by environmental stress. Expression levels of six NLPs were differentially regulated in response to imbibition, early plant growth and seed deterioration treatments (P ≤ 0.05). The imbibition and seedling specific expression of NLPs correlate with the nitrate signalling mediated degradation of ABA and developmental changes as reported in model plants.
Figure 4.
Expression analysis of NLP genes in response to nutrient (a) and abiotic stress (b) perturbations showing significant expression changes at P value ≤ 0.05 (b) using Genevestigator database.
Expression of nitrogen metabolism and NLP genes in rice genotypes with contrasting NUE
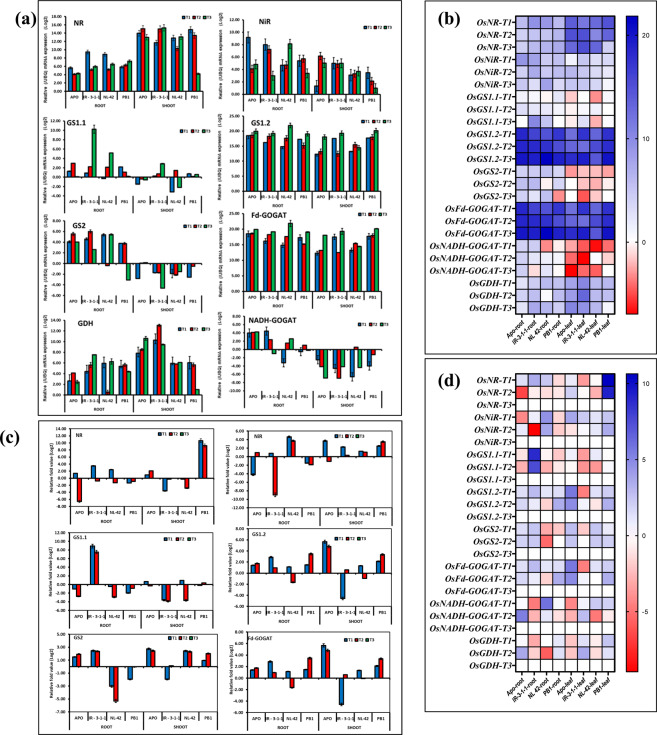
To further validate and analyze the expression of NLPs and other candidate genes, by qRT-PCR, experiments were conducted at seedling stage in hydroponics, in the selected four rice genotypes. Both mRNA expression (/UBQ) and relative expression w.r.to T3 as calibrator is presented. Expression of assimilation genes is represented as Fig. 5a–d. Expression analysis of OsNIA1, OsNIR, OsGS1.1, OsGS1.2, OsGS2, OsNADH-GOGAT, OsFd-GOGAT and OsGDH were studied in the leaf and root tissues of rice seedlings subjected to treatments T1, T2 and T3 for 24 h. Relative fold expression was computed by keeping T3 treatment (leaf or root) as calibrator in all the genotypes. N assimilatory gene expression was significantly higher in T1compared T2 irrespective of tissues. Relative mRNA expression of N assimilation genes showed conclusive trends. Genotype IR-3-1-1 showed high N assimilatory gene expression followed by APO and NL-42. PB1 showed very low expression of OsGS1.1, OsGS2, OsGDH and OsNADH-GOGAT. Plant growth under different N forms most notably impacted gene expression of N assimilation genes in leaves of rice plants. The transcription of primary nitrate assimilation genes, OsNIA1 and OsNIR, OsGS2 were specifically induced by nitrate supply (T1). Comparison of response to nitrate availability, ammonia supply (T2) also revealed a conspicuous re-programming of N assimilation, for example in IR-3-1-1, the transcript abundance of ammonia assimilation genes, OsGS1.2, OsNADH-GOGAT and OsGDH were up regulated.
Figure 5.
Relative mRNA expression (/UBQ) (a,b) and relative expression of (w.r.to T3 treatment) (c,d) of nitrogen assimilation genes in leaves and roots of rice genotypes Apo, IR-83929-B-B-291-3-1-1 (IR-3-1-1), and Nerica L-42 (NL-42), and Pusa Basmati 1 (PB1) in hydroponics and subjected to nitrogen treatments. High nitrate: Low ammonia ratio (T1: 6.5 mM Nitrate: 1 mM Ammonium), Low nitrate: High ammonia (T2: 6.5 mM Ammonium: 1 mM Nitrate), Low N (T3: 0.24 mM Ammonium Nitrate). Values are means (±SE) of 3 biological replicates.
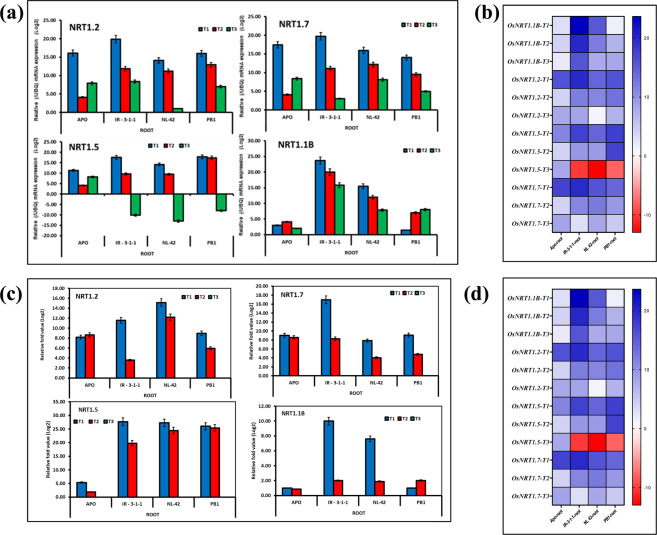
Expression analysis of LATS tansporters OsNRT1.1B, OsNRT1.2, OsNRT1.5 and OsNRT1.7 was studied in the root tissues of rice seedlings subjected to treatments T1, T2 and T3 for 24 hrs (Fig. 6a–d). mRNA expression (/UBQ) as well as relative fold expression depicts nitrate meditaed (T1) induction of LATS expression. The expression of all the LATS genes studied were highest in IR-3-1-1, most importantly, it showed highest expression of OsNRT1.1B gene which is one of the rice homologues of AtNRT1.1 and also plays a role in nitrate uptake, translocation and signaling in rice. PB1 also showed comparatively higher expression of LATS except in the case of OsNRT1.1B.
Figure 6.
Relative mRNA expression (/UBQ) (a,b) and relative expression of (w.r.to T3 treatment) (c,d) of nitrogen uptake genes in leaves and roots of rice genotypes Apo, IR-83929-B-B-291-3-1-1 (IR-3-1-1), and Nerica L-42 (NL-42), and Pusa Basmati 1 (PB1) in hydroponics and subjected to nitrogen treatments. High nitrate: Low ammonia ratio (T1: 6.5 mM Nitrate: 1 mM Ammonium), Low nitrate: High ammonia (T2: 6.5 mM Ammonium: 1 mM Nitrate), Low N (T3: 0.24 mM Ammonium Nitrate). Values are means (±SE) of 3 biological replicates.
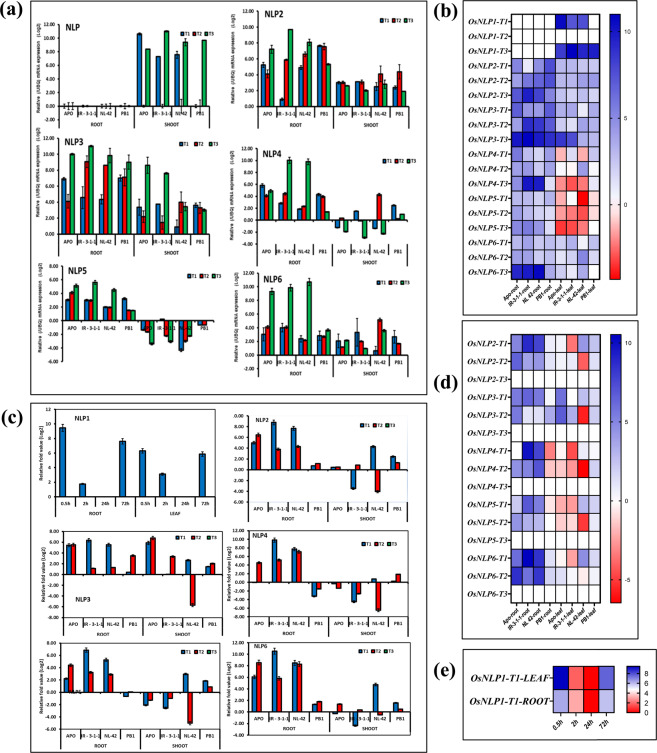
Expression analysis of all 6 NLPs were also studied in leaf and root tissues of rice seedlings subjected to treatments T1, T2 and T3 for 24 hrs (Fig. 7a–e). Expression of NLP1 in shoots showed no or negligible expression at 24 h, whereas in roots the expression was up regulated by T1 and T3 treatments. Time course expression analysis of NLP1 in IR-3-1-1 after 0.5, 2, 24 and 72 hr of exposure to T1 treatment showed that NLP1 expression was maximum in roots after 30 min exposure to nitrate, and thus NLP1 may have probable potential role in primary nitrate response (Fig. 7e). This kind of temporal variation in expression can be expected in other NLPs also. In general, NLPs were expressed more in roots. as depicted in Fig. 7a and heatmap Fig. 7b, mRNA expression of NLPs were highest in response to by N deficiency (T3) followed by nitrate treatment (T1), in root tissues. However there were genotypic differences in the pattern of gene expression, for example IR-3-1-1 did not follow upregulation in expression of NLP 2/4/5 by N deficiency. Relative fold expression (Fig. 7c,d) depicts that T1 treatment resulted in highest expression of NLPs, indicating nitrate regulated expression of NLPs. Genotype IR-3-1-1 showed a general up regulation of NLP expression which is correlated with higher uptake gene transcript abundance. NLP3 and NLP4 expression were highly up regulated in roots of IR-3-1-1 and NL-42 plants receiving T1 treatment.
Figure 7.
Relative mRNA expression (/UBQ) (a,b) and relative expression of (w.r.to T3 treatment) (c,d) NLP genes in leaves and roots of rice genotypes Apo, IR-83929-B-B-291-3-1-1 (IR-3-1-1), and Nerica L-42 (NL-42), and Pusa Basmati 1 (PB1) in hydroponics and subjected to nitrogen treatments. High nitrate: Low ammonia ratio (T1: 6.5 mM Nitrate: 1 mM Ammonium), Low nitrate: High ammonia (T2: 6.5 mM Ammonium: 1 mM Nitrate), Low N (T3: 0.24 mM Ammonium Nitrate) and (e) time course of NLP1 relative expression in roots of IR-3-1-1 exposed to T1 treatment.Values are means (±SE) of 3 biological replicates.
As the data on N responsive expression of genes was is limited in Genevestigator, we searched NCBI GEO for relevant experiments (Fig. 8a,b). Expression data of experiments GSE61370 (rice roots exposed to 0.005 and 1 mM NH4Cl for 10 days) and GSE66807 (low N adapted rice plants (0.3 mM NO3−) transferred to normal (high, 3 mM NO3−) N condition (induction treatment) and plants grown with normal N conditions moved to a low N system (reduction), were downloaded from NCBI-GEO (https://www.ncbi.nlm.nih.gov/gds) and analysed to study NLP gene expression. In agreement with our experimental results, expression of OsNLP3 and OsNLP4 were responsive to N availability, though the pattern showed by OsNLP3 was more consistent.
Figure 8.
Expression heat map of (a) experiment GSE61370 (rice roots exposed to 0.005 and 1 mM NH4Cl for 10 days) (b) GSE66807 (low N adapted rice plants (0.3 mM NO3) transferred to normal (high, 3 mM NO3) N condition (induction treatment) and plants grown with normal N conditions moved to a low N system (reduction). Retrieved from publically open database (https://www.ncbi.nlm.nih.gov/gds).
Protein interaction networks of the NLP proteins were predicted by STRING bioinformatics tool (Supplementary Fig. 8). Among all the NLPs, OsNLP1, OsNLP3 and OsNLP4 showed potential interactions with annotated putative target proteins from the rice genome. The cut-off for the protein interaction networks were,2.31e-25 for PB1 NLP1, 1.05e-23 for PB1 NLP3, 1.94e-18 for PB1 NLP4 respectively. OsNLP1 displayed major interaction with, Putative transcription factor PCF3 (Os01g0924400), Protein kinase, domain containing protein, Nitrate transporter NTL1. CBL-interacting protein kinase 8, Putative nodulation receptor kinase etc. OsNLP3 showed potential interactions with Nitrate/chlorate transporter, Putative leucine zipper protein (Os10g0562700), High-affinity nitrate transporter 2.1, Nitrate/chlorate transporter (Os10g0554200), High affinity nitrate transporter (Os02g0112100), Nitrite reductase etc. OsNLP4 has potential interactions with, protein kinase (Os10g0571300), Transcription factor-like (Os02g0739700) protein; Myb-like DNA-binding domain containing protein (Os12g0238000 protein), Leucine-rich repeat transmembrane protein kinase (Os10g0389800), Growth-regulating factor 8 etc. Iinteraction network involving Os NLP3, OsJ_19340 and OS10T0554200-02 (Nitrate/chlorate transporter), NRT2.1 (HATS nitrate transporter 2.1) and nitrite reductase provide evidence that NLP3 act as central regulator associated with N responses in coordination with NRT1.1B and NRT1.1 A. Summary of the gene expression regulation by N in rice plants is represented as a schematic model in Fig. 9.
Discussion
Nitrogen is the most important mineral nutrient essential for crop production and al also function as a nutrient signal determining plant growth and metabolism38. NUE of cereal crops including rice is approximately 40%, and the loss of 60% applied N incurs economic and environmental shortfalls39. NLPs are transcription factors that recognises NRE motifs in the promoter region of nitrate regulated genes14. The study aims at 1) analysing rice genotypes for traits determining NUE 2) genome-wide identification, insilico and expression analysis of NLP genes family in selected rice genotypes.
We found significant variation in N accumulation of culm and grains. Grain N accumulation is a key indicator of NUE and both NUpE and NUtE (uptake or N utilization efficiencies), in crops40,41,1. The present research also showed considerably higher N uptake by both grain and straw in case of high NUE genotype IR-3-1-1 when compared with low NUE genotype PB1. NUpE is influenced by mass flow of soil water to the root, root morphology, transporter activity on the root surface, timing of N application, and microbial competition42. Significant difference was observed in root traits like total root length, total root volume, total root surface area, with N treatments and varieties. Irrespective of genotype, nitrate nutrition (T1) significantly improved the root growth, root surface area and number of fine root hairs in rice. N deficiency also significantly promoted root proliferation. Genotypic variation in root traits was also significant. Root proliferation also has significant correlation with nitrate uptake kinetics. Barber (1984)43 reported that rate of nitrate uptake was a critical parameter for nitrate uptake and the plants having higher rate of uptake can use nitrogen more efficiently. Comparison of rate of uptake revealed that most efficient uptake system was in IR-3-1-1 followed by APO when nitrate supply was low. Whereas at higher N supply rate of nitrate uptake was highest in IR-3-1-1 and PB1. Nitrate uptake at high external concentrations are mediated by NRT1/NPF family of nitrate transporters falling into the category of low affinity nitrate transport system (LATS). The fore most member of LATS family in rice is OsNRT1/OsNPF8.944 and transgenic plants overexpressing of OsNPF8.9 improved N uptake efficiency under high N conditions45. Another prominent member of rice LATS family, OsNPF6.5/OsNRT1.1B is involverd in root nitrate uptake and translocation to shoot. Specific OsNRT1.1B allele present in indica rice could impart improvement in yield (by 30-33%) and NUE in japonica NILs46 under low N conditions. To understand the varietal differences nitrate uptake, expression analysis of LATS transporters NRT1.1B, NRT1.2, NRT1.5 and NRT1.7 was also studied. Interestingly, the expression of all the LATS genes studied were highest in IR-3-1-1, contributing to the high uptake rate. The high expression of NRT1.1B gene which is one of the rice homologue of AtNRT1.1 and also plays a role in nitrate uptake, translocation and signaling in rice. PB1 also showed comparatively higher relative fold change w.r.to other genotypes APO and NL-42.
Transcription factors such as NLPs, TGA, bZIP1, LBDs, TCPs, and NAC4 are involved in nitrate signalling in Arabidopsis47. The altered transcript levels of these genes regulate downstream NO3− responsive genes. In Arabidopsis, the presence and abundance of nitrate is perceived by NPF6.3, in turn activating phospholipase-C to induce a transient increase in cytoplasmic calcium. This oscillation in calcium level induces the expression of TGA1/TGA4. Expression of the auxin receptor, AFB3 is also responsive to nitrate level via a calcium-independent mechanism. AFB3 up regulates the expression of NAC4 and OBP4. TabHLH1, a bHLH-type transcription factor gene in wheat, improves tolerance to N deprivation via regulation of nutrient transporter gene transcription48. The current understanding of NLPs is mainly based on the studies in Arabidopsis and legumes. Apart from the bioinformatic analysis14,49, functional studies on rice NLPs are lacking, except the finding that OsNRT1;1 affect the subcellular localization of OsNLP450.
We found six NLPS in rice based on protein BLAST for presence of conserved RWP-RK and PB1 domains (RWP-RK. hmm, PF02042; PB1.hmm, PF00564). The genes were named as NLP1 to NLP 6 based on similarity to Arabidopsis NLPs and previous reports in Rice14. All the other NLPs are localised to cellular locations other than mitochondria (mTP) and chloroplast (cTP)- cytosolic and or nuclear localisation as reported in AtNLPs17. The sequence information of 11 rice species, Oryza barthii, Oryza brachyantha, Oryza glaberrima, Oryza glumipatula, Oryza longistaminata,, Oryza meridionalis, Oryza punctata, Oryza rufipogon, Oryza sativa Indica Group, Oryza sativa Japonica Group, Oryza nivara were retrieved and the information on NLP homologues, the chromosome localization, protein Length (aa) and molecular weight (kDa) were deciphered. Evolutionary analysis of NLPs indicated three origins of this gene family, where Group 3 has the most ancestral genes originating from green algae. The well-known AtNLP6 and AtNLP7 genes belong to Group 323. In the current study also, rice and Arabidopsis NLPs were classified in to 3 groups. Group III consisted of forty two rice (NLP2, NLP3, NLP5, NLP6) and four Arabidopsis (NLP6, NLP7, NLP8 and NLP9) NLPs. Hence in agreement with recent reports in rice, OsNLP3 could be the closest homologue of AtNLP7. Spatial and temporal expression pattern NLPs in rice development and stress response were analysed by using Genevestigator. High expression of NLPs coincides with early vegetative stage, early and late reproductive phases. Stage specificity of expression implies role in crop establishment and reproductive success in response to nutritional and environmental cues. Expression of five NLPs (except NLP6) was differentially regulated in response to ABA, salicylic acid and Trans-zeatin treatments (P ≤ 0.05). The presence of the respective putative promoter elements in NLP promoter region reinstates the expression data. The hormone responsive expression also indicates the role of NLPs in sensing and transducing hormone signals to modulate downstream metabolism. Expression of six in was differentially regulated in response to imbibition, early plant growth and seed deterioration treatments (P ≤ 0.05). The imbibition and seedling specific expression of NLPs correlate with the nitrate signalling mediated degradation of ABA and developmental changes as reported in model plants. In Arabidopsis, nuclear localisation of NLP8 stimulate seed germination by activating abscisic acid (ABA) catabolic enzyme and reducing seed ABA level in a nitrate dependent manner51,52.
A seedling stage hydropincs experiment was conducted to understand N regulated expression changes of NLP genes (Fig. 7). Recently it was found that rice NLP1 protein is nuclear localized localizes in nucleus and the gene expression is N regulated unlike AtNLPs. Trangenic manipulation of OsNLP1 expression correlated that higher expression OsNLP1 enhances NUE, by regulating both N uptake and assimilation, whereas knocking out of OsNLP1 reduces NUE and yield under low N conditions53. Expression of NLP1 in shoot showed no or negligible expression at 24 hr, however a time course of NLP1 expression analysis indicated, the maximum expression was observed in roots after 30 min exposure to nitrate. Primary nitrate response (PNR) is the earliest (peaks at 30 min after exposure) nitrate response observed in higher plants, followed by transcriptional up regulation of nitrate assimilation (NR, NIR) genes and nitrate transporters and other related genes. Despite intensive efforts to identify components involved in PNR for last two decades54, no significant progress was made until recently after the identification of AtCIPK8, a CBL interacting protein kinase which regulate low affinity phase of PNR55 nitrate modulated primary root growth in Arabidopsis. Protein interaction networks predicted by STRING bioinformatics tool (Supplementary Fig. 8) predicts that OsNLP1 has an interaction network involving CIPK8, which along with the expression peak at 30 min of nitrate exposure makes OsNLP1 a probable candidate for PNR in rice. There is possibility of involvement of other NLPs in PNR as temporal variation in expression is expected in other NLPs also. In general, NLP1, 2, 3 and 6 was expressed more in roots (Supplementary Fig. 8). More over recently showed, under different N regimes of field trials consistently showed that loss-of-OsNLP4 dramatically reduced yield and NUE compared with wild type. In contrast, the OsNLP4 overexpression lines remarkably increased yield by 30% and NUE by 47% under moderate N level compared with wild type. It clearly showed that OsNLP4 orchestrates the expression of a majority of known N uptake, assimilation and signaling genes by directly binding to the nitrate-responsive cis-element in their promoters to regulate their expression by transcriptomic analyses. Moreover, overexpression of OsNLP4 can recover the phenotype of Arabidopsis nlp7 mutant and enhance its biomass56.N deficiency (T3) and Nitrate (T1) treatments resulted in highest expression of NLPs – indicating N deficiency vis-a-vis nitrate regulated expression of NLPs. Genotype IR-3-1-1 showed a general up regulation of NLP expression which is correlated with higher transcript abundance of nitrate uptake genes. NLP3 expression was highly up regulated in roots of IR-3-1-1 and NL-42 plants receiving T1 treatment.
OsNLP3 and OsNLP4 are interesting candidate genes because of the evolutionary relationship, they are the best rice homologue of AtNLP6 and AtNLP714. Recently, it was shown that the in the transgenic lines expressing OsNRT1.1 A/OsNPF6.3 showed higher nuclear retention of OsNLP3 and OsNLP4 and thereby enhanced the expression levels nitrate metabolism genes and promoted early maturation in rice44. These transgenic lines also had higher economic production and NUE: demonstrating the major role of OsNRT1.1 A/ OsNPF6.3-OsNLP3/4 in regulating the uptake and assimilation of nitrate to improve crop yields.
Another important candidate is OsNRT1.1B which was found to be highly up regulated in high NUE genotype, IR-3-1-1 (Fig. 6). OsNRT1.1B was identified by screening genotypes for resistance to chlorate the toxic analogue of nitrate57. The OsNRT1.1B allele found in indica rice is thought to impart high NUE and is part of a critical QTL contributing to NUE divergence between rice subspecies. Transgenic over expression of OsNRT1.1B improved grain yield and NUE of japonica rice57. OsNRT1.1B also has an important role in regulating nitrate signalling and can contribute in regulating NUpE and NUtE, which both contribute to NUE improvement in rice57. OsNRT1.1B and OsNRT1.1 A are both functional paralogs of AtNRT1.1 which functions as a sensor to trigger the PNR58 and nitrate signaling in Arabidopsis. The increase in transcript abundance of OsNRT1.1B in high NUE genotype also confirms its probable involvement in determining NUpE and NUtE.
As the data on N responsive expression of genes was is limited in Genevestigator, we searched NCBI GEO for relevant experiments (Fig. 8a,b). Expression data of experiments GSE61370 (rice roots exposed to 0.005 and 1 mM NH4Cl for 10 days) and GSE66807 (low N adapted rice plants (0.3 mM NO3) transferred to normal (high, 3 mM NO3) N condition (induction treatment) and plants grown with normal N conditions moved to a low N system (reduction), were downloaded from NCBI-GEO (https://www.ncbi.nlm.nih.gov/gds) and analysed to study NLP gene expression. In agreement with our experimental results, expression of OsNLP3 and OsNLP4 were responsive to N availability, though the pattern showed by OsNLP3 was more consistent.
Hu et al.57 showed that the nitrate sensor NRT1.1B interacts with a phosphate signalling repressor SPX4 which in turn interacts with NLP3. They demonstrated that SPX4 connects nitrate signal perception through NRT1.1B and downstream nitrate response activation via NLP3 in the nitrate signal transduction pathway. Protein interaction networks predicted by STRING bioinformatics tool (Supplementary Fig. 8) predicts that OsNLP3 has an interaction network involving OsJ_19340 and OS10T0554200-02 (Nitrate/chlorate transporter) and NRT2.1 (HATS nitrate transporter 2.1) provide evidence that NLP3 act as central regulator associated with N responses in coordination with NRT1.1B and NRT1.1 A.
Conclusions
Based on physiological analysis of rice germplasm four genotypes with contrasting NUE were identified. The low NUE variety PB1 showed an overall decline in photosynthetic pigment content in both field and hydroponic condition. Based on the activity and expression of N assimilatory and low affinity nitrate uptake (LATS) genes, we found that PB1 being N responsive genotype was efficient in uptake when N availability is high, IR3-1-1 has high uptake efficiency, translating into high NUE. The expression of all the LATS genes studied were highest in IR3-1-1, most importantly, it showed highest expression of OsNRT1.1B gene which is one of the rice homologues of AtNRT1.1 and also plays a role in nitrate uptake, translocation and signaling in rice. Based on insilico analysis, 6 NLP genes (including alternative splice forms 11NLPs) were identified from rice. Insilico expression profiling, chromosomal localization, putative promoter elements, protein localization, co-expression net-work, were also conducted. Expression of NLPs was promoted by nitrate supply as well as N deficiency (NLP1, NLP4, NLP5). The effective and coordinated signal transduction network involving the rice homologue of nitrate transceptor OsNRT1.1B, the probable PNR regulator OSNLP1 and the master response regulator OsNLP3, a homologue of AtNLP6/7 renders high NUE in rice (Fig. 9).
Supplementary information
Acknowledgements
Authors are thankful to the ICAR-Indian Agricultural Research Institute for funding (institute project-CRSCIARISIL20144047279) and providing the necessary facilities. Authors also acknowledge the financial support received from NAHEP-CAAST, ICAR-IARI (Grant No. NAHEP/CAAST/2018-19/07). JB acknowledge ICAR- Junior research fellowship support received during the course of the study.
Author contributions
J.B. and L.S. conducted the experiments. L.S. designed primers, did the expression profiling and statistical analysis and finalised the figures. L.S. and J.B. together prepared the first draft. H.S. assisted in growing of plants and taking physiological and biochemical observations. L.S. and V.C. contributed resources. L.S., V.C., S.K.J., S.K. and A.K. finalised experiments, contributed resources and finalised the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: B. Jagadhesan and Lekshmy Sathee.
Contributor Information
Lekshmy Sathee, Email: lekshmyrnair@gmail.com.
Viswanathan Chinnusamy, Email: viswa_iari@hotmail.com.
Supplementary information
is available for this paper at 10.1038/s41598-020-66338-6.
References
- 1.Havé, M., Marmagne, A., Chardon, F. & Masclaux-Daubresse, C. Nitrogen remobilization during leaf senescence: Lessons from Arabidopsis to crops. Journal of Experimental Botany vol. 68 (2017). [DOI] [PubMed]
- 2.Tegeder M, Masclaux-Daubresse C. Source and sink mechanisms of nitrogen transport and use. New Phytol. 2018;217:35–53. doi: 10.1111/nph.14876. [DOI] [PubMed] [Google Scholar]
- 3.Crawford NM, Glass AD. Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci. 1998;3:389–395. doi: 10.1016/S1360-1385(98)01311-9. [DOI] [Google Scholar]
- 4.FAo, I., FAO, U. food security., fao, R. U. http://www. & 2017, undefined. WFP, WHO (2017) The state of food security and nutrition in the world 2017.
- 5.Mueller ND, et al. A tradeoff frontier for global nitrogen use and cereal production. Environ. Res. Lett. 2014;9:054002. doi: 10.1088/1748-9326/9/5/054002. [DOI] [Google Scholar]
- 6.Zhang, X. et al. Managing nitrogen for sustainable development. Nature vol. 528 (2015). [DOI] [PubMed]
- 7.Hirel B, Tétu T, Lea PJ, Dubois F. Improving Nitrogen Use Efficiency in Crops for Sustainable Agriculture. Sustainability. 2011;3:1452–1485. doi: 10.3390/su3091452. [DOI] [Google Scholar]
- 8.Xuan W, Beeckman T, Xu G. Plant nitrogen nutrition: sensing and signaling. Curr. Opin. Plant Biol. 2017;39:57–65. doi: 10.1016/j.pbi.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 9.Chen, C. et al. Nitrogen forms affect root growth, photosynthesis, and yield of tomato under alternate partial root-zone irrigation. J Plant Nutr Soil Sci179 (2016).
- 10.Undurraga SF, Ibarra-Henríquez C, Fredes I, Álvarez JM, Gutiérrez RA. Nitrate signaling and early responses in Arabidopsis roots. J. Exp. Bot. 2017;68:2541–2551. doi: 10.1093/jxb/erx041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y-Y, Cheng Y-H, Chen K-E, Tsay Y-F. Annual Review of Plant Biology Nitrate Transport. Signaling, and Use Efficiency. 2018 doi: 10.1146/annurev-arplant-042817. [DOI] [PubMed] [Google Scholar]
- 12.Camargo A, et al. Nitrate Signaling by the Regulatory Gene NIT2 in Chlamydomonas. Plant Cell Online. 2007;19:3491–3503. doi: 10.1105/tpc.106.045922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schauser, L., Wieloch, W. & Stougaard, J. Evolution of NIN-like proteins in Arabidopsis, rice, and Lotus japonicus. J. Mol. Evol. 60, 229–237. [DOI] [PubMed]
- 14.Chardin C, Girin T, Roudier F, Meyer C, Krapp A. The plant RWP-RK transcription factors: key regulators of nitrogen responses and of gametophyte development. J. Exp. Bot. 2014;65:5577–87. doi: 10.1093/jxb/eru261. [DOI] [PubMed] [Google Scholar]
- 15.Kumar A, et al. Genome-wide identification and characterization of gene family for RWP-RK transcription factors in wheat (Triticum aestivum L.) PLoS One. 2018;13:e0208409. doi: 10.1371/journal.pone.0208409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guan, P. Dancing with Hormones: A Current Perspective of Nitrate Signaling and Regulation in Arabidopsis. Front. Plant Sci. 8 (2017). [DOI] [PMC free article] [PubMed]
- 17.Gaudinier A, et al. Transcriptional regulation of nitrogen-associated metabolism and growth. Nature. 2018;563:259–264. doi: 10.1038/s41586-018-0656-3. [DOI] [PubMed] [Google Scholar]
- 18.Konishi M, Yanagisawa S. Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nat. Commun. 2013;4:1617–1619. doi: 10.1038/ncomms2621. [DOI] [PubMed] [Google Scholar]
- 19.Marchive C, et al. Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat. Commun. 2013;4:1713. doi: 10.1038/ncomms2650. [DOI] [PubMed] [Google Scholar]
- 20.Liu K, et al. Discovery of nitrate–CPK–NLP signalling in central nutrient–growth networks. Nature. 2017;545:311–316. doi: 10.1038/nature22077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vidal EA, Álvarez JM, Moyano TC, Gutiérrez RA. Transcriptional networks in the nitrate response of Arabidopsis thaliana. Curr. Opin. Plant Biol. 2015;27:125–132. doi: 10.1016/j.pbi.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y-Y, Cheng Y-H, Chen K-E, Tsay Y-F. Nitrate Transport, Signaling, and Use Efficiency. Annu. Rev. Plant Biol. 2018;69:85–122. doi: 10.1146/annurev-arplant-042817-040056. [DOI] [PubMed] [Google Scholar]
- 23.Mu X, Luo J. Evolutionary analyses of NIN-like proteins in plants and their roles in nitrate signaling. Cell. Mol. Life Sci. 2019 doi: 10.1007/s00018-019-03164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hakeem, K., Chandna, R., Ahmad, A., science, M. I.-R. & 2012, undefined. Physiological and molecular analysis of applied nitrogen in rice genotypes. Elsevier.
- 25.Lim SS, et al. Soil and plant nitrogen pools in paddy and upland ecosystems have contrasting δ15N. Biol. Fertil. Soils. 2015;51:231–239. doi: 10.1007/s00374-014-0967-y. [DOI] [Google Scholar]
- 26.Kawahara, Y., Bastide, M. de la, Hamilton, J., Rice, H. K.- & 2013, undefined. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Springer. [DOI] [PMC free article] [PubMed]
- 27.Verma G, et al. Genome-wide analysis of rice dehydrin gene family: Its evolutionary conservedness and expression pattern in response to PEG induced dehydration stress. PLoS One. 2017;12:1–22. doi: 10.1371/journal.pone.0176399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 29.Hruz, T. et al. Genevestigator V3: A Reference Expression Database for the Meta-Analysis of Transcriptomes. Adv. Bioinforma.2008, 420747 (2008). [DOI] [PMC free article] [PubMed]
- 30.Jones JM, Richards BN. Effect of reforestation on turnover of 15N-labelled nitrate and ammonium in relation to changes in soil microflora. Soil Biol. Biochem. 1977;9:383–392. doi: 10.1016/0038-0717(77)90016-5. [DOI] [Google Scholar]
- 31.Lim, S.-S. et al. Ammonia Volatilization from Rice Paddy Soils Fertilized with 15 N-Urea Under Elevated CO 2 and Temperature. Korean J. Environ. Agric. 28, 233–237 (2009).
- 32.Feng Y, et al. Mapping QTLs for nitrogen-deficiency tolerance at seedling stage in rice (Oryza sativa L.) Plant Breed. 2010;129:652–656. doi: 10.1111/j.1439-0523.2009.01728.x. [DOI] [Google Scholar]
- 33.S, L., Jain, V., Khetarpal, sangeeta, Pandey, R. & Singh, R. Effect of elevated carbon dioxide on kinetics of Nitrate uptake in wheat roots. (2009).
- 34.Downes MT. An improved hydrazine reduction method for the automated determination of low nitrate levels in freshwater. Water Res. 1978;12:673–675. doi: 10.1016/0043-1354(78)90177-X. [DOI] [Google Scholar]
- 35.Lekshmy S, Jain V, Khetarpal S, Pandey R. Inhibition of nitrate uptake and assimilation in wheat seedlings grown under elevated CO2. Indian J. Plant Physiol. 2013;18:23–29. doi: 10.1007/s40502-013-0010-6. [DOI] [Google Scholar]
- 36.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 37.Abdi, H. Bonferroni and Šidák corrections for multiple comparisons. Encycl. Meas. Stat. 103–107 (2007).
- 38.Undurraga, S. F., Ibarra-Henríquez, C., Fredes, I., Álvarez, J. M. & Gutiérrez, R. A. Nitrate signaling and early responses in Arabidopsis roots. J. Exper. Botany68(10), 2541–2551 (2017). [DOI] [PMC free article] [PubMed]
- 39.Hu B, et al. Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat. Genet. 2015;47:834–838. doi: 10.1038/ng.3337. [DOI] [PubMed] [Google Scholar]
- 40.Moose, S. & Below F. E. Biotechnology Approaches to Improving Maize Nitrogen Use Efficiency. In: Kriz A.L., Larkins B.A. (eds) Molecular Genetic Approaches to Maize Improvement. Biotechnology in Agriculture and Forestry, vol 63. Springer, Berlin, Heidelberg (2009).
- 41.Vijayalakshmi P, et al. Biochemical and physiological characterization for nitrogen use efficiency in aromatic rice genotypes. F. Crop. Res. 2015;179:132–143. doi: 10.1016/j.fcr.2015.04.012. [DOI] [Google Scholar]
- 42.Garnett T, Plett D, Heuer S, Okamoto M. Genetic approaches to enhancing nitrogen-use efficiency (NUE) in cereals: Challenges and future directions. Funct. Plant Biol. 2015;42:921–941. doi: 10.1071/FP15025. [DOI] [PubMed] [Google Scholar]
- 43.Barber, S. Soil nutrient bioavailability: a mechanistic approach. (1995).
- 44.Wang, W. et al. Expression of the Nitrate Transporter Gene OsNRT1.1A/OsNPF6.3 Confers High Yield and Early Maturation in Rice. Plant Cell30 (2018). [DOI] [PMC free article] [PubMed]
- 45.Fan X, et al. A putative 6-transmembrane nitrate transporter OsNRT1.1b plays a key role in rice under low nitrogen. J. Integr. Plant Biol. 2016;58:590–599. doi: 10.1111/jipb.12382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lekshmy S, Jha SKK. Selection of reference genes suitable for qRT-PCR expression profiling of biotic stress, nutrient deficiency and plant hormone responsive genes in bread wheat. Indian J. Plant Physiol. 2017;22:101–106. doi: 10.1007/s40502-017-0282-3. [DOI] [Google Scholar]
- 47.O’Brien J, et al. Nitrate Transport, Sensing, and Responses in Plants. Mol. Plant. 2016;9:837–856. doi: 10.1016/j.molp.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 48.Jiang S, et al. Root extension and nitrate transporter up-regulation induced by nitrogen deficiency improves nitrogen status and plant growth at the seedling stage of winter wheat (Triticum aestivum L.) Environ. Exp. Bot. 2017;141:28–40. doi: 10.1016/j.envexpbot.2017.06.006. [DOI] [Google Scholar]
- 49.Schauser L, Wieloch W, Stougaard J. Evolution of NIN-like proteins in Arabidopsis, rice, and Lotus japonicus. J. Mol. Evol. 2005;60:229–237. doi: 10.1007/s00239-004-0144-2. [DOI] [PubMed] [Google Scholar]
- 50.Wang, W. et al. Expression of the Nitrate Transporter Gene OsNRT1.1A/ OsNPF6.3 Confers High Yield and Early Maturation in Rice OPEN, 10.1105/tpc.17.00809. [DOI] [PMC free article] [PubMed]
- 51.Okamoto M. CYP707A1 and CYP707A2, Which Encode Abscisic Acid 8’-Hydroxylases, Are Indispensable for Proper Control of Seed Dormancy and Germination in Arabidopsis. PLANT Physiol. 2006;141:97–107. doi: 10.1104/pp.106.079475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yan, D. et al. NIN-like protein 8 is a master regulator of nitrate-promoted seed germination in Arabidopsis. Nat. Commun. 7 (2016). [DOI] [PMC free article] [PubMed]
- 53.Alfatih, A. et al. Rice NIN-LIKE PROTEIN 1 Rapidly Responds to Nitrogen Deficiency and Improves Yield and Nitrogen Use Efficiency. bioRxiv. (2020). [DOI] [PubMed]
- 54.Deng M, Moureaux T, Caboche M. Tungstate, a Molybdate Analog Inactivating Nitrate Reductase, Deregulates the Expression of the Nitrate Reductase Structural Gene. PLANT Physiol. 1989;91:304–309. doi: 10.1104/pp.91.1.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang C, et al. FIP1 plays an important role in nitrate signaling and regulates CIPK8 and CIPK23 expression in arabidopsis. Front. Plant Sci. 2018;9:1–14. doi: 10.3389/fpls.2018.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu, J. et al. Rice NIN-LIKE PROTEIN 4 is a master regulator of nitrogen use efficiency. bioRxiv. (2020).
- 57.Hu B, et al. Nitrate–NRT1.1B–SPX4 cascade integrates nitrogen and phosphorus signalling networks in plants. Nat. Plants. 2019;5:401–413. doi: 10.1038/s41477-019-0384-1. [DOI] [PubMed] [Google Scholar]
- 58.Ho C-H, Lin S-H, Hu H-C, Tsay Y-F. CHL1 Functions as a Nitrate Sensor in Plants. Cell. 2009;138:1184–1194. doi: 10.1016/j.cell.2009.07.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.