Abstract
Carbonic anhydrase (CA) is a zinc enzyme that catalyzes the reversible conversion of carbon dioxide to bicarbonate and proton. Currently, CA inhibitors are widely used as antiglaucoma, anticancer, and anti-obesity drugs and for the treatment of neurological disorders. Recently, the potential use of CA inhibitors to fight infections caused by protozoa, fungi, and bacteria has emerged as a new research line. In this article, the X-ray crystal structure of β-CA from Burkholderia pseudomallei was reported. The X-ray crystal structure of this new enzyme was solved at 2.7 Å resolution, revealing a tetrameric type II β-CA with a “closed” active site in which the zinc is tetrahedrally coordinated to Cys46, Asp48, His102, and Cys105. B. pseudomallei is known to encode at least two CAs, a β-CA, and a γ-CA. These proteins, playing a pivotal role in its life cycle and pathogenicity, offer a novel therapeutic opportunity to obtain antibiotics with a different mechanism of action. Furthermore, the new structure can provide a clear view of the β-CA mechanism of action and the possibility to find selective inhibitors for this class of CAs.
Keywords: β-Carbonic Anhydrase, Burkholderia pseudomallei, crystal structure, type II CA
1. Introduction
Burkholderia pseudomallei is the etiologic agent of a severe and often fatal syndrome known as melioidosis, or Whitmore’s disease [1]. Melioidosis is a severe disease of humans and animals, causing an estimated 165,000 cases per year, resulting in a predicted 89,000 deaths [2,3]. Infection with B. pseudomallei was usually associated with environmental exposure and can occur through breaks in the skin, inhalation, or ingestion [4]. In addition, B. pseudomallei is one of the prominent opportunistic pathogens classified as a bioterrorism agent by both the UK government and the US Centers for Disease Control and Prevention [4,5]. Finally, in recent years, the tolerance to antimicrobials has increased considerably [6,7].
In this scenario, a novel and promising approach for fighting antibiotic resistance is represented by the inhibition of carbonic anhydrases (CAs, EC 4.2.1.1 [8,9,10,11,12], a superfamily of metalloenzymes which catalyzes the simple but physiologically crucial reaction of carbon dioxide hydration to bicarbonate and protons [13,14,15]. These enzymes are present in all life kingdoms and, to date, are divided into eight distinct classes which exhibit no significant sequence or structural similarities, known as the α, β, δ, γ, ζ, θ, η, and the recently discovered ι [16,17]. All the catalytically active CAs contain, independently of the genetic groups, a metal ion cofactor, which is necessary for enzyme catalysis [13,14,15,16,17]. The α-, β-, δ-, γ-CAs use the Zn2+ ion as a catalytic metal, in addition, γ-CAs use Fe2+ or Co2+ ions too [13,14,15]. ζ-CA is cambialistic enzymes, which are active with Cd2+ or Zn2+ [15,16]. Unexpectedly, the last identified ι-CA, which is encoded in the genome of the marine diatom, Thalassiosira pseudonana, prefers Mn2+ to Zn2+ as a cofactor [17,18]. In addition, in many bacteria, these enzymes are known to be essential for their life cycle, whereas several essential metabolic pathways require either CO2 or bicarbonate as a substrate [19,20]. It was demonstrated in vivo that the bacterial growth at an ambient CO2 concentration was dependent on CA activity for several species.
The genome of B. pseudomallei encodes for β- and γ-CAs. Recently, a gene encoding for the ι-CA was found in the genome of another genus of Burkholderia (Burkholderia territorii) [18]. However, neither of the two species had genes encoding for the α-class [18]. This feature is of great interest, because these three classes are not expressed in humans, giving the opportunity to inhibit these classes preferentially. Our group recently reported the catalytic activity and the sulfonamide and anion inhibition profiles of the recombinant β- and γ-CAs from B. pseudomallei, named BpsβCA and BpsγCA, respectively [21,22,23]. In the last ten years, numerous results concerning the inhibition profile of the three bacterial CA classes (α, β, and γ) were reported using anions and sulfonamides. Most of these studies were carried out on bacterial CAs from pathogenic bacteria, such as Francisella tularensis, Burkholderia pseudomallei, Vibrio cholerae, Streptococcus mutans, Porphyromonas gingivalis, Legionella pneumophila, Clostridium perfringens, and Mycobacterium tuberculosis [20,21,24,25,26]. The results indicated that certain CA inhibitors were able to highly inhibit most of the CAs identified in the genome of the aforementioned bacteria. Moreover, certain CA inhibitors, such as acetazolamide and methazolamide, were shown to effectively inhibit bacterial growth in cell cultures [27].
Here, we reported for the first time the crystallographic structure of BpsβCA that was solved in order to understand its function, and laid down the foundation for developing inhibitors that were more potent and selective towards this isoform.
Previous works on the β-CAs class revealed two distinct subtypes of this enzyme called type I or type II β-Cas, according to their active-site organization. [28] Type I presents in the active site the zinc ion coordinated with one histidine, two cysteine residues, and a fourth coordination site occupied by water or a substrate analogue (the so-called open conformation). This particular conformation was reported for the β-CAs from the bacteria, such as Pisum sativum [29], Methanobacterium thermoautotrophicum [30] and M. tuberculosis (Rv1284) [28]. On the other hand, the Type II subclass of β-CAs has a unique zinc-coordination geometry, in which the water molecule is replaced by an aspartate side chain, forming a non-canonical CA active site (the closed conformation), as observed in Haemophilus influenza [31,32], Escherichia coli [33], Porphyridium purpureum [34], and M. tuberculosis (Rv3588c) [28]. This subtype is characterized by little or no CO2 hydration activity at pH values less than 8.0. Therefore, it was hypothesized that the closed conformation (called T state) observed in the structures of type II β-CAs is an allosteric form of the enzyme and, is the inactive form at pH values below 8.0. However, at pH values larger than 8.3, the closed active site is converted to an open one, with an incoming water molecule replacing the carboxylate moiety of the Asp residue, thus generating the nucleophile required in the catalytic cycle. This was demonstrated by X-ray crystallography (and kinetic studies) in an elegant work by Jones and coworkers [28]. Indeed, at this pH value, the carboxylate of the Asp has a strong interaction with the guanidine/guanidinium moiety of a conserved Arg residue present in all β-CAs investigated so far [28].
2. Results
First of all, the catalytic efficiency of recombinant BpsβCA for the physiologic reaction, CO2 hydration to bicarbonate and protons, was measured and its kinetic parameters were compared with those of γ-CA and ι-CA classes, CAs from the same gram-negative genus (Table 1).
Table 1.
Kinetic parameters for the CO2 hydration reaction catalyzed by the β- and γ-CAs from B. pseudomallei and ι-CA from B. territorii measured at 20 °C, pH 8.3 in 20 mM TRIS buffer, and 20 mM NaClO4 [20,21,35]. Acetazolamide inhibition data are also shown.
| Enzyme | Activity Level | Class | kcat (s−1) | kcat/Km (M−1 s−1) | Ki (Acetazolamide) (nM) |
|---|---|---|---|---|---|
| BpsβCA | Moderate | β | 1.6 × 105 | 3.4 × 107 | 745 |
| BpsγCA | Moderate | γ | 5.3 × 105 | 2.5 × 107 | 149 |
| BteCAι | Moderate | ι | 3.0 × 105 | 9.7 × 107 | 64.9 |
Data of Table 1 shows similar activities among the different classes of CAs from the Burkholderia genus, possessing a moderate but significant CO2 hydrase activity with kinetic parameters (kcat) spanning between 1.6 to 5.3 × 105. Furthermore, the activity of BpsβCA is only moderately inhibited (Ki of 745 nM) by the clinically used sulfonamide inhibitor acetazolamide (5-acetamido-1,3,4-thiadiazole-2-sulfonamide), which was a much better inhibitor of the other two enzymes belonging to different classes [18,21,22].
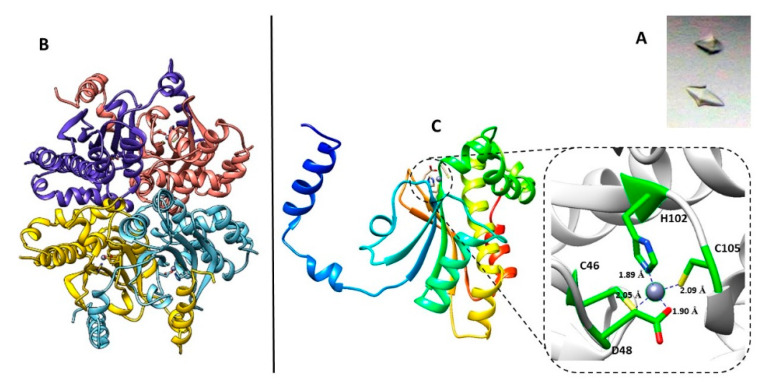
Then, the crystal structure of the recombinant type II β-CA from B. pseudomallei was determined at a resolution of 3.1 Å (Table 2). Enzyme crystals were obtained by the sitting-drop vapor diffusion method. They belong to the space group P6422, with one molecule per asymmetric unit. Among the β-CAs of the known structure, the highest level of sequence homology of Bpsβ-CA was observed with the β-CAs from Pseudomonas aeruginosa (57.4% identity), Porphyridium purpureum (55.3% identity), Salmonella typhimurium (49.5% identity) and, finally, from Vibrio cholerae (49.3% identity). The structure was solved by molecular replacement using the β-CA from Pseudomonas aeruginosa (PsCA3, 57.4% sequence identity, PDB code: 4rxy) as the initial model [36]. The biological assembly was investigated by the PISA (Protein Interfaces, Surfaces and Assemblies) software application, that confirmed a tetrameric organization of the enzyme which strictly resembles that of the other structurally characterized β–CAs, which have a dimer, a tetramer, or an octamer arrangement (Figure 1). The active site is located in a cleft at the interface of one dimer, and contains a zinc ion at the bottom, coordinated by three protein residues, namely Cys46, His102, and Cys105. Furthermore, Asp48 is visible in the fourth coordination position instead of the typical water molecule (Figure 1), revealing a “closed” configuration of the active site. Therefore, the enzyme can be classified as a type II β-CA that, as expected, assumes a “closed” conformation, considering the pH of the crystallization condition (pH 7.5).
Table 2.
Summary of Data Collection and Atomic Model Refinement Statistics.
| BpsβCA pH 7.5 | BpsβCA pH 8.5 | |
|---|---|---|
| PDB ID | 6YL7 | 6YJN |
| Wavelength (Å) | 1.0399 | 1.0000 |
| Space Group | P6422 | P6422 |
| Unit cell (a, b, c, α, β, γ) (Å,°) | 88.74;88.74;112.43; | 88.03;88.03;111.64; |
| 90.0;90.0;120.0 | 90.00;90.00;120.00 | |
| Limiting resolution (Å) | 45.37–3.16 (3.38–3.16) | 45.04–2.70 (2.83–2.70) |
| Unique reflections | 4858 (743) | 13330 (2141) |
| Rmerge (%) | 27.4 (265.4) | 26.0 (789.6) |
| Rmeas (%) | 28.08 (271.5) | 26.7 (811.5) |
| Redundancy | 24.7 (23.4) | 18.4 (18.6) |
| Completeness overall (%) | 99.7 (98.7) | 99.9 (99.6) |
| <I/σ(I)> | 10.42 (1.15) | 10.93 (0.34) |
| CC (1/2) | 99.8 (59.4) | 99.9 (32.1) |
| Refinement statistics | ||
| Resolution range (Å) | 45.412–3.166 | 45.080–2.701 |
| Unique reflections, working\free | 4605\3440 | 7508\7093 |
| Rfactor (%) | 19.26 | 21.4 |
| Rfree(%) | 29.62 | 32.4 |
| r.m.s.d. bonds(Å) | 0.0060 | 0.0052 |
| r.m.s.d. angles (°) | 1.5874 | 1.4987 |
| Ramachandran statistics (%) | ||
| Most favored | 79.3 | 82.9 |
| Additionally allowed | 17.8 | 11.4 |
| Outlier regions | 2.9 | 5.7 |
| Average B factor (Å2) | ||
| Solvent | 78.811 | 83.801 |
Figure 1.
(A) The shape of BpsβCA crystals, under bright field illumination. (B) Ribbon diagram showing the tetrameric arrangement of BpsβCA. (C) Crystal structure of BpsβCA. Ribbon diagram of the BpsβCA structure, asymmetric unit content, and active site of BpsβCA. The detailing insert shows the enzyme active site with the zinc ion (gray sphere) and its ligands (Cys46, His102, Cys105, and Asp48).
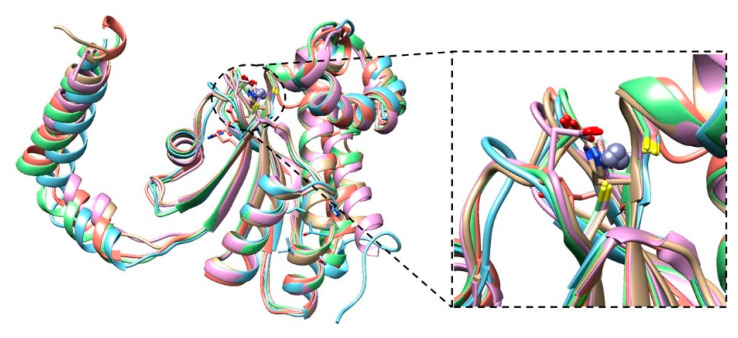
The structural comparison of BpsβCA with other β-CAs belonging to different bacterial species, shows substantial conservation of the BpsβCA three-dimensional structure (Figure 2), which is the highest with the Pseudomonas aeruginosa (PsCA3).
Figure 2.
Superposition of the BpsβCA structure (brown), with the previously determined type II β-CAs from Pseudomonas aeruginosa (cyan, r.m.s.d of 0.784 Å), Porphyridium purpureum (violet r.m.s.d of 0.749 Å), Salmonella typhimurium (green r.m.s.d of 0.785 Å), and Vibrio cholerae (red r.m.s.d of 0.952 Å). The gray sphere represents the zinc atom in the active site. The right panel highlights the active site of type II β-CAs.
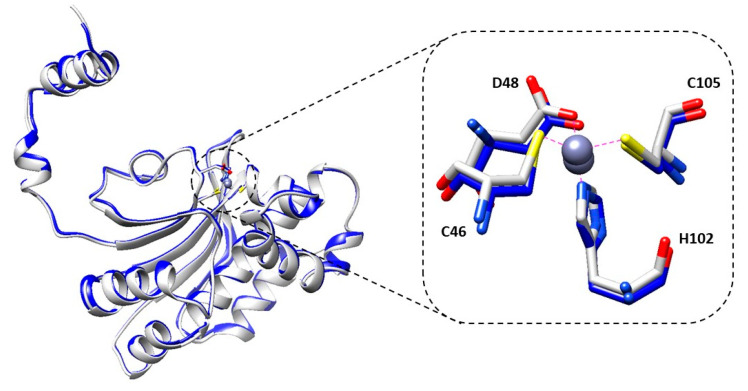
The active site is also well conserved compared to the other type II β-CAs. Arg50 is supposed to interact through two hydrogen bonds with Asp48 in the “open” conformation, as inferred from the structures showing the active site of the type I β-CAs. Unfortunately, the electron density maps did not show any density at the side chain of Arg50 that was not included in the model. It was hypothesized that a Tyrosine was necessary for efficient proton transfer inside the mechanism of the β-CAs, for example Tyr212 in Arabidopsis Thaliana and Tyr83 in Vibrio cholerae β-CA. BpsβCA has phenylalanine (Phe87) at that position and other residues in the active site may play an essential role for catalysis. Finally, to gain information on the open conformation of the enzyme active site, we solved the structure of the enzyme from one crystal obtained in the same crystallization condition, except pH was increased to 8.5, a value at which the enzyme was shown to possess catalytic activity. Although the structure was determined at quite a low resolution (maximum resolution 2.7 Å), we could observe that in the active site, the Asp48 is in the same position as in the structure at pH 7.5, interacting directly with the zinc ion as reported above (see Figure 3).
Figure 3.
A comparison of the BpsβCA active site arrangements in two different pH conditions: 7.5 and 8.5. In gray: BpsβCA crystalized at pH 7.5. In blue: BpsβCA crystalized at pH 8.5.
Nevertheless, the active form was never observed in a β-CA structure, regardless of the crystallization pH, except for in two mutants of H. influenzae CAs [26] and a thiocyanate inhibitor complex of M. tuberculosis CA [28].
3. Materials and Methods
3.1. Enzyme Preparation
The identification of the gene encoding for B. pseudomallei β-CA (BpsβCA) was performed, as described by Del Prete et al. [22] Briefly, The β-CA gene of B. pseudomallei (accession number: WP_004189176.1) was identified by running the Basic Local Alignment Search Tool (BLAST) software application, using the nucleotide sequences of bacterial β-CAs as a query sequence. The GeneArt Company (Invitrogen), specializing in gene synthesis, designed the synthetic BpsβCA gene (BpsβCA-DNA) encoding for the BpsβCA (a protein made of 256 amino acid residues) containing four base-pair sequences (CACC) necessary for directional cloning at the 50 end of the PfCAdom gene. The recovered BpsβCA gene and the linearized expression vector (pET-100/D-TOPO) were ligated by T4 DNA ligase to form the expression vector pET-100/BpsβCA. BL21 DE3 codon plus competent cells (Agilent) were transformed with pET-100/BpsβCA, grown at 37 °C, and induced with 1 mM IPTG. After 30 min, ZnSO4 (0.5 mM) was added to the culture medium (2 L), and cells were grown for additional 3 h. Subsequently, cells were harvested and resuspended in the following buffer: 50 mM Tris/HCl, pH 8.0, 0.5 mM PMSF, and 1 mM benzamidine. Cells were then disrupted by sonication at 4 °C. After centrifugation at 12,000× g for 45 min, the supernatant was incubated with His Select HF nickel affinity gel resin (Sigma) equilibrated in lysis buffer for 30 min. Following centrifugation at 2000 g, the resin was washed in wash buffer (50 mM Tris/HCl, pH 8.3, 500 mM KCl, and 20 mM imidazole). The protein was eluted with the wash buffer containing 300 mM imidazole. Collected fractions were dialyzed against 50 mM Tris/HCl, pH 8.3. At this stage of purification, the protein was at least 95% pure, and the obtained recovery was about 20 mg of the recombinant protein.
3.2. Crystallization and Data Collection
The enzyme was crystallized at 296 K using the sitting-drop vapor-diffusion method in 96-well plates (CrystalQuick, Greiner Bio-One, Maulbronn, Germany). Drops were prepared using 1 µL of protein solution mixed with 1 µL of reservoir solution and were equilibrated against 100 µL precipitant solution. The concentration of the protein was 10 mg mL−1 in 50 mM Tris pH 7.2. Initial crystallization condition was found using the JCSG plus screen kit (Molecular Dimensions) and were optimized. Diffraction-quality crystals grew within four months from a solution consisting of 22% PEG 4000, 10% isopropanol, 100 mM HEPES pH 7.5 or 8.5, and 3% v/v 1,5-Diaminopentene di-HCl. The crystals belonged to the primitive hexagonal space group P6422. Data obtained from crystals at pH 8.5 were collected on the XRD2 beamline at Elettra, Trieste, Italy, using a Pilatus3_6M Dectris CCD detector and a wavelength of 1.000 Å. Data obtained from crystals at pH 7.5 was collected on the ID-29 beamline at ESRF (Grenoble, France) with a wavelength of 1.0399 Å and a Pilatus3_6M Dectris CCD detector. For data collection, a crystal of the enzyme was cooled to 100 K using a solution consisting of 22% PEG 4000, 10% isopropanol, 100 mM HEPES pH 7.5 or 8.5, 3% v/v 1,5-Diaminopentene di-HCl, and 15% ethylene glycol, as cryoprotectant. The data were processed with an XDS program package [37].
3.3. Structure Determination and Refinement
The structure was solved by the molecular-replacement technique using the MOLREP program for molecular replacement [35] using the coordinates of the structure of β-carbonic anhydrase from P. aeruginosa (PDB entry 4rxy) as a starting model. The model was refined using the REFMAC5 program [38] from the CCP4 suite [39]. Manual rebuilding of the model was performed using the Crystallographic Object-Oriented Toolkit (Coot) [40]. Solvent molecules were introduced automatically using the ARP/wARP software suite [41]. Data processing and refinement statistics are summarized in Table 1. Protein coordinates were deposited in the Protein Data Bank (PDB entry 6YL7; 6YJN). Structural figures were generated with the UCSF Chimera package [42].
3.4. Kinetic and Inhibition Assay
An Applied Photophysics stopped-flow instrument was used for assaying the CA catalyzed CO2 hydration activity. [43] Phenol red (at a concentration of 0.2 mM) was used as an indicator, working at the absorbance maximum of 557 nm, with 20 mM TRIS (pH 8.3) as buffer, and 20 mM NaClO4 (for maintaining constant the ionic strength), following the initial rates of the CA-catalyzed CO2 hydration reaction for a period of 10–100 s. The CO2 concentrations ranged from 1.7 to 17 mM for the determination of the kinetic parameters (by Lineweaver-Burk plots) and inhibition constants. For each inhibitor, at least six traces of the initial 5–10% of the reaction were used for determining the initial velocity. The uncatalyzed rates were determined in the same manner and subtracted from the total observed rates. Stock solutions of the inhibitor (10–100 mM) were prepared in distilled-deionized water and dilutions up to 0.01 mM were done thereafter with the assay buffer. Inhibitor and enzyme solutions were preincubated together for 15 min at room temperature prior to assay, in order to allow for the formation of the E-I complex, or for the eventual active site mediated hydrolysis of the inhibitor. The inhibition constants were obtained by non-linear least-squares methods using PRISM 3 and the Cheng–Prusoff equation, as reported earlier, and represent the mean from at least three different determinations. All CA isoforms were recombinant ones obtained in-house. All salts and small molecules were of the highest purity available, from Sigma-Aldrich (Milan, Italy).
4. Conclusions
The X-ray crystal data of the recombinant β-CA from Burkholderia pseudomallei (BpsβCA) are reported in this paper. The X-ray crystal structure of the enzyme was solved at 2.7 Å resolution and two different pH levels (7.5 and 8.5). BpsβCA was revealed to be a tetrameric type II β-CA with a closed active site in which the zinc is tetrahedrally coordinated to Cys46, Asp48, His102, and Cys105. The X-ray structure solved at two pH levels (7.5 and 8.5) showed the same “close” conformation at the active site. The genome of B. pseudomallei encodes for different classes of CAs (β and γ). Besides, the bacterial CAs play a pivotal role in the life cycle and pathogenicity of the microorganism, balancing their endogenous equilibrium between CO2 and HCO3−. The resolution of the BpsβCA structure provides new insights for the understanding of the enzyme catalytic site, as well as the possibility of finding selective inhibitors for β-CAs. These findings offer the opportunity to obtain new antibiotics that are able to impair the growth or the virulence of the microorganism, with a mechanism of action different to that of the existing drugs.
Acknowledgments
We acknowledge Elettra-Sincrotrone for the provision of synchrotron radiation facilities, and we would like to thank Nicola Demitri for her assistance in using the beamline XRD2. We also acknowledge the European Synchrotron Radiation Facility for the provision of synchrotron radiation facilities (ID-29).
Author Contributions
Writing—original draft preparation, A.A., M.F.; writing—review and editing, F.C., S.S.M., B.C.S., S.D.P., C.C.; supervision, C.T.S.; date curation, M.P. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by a grant from the Romanian Ministry of Research and Innovation, CNCS–UEFISCDI, project number PN-III-P4-ID-PCCF-2016–0050, within PNCDI II.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Samples of the compounds are not available from the authors.
References
- 1.Limmathurotsakul D., Peacock S.J. Melioidosis: A clinical overview. Br. Med. Bull. 2011;99:125–139. doi: 10.1093/bmb/ldr007. [DOI] [PubMed] [Google Scholar]
- 2.Limmathurotsakul D., Golding N., Dance D., Messina J.P., Pigott D.M., Moyes C., Rolim D.B., Bertherat E., Day N.P., Peacock S.J., et al. Predicted global distribution of Burkholderia pseudomallei and burden of melioidosis. Nat. Microbiol. 2016;1:15008. doi: 10.1038/nmicrobiol.2015.8. [DOI] [PubMed] [Google Scholar]
- 3.Wiersinga W.J., van der Poll T., White N.J., Day N.P., Peacock S.J. Melioidosis: Insights into the pathogenicity of Burkholderia pseudomallei. Nat. Rev. Genet. 2006;4:272–282. doi: 10.1038/nrmicro1385. [DOI] [PubMed] [Google Scholar]
- 4.Cheng A., Currie B.J. Melioidosis: Epidemiology, pathophysiology, and management. Clin. Microbiol. Rev. 2005;18:383–416. doi: 10.1128/CMR.18.2.383-416.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rotz L.D., Khan A.S., Lillibridge S.R., Ostroff S.M., Hughes J.M. Public health assessment of potential biological terrorism agents. Emerg. Infect. Dis. 2002;8:225–230. doi: 10.3201/eid0802.010164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balaban N.Q., Gerdes K., Lewis K., McKinney J.D. A problem of persistence: Still more questions than answers? Nat. Rev. Genet. 2013;11:587–591. doi: 10.1038/nrmicro3076. [DOI] [PubMed] [Google Scholar]
- 7.Monroe D. Looking for chinks in the armor of bacterial biofilms. PLoS Biol. 2007;5:e307. doi: 10.1371/journal.pbio.0050307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Supuran C.T., Capasso C. An overview of the bacterial carbonic anhydrases. Metabolites. 2017;7:56. doi: 10.3390/metabo7040056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Supuran C.T., Capasso C. Biomedical applications of prokaryotic carbonic anhydrases. Expert Opin. Ther. Pat. 2018;28:745–754. doi: 10.1080/13543776.2018.1497161. [DOI] [PubMed] [Google Scholar]
- 10.Capasso C., Supuran C.T. Inhibition of bacterial carbonic anhydrases as a novel approach to escape drug resistance. Curr. Top Med. Chem. 2017;17:1237–1248. doi: 10.2174/1568026617666170104101058. [DOI] [PubMed] [Google Scholar]
- 11.Capasso C., Supuran C.T. Bacterial, fungal and protozoan carbonic anhydrases as drug targets. Expert Opin. Ther. Targets. 2015;19:1689–1704. doi: 10.1517/14728222.2015.1067685. [DOI] [PubMed] [Google Scholar]
- 12.Capasso C., Supuran C.T. An overview of the alpha-, beta- and gamma-carbonic anhydrases from Bacteria: Can bacterial carbonic anhydrases shed new light on evolution of bacteria? J. Enzyme Inhib. Med. Chem. 2015;30:325–332. doi: 10.3109/14756366.2014.910202. [DOI] [PubMed] [Google Scholar]
- 13.Supuran C.T. Structure-based drug discovery of carbonic anhydrase inhibitors. J. Enzyme Inhib. Med. Chem. 2012;27:759–772. doi: 10.3109/14756366.2012.672983. [DOI] [PubMed] [Google Scholar]
- 14.Supuran C.T. Carbonic anhydrase inhibitors and activators for novel therapeutic applications. Future Med. Chem. 2011;3:1165–1180. doi: 10.4155/fmc.11.69. [DOI] [PubMed] [Google Scholar]
- 15.Bozdag M., Altamimi A.S.A., Vullo D., Supuran C.T., Carta F. State of the art on carbonic anhydrase modulators for biomedical purposes. Curr. Med. Chem. 2019;26:2558–2573. doi: 10.2174/0929867325666180622120625. [DOI] [PubMed] [Google Scholar]
- 16.Supuran C.T. Structure and function of carbonic anhydrases. Biochem. J. 2016;473:2023–2032. doi: 10.1042/BCJ20160115. [DOI] [PubMed] [Google Scholar]
- 17.Akocak S., Supuran C.T. Activation of α-, β-, γ- δ-, ζ- and η- class of carbonic anhydrases with amines and amino acids: A review. J. Enzyme Inhib. Med. Chem. 2019;34:1652–1659. doi: 10.1080/14756366.2019.1664501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Del Prete S., Nocentini A., Supuran C.T., Capasso C. Bacterial ι-carbonic anhydrase: A new active class of carbonic anhydrase identified in the genome of the Gram-negative bacterium Burkholderia territorii. J. Enzyme Inhib. Med. Chem. 2020;35:1060–1068. doi: 10.1080/14756366.2020.1755852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Capasso C., Supuran C.T. Anti-infective carbonic anhydrase inhibitors: A patent and literature review. Expert Opin. Ther. Pat. 2013;23:693–704. doi: 10.1517/13543776.2013.778245. [DOI] [PubMed] [Google Scholar]
- 20.Supuran C.T. Inhibition of bacterial carbonic anhydrases and zinc proteases: From orphan targets to innovative new antibiotic drugs. Curr. Med. Chem. 2012;19:831–844. doi: 10.2174/092986712799034824. [DOI] [PubMed] [Google Scholar]
- 21.Del Prete S., Vullo D., Di Fonzo P., Osman S.M., Alothman Z., Donald W.A., Supuran C.T., Capasso C. Sulfonamide inhibition profile of the γ-carbonic anhydrase identified in the genome of the pathogenic bacterium Burkholderia pseudomallei the etiological agent responsible of melioidosis. Bioorg. Med. Chem. Lett. 2017;27:490–495. doi: 10.1016/j.bmcl.2016.12.035. [DOI] [PubMed] [Google Scholar]
- 22.Vullo D., Del Prete S., Di Fonzo P., Carginale V., Donald W.A., Supuran C.T., Capasso C. Comparison of the sulfonamide inhibition profiles of the β- and γ-carbonic anhydrases from the pathogenic bacterium Burkholderia pseudomallei. Molecules. 2017;22:421. doi: 10.3390/molecules22030421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stefanucci A., Angeli A., Dimmito M.P., Luisi G., Del Prete S., Capasso C., Donald W.A., Mollica A., Supuran C.T. Activation of β- and γ-carbonic anhydrases from pathogenic bacteria with tripeptides. J. Enzyme Inhib. Med. Chem. 2018;33:945–950. doi: 10.1080/14756366.2018.1468530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Del Prete S., Vullo D., De Luca V., Carginale V., Osman S.M., Alothman Z., Supuran C.T., Capasso C. Comparison of the sulfonamide inhibition profiles of the α-, β- and γ-carbonic anhydrases from the pathogenic bacterium Vibrio cholerae. Bioorg. Med. Chem. Lett. 2016;26:1941–1946. doi: 10.1016/j.bmcl.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 25.Del Prete S., Vullo D., Osman S.M., Alothman Z., Supuran C.T., Capasso C. Sulfonamide inhibition profiles of the β-carbonic anhydrase from the pathogenic bacterium Francisella tularensis responsible of the febrile illness tularemia. Bioorg. Med. Chem. 2017;25:3555–3561. doi: 10.1016/j.bmc.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 26.Dedeoglu N., De Luca V., Isik S., Yildirim H., Köçkar F., Capasso C., Supuran C.T. Cloning, characterization and anion inhibition study of a β-class carbonic anhydrase from the caries producing pathogen Streptococcus mutans. Bioorg. Med. Chem. 2015;23:2995–3001. doi: 10.1016/j.bmc.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 27.Shahidzadeh R., Opekun A., Shiotani A., Graham D.Y. Effect of the carbonic anhydrase inhibitor, acetazolamide, on Helicobacter pylori infection in vivo: A pilot study. Helicobacter. 2005;10:136–138. doi: 10.1111/j.1523-5378.2005.00306.x. [DOI] [PubMed] [Google Scholar]
- 28.Covarrubias A.S., Bergfors T., Jones T.A., Högbom M. Structural mechanics of the pH-dependent activity of beta-carbonic anhydrase from Mycobacterium tuberculosis. J. Biol. Chem. 2006;281:4993–4999. doi: 10.1074/jbc.M510756200. [DOI] [PubMed] [Google Scholar]
- 29.Kimber M.S., Pai E.F. The active site architecture of Pisum sativum beta-carbonic anhydrase is a mirror image of that of alpha-carbonic anhydrases. EMBO J. 2000;19:1407–1418. doi: 10.1093/emboj/19.7.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strop P., Smith K.S., Iverson T.M., Ferry J.G., Rees D.C. Crystal structure of the “cab”-type β class carbonic anhydrase from the archaeon Methanobacterium thermoautotrophicum. J. Biol. Chem. 2000;276:10299–10305. doi: 10.1074/jbc.M009182200. [DOI] [PubMed] [Google Scholar]
- 31.Cronk J.D., Rowlett R., Zhang K.Y., Tu C., Endrizzi J.A., Lee J., Gareiss P.C., Preiss J.R. Identification of a novel noncatalytic bicarbonate binding site in eubacterial β-carbonic anhydrase. Biochemistry. 2006;45:4351–4361. doi: 10.1021/bi052272q. [DOI] [PubMed] [Google Scholar]
- 32.Hoffmann K.M., Million-Perez H.R., Merkhofer R., Nicholson H.E., Rowlett R. Allosteric reversion of Haemophilus influenza β-Carbonic anhydrase via a proline shift. Biochemistry. 2014;54:598–611. doi: 10.1021/bi501116e. [DOI] [PubMed] [Google Scholar]
- 33.Cronk J.D., Endrizzi J.A., Cronk M.R., O’Neill J.W., Zhang K.Y. Crystal structure of E. coli β-carbonic anhydrase, an enzyme with an unusual pH-dependent activity. Protein Sci. 2001;10:911–922. doi: 10.1110/ps.46301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitsuhashi S., Mizushima T., Yamashita E., Yamamoto M., Kumasaka T., Moriyama H., Ueki T., Miyachi S., Tsukihara T. X-ray structure of beta-carbonic anhydrase from the red alga, Porphyridium purpureum, reveals a novel catalytic site for CO(2) hydration. J. Biol. Chem. 2000;275:5521–5526. doi: 10.1074/jbc.275.8.5521. [DOI] [PubMed] [Google Scholar]
- 35.Vagin A., Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr Sect. D Biol Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 36.Pinard M.A., Lotlikar S.R., Boone C.D., Vullo D., Supuran C.T., Patrauchan M.A., McKenna R. Structure and inhibition studies of a type II beta-carbonic anhydrase psCA3 from Pseudomonas aeruginosa. Bioorg. Med. Chem. 2015;23:4831–4838. doi: 10.1016/j.bmc.2015.05.029. [DOI] [PubMed] [Google Scholar]
- 37.Kabsch W. XDS. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murshudov G.N., Skubak P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R., Winn M.D., Long F., Vagin A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hough M.A., Wilson K. From crystal to structure with CCP4. Acta Crystallogr. Sect. D Struct. Biol. 2018;74:67. doi: 10.1107/S2059798317017557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lamzin V.S., Perrakis A., Wilson K.S. The ARP/wARP suite for automated construction and refinement of protein models. In: Rossmann M.G., Arnold E., editors. International Tables for Crystallography: Volume F. Crystallography of Biological Macromolecules, Kluwer Academic Publishers; Dordrecht, The Netherlands: 2001. pp. 720–722. [Google Scholar]
- 42.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 43.Khalifah R.G. The carbon dioxide hydration activity of carbonic anhydrase. I. Stop-flow kinetic studies on the native human isoenzymes B and C. J. Biol. Chem. 1971;246:2561. [PubMed] [Google Scholar]