Abstract
The putative synergistic action of target-site mutations and enhanced detoxification in pyrethroid resistance in insects has been hypothesized as a major evolutionary mechanism responsible for dramatic consequences in malaria incidence and crop production. Combining genetic transformation and CRISPR/Cas9 genome modification, we generated transgenic Drosophila lines expressing pyrethroid metabolizing P450 enzymes in a genetic background along with engineered mutations in the voltage-gated sodium channel (para) known to confer target-site resistance. Genotypes expressing the yellow fever mosquito Aedes aegypti Cyp9J28 while also bearing the paraV1016G mutation displayed substantially greater resistance ratio (RR) against deltamethrin than the product of each individual mechanism (RRcombined: 19.85 > RRCyp9J28: 1.77 × RRV1016G: 3.00). Genotypes expressing Brassicogethes aeneus pollen beetle Cyp6BQ23 and also bearing the paraL1014F (kdr) mutation, displayed an almost multiplicative RR (RRcombined: 75.19 ≥ RRCyp6BQ23: 5.74 × RRL1014F: 12.74). Reduced pyrethroid affinity at the target site, delaying saturation while simultaneously extending the duration of P450-driven detoxification, is proposed as a possible underlying mechanism. Combinations of target site and P450 resistance loci might be unfavourable in field populations in the absence of insecticide selection, as they exert some fitness disadvantage in development time and fecundity. These are major considerations from the insecticide resistance management viewpoint in both public health and agriculture.
Keywords: toxicology, malaria, synergism, CRISPR/Cas9, insecticide resistance management, Drosophila
1. Introduction
The prevention of vector-borne diseases and the protection of agricultural production largely relies on the control of pest insects through the use of insecticides. However, insects display a striking ability to develop resistance, an intriguing evolutionary adaptation to a very fast environmental change, with dramatic consequences. For example, the number of malaria cases increased in 2018 after many years of decline, indicative of a failure of pyrethroid based intervention strategies [1].
To mitigate against the failure of insecticide-based control tools, the mechanisms by which insects have evolved resistance must be elucidated and understood. Mutations at the insecticide target site which reduce insecticide binding affinity, and metabolic detoxification which inactivates and sequesters insecticidal active ingredients, are the most common mechanisms of insecticide resistance [2]. However, it has been widely hypothesized that it is only the synergism of different mechanisms in the same insect population that causes a real operational control failure in many cases [3–6]. This has important ramifications on insecticide resistance management (IRM) strategies. For example, the synergist piperonyl butoxide when incorporated into bednets seems to restore their efficacy even in areas with fixed target site resistance alleles [7], while the value of molecular diagnostics for IRM might be different depending on the presence or absence of additional mechanisms in the same mosquito population [8,9]. Although a putative synergistic epistasis of the metabolic and target site resistance loci has been considered [6], it has only been tested by crossing lines with different resistance factors together in order to see their effect. However, this process introduces a large amount of unrelated genetic variation, which complicates inferences drawn about the specific loci being studied. While there have been several efforts to isolate the contribution of resistance alleles by introducing them into model organisms like Drosophila, many of these studies have only managed to recapitulate a fraction of the total resistance levels observed in the field [10,11].
An additional factor in IRM strategies are the evolutionary fitness costs imposed by resistance alleles. Alleles that pose high costs will tend to revert back to their susceptible form once the selective pressure (pesticide) is removed. Variants that do not pose such a cost can persist indefinitely. Fitness costs related to drug resistance have long been the subject of investigation in clinical settings with resistant bacteria and cancer lines [12,13], and have also been subjected to investigation in insects [14]. From these studies it has become clear that the severity, and indeed presence, of a fitness cost brought about by a given resistance allele depends on the particular allele and the genetic background in which it is observed. However, the understanding of the costs of each variant and their epistatic effects are poorly understood.
Of particular interest to the insecticide community are the mechanisms underpinning pyrethroid resistance and their resulting evolutionary implications. This large class of structurally related insecticides targets the voltage-gated sodium channel (the orthologues to the para gene in Drosophila), and representative pyrethroids such as deltamethrin have been widely used in both agricultural and public health related pest control since the 1970s. Widespread use was followed by the appearance of several independent resistance mechanisms. Among several examples, the P450 Cyp6BQ23 was found to be overexpressed in the pollen beetle Brassicogethes aeneus [15], while other resistant strains carried the kdr (L1014F) substitution in para [16]. In the mosquito Aedes aegypti, the main vector of yellow fever worldwide, a similar array of mechanisms have been identified including the overexpression of Cyp9J28; [17] and another mutation in para (V1016G). However, the interaction of these alleles in vivo has not, to our knowledge, been studied, neither in terms of contribution to pyrethroid resistance nor any resulting fitness cost.
Here, we report the generation of transgenic Drosophila melanogaster lines expressing pyrethroid metabolizing cytochrome P450 enzymes from major mosquito vectors (A. aegypti) and agricultural pests (B. aeneus) in a genetic background where we have engineered by CRISPR/Cas9 specific homozygous target-site resistance mutations in the voltage-gated sodium channel (para), also found in these insects. This strategy enabled us to directly measure the resulting resistance phenotypes, encountering either the contribution of both mechanisms or each one separately, with very limited confounding genetic effects.
2. Material and methods
(a). Drosophila strains
Drosophila strains used in this study are shown in the electronic supplementary material, table S1. Strain yw nos int; attP40 [18] was a gift by Pawel Piwko and Christos Delidakis (IMBB/FORTH). CRISPR/Cas9 genome modification was performed at strain y1 M{nos-Cas9.P}ZH-2A w*, where Cas9 is expressed under the control of nanos promoter [19] (herein referred as nos.Cas9, #54591 in the Bloomington Drosophila stock centre). Background strain yellow white (yw) and several balancer lines (see the electronic supplementary material, table S1) are part of the IMBB/FORTH facility fly collection (kindly provided by Prof. Christos Delidakis, IMBB and University of Crete). The HR-GAL4 driver line is previously described [20], while the responder line UAS.AaegCYP9J28 (herein referred as UAS-CYP9J28) was generated also as described previously [17]. All flies were kept at a temperature of 25°C, humidity 60–70% and 12 : 12 h photoperiod on a standard fly diet.
(b). Amplification and sequencing of para target regions
DNA from nos.Cas9 Drosophila adults was extracted with DNAzol (MRC, Cincinnati, OH) following the manufacturer's instructions. Several primers (paraInF, paraInR, kdrF, kdrR, exoF, exoR, electronic supplementary material, table S2) were designed based on the para gene sequence in order to amplify and sequence overlapping fragments that correspond to a 2585 bp sequenced genomic region of strain nos.Cas9 (X: 16485234:16487819, numbering according to BDGP6.22 genome assembly) which contains the exons that harbour positions L1014 and V1016, respectively. The amplification reactions were performed using KapaTaq DNA polymerase (Kapa Biosystems, Wilmington, MA). The conditions were 95°C for 2 min for initial denaturation followed by 30–35 cycles of denaturation at 95°C for 30 s, annealing at 54–60°C for 15 s, extension at 72°C for 90 s and a final extension step for 2 min. The polymerase chain reaction (PCR) products were purified with a PCR clean-up kit (Macherey-Nagel, Düren, Germany) according to manufacturer's instructions. Sequencing of the products was performed from both ends at Macrogen (Amsterdam, The Netherlands).
(c). Strategy for genome editing
An ad hoc CRISPR–Cas9 strategy was implemented in order to generate Drosophila strains bearing either mutation (equivalent to L1014F and V1016G according to housefly Musca domestica numbering) in the para gene. Based on the genomic sequence of para obtained for strain nos.Cas9 several CRISPR targets in the desired region were identified using the Optimal Target Finder online tool [21] (http://targetfinder.flycrispr.neuro.brown.edu/). We selected three CRISPR targets in total, which had minimal predicted off-target effects. Targets para935 and para406 were used to obtain L1014F, while targets para406 and para205 were used for V1016G (figure 1). In order to generate single guide RNAs (sgRNAs) targeting those sequences, three different RNA-expressing plasmids were generated (sgRNA935, sgRNA406 and sgRNA205, respectively) based on the vector pU6-BbsI chiRNA [22] following digestion with BbsI and ligation of three relevant double-stranded oligonucleotides, which were generated by annealing single stranded oligonucleotides 935F/935R, 406F/406R and 205F/205R, respectively (electronic supplementary material, table S2). Following ligation and transformation, single colonies for each construct were picked and checked for the correct insert by performing colony PCR using T7 universal primer and the reverse primer for each double stranded DNA. The sequence of each sgRNA expressing plasmid was verified by sequencing (Macrogen).
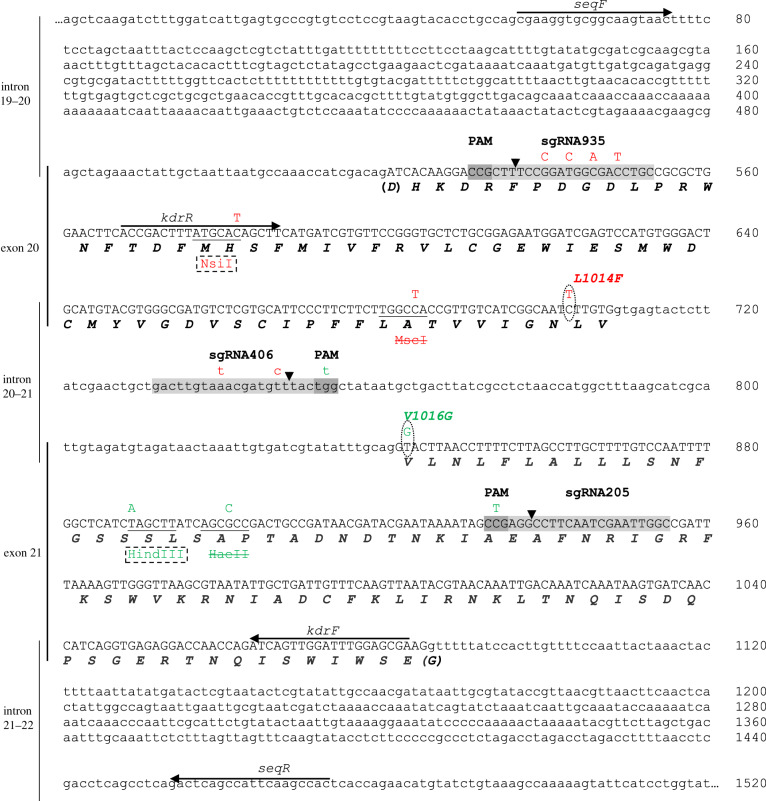
Figure 1.
CRISPR/Cas9 strategies for the generation of genome modified flies bearing mutations L1014F and V1016G. Nucleotide and deduced amino acid sequence of a 1520 bp fragment of para, encompassing exons 20 and 21 that contain positions 1014 and 1016, respectively (M. domestica numbering) of the Drosophila melanogaster amino acid sequence. Light grey areas indicate the CRISPR/Cas9 targets selected (sgRNA935, sgRNA406 and sgRNA2015), while dark grey areas indicate the corresponding PAM (-NGG) triplets. Vertical arrows denote break points for CRISPR/Cas9-induced double-stranded breaks. Red lettering indicates the differences introduced in exon 20 for the generation of L1014F, while green lettering indicates the differences introduced in exon 21 for the generation of V1016G. Ovals mark non-synonymous differences between the target (wild-type) and donor (genome modified) sequences. Synonymous mutations incorporated for diagnostic purposes, as well as to avoid cleavage of the donor plasmid by the CRISPR/Cas9 machinery, are shown above the nucleotide sequence. Restriction sites abolished because of the genome modification are shown with strikethrough letters and the corresponding sequence is underlined. Restriction sites introduced because of the genome modification are shown in dashed boxes and the corresponding sequence is also underlined. Horizontal arrows indicate the positions of primer pairs kdrF/kdrR and seqF/seqR (electronic supplementary material, table S2) used for sequencing of the genome modified alleles. (Online version in colour.)
Two different donor plasmids, vgscL1014F and vgscV1016G were synthesized de novo (Genscript, Piscataway, NJ) to facilitate homologous directed repair for the generation of strains L1014F and V1016G, respectively (newly synthesized sequences were subcloned in the pUC57 vector EcoRV site; relevant insert sequences for each donor plasmid are shown in the electronic supplementary material, figure S1). Each plasmid contained two approximately 900 bp homology arms flanking the target region between sgRNA targets para935 and para406 (for L1014F) or para406 and para205 (for V1016G) (figure 1). The target regions were specifically designed in order to contain the desired mutations along with certain additional synonymous mutations (figure 1) serving either as molecular markers (to facilitate molecular screening of CRISPR events), or to prevent unwanted CRISPR digestion of the donor itself.
(d). Molecular screening and establishment of genome modified lines
Injection of nos.Cas9 pre-blastoderm embryos was performed at the IMBB/FORTH facility with injection mixes containing 75 ng µl−1 of each sgRNA plasmid vector and 100 ng µl−1 of donor template as previously described [23]. Hatched larvae were transferred into standard fly artificial diet and after 9–13 days G0 (generation zero) surviving adults were collected and individually backcrossed with nos.Cas9 flies. In order to screen for CRISPR events, G1 progeny from each cross were pooled into batches of approximately 30 and genomic DNA extraction was performed en masse in order to be screened with two different strategies. Initially, 2 µg of pooled genomic DNA (gDNA) were digested with MscI (for L1014F crosses) or HaeII (for V1016G crosses); these enzymes cut only the wild-type alleles but not potential mutant alleles in each DNA pool. Then, the strategy for screening for L1014F mutants consists of amplification with specific primers 1014UP/1014DOWN (electronic supplementary material, table S2) that were designed taking into account the synonymous mutations introduced in the relevant target sequences in donor template vgscL1014F, in order to generate a 234 bp diagnostic fragment that is specific to genome modified alleles, but not wild-type ones (electronic supplementary material, figure S2A). PCR was performed with Kapa Taq polymerase as previously described using approximately 60 ng of digested template DNA mix. For screening of V1016G mutants, an alternative strategy was used, which consists of PCR amplification with the ‘generic’ primer pair kdrF/kdrR (electronic supplementary material, table S1) which was designed in order to amplify a 516 bp fragment that may be derived by either wild-type (if still present, given the initial enzymatic cleavage of the template DNA mix) or genome modified alleles. Following PCR amplification, the product was digested with the diagnostic enzyme HindIII introduced in the vgscV1016G donor plasmid sequence, producing two diagnostic fragments of 324 and 192 bp (electronic supplementary material, figure S2B). Crosses that proved positive for genome modified alleles were further explored in order to identify individual flies bearing mutant alleles and establish homozygous lines. DNA was extracted from several homozygous female and hemizygous male adults, amplified by using primers kdrF/kdrR or seqF/seqR (electronic supplementary material, table S2) and the relevant amplification fragments were sequence verified (Macrogen) for the presence of the desired mutations (electronic supplementary material, figure S2C).
(e). Generation of transgenic Drosophila expressing Cyp6BQ23
In order to generate a transgenic D. melanogaster strain conditionally expressing Cyp6BQ23, a GAL4/UAS strategy was employed. The responder strain, UAS-CYP6BQ23, was generated by PhiC31 integrase mediated attB insertion at an attP40 landing site [24]. An ad hoc integration vector, dPelican-attB-UAS_CYP6BQ23 was generated by replacing the insert of plasmid dPelican-attB-UAS_CYP6A51 we had previously generated [25]. We performed de novo synthesis (Genescript) of the CYP6BQ23 coding sequence (GenBank acc. no. KC840055.1) with some modifications in order to optimize for expression in Drosophila, i.e. introducing a CACC Kozac-consensus sequence just upstream of the initiation codon and taking into account codon usage optimal for Drosophila (full construct sequence shown in the electronic supplementary material, figure S1). An MluI/XhoI fragment encompassing the CYP6BQ23 coding sequence was subcloned into dPelican-attB-UAS_CYP6A51 [25] plasmid backbone that had been digested with MluI and XhoI so that the existing CYP6A51 expression cassette was removed and replaced by the CYP6BQ23 fragment downstream of 5xUAS and just upstream of an SV40 polyadenylation sequence, to produce the final recombinant plasmid dPelican.attB.UAS_CYP6BQ23. This plasmid also contains a mini-white marker gene for Drosophila. The sequence was verified using sequencing primers pPel_uas F and pPel_sv40 R [26] and the recombinant plasmid was used to inject pre-blastoderm embryos of the D. melanogaster strain yw nos int; attP40. Injected G0 flies were outcrossed with yw background flies and G1 progeny was screened for w+ phenotypes (red eyes) indicating integration of the recombinant plasmid. Independent transformed lines were crossed with a strain bearing a balancer for the second chromosome (yw; CyO/Sco), and G2 flies with red eyes and Cy phenotype were selected and crossed among themselves to generate homozygous UAS-CYP6BQ23 flies used to establish the transgenic responder line population.
(f). Generation of null background strain yw;attP40
In order to generate a Drosophila line that is fully equivalent to the UAS-CYP6BQ23 strain in terms of genetic background and can be used as null control in downstream experiments, male non-injected flies of the D. melanogaster strain yw nos int; attP40 were outcrossed with female yw background flies, and male G1 progeny (not carrying the yw nos int chromosome) was crossed with females bearing a balancer for the second chromosome (yw; CyO/Sco). G2 flies with Cy phenotype were selected and crossed among themselves to generate homozygous yw; attP40 flies that have essentially the same genetic background with the transgenic responder line population, apart from the UAS-CYP6BQ23 expression cassette.
(g). Generation of homozygous recombinant yw; HR-GAL4>UAS-CYP9J28(2N) strain
We generated a strain bearing both HR-GAL4 and UAS-CYP9J28 in the second chromosome by genetic recombination, as shown in the electronic supplementary material, figure S3. This was performed via a cross between lines HR-GAL4 [20] and UAS-CYP9J28 [17] that produces a heterozygous genotype (y)w;HR-GAL4>UAS-CYP9J28. However, while this genotype produces a detectable resistant phenotype in contact bioassays [17], preliminary topical application bioassays indicated that only marginal (i.e. not always significant) changes in resistance were detected (data not shown). Thus, heterozygous (y)w;HR-GAL4>UAS-CYP9J28 females (where chromosomal crossover is feasible) were crossed to yw; CyO/Sco balancer flies and the progeny screened for genetic recombination events as shown in the electronic supplementary material, figure S3. We identified heterozygous yw; [HR-GAL4_UAS-CYP9J28]/CyO recombinant flies bearing both HR-GAL4 and UAS-CYP9J28 transgenes in the second chromosome and these were intercrossed to generate the homozygous yw; HR-GAL4>UAS-CYP9J28(2N) strain, which contains two copies of both driver and responder transgenes.
(h). Generation of driver and responder lines in genome modified (mutant) background
Taking into account that the voltage-gated sodium channel (para) gene in Drosophila is located at the X chromosome, while the HR-GAL4 driver strain used in our laboratory, the Cyp9J28 insertion site, as well as the attp40 insertion site bearing the Cyp6BQ23 transgene, are all located in the second chromosome, we devised a simple genetic crosses strategy (electronic supplementary material, figure S4) to: (i) introduce both the HR-GAL4 driver and the UAS-CYP6BQ23 responder transgene in a paraL1014F genetic background in order to generate strain paraL1014F; HR-GAL4>UAS-CYP6BQ23 that represents the ‘beetle’ allele combination, and (ii) generate a strain bearing the linked [HR-GAL4_UAS-CYP9J28] chromosome 2 (see (g) above) in a paraV1016G genetic background (strain paraV1016G;HR-GAL4>UAS-CYP9J28(2N)) that represents the ‘mosquito’ allelic combination. As shown in the electronic supplementary material, figure S4, the crossing scheme results in lines where the X chromosome is derived from nos.Cas9 strain, the second chromosome from the respective transgenic line (HR-GAL4, yw nos int; attP40 or UAS-CYP9J28) and the other chromosomes from yw (note that all balancer lines as well as nos.Cas9, yw nos int; attP40 and UAS-CYP9J28 had been originally generated in yw or extensively outcrossed to it in the IMBB fly facility).
(i). GAL4/UAS-expression in Drosophila melanogaster
In order to drive conditional expression of Cyp9J28 or Cyp6BQ23 in wild-type or mutant genetic backgrounds we used the HR-GAL4 driver [20] which drives expression in specific tissues related to detoxification (malpighian tubules, midgut and fat body). Transgenic responder virgin females were crossed with HR-GAL4 males and the progeny was used in toxicity bioassays with deltamethrin in order to validate the potential of each line to confer insecticide resistance. Crosses of yw; attP40 or nos.Cas9 virgin females with HR-GAL4 males were used as negative controls.
(j). Extraction of RNA, complementary DNA synthesis, reverse transcription polymerase chain reaction and quantitative polymerase chain reaction
Reverse transcription PCR was performed in order to confirm expression of Cyp6bq23 or Cyp9j28 in the progeny. Total RNA was extracted from pools of 20 adult Drosophila flies (1–3 day old) using Trizol reagent (MRC), according to the manufacturer's instructions. Extracted RNA samples were treated with Turbo DNase (Ambion, Foster City, CA) to remove genomic DNA and 2 µg of treated RNA was used to generate first strand complementary DNA (cDNA) using oligo-dT20 primers with Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA). One microliter of cDNA was used in the PCR reaction using specific primers for each transgene and for rpl11 (ribosomal protein L11) which served as a reference gene (electronic supplementary material, table S1). The conditions of the reactions were 95°C for 5 min followed by 35 cycles of 95°C for 30 s, 55°C for 30 s, 72°C for 30 s and final extension for 2 min.
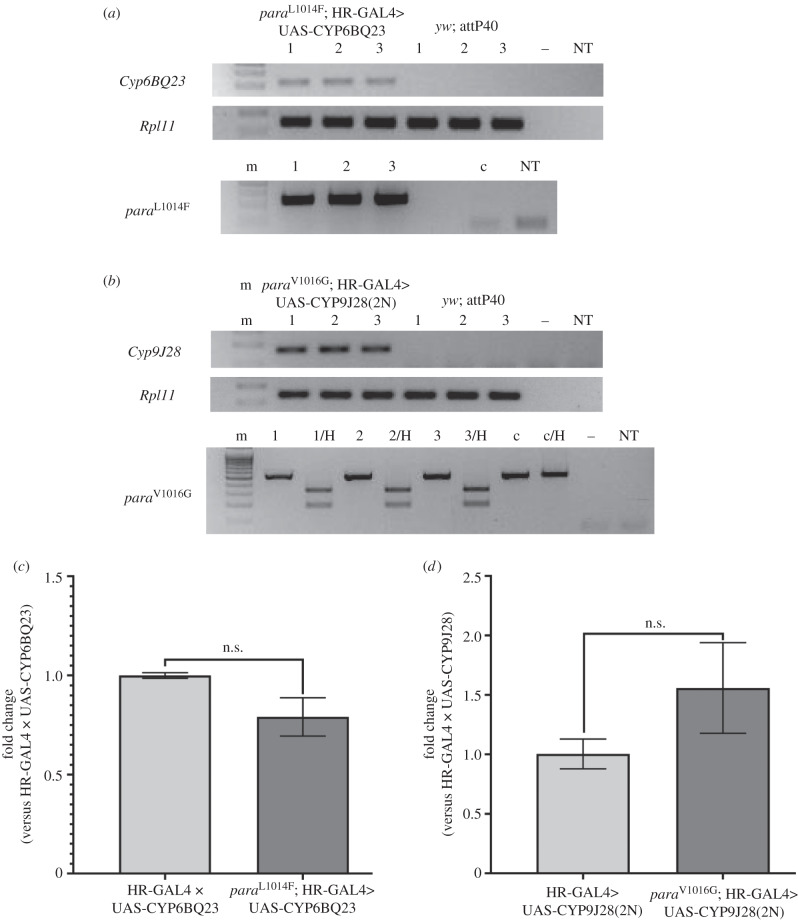
A two-step quantitative-reverse transcriptase PCR was performed in order to analyse the expression levels of Cyp6BQ23 and Cyp9j28 between genotypes bearing the relevant expression trangenes in either wild-type or genome modified para background (figure 2c,d). Quantitative PCR (qPCR) was conducted using the Kapa SYBR Fast qPCR Master Mix kit (Kapa Biosystems) and the reactions were carried out in the Bio-Rad CFX Connect using the following conditions: 95°C for 2 min, followed by 40 cycles at 95°C for 5 s and 60°C for 30 s. The efficiency of the qPCR reaction for each primer pair was assessed in 10-fold dilution series of pooled cDNA samples. The experiment was performed using three biological and two technical replicates. Relative expression was normalized to the reference genes rpl11 and rpl32, while the analysis was conducted as previously described [27]. All primer sequences are shown in the electronic supplementary material, table S2.
Figure 2.
P450 overexpression in the flies bearing the kdr mutations. (a) (top) Cyp6bg23 expression is confirmed by reverse transcription and PCR amplification of cDNAs. Lanes paraL1014F;HR-GAL4>UAS-CYP6BQ23 (1–3) indicate three biological replicates of the flies tested for the overexpression of the transgene. Lanes yw; attP40 (1–3) indicate the three biological replicates of the control line. The same cDNAs were used to amplify the housekeeping gene rpl11 as a reference gene. —: no reverse transcription control (to monitor for genomic DNA contamination); NT: no template control. (bottom): The presence of L1014F mutation in the same flies is tested by PCR of genomic DNA with allele-specific primers. c: yw; attP40 negative control DNA. (b) (top) Cyp9j28 expression is similarly confirmed. Lanes paraV1016G;HR-GAL4>UAS-CYP9J28(2N) (1–3) indicate three biological replicates of the flies tested for the overexpression of the transgene, while lanes yw; attP40 (1–3) indicate the three biological replicates of the control line. The same cDNAs were used to amplify the housekeeping gene rpl11 as a reference gene. —: no reverse transcription; NT: no template. (bottom): The presence of V1016G mutation in the same flies is tested by PCR of genomic DNA with generic primers and subsequent digestion of the product with HindIII (/H). c: yw; attP40 negative control DNA. (c,d) qRT-PCR for evaluation of P450 expression levels in different strains. The Ct values of strains expressing CYP6BQ23 (c) and CYP9J28 (d) were calculated in the absence or presence of the relevant para mutations. No significant difference in expression was observed (p = 0.0618 for CYP6BQ23, p = 0.1161 for CYP9J28).
(k). Toxicity bioassays
Deltamethrin (99.6%) of technical grade was purchased from Sigma Aldrich (Darmstadt, Germany) and used in contact assays and topical applications. Contact assays were performed as described previously [28]. More specifically, 20 adult female flies (1–3 days old, as per [20,28]) were used for each toxicity assay. Flies were collected in plastic vials and transferred in scintillation vials coated with insecticide. Serial dilutions of 6–7 concentrations of technical grade deltamethrin in acetone were used for dose response bioassays, while vials coated only with acetone served as control. The vials were plugged with cotton that was kept moist with 5% sucrose solution. Each deltamethrin concentration was assayed in three replicates. Knockdown was scored for 180 min with 15–30 min intervals and mortality was scored after 24 h. Topical application of deltamethrin was performed on 1–3 day old female flies. Deltamethrin was dissolved in acetone and serial dilutions were made to generate the appropriate concentrations. Each insecticide concentration (or acetone as negative control) was applied in a dose of 1 µl per fly using a 10 µl Hamilton syringe. Flies were immobilized by keeping them on an ice cold slide. For each concentration 40 flies were tested. Following insecticide application the flies were transferred into glass scintillation vials covered with cotton moisturized with 5% sucrose solution. The vials were maintained in a 25°C incubator while mortality was scored after 24 h.
(l). Life table parameters
Determination of life table parameters was performed as previously described [23]. To determine developmental time and sex ratio, cages with 50 virgins (1–3 day old) and 20 males were capped with cherry juice-agar plates supplemented with yeast, left to cross overnight and after plate replacement the flies were left to lay eggs for 4–5 h. Eggs were transferred into vials with standard fly food in batches of ca 50 (10 replicates for each genotype). Pupation, adult emergence time and total number of males/females were scored daily from day 8. To monitor daily and total fecundity, c. 15 females from each genotype were mated, transferred in small cages capped with 35 mm yeast-supplemented cherry agar plates, and the number of eggs laid by each female was counted daily.
(m). Statistical analyses
Concentration-response data of each bioassay setup were collected and analysed with ProBit analysis using PoloPlus (LeOra Software, Berkeley, CA) in order to calculate lethal concentrations of the 50% of the population subjected to the experiment (LC50 values), 95% fiducial limits (FL), linearity of the dose-mortality response and statistical significance of the results. Α χ2-test was used to assess how well the individual LC50 values observed in the bioassays agree with the calculated linear regression lines. The LC50 values and resistance ratio (RR) are considered significant if the 95% FL did not include 1 [29]. Life table parameter data (electronic supplementary material, dataset 1) were analysed for significant differences between strains with one-way ANOVA using the software GraphPad Prism 8.0.2. A two-tailed unpaired student's t-test (also using GraphPad Prism) was carried out in order to compare relative expression in qPCR data.
3. Results
(a). Generation of Drosophila lines expressing detoxification enzymes in a genetic background bearing target-site resistance mutations in para
We used CRISPR/Cas9 genome engineering (figure 1) to generate strains bearing homozygous target-site resistance mutations in the voltage-gated sodium channel (para) of D. melanogaster (strain paraL1014F bearing mutation L1014F (kdr) and strain paraV1016G bearing mutation V1016G). Additionally, we employed GAL4/UAS for the expression of known detoxification enzyme CYP6BQ23 from the pollen beetle B. aeneus. We also used a previously generated strain for expression of CYP9J28 from the mosquito A. aegypti under the control of HR-GAL4 driver. We used genetic recombination to bring HR-GAL4 and UAS-CYP9J28 in the same chromosome, resulting in a strain bearing two copies of each transgene (yw; HR-GAL4>UAS-CYP9J28(2N)) in order to obtain higher expression levels providing a readily detectable effect in topical application assays (see below). Standard genetic crosses enabled Cyp6BQ23 transgenic expression in paraL1014F genetic background and Cyp9J28 transgenic expression in paraV1016G background so that both mechanisms were combined (figure 2).
(b). Toxicity bioassays in Drosophila indicate synergistic action of different resistance mechanisms
Slow uptake contact bioassay experiments (all results shown in the electronic supplementary material, table S3) showed that resistance levels were extremely high in paraL1014F;HR-GAL4>UAS-CYP6BQ23 flies (totally insensitive at deltamethrin doses exceeding 5000 µg vial−1), compared to the control flies nos.Cas9 (LC50 5.45 (2.40–8.57) µg vial−1) or the flies bearing any of the resistant mechanisms alone (RRL1014F: 158.9; RRCyp6BQ23: 9.68 compared to relevant control flies nos.Cas9 and HR-GAL4 × yw; attP40, respectively).
Thus, in order to more precisely quantify the intensity of resistance phenotype, topical application bioassays were used. The results are shown in table 1. paraL1014F flies exhibit 12.74-fold resistance to deltamethrin, while flies expressing Cyp6BQ23 in wild-type para background exhibit 5.74-fold resistance compared to controls (nos.Cas9 and HR-GAL4 × yw; attP40), which had an absolutely similar response to deltamethrin toxicity.
Table 1.
Topical application deltamethrin bioassay responses of transgenic flies expressing pyrethroid metabolizing P450s alone or along engineered target-site resistance mutations in their voltage-gated sodium channel (para).
| strain/cross | LD50 (ng fly−1) | (95% FL) | slope (±s.e.) | RR |
|---|---|---|---|---|
| HR-GAL4 × yw; attP40 | 3.10 | (2.65–3.65) | 3.59 (±0.47) | 1 |
| HR-GAL4 × UAS-CYP6BQ23 | 17.8 | (12.50–21.65) | 4.27 (±0.87) | 5.74 |
| yw;HR-GAL4>UAS-CYP9J28(2N)a | 5.49 | (4.051–6.60) | 4.4 (±1.09) | 1.77 |
| nos.Cas9 | 3.33 | (1.3–5.1) | 2.259 (±0.39) | 1.07 |
| paraL1014F | 39.49 | (23.1–53.95) | 2.949 (±0.39) | 12.74 |
| paraV1016G | 9.30 | (4.98–14.55) | 1.696 (±0.36) | 3.00 |
| paraL1014F; HR-GAL4>UAS-CYP6BQ23b | 233.08 | (161.70–333.85) | 1.508 (±0.21) | 75.19 |
| paraV1016G; HR-GAL4>UAS-CYP9J28(2N)c | 61.53 | (47.48–78.50) | 4.851 (±0.80) | 19.85 |
ahomozygous recombinant yw; HR-GAL4>UAS-CYPJ28(2N) contains two copies of driver and responder.
bparaL1014F; HR-GAL4 × paraL1014F; UAS-CYP6BQ23.
cparaV1016G; HR-GAL4>UAS-CYPJ28(2N) contains two copies of driver and responder in paraV1016G X-chromosome background.
Flies expressing Cyp6BQ23 in paraL1014F background displayed an almost multiplicative RR compared to the control (RRcombined: 75.19 ≥ RRCyp6BQ23: 5.74 × RRL1014F: 12.74). In the case of the resistance alleles known from A. aegypti an even more striking effect was found: paraV1016G flies show modest levels of resistance (RR: 3.00), while flies stably expressing Cyp9J28 following genetic recombination between the UAS-CYP9J28 responder with the HR-GAL4 driver (bearing two copies of each) also exhibit modest resistance (RR: 1.77) compared to controls. However, the paraV1016G;HR-GAL4>UAS-CYP9J28(2N) flies displayed a significantly greater RR than the product of the individual RRs (RRcombined: 19.85 ≫ RRCyp9J28: 1.77 × RRV1016G: 3.00).
(c). The presence of multiple resistance alleles may be associated with some fitness disadvantage
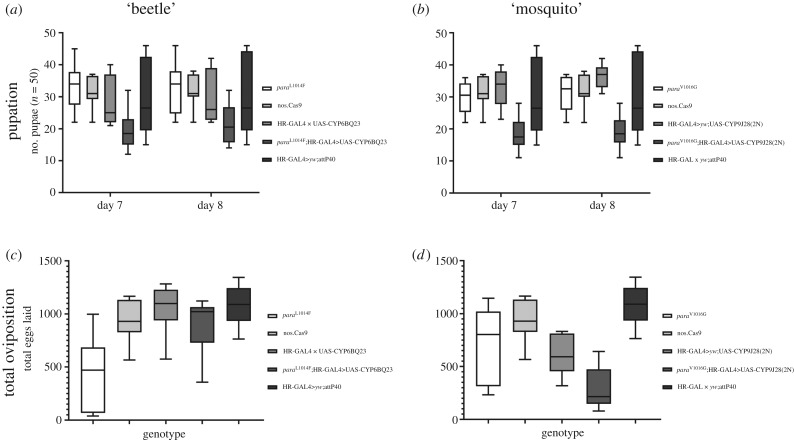
By contrast to the lines bearing only one resistant mechanism, which (with a possible exception on paraL1014F fecundity) exhibited no statistically significant difference compared to the control lines (electronic supplementary material, dataset 1), both the ‘super-resistant’ lines, paraL1014F;HR-GAL4>UAS-CYP6BQ23 and paraV1016G;HR-GAL4>UAS-CYP9J28(2N) showed a significant cost in development time as indicated by one-way ANOVA of pupation time (figure 3 and electronic supplementary material, table S4, pupation after 7–8 days). For the ‘beetle’ allelic combination, paraL1014F;HR-GAL4>UAS-CYP6BQ23 flies exhibit some developmental delay compared to nos.Cas9 controls (pd7 = 0.0249), as well as to paraL1014F (pd7 = 0.0044 and pd8 = 0.0274). Regarding the ‘mosquito’ combination, this is also evident in comparisons of paraV1016G;HR-GAL4>UAS-CYP9J28(2N) flies against nos.Cas9 (pd7 = 0.0039, pd8 = 0.0045), paraV1016G(pd7 = 0.0110, pd8 = 0.0048), yw;HR-GAL4>UAS-CYP9J28 (pd7 = 0.0004, pd8 < 0.0001) and HR-GAL4>yw;attP40 (pd7 = 0.0093, pd8 = 0.0089). Furthermore, paraV1016G;HR-GAL4>UAS-CYP9J28(2N) flies also exhibit a significant cost in total fecundity after 20 days, compared to all controls tested (figure 3 and electronic supplementary material, table S5).
Figure 3.
Evaluation of ‘super-resistant’ fly lines fitness. (a,b): one-way ANOVA of pupation after 7–8 days in strains bearing both resistance alleles for the ‘beetle’ (a) and ‘mosquito’ (b) allele combinations compared to controls, indicating an extended developmental time when both alleles are present. All p-values are shown in the electronic supplementary material, table S4. (c,d): one-way ANOVA of total oviposition in strains bearing both resistance alleles for the ‘beetle’ (c) and ‘mosquito’ (d) allele combinations. Oviposition is significantly reduced in paraV1016G; HR-GAL4>UAS-CYP9J28(2N) flies against all controls, indicating reduced overall fecundity. All p-values are shown in the electronic supplementary material, table S5. For the full life table parameters dataset, see the electronic supplementary material, dataset 1.
4. Discussion
An old enigma in insect toxicology, the putative synergistic action of target site resistance mutations and upregulated cytochrome P450s in pyrethroid resistance of major disease vectors and agricultural pests, has been functionally resolved. Specifically, field evolved P450s conferring pyrethroid resistance and target site resistance alleles were individually introduced in a single in vivo system. Genotypes overexpressing P450s in addition to the target-site resistance displayed a multiplicative RR equal or greater than the product of the RRs obtained for the individual resistance mechanisms. This is in line with previous studies focusing on synergism associated with insecticide resistance alleles (reviewed in [6]), which found that the combination of resistance alleles was mostly multiplicative.
While the work presented here then largely agrees with prior estimations of P450 target site synergism, the use of D. melanogaster in this study provides several advantages. Most critically Drosophila reduces confounding genetic factors arising from different backgrounds which are known to cause substantial variation in ‘wild-type’ lines [30]. While the flies used in this study were not completely isogenic, the backgrounds were much more similar compared to previous studies considering synergism in model organisms [6] and provide similar levels of resistance in topical application bioassays (table 1). When genetic background was controlled for in pest species, it has only been done by backcrossing individual variants such as the 1016 mutation in A. aegypti [31]. Drosophila provides an easier alternative, but caution must be taken between interpreting findings across species, taking into account not only the powers but also the limitations of this system (for a detailed relevant discussion see [32,33]).
The mechanism for the apparent synergism between P450s and target site mutations is still not fully understood. It may involve only the parent compound, i.e. reduced binding affinity for the target site could simply give P450s additional time to perform insecticide metabolism and avoid saturation. This is thought to be the case with P-glycoprotein CYP3A4 synergism in humans, whereby the former increases the time a compound spends in the intestine and thereby increases its chance of being metabolized by the latter [34]. The higher RRcombined in the slow uptake compared to the fast-uptake topical application seems to corroborate this hypothesis (electronic supplementary material, table S3 and table 1, respectively). Alternatively, the less toxic P450-generated metabolites might bind the mutant target receptors disproportionately less effectively, thus manifesting in a synergistic phenotype. It is currently unknown what, if any, affinity the P450 derived metabolites of deltamethrin would have for para, but it has been suggested that their accumulation is likely to be detrimental in mosquitoes and that their metabolism by certain P450s, such as the Anopheles gambiae CYPZ8 and CYP6Z2, is a secondary but important mechanism of insecticide resistance [35]. In addition, examples from other compounds may provide some guidance. Imidacloprid is ‘detoxified’ by P450s into a variety of metabolites which still show levels of toxicity that would probably be relevant in vivo [36,37]. Nevertheless, there is currently no data regarding the binding of various pyrethroid metabolites for different alleles of para, so such a hypothesis awaits functional validation.
In addition to differences in resistance level, fitness costs were also observed for the various genotypes used in this study. While genotypes carrying a single resistance allele behaved similarly to the control lines (with a possible exception regarding paraL1014F fecundity), the combination of these alleles significantly increased the developmental time both in the ‘beetle’ and ‘mosquito’ genotype combinations and significantly reduced fecundity in the ‘mosquito’ combination against multiple controls contributing to the genetic background (figure 3). It thus seems possible that these alleles, may exert a fitness cost only in certain backgrounds, an implication which has far reaching ramifications for IRM. This hypothesis is also supported by studies done directly on these pest species. The combination of high level Cyp6BQ23 expression and kdr mutations is extremely rare in pollen beetle populations, supporting the fitness cost theory [16], but the mechanism underpinning this phenomenon is not known. By contrast, the results presented here on the V1016G mutation contradict those obtained by backcrossing the mutation into a susceptible A. aegypti background, although the precise mutation in that study was different [31]. These data collectively suggest epistatic effects between different resistance mechanisms and highlight the need for fitness cost assessment to be done in multiple backgrounds. Further work will thus be needed to establish and characterize the evolutionary significance of these resistance alleles in the field.
Several groups are currently developing and applying DNA-based technologies for insecticide resistance monitoring [8,9]. Our study shows that these molecular diagnostics need careful calibration, integration and interpretation, as an apparent epistasis (i.e. different phenotype depending on the genetic background) is present, and thus they may or may not diagnose the importance of resistance in the field. The reconstruction of complex resistance phenotypes by reverse-genetics based simultaneous introduction of individual mechanisms in a susceptible genetic background enhances our ability to elucidate the contribution of each individual molecular mechanism in the resistance phenotype; a concept that is perhaps best represented by the famous quote found at the blackboard of R. P. Feynman (1918–1988) at the time of his death (‘What I cannot create, I do not understand’). Although significant research effort remains to be done, the present study provides a ‘proof of principle’ of the applicability of such a reconstructed resistance ‘network of interactions' within a model Drosophila ‘test tube’ that enables the validation of hypotheses regarding the molecular mechanisms contributing to insecticide resistance phenotypes in field populations; in other words, to (re)create the interactions among different mechanisms, so that we can gain insight on their specific role.
Supplementary Material
Supplementary Material
Acknowledgements
We thank Ioannis Livadaras (IMBB/FORTH) for help in the generation of Drosophila lines, and Prof. Christos Delidakis (University of Crete and IMBB/FORTH) for providing balancer stocks and useful tips.
Data accessibility
All data generated or analysed during this study are available in this manuscript and its electronic supplementary material.
Authors' contributions
V.D. and J.V. conceived the project and designed the experiments; G.-R.S., R.P, V.D., S.K. and I.C. generated fly lines; G.-R.S. and R.P. performed toxicity bioassays and qPCR; G.-R.S. performed fitness cost experiments; V.D., S.D., R.N. and J.V. analysed the data, contributed to scientific discussion and wrote the manuscript, which has been revised and approved by all authors.
Competing interests
The authors declare no competing interests.
Funding
The research has been supported by the European Union Horizon 2020 Framework Programme (688207-SuperPests) grant to J.V., by a Fontation Santé research grant to V.D., by the Hellenic Foundation for Research and Innovation (HFRI) under the HFRI PhD fellowship grant (fellowship no. 10448) to R.P., and by Greece and the European Union (European Social Fund-ESF) through the Operational Programme ‘Human Resources Development, Education and Lifelong Learning’ in the context of the project ‘Strengthening Human Resources Research Potential via Doctorate Research’ (MIS-5000432), implemented by the Greek State Scholarships Foundation (ΙΚΥ) as a scholarship to G.-R.S.
References
- 1.WHO. 2018. WHO world malaria report 2018. Geneva, Switzerland: WHO.
- 2.Liu N. 2015. Insecticide resistance in mosquitoes: impact, mechanisms, and research directions. Annu. Rev. Entomol. 60, 537–559. ( 10.1146/annurev-ento-010814-020828) [DOI] [PubMed] [Google Scholar]
- 3.Strycharz JP, et al. 2013. Resistance in the highly DDT-resistant 91-R strain of Drosophila melanogaster involves decreased penetration, increased metabolism, and direct excretion. Pestic. Biochem. Physiol. 107, 207–217. ( 10.1016/j.pestbp.2013.06.010) [DOI] [Google Scholar]
- 4.Kasai S, Komagata O, Itokawa K, Shono T, Ng LC, Kobayashi M, Tomita T. 2014. Mechanisms of pyrethroid resistance in the dengue mosquito vector, Aedes aegypti: target site insensitivity, penetration, and metabolism. PLoS Negl. Trop. Dis. 8, e2948 ( 10.1371/journal.pntd.0002948) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ranson H, Edi CVA, Koudou BG, Jones CM, Weetman D. 2012. Multiple-insecticide resistance in Anopheles gambiae mosquitoes, southern Côte D'ivoire. Emerg. Infect. Dis. 18, 1508–1511. ( 10.3201/eid1809.120262) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hardstone MC, Scott JG. 2010. A review of the interactions between multiple insecticide resistance loci. Pestic. Biochem. Physiol. 97, 123–128. ( 10.1016/J.PESTBP.2009.07.010) [DOI] [Google Scholar]
- 7.Tiono AB, et al. 2018. Efficacy of Olyset Duo, a bednet containing pyriproxyfen and permethrin, versus a permethrin-only net against clinical malaria in an area with highly pyrethroid-resistant vectors in rural Burkina Faso: a cluster-randomised controlled trial. Lancet 392, 569–580. ( 10.1016/S0140-6736(18)31711-2) [DOI] [PubMed] [Google Scholar]
- 8.Donnelly MJ, Isaacs AT, Weetman D. 2016. Identification, validation, and application of molecular diagnostics for insecticide resistance in malaria vectors. Trends Parasitol. 32, 197–206. ( 10.1016/j.pt.2015.12.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vontas J, Mavridis K. 2019. Vector population monitoring tools for insecticide resistance management: myth or fact? Pestic. Biochem. Physiol. 161, 54–60. ( 10.1016/j.pestbp.2019.08.005) [DOI] [PubMed] [Google Scholar]
- 10.Daborn PJ, et al. 2012. Using Drosophila melanogaster to validate metabolism-based insecticide resistance from insect pests. Insect Biochem. Mol. Biol. 42, 918–924. ( 10.1016/j.ibmb.2012.09.003) [DOI] [PubMed] [Google Scholar]
- 11.Zimmer CT, Garrood WT, Puinean AM, Eckel-Zimmer M, Williamson MS, Davies TGE, Bass C. 2016. A CRISPR/Cas9 mediated point mutation in the α6 subunit of the nicotinic acetylcholine receptor confers resistance to spinosad in Drosophila melanogaster . Insect Biochem. Mol. Biol. 73, 62–69. ( 10.1016/j.ibmb.2016.04.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melnyk AH, Wong A, Kassen R. 2015. The fitness costs of antibiotic resistance mutations. Evol. Appl. 8, 273–283. ( 10.1111/eva.12196) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedman R. 2016. Drug resistance in cancer: molecular evolution and compensatory proliferation. Oncotarget 7, 11 746–11 755. ( 10.18632/oncotarget.7459) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.ffrench-Constant RH, Bass C. 2017. Does resistance really carry a fitness cost? Curr. Opin. Insect Sci. 21, 39–46. ( 10.1016/j.cois.2017.04.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zimmer CT, Bass C, Williamson MS, Kaussmann M, Wölfel K, Gutbrod O, Nauen R. 2014. Molecular and functional characterization of CYP6BQ23, a cytochrome P450 conferring resistance to pyrethroids in European populations of pollen beetle, Meligethes aeneus. Insect Biochem. Mol. Biol. 45, 18–29. ( 10.1016/j.ibmb.2013.11.008) [DOI] [PubMed] [Google Scholar]
- 16.Nauen R, et al. 2012. Target-site resistance to pyrethroids in European populations of pollen beetle, Meligethes aeneus F. Pestic. Biochem. Physiol. 103, 173–180. ( 10.1016/j.pestbp.2012.04.012) [DOI] [Google Scholar]
- 17.Pavlidi N, Monastirioti M, Daborn P, Livadaras I, Van Leeuwen T, Vontas J.. 2012. Transgenic expression of the Aedes aegypti CYP9J28 confers pyrethroid resistance in Drosophila melanogaster. Pestic. Biochem. Physiol. 104, 132–135. ( 10.1016/j.pestbp.2012.07.003) [DOI] [Google Scholar]
- 18.Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N. 2008. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat. Genet. 40, 476–483. ( 10.1038/ng.101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Port F, Chen H-M, Lee T, Bullock SL. 2014. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl Acad. Sci. USA 111, E2967–E2976. ( 10.1073/pnas.1405500111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chung H, Bogwitz MR, McCart C, Andrianopoulos A, Ffrench-Constant RH, Batterham P, Daborn PJ. 2007. Cis-regulatory elements in the Accord retrotransposon result in tissue-specific expression of the Drosophila melanogaster insecticide resistance gene Cyp6g1. Genetics 175, 1071–1077. ( 10.1534/genetics.106.066597) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM, Oconnor-Giles KM. 2014. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196, 961–971. ( 10.1534/genetics.113.160713) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, Wildonger J, O'Connor-Giles KM. 2013. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194, 1029–1035. ( 10.1534/genetics.113.152710) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Douris V, Steinbach D, Panteleri R, Livadaras I, Pickett JA, Van Leeuwen T, Nauen R, Vontas J.. 2016. Resistance mutation conserved between insects and mites unravels the benzoylurea insecticide mode of action on chitin biosynthesis. Proc. Natl Acad. Sci. USA 113, 14 692–14 697. ( 10.1073/pnas.1618258113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Groth AC, Fish M, Nusse R, Calos MP. 2004. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 166, 1775–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsakireli D, Riga M, Kounadi S, Douris V, Vontas J. 2019. Functional characterization of CYP6A51, a cytochrome P450 associated with pyrethroid resistance in the Mediterranean fruit fly Ceratitis capitata. Pestic. Biochem. Physiol. 157, 196–203. ( 10.1016/j.pestbp.2019.03.022) [DOI] [PubMed] [Google Scholar]
- 26.Kefi M, Balabanidou V, Douris V, Lycett G, Feyereisen R, Vontas J. 2019. Two functionally distinct CYP4G genes of Anopheles gambiae contribute to cuticular hydrocarbon biosynthesis. Insect Biochem. Mol. Biol. 110, 52–59. ( 10.1016/j.ibmb.2019.04.018) [DOI] [PubMed] [Google Scholar]
- 27.Pfaffl MW, Horgan GW, Dempfle L. 2002. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30, e36 ( 10.1093/nar/30.9.e36) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Samantsidis G-R, O'Reilly AO, Douris V, Vontas J. 2019. Functional validation of target-site resistance mutations against sodium channel blocker insecticides (SCBIs) via molecular modeling and genome engineering in Drosophila. Insect Biochem. Mol. Biol. 104, 73–81. ( 10.1016/J.IBMB.2018.12.008) [DOI] [PubMed] [Google Scholar]
- 29.Robertson JL, Russell RM, Preisler HK, Savin E. 2007. Bioassays with arthropods, 2nd edn Boca Raton, FL: CRC Press. [Google Scholar]
- 30.Chandler CH, Chari S, Dworkin I. 2013. Does your gene need a background check? How genetic background impacts the analysis of mutations, genes, and evolution. Trends Genet. 29, 358–366. ( 10.1016/j.tig.2013.01.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brito LP, Linss JGB, Lima-Camara TN, Belinato TA, Peixoto AA, Lima JBP, Valle D, Martins AJ. 2013. Assessing the effects of Aedes aegypti kdr mutations on pyrethroid resistance and its fitness cost. PLoS ONE 8, e60878 ( 10.1371/journal.pone.0060878) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scott JG, Buchon N. 2019. Drosophila melanogaster as a powerful tool for studying insect toxicology. Pestic. Biochem. Physiol. 161, 95–103 ( 10.1016/j.pestbp.2019.09.006) [DOI] [PubMed] [Google Scholar]
- 33.Douris V, Denecke S, Van Leeuwen T, Nauen R, Bass C, Vontas J.. In press Using CRISPR/Cas9 genome modification to understand the genetic basis of insecticide resistance: Drosophila and beyond. Pestic. Biochem. Physiol. ( 10.1016/j.pestbp.2020.104595) [DOI] [PubMed] [Google Scholar]
- 34.van Waterschoot RAB, Schinkel AH.. 2011. A critical analysis of the interplay between cytochrome P450 3A and P-glycoprotein: recent insights from knockout and transgenic mice. Pharmacol. Rev. 63, 390–410. ( 10.1124/pr.110.002584) [DOI] [PubMed] [Google Scholar]
- 35.Chandor-Proust A, et al. 2013. The central role of mosquito cytochrome P450 CYP6Zs in insecticide detoxification revealed by functional expression and structural modelling. Biochem. J. 455, 75–85. ( 10.1042/BJ20130577) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nauen R, Tietjen K, Wagner K, Elbert A. 1998. Efficacy of plant metabolites of imidacloprid against Myzus persicae and Aphis gossypii (Homoptera: Aphididae). Pestic. Sci. 52, 53–57. () [DOI] [Google Scholar]
- 37.Denecke S, Fusetto R, Martelli F, Giang A, Battlay P, Fournier-Level A, O’ Hair RA, Batterham P. 2017. Multiple P450s and variation in neuronal genes underpins the response to the insecticide imidacloprid in a population of Drosophila melanogaster. Sci. Rep. 7, 11338 ( 10.1038/s41598-017-11092-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are available in this manuscript and its electronic supplementary material.