Abstract
Communicable diseases are often virulent, i.e. they cause morbidity symptoms in those infected. While some symptoms may be transmission-enhancing, other symptoms are likely to reduce transmission potential. For human diseases, the reduction in transmission opportunities is commonly caused by reduced activity. There is limited data regarding the potential impact of virulence on transmission potential. We performed an exploratory data analysis of 324 influenza patients at a university health centre during the 2016/2017 influenza season. We classified symptoms as infectiousness-related or morbidity-related and calculated two scores. The scores were used to explore the relationship between infectiousness, morbidity (virulence), and activity level. We found a decrease in the activity level with increasing morbidity scores. There was no consistent pattern between an activity level and an infectiousness score. We also found a positive correlation between morbidity and infectiousness scores. Overall, we find that increasing virulence leads to increased infectiousness and reduced activity, suggesting a trade-off that can impact overall transmission potential. Our findings indicate that a reduction of systemic symptoms may increase host activity without reducing infectiousness. Therefore, interventions should target both systemic- and infectiousness-related symptoms to reduce overall transmission potential. Our findings can also inform simulation models that investigate the impact of different interventions on transmission.
Keywords: infectious diseases, influenza, trade-off, transmission, virulence
1. Introduction
Many infectious diseases cause symptoms in at least some infected hosts. Often, those symptoms increase the host's infectiousness and facilitate the transmission of the pathogen [1,2]. Coughing and sneezing for respiratory infections are prime examples. However, symptoms that are too severe may reduce host activity or, in extreme cases, cause host death, reducing transmission opportunities. The induction of symptoms by a pathogen is often called virulence, and the idea that some virulence is needed for efficient transmission but too much virulence reduces transmission is often called the trade-off hypothesis and has been studied extensively [3–5]. The trade-off hypothesis predicts that an intermediate level of virulence leads to maximum pathogen fitness (usually quantified by the reproductive number). The virulence level which optimizes fitness depends on both population-level and within-host-level processes, the implications of which have been theoretically explored previously [2,3,6–13].
The most studied trade-off is between increasing transmission potential due to increased host infectiousness and decreasing transmission potential due to host mortality [3]. For humans, malaria might fall into this category. It has been shown that as malaria parasite density increased within a host (considered a measure of virulence), per-contact transmission potential (quantified by gametocyte density) increased, as did the duration of infection and mortality [14,15]. The former two quantities are assumed to increase transmission potential, while the latter reduces it. It is, however, unclear if the increase in mortality is a strong enough effect to play an important role in the virulence trade-off in malaria. Host death is likely an important determining factor of transmission potential for many animal diseases and some human diseases (e.g. viral haemorrhagic diseases [16]). However, for many human pathogens mortality is low, and it is more likely that increased virulence leads to reduced host activity and thus reduced transmission potential. Transmission potential, T, is proportional to the product of per-contact transmission potential (i.e. infectiousness), p, the rate at which infected hosts have contact with other hosts, c, and the duration of the infectious period, d. All three quantities can potentially be impacted by virulence, v, leading to T ∼ p(v) × c(v) × d(v).
A previous study for HIV infection in humans showed that d and p are both impacted by virulence, namely increased virulence (quantified by increased pathogen load), was shown to increase per-contact transmission potential, p, and reduce the duration of infectiousness (i.e. time to AIDS/death), d. Maximum transmission potential was found to be at an intermediate level of pathogen load (virulence) [17,18]. A study of dengue infection showed similar results [19]. The viral production rate, which was considered as a proxy for virulence, led to both increased peak viral load (which is correlated with increased transmission potential) as well as increased clearance rate, i.e. shorter duration of infection. This led to a similar relationship as found for HIV, namely an intermediate peak viral load resulted in the highest transmission potential.
While there is some evidence that the virulence trade-off applies to other human diseases [20–23], there are, as far as we are aware, no other studies of human pathogens that directly address this topic. In addition, the studies we just described focused on the duration, d, and per-contact transmission potential, p. None were able to investigate the impact of virulence on contact behaviour, c.
Here, we investigate the trade-off hypothesis using data from influenza infections in humans, focusing on transmission potential (p) and contact rate (c). Influenza induces symptoms in around 84% of infected individuals [24]. Some of the symptoms, such as coughing and sneezing, likely enhance transmission by increasing the infectiousness of a host (p). A recent study provided estimates for the transmission potential of symptomatic versus asymptomatic individuals and found that individuals with symptomatic infections are about 3–12 times as infectious as persons with asymptomatic infections [25]. Other symptoms, such as fever, body aches, and general malaise, are more likely to lead to a reduction in transmission by reducing host activity (c). A previous study on influenza in 146 adults and children in the UK found that individuals without symptoms had a mean of 12.72 contacts per day, while those individuals with symptoms only had 3.58 [26]. The same study showed that the number of contacts decreased as the number of symptoms increased. These studies suggest that there might be a trade-off between infectiousness and activity for influenza, which together are important factors in the overall transmission potential. Here, we investigate this relationship further using individual patient data. While it is quite likely that virulence can impact the duration of infectiousness for influenza, we, unfortunately, do not have data for this component and thus focus on the other two quantities. We expect that virulence leads to increasing p while negatively affecting c. The transmission potential due to p and c can be considered as proportional to the product of those quantities, i.e. Tpc ∼ p(v) × c(v). The lines in figure 2 illustrate our hypothesized relation for contact rate, per-contact transmission potential, and overall transmission potential, Tpc, as a function of virulence. Our data (symbols in figure 2, discussed in detail next) show overall support for this relation.
Figure 2.
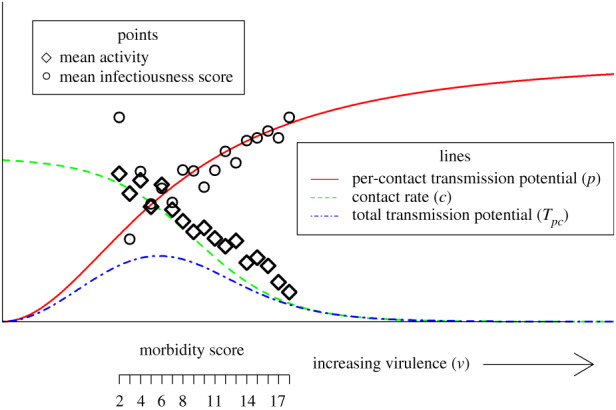
Theoretical framework and data for virulence-mediated transmission trade-off. A morbidity score is a proxy for virulence, an infectiousness score is a proxy for per-contact transmission potential, and the activity level is a proxy for a contact rate. The values for infectiousness and activity are re-scaled to allow better visualization. Lines are adjusted to pass through data. Thus, this figure does not show a fit but instead a conceptual framework in which our data can be placed and interpreted. (Online version in colour.)
2. Material and methods
(a). Data collection
Students with a primary complaint related to a respiratory infection who made an appointment at the health centre of a large research university from December 2016 to February 2017 filled out an electronic questionnaire (see [27] for more details). The questionnaire collected data about their current symptoms and activity level. A response was required for all symptom-related questions when they scheduled their appointments. We included all symptoms collected by the questionnaire in this analysis. The complete questionnaire is available in the electronic supplementary material.
For the symptoms of cough, weakness, and body aches, the patient graded the severity of the symptom as none, mild, moderate, and severe. The patient recorded all other symptom data as present or absent. The patient also reported any changes in their normal behaviour. Patients described their activity level as a number between 0 and 10, with 10 indicating no change in regular activity and 0 being bedridden.
The study population includes all patients with a diagnosis of influenza. The data and results presented in the main text include patients diagnosed with a rapid antigen or rapid polymerase chain reaction (PCR) test. To address the impact of the influenza diagnosis method, we performed the same analyses for all patients diagnosed with influenza regardless of the method used. These results are shown in the electronic supplementary material.
The institutional review board approved the study protocol.
(b). Data cleaning
We cleaned the data to format the variables and to check for variables with potential errors or missing entries. During the cleaning process, we removed uninformative variables, which we defined as any symptoms found to occur in less than 5% of patients. The symptoms of blurred vision and hearing loss both had a prevalence of less than 5%, so they were not considered for further analysis. To allow easy comparison of morbidity symptoms, we dichotomized weakness and body aches to ‘absent’ or ‘present’. Patients reported cough as present or absent as well as severity, but only cough absent or present was considered for the main analysis.
(c). Analysis
We assessed the univariate relationships between activity and each symptom using the linear regression-treating activity level as a continuous variable. We also performed multiple linear regressions. We determined the variables to include in our final model with a sequential forward floating selection, minimizing the root-mean-square error (RMSE) on test data through a fivefold cross-validation (20 times repeated) [28].
Next, we constructed two cumulative scores, one for overall infectiousness and one for overall morbidity. To that end, we divided all symptoms into those related to infectiousness and those related to morbidity (electronic supplementary material, table S1). We defined morbidity symptoms as symptoms that influence overall feelings of well-being but are not associated with infectiousness. Infectiousness symptoms are any symptoms that could plausibly contribute to passing the virus from an infected host to a susceptible host [29,30]. Importantly, the grouping of variables into either the morbidity or infectiousness symptom categories was based on a priori medical and biological considerations, independently of any observed correlation with the activity level. Doing so prevents any circular reasoning since only including symptoms correlated with activity would, of course, generate a score which would match the impact on the activity level. These scores are similar to systemic and respiratory scores used in past studies of influenza infection [29,31,32]. The scores are computed as a sum of the symptoms that are present. Our data did not allow us to take into account symptom severity, though a comparison of our scores with cough, weakness, and body aches, for which we have severity, shows that there is a good positive correlation between strength and the number of symptoms (see electronic supplementary material). To test the robustness of our results, we completed several sensitivity analyses using different approaches to generate the scores. Results from these analyses are shown in the electronic supplementary material.
Correlations between the infectiousness score, morbidity score, and activity were assessed using Spearman correlation [33] and the generalized Mantel–Haenszel procedure [34]. Linear regression is used to estimate the average change in activity, and the lines are included in the plots to help visualize the relationships. All analyses were completed using R (v. 3.5.3) [35]. We used the mlr package for cross-validation [36], vcdExtra to compute Yule's Q, and the Cochrane-Mantel-Haenszel trend test [37] and DescTools to compute Spearman's rank correlation coefficient and corresponding confidence intervals [38]. All of the code and data required to reproduce the results are available through Dryad.
3. Results
(a). Study population
During the study period, 2380 patients had a respiratory complaint and filled out the questionnaire. Among those, 324 had a lab-based diagnosis of influenza (PCR or rapid antigen). The following analyses focus on those patients since they are most likely infected with influenza. For analyses of patients who received a flu diagnosis based on lab tests or empirically from a physician, see the electronic supplementary material. Patients with influenza reported activity levels ranging from 0 to 10, with a median of 4 (electronic supplementary material, figure S1). All of the patients reported symptoms, with only 14% reporting 9 or fewer (maximum possible 25). The most common symptoms were coughing and weakness, while the least common was abdominal pain (electronic supplementary material, table S1).
(b). Univariate and subset selection
We assessed correlations between the activity level and each symptom in a univariate linear analysis (electronic supplementary material, table S2). All statistically significant symptoms had a negative correlation with the activity level (electronic supplementary material, table S2). Next, we considered a multivariable regression model and performed a variable selection based on the cross-validated minimization of RMSE. We found that the best performing model was one that included chills/sweats, subjective fever, headache, weakness, sleeplessness, and vomiting (electronic supplementary material, table S2). While vomiting is not a common symptom of influenza, in those patients who did report vomiting it led to major reductions in their activity.
(c). Computation of infectiousness and morbidity scores
For the main analysis, we classified five symptoms as infectiousness-related: coughing, sneezing, runny nose, nasal congestion, and chest congestion. Twenty symptoms were classified as morbidity-related: subjective fever, having chills and or sweats, body aches, weakness, headache, fatigue, sleeplessness, breathlessness, wheezing, chest pain, sore throat, abdominal pain, diarrhoea, nausea, vomiting, ear pain, tooth pain, eye pain, itchy eyes, and swollen lymph nodes. Each symptom present in a patient contributed one point to their respective scores. For those symptoms for which we had severity, we investigated correlations with the number of symptoms and found total number of symptoms to be a good proxy (see the electronic supplementary material). Analyses using several alternative approaches for computing the score are shown in the electronic supplementary material and summarized below in the Sensitivity Analysis section.
The infectiousness score had a possible range of 0–5, and the morbidity score had a possible range of 0–20. The median infectiousness score was 4. Only two patients had an infectiousness score of 0, 20% had a score of 2 or less, and 28% of patients had the maximum possible score of 5. The mean morbidity score was 9.574, and no patients had a morbidity score of 0, 1, 19, or 20. The centred distribution is assumed to be the result of how patients were included in the study, that is all the patients felt sick enough to seek medical care, but none were sick enough to require urgent care or hospitalization. Results presented in the electronic supplementary material show plots of the score distributions (electronic supplementary material, figure S2A,B).
(d). Impact of infectiousness score on activity
Analysis of the association between the infectiousness score and the patient's self-reported activity level suggests that the value of this score has a small impact on the activity level of a patient, with higher infectiousness correlating with reduced activity. Spearman's rank correlation indicates a negative relationship (r = −0.15 (95% CI: −0.25, −0.04)), and the Cochran–Mantel–Haenszel trend test is statistically significant (χ2 = 6.363, d.f. = 1, p < 0.01) (figure 1a). Note, however, that the data suggest a nonlinear relationship between infectiousness and activity. We cannot think of a biological mechanism that might lead to this pattern. Given that the observed negative trend is small and does not show a monotone decline, it is most reasonable to assume based on this data that there is no meaningful relationship between the infectiousness score and the activity level. Results presented in the electronic supplementary material show that the overall pattern remains the same if details of the analysis approach are changed (electronic supplementary material, figures S6–S8B and S13).
Figure 1.
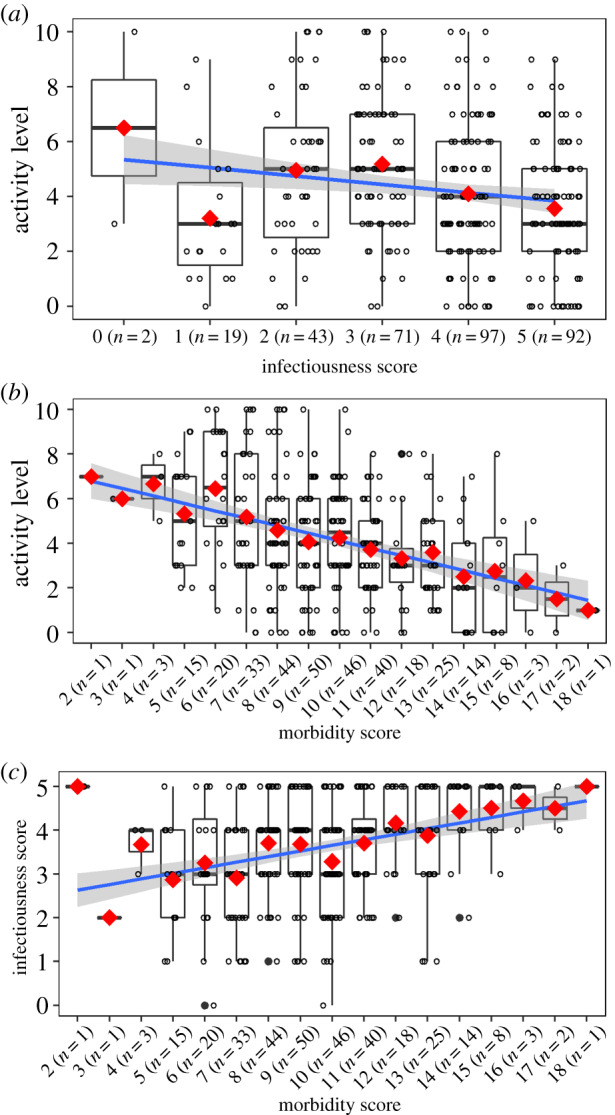
For all plots, the red diamonds indicate the mean and the solid blue line is the linear regression fit. The shaded area is the 95% confidence interval for the linear regression. (a) Activity level for each level of the infectiousness score. (b) Activity level for each level of the morbidity score. There are no patients with a morbidity score of 0, 1, 19, and 20. (c) Infectiousness score for each level of the morbidity score. (Online version in colour.)
(e). Impact of morbidity score on activity
Analysis of the association between the morbidity score and the patient's self-reported activity level suggests that the higher morbidity score is associated with reduced activity levels. Spearman's rank correlation indicates a negative relationship (r = −0.32 (95% CI: −0.42, −0.22)), and the Cochran–Mantel–Haenszel trend test is statistically significant (χ2 = 38.577, d.f. = 1, p < 0.01) (figure 1b). The observed pattern is consistent and clear, with a mean 4.7-fold reduction in the activity level going from the lowest to the highest morbidity score. The strong negative relationship is preserved if details of the analysis approach are changed and results are shown in electronic supplementary material, figures S9B, S10B and S14.
(f). Impact of morbidity score on the infectiousness score
Analysis of the relationship between the morbidity and infectiousness scores shows a positive correlation. Spearman's rank correlation indicates a positive relationship (r = 0.28 (95% CI: 0.18, 0.38)), and the Cochran–Mantel–Haenszel trend test is statistically significant (χ2 = 26.093, d.f. = 1, p < 0.01) (figure 1c). Apart from the mean activity levels for very low morbidity score values (with very small sample sizes), the pattern is consistent and clear, with a mean 1.8-fold increase in the infectiousness score going from the lowest to the highest morbidity score. A positive relationship is observed regardless of how the infectiousness or morbidity scores are calculated (electronic supplementary material, figures S6–S10C) and in the analysis of empirically diagnosed patients (electronic supplementary material, figure S15).
(g). Sensitivity analysis
We performed several sensitivity analyses of our results. In one type of analysis, we computed both the morbidity and infectiousness scores in different ways and showed that overall results remain the same. In a different analysis, we considered all patients diagnosed with influenza, not just those that had a positive lab test. Again, overall results remained robust. Details of all these analyses are presented in the electronic supplementary material.
(h). Conceptualizing our results
We can place our data into the conceptual framework introduced in the introduction with the morbidity score as a proxy of virulence, v, the infectiousness score as a proxy of per-contact transmission potential, p, and patient-reported activity as a proxy for the contact rate, c (figure 2). Since our data are measured in units with indirect and uncertain mapping to actual per-contact transmission potential and actual contact rate, we standardized the data and manually placed it on top of the conceptual lines. This should not be considered a quantitative mapping.
Our study population consisted of individuals who felt sick enough to seek medical care, but none were ill enough to require emergency care. It is thus a reasonable assumption to expect them to be somewhere in the middle of the virulence spectrum. Based on our data, this range is characterized by infectiousness levels that are increasing as morbidity (virulence) increases (figure 2). Activity is more strongly impacted and decreases as morbidity increases. We speculate that a study population that included asymptomatic and mildly symptomatic-infected persons would be on the left side of our data, while severely ill and hospitalized individuals would fall to the right side of our data.
4. Discussion
We believe this is the first study that investigates a trade-off between contact rate and per-contact transmission potential for influenza in humans. We showed that for our population, activity decreased as both morbidity and infectiousness scores increased, and we found a positive association between morbidity and infectiousness symptoms. This is a secondary analysis of data that was not specifically collected to address our question. As such, results should be considered exploratory instead of confirmatory. All data were self-reported, and most symptoms were only reported as present or absent. Patient-reported symptoms can be very subjective [39]. Due to limitations in the available data, we had to make certain assumptions. We only used the absence or presence of symptoms for our scores. More detailed data would have allowed for a more accurate score taking into account severity/frequency of symptoms. We only collected data on individuals who were experiencing enough symptoms to seek care, but not symptoms so severe that emergency care was required. As a result, we do not have data on individuals with low or high virulence infections. Such data would allow for a complete exploration across the full range of virulence and determine relationships between transmission, morbidity, and infectiousness. Finally, our study population was made up of college students, i.e. generally young and healthy individuals. As such, their symptoms, infectiousness, and activity behaviour distributions might not fully apply to a more general population. Our data also does not contain information on the influenza strain with which individuals were infected, which may impact disease outcomes [40].
The mapping of our infectiousness score on actual transmission potential is unclear. Different symptoms likely contribute to the infectiousness of a patient in different amounts. Coughing and sneezing create both droplets and aerosols that have varying concentrations of virus and environmental persistence and can transmit the disease directly and indirectly [41–47]. While a runny nose does not create aerosols or droplets without proper hygiene, it can lead to contaminated objects or self-inoculation of contacts [42,48]. Our score is limited but similar to scores used in previous studies to quantify infectiousness [29,31,32].
Despite these potential limitations, our study can help inform current and future interventions targeting influenza. For example, if our results hold up, it suggests that any anti-influenza treatment that reduces morbidity-related symptoms, without affecting infectiousness-related symptoms, could lead to increased transmission. While from the perspective of a patient or a clinician, a reduction in any symptom may be viewed as a positive, such an intervention might lead to worse outcomes on the population level [49]. The current Food and Drug Administration (FDA) approval of anti-influenza drugs relies on showing an impact on the symptoms, with a focus on more severe and systemic (i.e. morbidity) symptoms [50–52]. From a population perspective, it is essential that such drugs also reduce host infectiousness [52–55]. Some evidence for this has been found in previous studies [54,56–58] as well as being explored in mathematical models [49,59,60].
Population-level control of infectious diseases makes increasing use of mathematical models [61]. The need for these models to be accurate is critical. Researchers have increasingly recognized that capturing human behaviour changes during an infectious disease outbreak, both for uninfected and infected individuals, is relevant [62,63]. As far as we are aware, only a few previous modelling studies for influenza have tried to capture the impact of infection on contact rates [64–67]. Previous studies have shown that symptoms aid infectiousness and impact the number of contacts [25,26]. In our analysis, we found a strong reduction in mean activity as a result of increased morbidity. Using data from our study and past studies [25,26] can be a starting point for future models to explore the impacts of infectiousness and contact behaviour of infected hosts [68].
Supplementary Material
Ethics
The University of Georgia's Institutional Review Board (IRB) approved the study protocol.
Data accessibility
All of the data and code required to complete the analysis are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.51c59zw4v [69].
Authors' contributions
B.M. aided in data collection, data analysis, and drafting the manuscript; M.E. designed and facilitated the data collection, reviewed and revised the manuscript. A.P.D. aided in data collection and reviewed the manuscript; Y.S. helped develop the analytical strategy and the methods and results section of the manuscript; A.H. was responsible for the conceptual design, analysis, and revised the manuscript. All authors gave final approval for publication and agree to be held accountable for the work performed therein.
Competing interests
We declare we have no competing interests.
Funding
A.H. and B.M. were partially supported by the NIH grant no. U19AI117891. This is a secondary analysis of data that had been previously collected with support from Roche diagnostics.
References
- 1.Tellier R. 2009. Aerosol transmission of influenza a virus: a review of new studies. J. R. Soc. Interface 6, S783–S790. ( 10.1098/rsif.2009.0302.focus) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lipsitch M, Moxon ER. 1997. Virulence and transmissibility of pathogens: what is the relationship? Trends Microbiol. 5, 31–37. ( 10.1016/S0966-842X(97)81772-6) [DOI] [PubMed] [Google Scholar]
- 3.Alizon S, Hurford A, Mideo N, Van Baalen M. 2009. Virulence evolution and the trade-off hypothesis: history, current state of affairs and the future. J. Evol. Biol. 22, 245–259. ( 10.1111/j.1420-9101.2008.01658.x) [DOI] [PubMed] [Google Scholar]
- 4.Cressler CE, McLeod DV, Rozins C, Van Den Hoogen J, Day T. 2016. The adaptive evolution of virulence: a review of theoretical predictions and empirical tests. Parasitology 143, 915–930. ( 10.1017/S003118201500092X) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bull JJ, Lauring AS. 2014. Theory and empiricism in virulence evolution. PLoS Pathog. 10, e1004387 ( 10.1371/journal.ppat.1004387) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson RM, May RM. 1979. Population biology of infectious diseases: Part I. Nature 280, 361 ( 10.1038/280361a0) [DOI] [PubMed] [Google Scholar]
- 7.Antia R, Levin BR, May RM. 1994. Within-host population dynamics and the evolution and maintenance of microparasite virulence. Am. Nat. 144, 457–472. ( 10.1086/285686) [DOI] [Google Scholar]
- 8.Sofonea MT, Alizon S, Michalakis Y. 2017. Exposing the diversity of multiple infection patterns. J. Theor. Biol. 419, 278–289. ( 10.1016/j.jtbi.2017.02.011) [DOI] [PubMed] [Google Scholar]
- 9.Brown SP, Cornforth DM, Mideo N. 2012. Evolution of virulence in opportunistic pathogens: generalism, plasticity, and control. Trends Microbiol. 20, 336–342. ( 10.1016/j.tim.2012.04.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilchrist MA, Coombs D. 2006. Evolution of virulence: interdependence, constraints, and selection using nested models. Theor. Popul. Biol. 69, 145–153. ( 10.1016/j.tpb.2005.07.002) [DOI] [PubMed] [Google Scholar]
- 11.Mideo N, Alizon S, Day T. 2008. Linking within- and between-host dynamics in the evolutionary epidemiology of infectious diseases. Trends Ecol. Evol. 23, 511–517. ( 10.1016/j.tree.2008.05.009) [DOI] [PubMed] [Google Scholar]
- 12.Antolin MF. 2008. Unpacking β: within-host dynamics and the evolutionary ecology of pathogen transmission. Ann. Rev. Ecol. Evol. Syst. 39, 415–437. ( 10.1146/annurev.ecolsys.37.091305.110119) [DOI] [Google Scholar]
- 13.Brown NF, Wickham ME, Coombes BK, Finlay BB. 2006. Crossing the line: selection and evolution of virulence traits. PLoS Pathog. 2, e42 ( 10.1371/journal.ppat.0020042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mackinnon MJ, Read AF. 2004. Virulence in malaria: an evolutionary viewpoint. Proc. R. Soc. B 359, 965–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mackinnon MJ, Gandon S, Read AF. 2008. Virulence evolution in response to vaccination: the case of malaria. Vaccine 26, C42–C52. ( 10.1016/j.vaccine.2008.04.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sofonea MT, Aldakak L, Boullosa LV, Alizon S. 2018. Can ebola virus evolve to be less virulent in humans? J. Evol. Biol. 31, 382–392. ( 10.1111/jeb.13229) [DOI] [PubMed] [Google Scholar]
- 17.Fraser C, Hollingsworth TD, Chapman R, de Wolf F, Hanage WP. 2007. Variation in HIV-1 set-point viral load: epidemiological analysis and an evolutionary hypothesis. Proc. Natl Acad. Sci. USA 104, 17 441–17 446. ( 10.1073/pnas.0708559104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fraser C, Lythgoe K, Leventhal GE, Shirreff G, Hollingsworth TD, Alizon S, Bonhoeffer S. 2014. Virulence and pathogenesis of HIV-1 infection: an evolutionary perspective. Science 343, 1243727 ( 10.1126/science.1243727) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ben-Shachar R, Koelle K. 2018. Transmission-clearance trade-offs indicate that dengue virulence evolution depends on epidemiological context. Nat. Commun. 9, 2355 ( 10.1038/s41467-018-04595-w) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ewald PW. 1983. Host-parasite relations, vectors, and the evolution of disease severity. Annu. Rev. Ecol. Syst. 14, 465–485. ( 10.1146/annurev.es.14.110183.002341) [DOI] [Google Scholar]
- 21.Levin S, Pimentel D. 1981. Selection of intermediate rates of increase in parasite-host systems. Am. Nat. 117, 308–315. ( 10.1086/283708) [DOI] [Google Scholar]
- 22.Levin BR. 1996. The evolution and maintenance of virulence in microparasites. Emerg. Infect. Dis. 2, 93 ( 10.3201/eid0202.960203) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bull JJ. 1994. Virulence. Evolution 48, 1423–1437. [DOI] [PubMed] [Google Scholar]
- 24.Leung NH, Xu C, Ip DK, Cowling BJ. 2015. The fraction of influenza virus infections that are asymptomatic: a systematic review and meta-analysis. Epidemiology 26, 862 ( 10.1097/EDE.0000000000000340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Kerckhove K, Hens N, Edmunds WJ, Eames KT. 2013. The impact of illness on social networks: implications for transmission and control of influenza. Am. J. Epidemiol. 178, 1655–1662. ( 10.1093/aje/kwt196) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eames K, Tilston N, White P, Adams E, Edmunds W. 2010. The impact of illness and the impact of school closure on social contact patterns. Health Technol. Assess. 14, 267–312. ( 10.3310/hta14340-04) [DOI] [PubMed] [Google Scholar]
- 27.Dale AP, Ebell M, McKay B, Handel A, Forehand R, Dobbin K. 2019. Impact of a rapid point of care test for influenza on guideline consistent care and antibiotic use. J. Am. Board Fam. Med. 32, 226–233. ( 10.3122/jabfm.2019.02.180183) [DOI] [PubMed] [Google Scholar]
- 28.Kohavi R, John GH. 1997. Wrappers for feature subset selection. Artif. Intellig. 97, 273–324. ( 10.1016/S0004-3702(97)00043-X) [DOI] [Google Scholar]
- 29.Yan J, et al. 2018. Infectious virus in exhaled breath of symptomatic seasonal influenza cases from a college community. Proc. Natl Acad. Sci. USA 115, 1081–1086. ( 10.1073/pnas.1716561115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Killingley B, Nguyen-Van-Tam J. 2013. Routes of influenza transmission. Influenza Other Respir. Virus. 7, 42–51. ( 10.1111/irv.12080) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carrat F, Vergu E, Ferguson NM, Lemaitre M, Cauchemez S, Leach S, Valleron A-J. 2008. Time lines of infection and disease in human influenza: a review of volunteer challenge studies. Am. J. Epidemiol. 167, 775–785. ( 10.1093/aje/kwm375) [DOI] [PubMed] [Google Scholar]
- 32.Hayden FG, Fritz R, Lobo MC, Alvord W, Strober W, Straus SE. 1998. Local and systemic cytokine responses during experimental human influenza a virus infection. Relation to symptom formation and host defense. J. Clin. Invest. 101, 643–649. ( 10.1172/JCI1355) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conover W. 1999. Practical nonparametric statistics. New York, NY: John Wiley & Sons. [Google Scholar]
- 34.Kuritz SJ, Landis JR, Koch GG. 1988. A general overview of Mantel-Haenszel methods: applications and recent developments. Annu. Rev. Public Health 9, 123–160. ( 10.1146/annurev.pu.09.050188.001011) [DOI] [PubMed] [Google Scholar]
- 35.R Core Team. 2016. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; See https://www.R-project.org/. [Google Scholar]
- 36.Bischl B, Lang M, Kotthoff L, Schiffner J, Richter J, Studerus E, Casalicchio G, Jones ZM. 2016. mlr: Machine learning in r. J. Mach. Learn. Res. 17, 1–5. [Google Scholar]
- 37.Meyer D, Zeileis A, Hornik K.2017. Vcd: visualizing categorical data. R package version 1.4-7.
- 38.Signorell A.2019. DescTools: tools for descriptive statistics. See https://cran.r-project.org/package=DescTools .
- 39.McCoul ED, Mohammed AE, Debbaneh PM, Carratola M, Patel AS. 2019. Differences in the intended meaning of congestion between patients and clinicians. JAMA Otolaryngol. Head Neck Surg. 145, 634–640. ( 10.1001/jamaoto.2019.1023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cowling BJ, et al. 2010. Comparative epidemiology of pandemic and seasonal influenza A in households. New Engl. J. Med. 362, 2175–2184. ( 10.1056/NEJMoa0911530) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicas M, Nazaroff WW, Hubbard A. 2005. Toward understanding the risk of secondary airborne infection: emission of respirable pathogens. J. Occup. Environ. Hyg. 2, 143–154. ( 10.1080/15459620590918466) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nicas M, Sun G. 2006. An integrated model of infection risk in a health-care environment. Risk Anal. 26, 1085–1096. ( 10.1111/j.1539-6924.2006.00802.x) [DOI] [PubMed] [Google Scholar]
- 43.Gralton J, Tovey E, McLaws M-L, Rawlinson WD. 2011. The role of particle size in aerosolised pathogen transmission: a review. J. Infect. 62, 1–13. ( 10.1016/j.jinf.2010.11.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weber TP, Stilianakis NI. 2008. Inactivation of influenza A viruses in the environment and modes of transmission: a critical review. J. Infect. 57, 361–373. ( 10.1016/j.jinf.2008.08.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nicas M, Jones RM. 2009. Relative contributions of four exposure pathways to influenza infection risk. Risk Anal. 29, 1292–1303. ( 10.1111/j.1539-6924.2009.01253.x) [DOI] [PubMed] [Google Scholar]
- 46.Atkinson MP, Wein LM. 2008. Quantifying the routes of transmission for pandemic influenza. Bull. Math. Biol. 70, 820–867. ( 10.1007/s11538-007-9281-2) [DOI] [PubMed] [Google Scholar]
- 47.Lindsley WG, et al. 2010. Measurements of airborne influenza virus in aerosol particles from human coughs. PLoS ONE 5, e15100 ( 10.1371/journal.pone.0015100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brankston G, Gitterman L, Hirji Z, Lemieux C, Gardam M. 2007. Transmission of influenza A in human beings. Lancet Infect. Dis. 7, 257–265. ( 10.1016/S1473-3099(07)70029-4) [DOI] [PubMed] [Google Scholar]
- 49.Earn DJ, Andrews PW, Bolker BM. 2014. Population-level effects of suppressing fever. Proc. R. Soc. B 281, 20132570 ( 10.1098/rspb.2013.2570) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ebell MH, Call M, Shinholser J. 2012. Effectiveness of oseltamivir in adults: a meta-analysis of published and unpublished clinical trials. Fam. Pract. 30, 125–133. ( 10.1093/fampra/cms059) [DOI] [PubMed] [Google Scholar]
- 51.Hayden FG, et al. 2018. Baloxavir marboxil for uncomplicated influenza in adults and adolescents. New Engl. J. Med. 379, 913–923. ( 10.1056/NEJMoa1716197) [DOI] [PubMed] [Google Scholar]
- 52.Jefferson T, Deeks J, Demicheli V, Rivetti D, Rudin M. 2004. Amantadine and rimantadine for preventing and treating influenza A in adults. Cochrane Database Syst. Rev. 3 ( 10.1002/14651858.CD001169.pub2) [DOI] [PubMed] [Google Scholar]
- 53.Nishiura H, Oshitani H. 2011. Household transmission of influenza (H1N1-2009) in Japan: age-specificity and reduction of household transmission risk by zanamivir treatment. J. Int. Med. Res. 39, 619–628. ( 10.1177/147323001103900231) [DOI] [PubMed] [Google Scholar]
- 54.Goldstein E, et al. 2010. Oseltamivir for treatment and prevention of pandemic influenza A/H1N1 virus infection in households, Milwaukee, 2009. BMC Infect. Dis. 10, 211 ( 10.1186/1471-2334-10-211) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Handel A, Ebell MH. 2015. Neuraminidase inhibitors for influenza: fully evaluating benefits and harms. Lancet Respir. Med. 3, e7–e8. ( 10.1016/S2213-2600(15)00066-1) [DOI] [PubMed] [Google Scholar]
- 56.Halloran ME, Hayden FG, Yang Y, Longini IM Jr, Monto AS. 2006. Antiviral effects on influenza viral transmission and pathogenicity: observations from household-based trials. Am. J. Epidemiol. 165, 212–221. ( 10.1093/aje/kwj362) [DOI] [PubMed] [Google Scholar]
- 57.Yang Y, Halloran ME, Longini IM Jr. 2009. A Bayesian model for evaluating influenza antiviral efficacy in household studies with asymptomatic infections. Biostatistics 10, 390–403. ( 10.1093/biostatistics/kxn045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsang TK, Lau LL, Cauchemez S, Cowling BJ. 2016. Household transmission of influenza virus. Trends Microbiol. 24, 123–133. ( 10.1016/j.tim.2015.10.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hozé N, Bonhoeffer S, Regoes R. 2018. Assessing the public health impact of tolerance-based therapies with mathematical models. PLoS Comput. Biol. 14, e1006119 ( 10.1371/journal.pcbi.1006119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Porco TC, Lloyd-Smith JO, Gross KL, Galvani AP. 2005. The effect of treatment on pathogen virulence. J. Theor. Biol. 233, 91–102. ( 10.1016/j.jtbi.2004.09.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lessler J, Cummings DA. 2016. Mechanistic models of infectious disease and their impact on public health. Am. J. Epidemiol. 183, 415–422. ( 10.1093/aje/kww021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Funk S, Bansal S, Bauch CT, Eames KT, Edmunds WJ, Galvani AP, Klepac P. 2015. Nine challenges in incorporating the dynamics of behaviour in infectious diseases models. Epidemics 10, 21–25. ( 10.1016/j.epidem.2014.09.005) [DOI] [PubMed] [Google Scholar]
- 63.Carrasco LR, Jit M, Chen MI, Lee VJ, Milne GJ, Cook AR. 2013. Trends in parameterization, economics and host behaviour in influenza pandemic modelling: a review and reporting protocol. Emerg. Themes Epidemiol. 10, 3 ( 10.1186/1742-7622-10-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Handel A, Longini IM Jr, Antia R. 2007. Neuraminidase inhibitor resistance in influenza: assessing the danger of its generation and spread. PLoS Comput. Biol. 3, e240 ( 10.1371/journal.pcbi.0030240) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen S-C, You S-H, Ling M-P, Chio C-P, Liao C-M. 2012. Use of seasonal influenza virus titer and respiratory symptom score to estimate effective human contact rates. J. Epidemiol. 22, 353–363. ( 10.2188/jea.JE20110146) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liao C-M, Yang S-C, Chio C-P, Chen S-C. 2010. Understanding influenza virus-specific epidemiological properties by analysis of experimental human infections. Epidemiol. Infect. 138, 825–835. ( 10.1017/S0950268809991178) [DOI] [PubMed] [Google Scholar]
- 67.Pawelek KA, Salmeron C, Valle SD. 2015. Connecting within and between-hosts dynamics in the influenza infection-staged epidemiological models with behavior change. J. Coupled Syst. Multiscale Dyn. 3, 233–243. ( 10.1166/jcsmd.2015.1082) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Handel A, Rohani P. 2015. Crossing the scale from within-host infection dynamics to between-host transmission fitness: a discussion of current assumptions and knowledge. Proc. R. Soc. B 370, 20140302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McKay B, Ebell M, Dale AP, Shen Y, Handel A.2020. Data from: Virulence-mediated infectiousness and activity trade-offs and their impact on transmission potential of influenza patients. Dryad Digital Repository. ( ) [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- McKay B, Ebell M, Dale AP, Shen Y, Handel A.2020. Data from: Virulence-mediated infectiousness and activity trade-offs and their impact on transmission potential of influenza patients. Dryad Digital Repository. ( ) [DOI]
Supplementary Materials
Data Availability Statement
All of the data and code required to complete the analysis are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.51c59zw4v [69].


