Abstract
We showed previously that inflammatory mediators, including IL8, in intervertebral disc tissues from patients with discogenic back pain may play a key role in back pain. To investigate the molecular mechanism of IL8 signaling in back pain, we generated a mouse model that conditionally expresses human (h) IL8. We hypothesized that hIL8 levels affect mouse activity and function. Briefly, hIL8 cDNA was inserted into the pCALL2 plasmid, linearized, and injected into mouse embryos. Resulting pCALL2–hIL8 mice were then bred with GDF5–Cre mice to express the transgene in cartilage and intervertebral disc (IVD) tissues. Functional capacities including nest-making and other natural behaviors were measured. Both male and female mice expressing hIL8 showed lower nesting scores than did littermates that did not express hIL8 (n = 14 to 16 per group). At 28 wk of age, mice expressing hIL8 (n = 35) spent more time immobile and eating during each night than littermate controls (n = 33). Furthermore, hIL8-expressing mice traveled shorter distances and at a lower average speed than littermate controls. Thus, in an initial effort to investigate the relationship between this chemokine and mouse behavior, we have documented changes in normal activities in mice conditionally expressing hIL8.
Abbreviations: GDF, growth differentiation factor; h, human; IVD, intervertebral disc tissues
Estimated to be discogenic in origin in 40% of patients,7,22 chronic lower back pain is a major public health and economic problem.17 Patients with back pain often have joint pain, commonly affecting knees, hips, and facet joints of the spine. Some patients with chronic back and joint pain develop pain behavior, including verbal (e.g., descriptions of the intensity, location, and quality of pain; vocalization of distress; moaning; complaining) and nonverbal (e.g., withdrawal from activities, use of pain medication, pain-related body postures and facial expressions) displays.9 These pain behaviors can disrupt work and normal social activities. To identify biologic markers that differentiate painful degenerative discs and asymptomatic discs, our group has collected intervertebral disc (IVD) tissues from patients undergoing surgery for discogenic back pain and has compared them with degeneration-matched IVD tissues from patients undergoing corrective surgery for scoliosis curvature.12,31 We have described elevated levels of inflammatory mediators in IVD tissues from patients with discogenic back pain that may play a key role in back pain etiology.12,31 In addition, other colleagues24,28,29 have reported elevated systemic inflammation, including elevated levels of cytokines and chemokines, in serum from patients with back pain. We have further shown that cultured IVD cells are fully capable of secreting inflammatory mediators,31 thus indicating that IVD cells account, at least in part, for the cytokine levels in serum. Infiltrating leukocytes in the injured32 or degenerative IVDs20 likely contribute to cytokine production as well. These previous studies firmly establish the clinical relevance of inflammation in discogenic back pain.
Among the chemokines identified in IVD tissues from patients with discogenic back pain, IL8 is the most highly inducible chemokine in cultured IVD cells.31 In healthy IVD tissue, IL8 is barely detectable, but it is rapidly induced in response to proinflammatory cytokines such as TNFα and IL1β, bacterial and viral products, and cellular stress.11 Our observation that IL1β readily induces IL8 in human IVD, with many-fold upregulation of mRNA expression and soluble protein levels,31 prompts further study of this chemokine in a transgenic mouse model.
Also known as CXCL8, IL8 is a member of a family of proinflammatory cytokines that are related by having a C-X-C motif, where X is any amino acid between 2 cysteines.15 Many cells produce IL8 in vitro, and it has been implicated in neutrophil and T-cell migration. IL8 binds to IL8 receptors (IL8RA/CXCRl and IL8RB/CXCR2).8,16 Although rodents lack an IL8 counterpart, human (h) IL8 can bind to rodent receptors for similar chemokines (CXCR1/2).8 One example is that expression of hIL8 in the cornea induces corneal ulcer formation in mice.21
Elevated levels of IL8 protein have been found in plasma and cerebral spinal fluids of patients with fibromyalgia,2 a chronic pain syndrome affecting multiple parts of the musculoskeletal system. Patients with chronic back and joint pain often exhibit symptoms that overlap with those of patients with fibromyalgia, including weight gain, chronic fatigue, and sleep disturbances.3 We hypothesize that elevated levels of IL8 in the IVD tissues have local and systemic effects in patients and in hIL8-expressing mice compared with control mice. To examine the effects of elevated IL8 in the mouse model, we used the Cre–lox system to establish a mouse line that allows tissue-specific expression of hIL8. We selected a mouse line that expresses Cre recombinase under the control of the promotor for growth differentiation factor (GDF) 5, which is active in both IVD and articular joint tissues.4,5,13,25 This line was then used to express hIL8 in both IVD and articular cartilage. Given that patients with disc degeneration and associated discogenic pain usually have ostheoarthitis in other articular joints,9 this system realistically models human disease. We then evaluated nest making, eating, drinking, and locomotion in these mice. The behavioral characterization of hIL8-expressing mice that we describe here is a preliminary effort to investigating the relationship between chemokine levels and behavior.
Materials and Methods
Mice.
All animal experimental procedures were approved by the IACUC of the Corporal Michael J Crescenz Veterans Affairs Medical Center in Philadelphia. Mice were housed (maximum, 5 per cage) in disposable cages (Innovive, San Diego, CA) with Alpha Dri bedding (Shepherd Specialty Papers, Watertown, TN) under SPF conditions with environmental enrichment (Nestlets, Ancare, Bellmore, NY; and Mouse Igloo Rodent Enrichment Device, Fisher Scientific, Hampton, NH). Mice are tested for pathogens quarterly by using the EAD Mouse Surveillance Plus PRIA series (Animal Diagnostic Services, Charles River Research, Wilmington, MA), which includes 11 common mouse viruses, 26 bacteria strains, and 7 parasitic strains. Our facility continues to be negative for all pathogens tested under this panel. Mice were fed PicoLab diet 5053 (PMI Nutrition International, St. Louis, MO) without restriction, provided acidified bottled water, and maintained on a 12:12-h light:dark cycle. Room temperature is kept at 70 to 76 °F (21.1 to 24.4 °C) and humidity between 30% and 70%.
Generation of transgenic mice.
pCALL2 plasmid (Miami Mice Research) was used to construct the hIL8 transgene (Figure 1). Specifically, cDNA of hIL8 was synthesized from cultured human AF cells stimulated with IL1β,31 by using random hexamers and polydT primers. To amplify the cDNA, primers were designed to include the Kozak consensus sequence to allow efficient translation.14 cDNA encoding the longest known IL8 protein (99 amino acids) and a small portion of 3′ untranslated region was included. A primer pair was designed based on published sequences (Ensembl transcript ID ENST00000307407.7)18,19,30 and included a BglII restriction endonuclease digestion site at the 5′ end and an XhoI site at the 3′ end of the amplicon. The primers (forward primer containing a BglII site, 5′ ATC GAT AGA TCT TGC ATA AGT TCT CTA GTA GGG 3′; reverse primer containing an XhoI site and FLAG tag, 5′ ACC TTA CTC GAG TTA CTT ATC GTC GTC ATC CTT GTA ATC TGA ATT CTC AGC CCT CTT C 3′) were synthesized by Invitrogen (Carlsbad, CA).
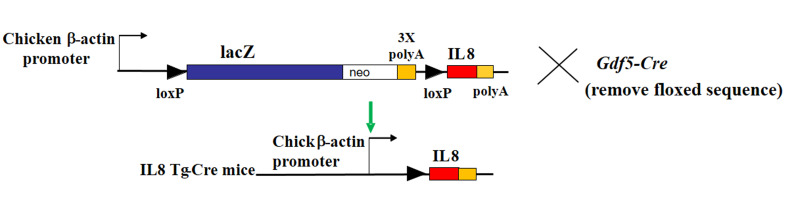
Figure 1.
Design for conditional expression of human IL8 in mice. Human IL8 cDNA was inserted into the pCALL2 plasmid polylinker region to generate IL8-transgenic mice that encode Cre-inducible IL8. IL8-transgenic mice were then bred with GDF5-Cre mice with inducible Cre activity in synovial joints and intervertebral discs.
Microinjection of the transgenic construct (linearized pCALL2–hIL8 cassette) into C57BL mice was performed by the University of Texas Health Science Center at Houston. Of the 17 mice generated, 5 were positive for the hIL8 insert (i.e., transgenic for hIL8) as verified by PCR analysis of DNA extracted from tail clips. Genotyping was conducted by using a nested primer pair (forward, 5′ TGG AAA GGT TTG GAG TAT GTC TTT A; reverse, 5′ CAG CCT TCC TGA TTT CTG CA 3′) and an annealing temperature of 56 °C.
The 5 founders (2 females and 3 males) were transported to the Corporal Michael J Crescenz Veterans Affairs Medical Center in Philadelphia and were bred with C57BL/6J mice (the Jackson Laboratories, Bar Harbor, ME). Four lines generated live offspring, and were tested for level of hIL8 expression. To verify and quantify hIL8 protein, mouse tail fibrocytes were cultured from these 4 founders and infected with adenovirus expressing Cre-recombinase (a generous gift from Dr Tong-Chuan He, University of Chicago, Chicago, IL) to induce hIL8 expression. HIL8 secreted into the supernatant was quantified through ELISA (R and D Systems, Minneapolis, MN). The pCALL2–hIL8 transgenic male mouse that expressed the highest level of hIL8 was bred with a female C57BL/6J mice (Jackson Laboratories, Bar Harbor, ME), followed by sibling-to-sibling inbreeding for at least 7 generations before being used in experiments. The Cre-inducible pCALL2-hIL8 transgenic mouse line will be available from The Jackson Laboratory as JAX no. 035378. We have acquired and are maintaining a mouse line expressing Cre-recombinase under the control of the GDF5 promoter (generously provided by Dr David Kinsley, Stanford University, CA).4,25 The hIL8 transgenic mice were bred with GDF5-Cre mice to conditionally express the transgene in GDF5-expressing tissues. hIL8 gene expression in various tissues was verified through real-time PCR analysis. For brevity, hIL8+;GDF5Cre mice are shown as hIL8+, and hIL8-;GDF5Cre mice are shown as controls.
RNA isolation and quantitative real-time PCR analysis.
IVD tissues were separated from their adjacent cartilaginous endplates and bone by using a scalpel under a dissecting microscope (VistaVision, VWR International, Radnor, PA), as previously described.26 Similarly, knee joint articular cartilage tissues were shaved off femoral condyls or tibial plateaus under the dissecting microscope. The isolated IVD or joint articular cartilage tissues were soaked in RNALater (Ambion, Foster City, CA) overnight and stored at –80 °C until extraction. On the day of RNA extraction, RNALater was removed, and the tissues were snap-frozen in liquid nitrogen and then transferred into Trizol (Invitrogen). Tissues were homogenized by using disposable OmniTip probes for hard tissue (Omni International, Kennesaw, GA). The aqueous component was extracted 3 time with phenol–chloroform. RNA was precipitated with 70% ethanol and further purified by using an RNeasy Micro Kit (Qiagen, Germantown, MD), according to the manufacturer's protocol. RNA concentration was determined by using a Synergy H4 Hybrid Reader (BioTek, Winooski, VT). To generate cDNA, all RNA from each IVD (7 to 20 ng/µL; total volume, 50 µL per sample) was used as template in a reverse-transcription reaction (SuperScript VILO cDNA synthesis kit, Life Technologies, Carlsbad, CA) containing random hexamers and added poly-dT primers (Invitrogen). cDNA sequences were retrieved from Ensembl.30 Primers for real-time PCR analysis were identical to the primer pair for genotyping (described earlier). For each PCR reaction, cDNA, SYBR Select master mix (Life Technologies), and primers (working concentration, 0.5 μM) were mixed, and deionized water was added to achieve a total volume of 20 μL per reaction. MicroAmp Optical 96-well reaction plates (Applied Biosystems, Foster City, CA) containing 20 µL of reaction mixture per well were sealed by using optical adhesive film (Life Technologies) and run in a ViiA7 real-time PCR system (Applied Biosystems) according to the following program: (1) 50 °C for 2 min, (2) 95 °C for 2 min, (3) 95 °C for 15 s, (4) 58 °C for 1 min, and (5) repeat steps 3 and 4 for a total of 40 cycles. Single products were confirmed by determining melting curves at the conclusion of the reaction. Relative expression was calculated by using the 2-ΔΔCt method and normalizing to gapdh (endogenous loading control).
Serum IL8 ELISA.
Immediately after euthanasia, mouse blood was collected via cardiocentesis. Blood was incubated at room temperature for 20 min, and then centrifuged at 5000 revolutions per minute for 20 min. The supernatant serum was isolated and stored at –80 °C until assayed. hIL8 was quantified by using the Human IL8/CXCL8 Quantikine ELISA Kit (R and D Systems) according to the manufacturer's instructions.
Mouse rectal temperature.
Body temperature was measured by using a rectal temperature probe (19 mm, model RET3, Thermoworks, Lindon, UT) connected to a thermometer (model TW2-193, MicroTherma, Thermoworks).
Mouse nesting behavior.
Mice were assigned to hIL8+ and littermate control groups according to their PCR-verified genotype. Littermates of the same sex were housed in the same cage, to a maximum of 5 mice per cage, regardless of genotype. Mice in the same cage usually were tested at the same time, regardless of their genotype. The mice were identified by using numbered ear tags; the operator of the behavioral testing was blind to mouse genotype. At 28 wk of age, each mouse was placed in a single cage and given a cotton square (Animal Specialties and Provisions, Quakertown, PA) on the night before evaluation. Mice and the nests they built were photographed, and nest quality was scored on a scale of 1 to 5.6 Next, the nesting material that was incorporated into nests and the unused material were weighed. Mice that made a nest of perfect shape, with all the material used, received a score of 5, and mice that did not make a nest at all received a score of 1. The procedure was repeated once for each mouse; the scores of 2 nests were averaged. The nests were scored by 3 independent reviewers.
Assessment of spontaneous behavior.
Each mouse was examined once at each time point (8, 12, 16, 20, 24, and 28 wk of age). Specifically, each mouse occupied a single cage, and as many as 6 animals were examined at each time point by using Laboras (Metris, Best, Netherlands).27 Laboras is a fully automatic and noninvasive system for recording spontaneous behaviors, including climbing, grooming, eating, drinking, rearing, and resting. Locomotive behaviors (distance traveled, climbing [hanging from a wire cage], rearing [standing on hind legs], and immobility) also were assessed overnight. Specifically, each mouse was tested for 16 h, between 1700 and 0900. Time spent in each activity and the distance traveled during the 16-h observation period were summed, and speeds of ambulation were averaged. The natural behavior at 28 wk of age was analyzed further by using MATLAB software (MathWorks, Natick, MA).
Statistics.
Mean mouse nesting scores were compared by using the Student t test. Interrater variability was calculated as the number of ratings in agreement divided by the total number of ratings. The ratings in agreement were defined as any whole-number rating and its half-number counterpart. For example, the scores 1 and 1.5 were considered to be in agreement. Similarly, intrarater variability was calculated by using the ratings from the same evaluator and nest image at 2 time points 10 mo apart.
The large data set on natural behaviors that was collected by using the Laboras platform was organized with MATLAB software. To assess between hIL8+ and control mice differences in time spent in each activity, distance traveled overnight, and average speed, 2-factor ANOVA for repeated measures was used, where type of mouse was a grouping factor and male or female was the repeated measure. Posthoc t tests using the ANOVA pooled variance were performed for sex-associated differences within each time period. A P value of less than 0.05 was considered statistically significant. All analyses were performed by using SAS statistical software (version 9.4, SAS Institute, Cary, NC).
CBC, Giemsa staining of blood films, and histologic analysis.
For CBC, approximately 200 μL of blood was collected by cardiocentesis immediately after euthanasia and stored in collection tubes containing K3 EDTA (Fisher Scientific, Waltham, MA). The samples in the tubes were mixed gently, placed in a 4 °C cooler, and analyzed (Genesis Veterinary Hematology Analyzer, Oxford Science, Oxford, CT) within 12 h after collection.
To examine blood cell morphology, approximately 10 µL of blood was used to make a film, which was fixed with methanol for 10 min. The blood films were then stained (Wright Giemsa Staining Pack, Sigma-Aldrich, St Louis, MO) according to the manufacturer's instructions.
For histologic analysis, IVD and portions of the adjacent bony vertebral bodies and knee joints were isolated immediately after euthanasia. Each IVD with its surrounding vertebral bodies was fixed with 4% paraformaldehyde for 24 h. The bone–disc–bone segments were decalcified in a solution consisting of 12.5% EDTA for approximately 1 wk, with shaking, until the bony portion was completely decalcified.26 The tissues were then dehydrated, embedded in paraffin, and sectioned to a thickness of 5 µm; sections were stained with hematoxylin and eosin.
In addition, mouse spleen and liver were harvested, weighed, fixed in 4% paraformaldehyde for 24 h, and stained with hematoxylin and eosin. The sections were imaged under a light microscope (Nikon, Tokyo, Japan).
Results
Blood cell morphology and CBC.
Neutrophil and lymphocyte morphology did not differ significantly between hIL8+ and control mice (Figure 2). In addition, total WBC, neutrophil, and lymphocyte counts were not significantly different between hIL8+ mice and their control littermates (n = 6 mice per group, P > 0.05). The lymphocyte percentage was higher in hIL8+ mice than their control littermates (n = 6 mice per group, P = 0.0250, Table 1).
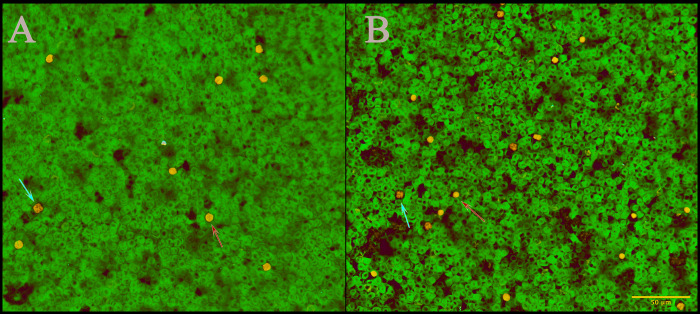
Figure 2.
No significant differences in lymphocyte or neutrophil morphology between (A) mice expressing human IL8 (hIL8+) and (B) control animals. Blood smears were stained with Giemsa; scale bar, 50 µm (B). Red arrows, neutrophils; blue arrows, lymphocytes.
Table 1.
CBC counts in mice (mean ± SEM; n = 6 male mice per group)
| WBC (×109/L) | Neutrophils (× 109/L) | Lymphocytes (× 109/L) | Neutrophils, % | Lymphocytes, % | |
| hIL8+ | 4.13 ± 0.85 | 0.95 ± 0.31 | 2.86 ± 0.65 | 25.3 ± 6.25 | 67.64 ± 5.38 |
| Control | 3.48 ± 0.46 | 1.06 ± 0.11 | 2.05 ± 0.41 | 32.98 ± 5.14 | 56.9 ± 4.36 |
| P | 0.5440 | 0.7121 | 0.3067 | 0.0515 | 0.0250 |
Serum hIL8 levels; rectal temperature; and body, spleen, and liver weights.
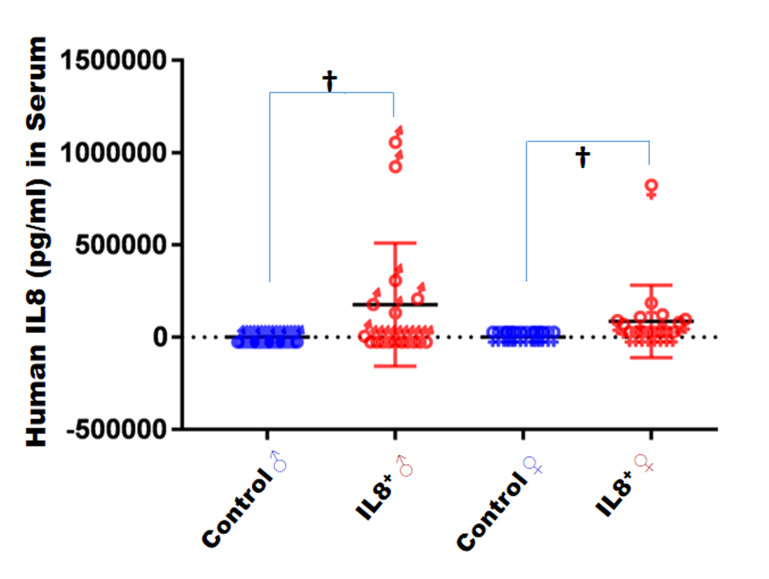
Immediately after euthanasia, mouse serum was collected and hIL8 levels determined by ELISA. The hIL8 protein level (mean ± 1 SD) in male hIL8+ mice (n = 17) was 176,313± 333,670 pg/mL and in female hIL8+ mice (n = 16) was 85,571 ± 195,753 pg/mL. Thus, the level of hIL8 varied greatly. Four male and 2 female hIL8+ mice did not have a detectable level of hIL8 protein in serum. Serum levels of hIL8 protein were not significantly different between male and female mice (P = 0.3461). None of the control mice had detectable hIL8 (males, n = 17; females, n = 16; Figure 3). Rectal temperature were not different between the hIL8+ mice and control littermates (n = 6 mice/group; P = 0.9128; Table 2).
Figure 3.
Serum levels (pg/mL; mean ± 1 SD) of human IL8 in mice, according to ELISA. †, P < 0.01.
Table 2.
Mouse body weight, spleen weight, liver weight, and body temperature.
| Body (g) | Spleen (g) | Liver (g) | Temperature (°C) | |
| hIL8+ | 28.10 ± 0.80 | 0.092 ± 0.006 | 1.365 ± 0.044 | 37.68 ± 0.10 |
| Control | 23.61 ± 0.80 | 0.082 ± 0.003 | 1.272 ± 0.077 | 37.67 ± 0.11 |
| P | 0.2807 | 0.1466 | 0.1490 | 0.9128 |
Data are given as mean ± SEM (n = 6 male mice per group).
Nesting.
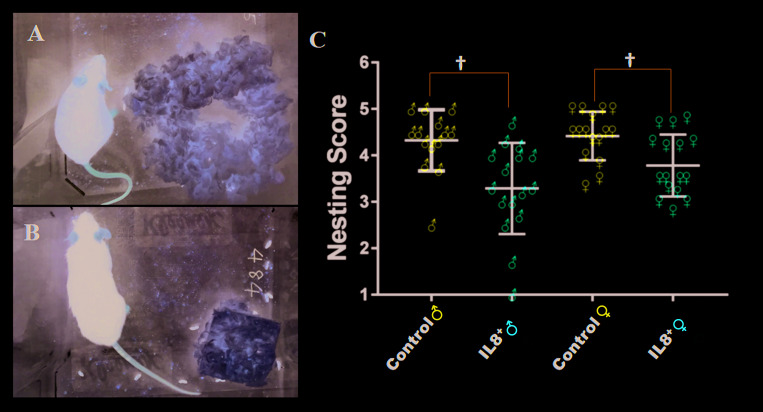
Three independent observers assessed nesting behavior in 28-wk-old mice. Each nest was assigned a score6 depending on the amount of cotton square used to build the nest and the nest shape (Figure 4 A and B). At 28 wk of age, hIL8+ mice built poorer-quality nests than their littermate controls (average score, 3.5 and 4.4, respectively; n = 32 and 29, respectively; P < 0.001). In particular, male hIL8+ mice had poorer nest scores than male littermate controls (average score, 3.3 and 4.3, respectively; n = 16 and 14, respectively; P = 0.0019). Female hIL8+ mice likewise showed poorer nest scores than their female littermate controls (average score, 3.8 and 4.4, respectively; n = 16 and 15, respectively; P = 0.0054; Figure 4 C), albeit to a lesser degree than male mice. The difference in nest-making scores between male and female mice was not statistically significant regardless of genotype. The 3 raters showed agreement of 65.6%. The crude intrarater agreement was 0.627, with a weighted κ of 0.644. The greatest disagreement occurred between nesting scores of 4 and 5. Nest score and serum hIL8 level were not correlated in hIL8+ mice (n = 26, R2 = 0.0003).
Figure 4.
Nesting in human IL8-expressing (hIL8+) and control littermates. (A) Nesting score 5. (B) Nesting score 1. (C) All scores (mean ± 1 SD) at 28 wk of age. †, P ≤ 0.01.
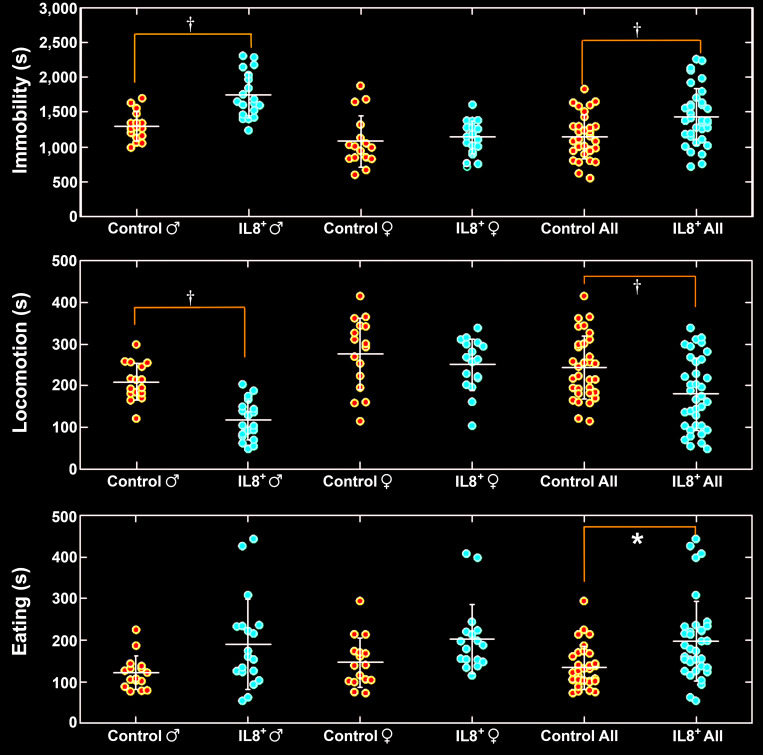
Time spent immobile, in locomotion, and eating.
We recorded natural behavior of hIL8+ mice and their control littermates overnight, for 16 h At 28 wk of age, hIL8+ mice spent more time immobile than controls (n = 35 and 33, respectively; P < 0.001). Interestingly, male hIL8+ mice spent more time immobile than male controls (n = 18 and 17, respectively; P < 0.001), whereas female hIL8+ mice spent similar amounts of time immobile as female controls (n = 17 and 16, respectively; P = 0.7220). Furthermore, male hIL8+ mice spent more time immobile than female hIL8+ mice (n = 18 and 17, respectively; P < 0.0001), and male control mice spent more time immobile than female control mice (n = 17 and 16, respectively; P < 0.0001; Figure 5, upper panel).
Figure 5.
Total time (s) spent immobile, in locomotion, and eating overnight. *, P < 0.05; †, P < 0.01.
At 28 wk of age, hIL8+ mice spent less time in locomotion than their littermate controls (n = 35 and 33, respectively; P = 0.001). Male hIL8+ mice spent less time in locomotion than male controls (n = 18 and 17, respectively; P = 0.001), whereas time spent in locomotion did not differ significantly between female hIL8+ and control mice (n = 17 and 16, respectively; P = 0.6133). Furthermore, male hIL8+ mice spent less time mice in locomotion than female hIL8+ mice (n = 18 and 17; P < 0.001). Similarly, male control mice spent less time in locomotion than female control mice (n = 17 and 16; P = 0.0001; Figure 5, middle panel).
HIL8+ mice spent more time eating than controls (n = 35 and 33, respectively; P = 0.0214). However, when comparing time spent eating between male hIL8+ mice and male controls (n = 18 and 17, respectively) or female hIL8+ mice and female controls (n = 17 and 16, respectively), the differences were not statistically significant (Figure 5, lower panel).
In contrast to eating, hIL8+ mice spent less time drinking than controls (n = 35 and 33, respectively; P = 0.0215). However, when comparing time spent drinking between male hIL8+ mice and male controls (n = 18 and 17) or female hIL8+ mice and female controls (n = 17 and 16), there was no statistically significant difference. The amount of food and water consumed was not measured.
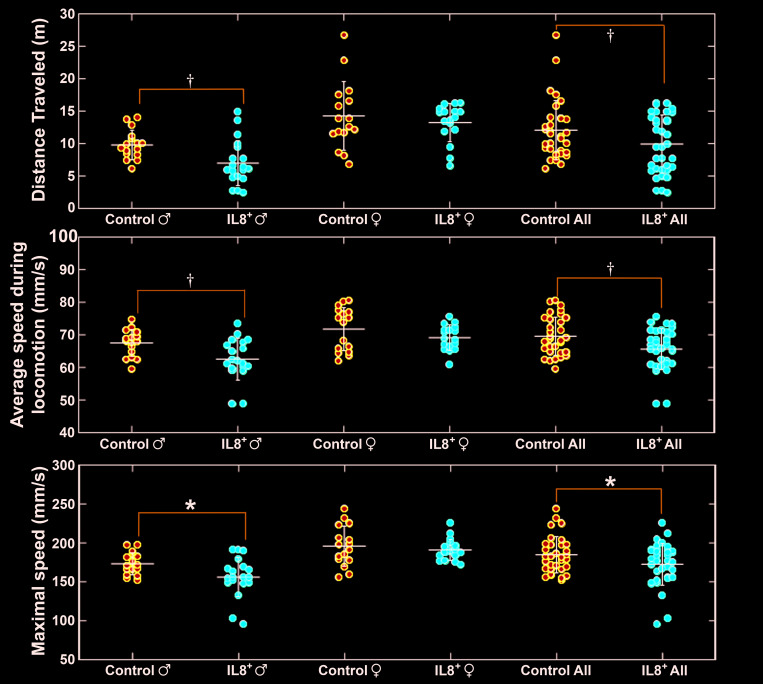
Distance and speed of travel.
At 28 wk of age, hIL8+ mice traveled a shorter distance during the 16-h observation period than their control littermates (n = 35 and 33, respectively; P = 0.0093). In particular, male hIL8+ mice traveled less distance than controls of the same sex (n = 18 and 17, respectively; P < 0.001), whereas female hIL8+ mice traveled a similar distance as female controls (n = 17 and 16, respectively; P = 0.8394). Furthermore, male hIL8+ mice traveled a shorter distance than female hIL8+ mice (n = 18 and 17, respectively; P < 0.0001), and male control mice traveled a shorter distance than female control mice (n = 17 and 16, respectively; P = 0.0079; Figure 6, upper panel).
Figure 6.
Distance traveled (m), average speed (mm/s), and maximal speed (mm/s) overnight. *, P < 0.05; †, P < 0.01.
Over the 16-h observation period, hIL8+ mice traveled at lower average and maximal speeds than their littermate controls (n = 35 and 33, respectively; P = 0.0013 and 0.0136, respectively). In particular, male IL8+ mice traveled at lower average and maximal speeds than male controls (n = 18 and 17, respectively; P = 0.0089 and 0.0208 respectively), whereas female hIL8+ mice traveled at similar average and maximal speeds as female controls (n = 17 and 16, respectively; P = 0.4419 and 0.1916, respectively; Figure 6, middle and lower panels). In summary, hIL8+ mice traveled shorter distances at slower speeds than their hIL8– controls, and this phenomenon is particularly evident in male mice.
Time spent climbing showed no difference between hIL8+ mice and their littermate controls (n = 35 or 33, respectively; P = 0.3686) or between male hIL8+ and male control mice (n = 18 and 17, respectively; P = 1.0000). Female IL8+ and female control mice spent similar amounts of time in climbing (n = 17 and 16, respectively; P = 0.5812). Female hIL8+ mice spent more time climbing than males of the same genotype (n = 17 and 18, respectively; P < 0.0001). Similarly, female control mice spent more time climbing than male controls (n = 16 and 17, respectively; P < 0.0001). Time spent rearing or grooming did not differ between hIL8+ mice of both sexes and control mice. In summary, compared with control mice, hIL8+ mice spent more time immobile and eating and less time in locomotion and drinking. These behavioral differences were more apparent in male than in female mice.
Histologic features of IVD, knee joint, spleen, and liver.
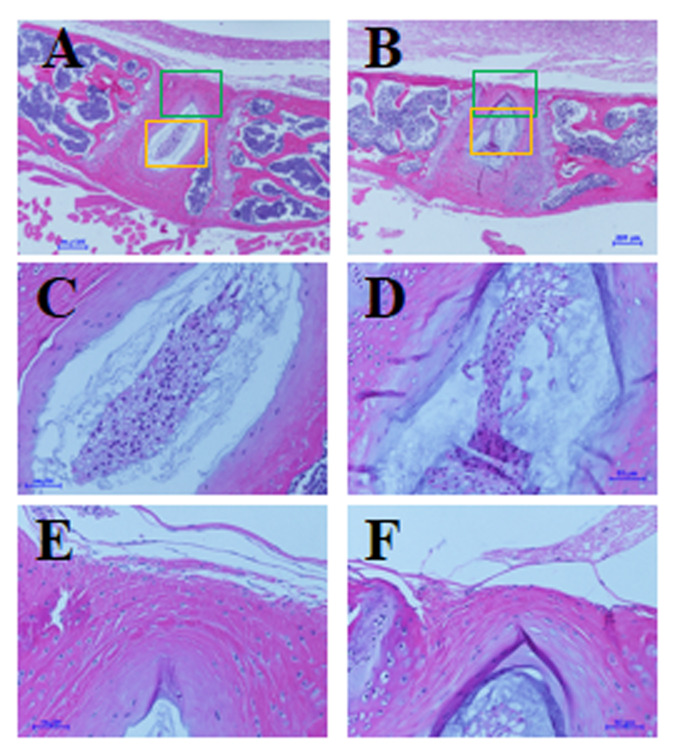
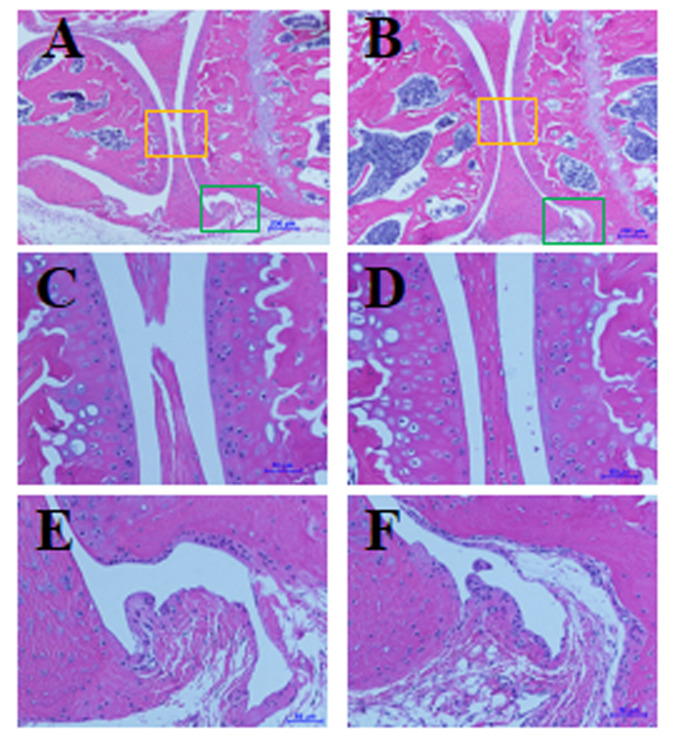
At the end of the study, mice (age, 28 wk) were euthanized, and histologic features of the IVDs and knee joints were evaluated as described previously.26 We found no significant differences in histologic features of lumbar spine IVD and knee joints between hIL8+ mice and controls, nor did we find an increase in leukocyte infiltration into the IVD or joint tissues (n = 7 per group, Figures 7 and 8). In addition, routine histologic examination of the spleen did not reveal any significant differences between hIL8+ and control littermates (n = 6 mice/group). Inflammatory cell infiltrations were present in the livers of hIL8+ mice but not control mice (n = 6 mice per group).
Figure 7.
No differences in histologic features of lumbar intervertebral discs (IVD) between (A, C, and E) human IL8-expressing (hIL8+) and (B, D, and F) control mice. Sagittal sections of IVD were stained with hematoxylin and eosin; C and D show nucleus pulposus, magnified from yellow squares in panels A and B, respectively; E and F show annulus fibrosus, magnified from green squares in images A and B, respectively. Scale bars, 200 μm (A and B); 50 μm (C through F).
Figure 8.
No differences in histologic features of knee joints between (A, C, and E) human IL8-expressing (hIL8+) and (B, D, and F) control mice. Sagittal sections of knee joints were stained with hematoxylin and eosin; C and D show knee articular cartilage and meniscus tissues, magnified from yellow squares in panels A and B, respectively; E and F show synovial tissues, magnified from green squares in panels A and B, respectively. Scale bars, 200 μm (A and B), 50 μm (C through F).
Discussion
The findings presented here are the first description of the effects of hIL8 on mouse activity and function. We previously found IL8 in IVD tissues from a patient with back pain and further showed that cultured cells isolated from human IVD produced massive amounts of IL8 in response to inflammation.31 To examine the effects of high hIL8 levels in a rodent model, we established an hIL8-transgenic mouse line. By breeding the hIL8Trasgenic line with GDF5Cre-recombinase-expressing mice, hIL8 gene expression was activated in the tissues that expressed GDF5, including IVD and synovial joints.5,13 Our data show that nest-making is impaired in both male and female IL8+ mice compared with littermate controls; the deficit is greater in male than in female mice. Furthermore, IL8+ mice tend to eat more and walk less than littermate controls.
The total lymphocyte percentage was higher in hIL8+ mice than their control littermates, but total WBC, neutrophil, and lymphocyte counts did not differ significantly between the 2 types of mice (F). We did not find leukocyte infiltration in IVD or synovial joint of hIL8+ mice (Figures 7 and 8). Our findings are consistent with the impaired migration of neutrophils into the inflamed peritoneal cavity of hIL8+ mouse.23 In addition, routine histologic examination of spleen did not reveal any significant differences between hIL8+ and control littermates, and only mild inflammatory cell infiltrations in the liver of hIL8+ mice were present.
Elevated systemic inflammation in patients with back pain, including elevated levels of cytokines and chemokines in serum, has been described.24,28,29 In the current study, we likewise found elevated levels of hIL8 in the serum of hIL8+ mice. Our findings of mouse behavior in the hIL8 expression model bear similarity to those of human patients with chronic inflammation or pain, in that such patients tend to be less active and eat more. Further work on the mouse brain will help to differentiate whether these behavioral deficits are due to depression-like mechanisms or to back or joint pain. However, we did not find any correlation between levels of serum IL8 and severity of behavior changes. The measurement was performed only once, at the end of the study, because we had to use cardiocentesis to collect sufficient serum for testing. We are currently developing a microchip to measure hIL8 levels in vivo in order to record potential fluctuations in hIL8 levels. This method will enhance our understanding of the relationship between hIL8 level and mouse behavior in future experiments.
Despite spending more time eating, IL8+ mice did not weigh more than their littermate controls (Table 2); despite spending more time eating, IL8+ mice may not have consumed extra enough food to cause a difference in weight. IL8+ mice also spent less time drinking water, which raises the possibility of dehydration. A detailed tracking of food and water consumption is needed in the future. The measurement of metabolic rate is clearly another important issue for the future. Changes between hIL8+ and control littermates were more pronounces at 28 wk of age than at earlier time points (data not shown). Body mass (i.e., amount of adipose tissue) changes with aging, potentially affecting the results.
In one study, mice that conditionally expressed hIL8 after birth had increased mobilization of immature myeloid cells to local inflammatory sites and accelerated carcinogenesis.1 The hIL8 expression model in that work1 was generated by using a bacterial artificial chromosome encompassing the entire hIL8 gene and various regulatory elements, thus allowing induction of hIL8 with a local pathophysiological stimulus, such as azoxymethane, in the gastrointestinal tract; that model could be useful to model musculoskeletal inflammation as well. In another study using hIL8 expression in the liver, hIL8 substantially limited hepatic apoptosis, reduced liver hemorrhage, and prolonged survival in mice treated with galactosamine and endotoxin.10 These findings suggest that hIL8 expression locally and systemically results in a variety of effects, in addition to leukocyte trafficking.
In summary, we have generated a mouse model that conditionally expresses hIL8 to investigate the effects of this expression in behavior. We have shown that nest-making is impaired in hIL8+ mice compared with littermate controls. Furthermore, hIL8+ mice tend to eat more and walk less than littermate controls, reminiscent of depression symptoms in humans with chronic back or joint pain. The mechanisms underlying IL8-associated behavioral changes are still unclear. In the future, we will examine various tissues, including brain, to differentiate central and peripheral mechanisms of functional deficits and altered behaviors.
Acknowledgments
This work is supported, in part, by research grants from the Department of Veterans Affairs Healthcare Network VISN 4, a Penn Center for Musculoskeletal Disorders (PCMD) pilot grant (P30-AR050950-10 Pilot), and a grant from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS, NIH R21 AR071623). The PCMD Histology Core Facility provided outstanding histology service (P30-AR050950-10). We gratefully thank Motomi Enomoto-Iwamoto, DDS, for valuable guidance and suggestions and Martin F Heyworth for critically editing the manuscript. We thank Yan Xiu and Lan Zhao for technical support and Debra Pawlowski and Pierre Conti, VMD, for care of the animals. We thank Dr James Olmsted Marx, for providing the rodent thermometer.
References
- 1.Asfaha S, Dubeykovskiy AN, Tomita H, Yang X, Stokes S, Shibata W, Friedman RA, Ariyama H, Dubeykovskaya ZA, Muthupalani S, Ericksen R, Frucht H, Fox JG, Wang TC. 2013. Mice that express human interleukin-8 have increased mobilization of immature myeloid cells, which exacerbates inflammation and accelerates colon carcinogenesis. Gastroenterology 144:155–166. 10.1053/j.gastro.2012.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bäckryd E, Tanum L, Lind AL, Larsson A, Gordh T. 2017. Evidence of both systemic inflammation and neuroinflammation in fibromyalgia patients, as assessed by a multiplex protein panel applied to the cerebrospinal fluid and to plasma. J Pain Res 10:515–525. 10.2147/JPR.S128508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett RM. 2009. Clinical manifestations and diagnosis of fibromyalgia. Rheum Dis Clin North Am 35:215–232. doi: 10.1016/j.rdc.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 4.Capellini TD, Chen H, Cao J, Doxey AC, Kiapour AM, Schoor M, Kingsley DM. 2017. Ancient selection for derived alleles at a GDF5 enhancer influencing human growth and osteoarthritis risk. Nat Genet 49:1202–1210. 10.1038/ng.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen H, Capellini TD, Schoor M, Mortlock DP, Reddi AH, Kingsley DM. 2016. Heads, shoulders, elbows, knees, and toes: Modular GDF5 enhancers control different joints in the vertebrate skeleton. PLoS Genet 12:1–27. 10.1371/journal.pgen.1006454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deacon RM. 2006. Assessing nest building in mice. Nat Protoc 1:1117–1119. 10.1038/nprot.2006.170. [DOI] [PubMed] [Google Scholar]
- 7.DePalma MJ, Ketchum JM, Saullo T. 2011. What is the source of chronic low back pain and does age play a role? Pain Med 12:224–233. 10.1111/j.1526-4637.2010.01045.x. [DOI] [PubMed] [Google Scholar]
- 8.Fan X, Patera AC, Pong-Kennedy A, Deno G, Gonsiorek W, Manfra DJ, Vassileva G, Zeng M, Jackson C, Sullivan L, Sharif-Rodriguez W, Opdenakker G, Van Damme J, Hedrick JA, Lundell D, Lira SA, Hipkin RW. 2006. Murine CXCR1 is a functional receptor for GCP-2/CXCL6 and interleukin-8/CXCL8. J Biol Chem 282:11658–11666. 10.1074/jbc.M607705200. [DOI] [PubMed] [Google Scholar]
- 9.Gellhorn AC, Katz JN, Suri P. 2013. Osteoarthritis of the spine: the facet joints. Nat Rev Rheumatol 9:216–224. doi: 10.1038/nrrheum.2012.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanson JC, Bostick MK, Campe CB, Kodali P, Lee G, Yan J, Maher JJ. 2006. Transgenic overexpression of interleukin-8 in mouse liver protects against galactosamine/endotoxin toxicity. J Hepatol 44:359–367. 10.1016/j.jhep.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 11.Hoffmann E, Dittrich-Breiholz O, Holtmann H, Kracht M. 2002. Multiple control of interleukin-8 gene expression. J Leukoc Biol 72:847–855. [PubMed] [Google Scholar]
- 12.Kepler CK, Markova DZ, Dibra F, Yadla S, Vaccaro AR, Risbud MV, Albert TJ, Anderson DG. 2013. Expression and relationship of proinflammatory chemokine RANTES/CCL5 and cytokine IL1β in painful human intervertebral discs. Spine (Phila Pa 1976) 38:873–880. 10.1097/BRS.0b013e318285ae08. [DOI] [PubMed] [Google Scholar]
- 13.Koyama E, Shibukawa Y, Nagayama M, Sugito H, Young B, Yuasa T, Okabe T, Ochiai T, Kamiya N, Rountree RB, Kingsley DM, Iwamoto M, Enomoto-Iwamoto M, Pacifici M. 2008. A distinct cohort of progenitor cells participates in synovial joint and articular cartilage formation during mouse limb skeletogenesis. Dev Biol 316:62–73. 10.1016/j.ydbio.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kozak M. 1987. An analysis of 5′–noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res 15:8125–8148. 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kraan MC, Patel DD, Haringman JJ, Smith MD, Weedon H, Ahern MJ, Breedveld FC, Tak PP. 2000. The development of clinical signs of rheumatoid synovial inflammation is associated with increased synthesis of the chemokine CXCL8 (interleukin-8). Arthritis Res 3:65–71. 10.1186/ar141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J, Cacalano G, Camerato T, Toy K, Moore MW, Wood WI. 1995. Chemokine binding and activities mediated by the mouse IL8 receptor. J Immunol 155:2158–2164. [PubMed] [Google Scholar]
- 17.Martin BI, Deyo RA, Mirza SK, Turner JA, Comstock BA, Hollingworth W, Sullivan SD. 2008. Expenditures and health status among adults with back and neck problems. JAMA 299:656–664. 10.1001/jama.299.6.656. [DOI] [PubMed] [Google Scholar]
- 18.Matsushima K, Morishita K, Yoshimura T, Lavu S, Kobayashi Y, Lew W, Appella E, Kung HF, Leonard EJ, Oppenheim JJ. 1988. Molecular cloning of a human monocyte-derived neutrophil chemotactic factor (MDNCF) and the induction of MDNCF mRNA by interleukin 1 and tumor necrosis factor. J Exp Med 167:1883–1893. 10.1084/jem.167.6.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mukaida N, Shiroo M, Matsushima K. 1989. Genomic structure of the human monocyte-derived neutrophil chemotactic factor IL8. J Immunol 143:1366–1371. [PubMed] [Google Scholar]
- 20.Nakazawa KR, Walter BA, Laudier DM, Krishnamoorthy D, Mosley GE, Spiller KL, Iatridis JC. 2018. Accumulation and localization of macrophage phenotypes with human intervertebral disc degeneration. Spine J 18:343–356. 10.1016/j.spinee.2017.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oka M, Norose K, Matsushima K, Nishigori C, Herlyn M. 2006. Overexpression of IL8 in the cornea induces ulcer formation in the SCID mouse. Br J Ophthalmol 90:612–615. 10.1136/bjo.2005.084525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwarzer AC, Aprill CN, Derby R, Fortin J, Kine G, Bogduk N. 1995. The prevalence and clinical features of internal disc disruption in patients with chronic low back pain. Spine (Phila Pa 1976) 20:1878–1883. 10.1097/00007632-199509000-00007. [DOI] [PubMed] [Google Scholar]
- 23.Simonet WS, Hughes TM, Nguyen HQ, Trebasky LD, Danilenko DM, Medlock ES. 1994. Long-term impaired neutrophil migration in mice overexpressing human interleukin-8. J Clin Invest 94:1310–1319. 10.1172/JCI117450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sowa GA, Perera S, Bechara B, Agarwal V, Boardman J, Huang W, Camacho-Soto A, Vo N, Kang J, Weiner D. 2014. Associations between serum biomarkers and pain and pain-related function in older adults with low back pain: A pilot study. J Am Geriatr Soc 62:2047–2055. 10.1111/jgs.13102. [DOI] [PubMed] [Google Scholar]
- 25.Storm EE, Kingsley DM. 1999. GDF5 coordinates bone and joint formation during digit development. Dev Biol 209:11–27. 10.1006/dbio.1999.9241. [DOI] [PubMed] [Google Scholar]
- 26.Tian Z, Ma X, Yasen M, Mauck RL, Qin L, Shofer FS, Smith LJ, Pacifici M, Enomoto-Iwamoto M, Zhang Y. 2018. Intervertebral disc degeneration in a percutaneous mouse tail injury model. Am J Phys Med Rehabil 97:170–177. 10.1097/PHM.0000000000000818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van de Weerd HA, Bulthuis RJ, Bergman AF, Schlingmann F, Tolboom J, Van Loo PL, Remie R, Baumans V, Van Zutphen LF. 2001. Validation of a new system for the automatic registration of behaviour in mice and rats. Behav Processes 53:11–20. 10.1016/S0376-6357(00)00135-2. [DOI] [PubMed] [Google Scholar]
- 28.Weber KT, Alipui DO, Sison CP, Bloom O, Quraishi S, Overby MC, Levine M, Chahine NO. 2016. Serum levels of the proinflammatory cytokine interleukin-6 vary based on diagnoses in individuals with lumbar intervertebral disc diseases. Arthritis Res Ther 18:1–14. 10.1186/s13075-015-0887-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weber KT, Satoh S, Alipui DO, Virojanapa J, Levine M, Sison C, Quraishi S, Bloom O, Chahine NO. 2015. Exploratory study for identifying systemic biomarkers that correlate with pain response in patients with intervertebral disc disorders. Immunol Res 63:170–180. 10.1007/s12026-015-8709-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Giron CG, Gil L, Gordon L, Haggerty L, Haskell E, Hourlier T, Izuogu OG, Janacek SH, Juettemann T, To JK, Laird MR, Lavidas I, Liu Z, Loveland JE, Maurel T, McLaren W, Moore B, Mudge J, Murphy DN, Newman V, Nuhn M, Ogeh D, Ong CK, Parker A, Patricio M, Riat HS, Schuilenburg H, Sheppard D, Sparrow H, Taylor K, Thormann A, Vullo A, Walts B, Zadissa A, Frankish A, Hunt SE, Kostadima M, Langridge N, Martin FJ, Muffato M, Perry E, Ruffier M, Staines DM, Trevanion SJ, Aken BL, Cunningham F, Yates A, Flicek P. 2018. Ensembl 2018. Nucleic Acids Res 46 D1:D754–D761. 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Chee A, Shi P, Adams SL, Markova DZ, Anderson DG, Smith HE, Deng Y, Plastaras CT, An HS. 2016. Intervertebral disc cells produce interleukins found in patients with back pain. Am J Phys Med Rehabil 95:407–415. 10.1097/PHM.0000000000000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, Chee A, Shi P, Wang R, Moss I, Chen EY, He TC, An HS. 2015. Allogeneic articular chondrocyte transplantation downregulates interleukin 8 gene expression in the degenerating rabbit intervertebral disk in vivo. Am J Phys Med Rehabil 94:530–538. 10.1097/PHM.0000000000000194. [DOI] [PMC free article] [PubMed] [Google Scholar]