SUMMARY
Most patients diagnosed with resected pancreatic adenocarcinoma (PDAC) survive less than 5-years, but a minor subset survives longer. Here, we dissect the role of the tumor microbiota and the immune system in influencing long-term survival. Using 16S rRNA gene sequencing, we analyzed the tumor microbiome composition in PDAC patients with short and long-term survival (STS, LTS). We found higher alpha-diversity in the tumor microbiome of LTS patients and identified an intra-tumoral microbiome signature (Pseudoxanthomonas/Streptomyces/Saccharopolyspora/Bacillus clausii) highly predictive of long term survivorship in both discovery and validation cohorts. Through human-into-mice Fecal Microbiota Transplantation (FMT) experiments from STS, LTS or control donors, we were able to differentially modulate the tumor microbiome and affect tumor growth as well as tumor immune infiltration. Our study demonstrates that PDAC microbiome composition, which cross-talks to the gut microbiome, influences the host immune response and natural history of the disease.
Graphical Abstract
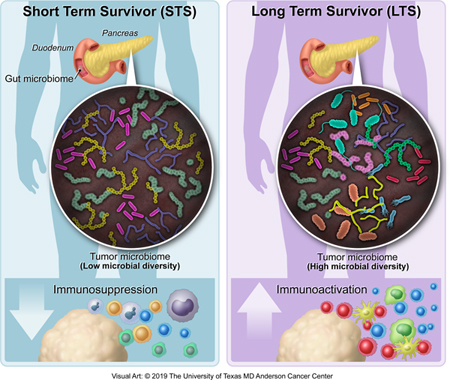
In Brief
The distinct tumor microbiome from pancreatic cancer long-term survivors can be used to predict PDAC survival in humans, and transfer of long-term survivor gut microbiomes can alter the tumor microbiome and tumor growth in mouse models.
INTRODUCTION
Pancreatic ductal adenocarcinoma (PDAC) is a disease of near uniform mortality (Hidalgo, 2010; Kamisawa et al., 2016; Miller et al., 2016). Most patients present with advanced stage disease and the prognosis is dismal, with a 5-year overall survival of 9% (Siegel et al., 2018). Even when patients can undergo surgical resection, the recurrence rate is very high and median overall survival varies between 24 to 30 months (Siegel et al., 2018). Despite this, a minor subset of patients survive more than 5-years post-surgery (Dal Molin et al., 2015; DeSantis et al., 2014). The factors that determine such enigmatic long-term survival are unknown. It is widely accepted that the cancer genomic landscape can be predictive of overall survival and response to therapy (Le et al., 2015; Ock et al., 2017; Vogelstein et al., 2013). However, analysis of stage-matched PDAC long term survivors has not demonstrated significant genomic differences compared to those patients with shorter survival (Dal Molin et al., 2015; Makohon-Moore et al., 2017). Recently, Balachandran et al. have used an in silico prediction approach to demonstrate that PDAC tumors from long term survivors have high quantity and quality of neoantigens, and stronger infiltration and activation of CD8+ T cells. Interestingly, the neoantigens exhibited homology to infectious disease-derived peptides, suggesting a neoantigen molecular mimicry with microbial epitopes (Balachandran et al., 2017). These data suggest that microbial host factors, independent of the genomic composition of the tumor, may determine tumor behavior and patient outcomes.
The key role of the gut microbiota as a host factor mediating tumor responses to chemotherapy and immunotherapy in patients with melanoma and lung cancers has been recently highlighted (Gopalakrishnan et al., 2018; Matson et al., 2018; Routy et al., 2018). These studies suggest that gut bacteria influences the activation of the immune system, promoting cancer-associated inflammation and ultimately affecting tumor responses to therapies. This information has allowed the stratification of patients into responders and non-responders using the microbiota composition as a predictive biomarker of response to immunotherapy.
Recently, Geller et al. reported the presence of Gammaproteobacteria (GP), besides other bacteria, in human PDAC. Importantly, they reported that GP is able to metabolize gemcitabine (2′,2′-difluorodeoxycytidine) into its inactive form (2′,2′-difluorodeoxyuridine), suggesting that the presence of this bacteria in PDAC may be responsible for the tumor resistance to gemcitabine (Geller et al., 2017). Additionally, Pushalkar et al. detected specific gut and tumor microbiome in murine models of PDAC, suggesting potential bacterial translocation from the intestinal tract into the peritumoral milieu (Pushalkar et al., 2018). Bacterial ablation with antibiotics in a PDAC orthotopic mouse model reshapes the tumor microenvironment, inducing T-cell activation, improving immune surveillance, and increasing sensitivity to immunotherapy. Together, these data suggest that modulating the gut and/or tumor microbiome could emerge as a novel strategy to sensitize tumors to therapeutics.
Despite all this knowledge, the composition of the human PDAC microbiome that contributes favorably or adversely to the natural history of pancreatic cancer remains incompletely studied. This represents a significant unmet need, as most chemotherapeutic and immunotherapeutic agents that have proven efficacy in other malignancies have limited efficacy in PDAC (Garrido-Laguna and Hidalgo, 2015; Manji et al., 2017). To help address this, we designed a study focused on dissecting the tumor microbiota of independent cohorts of PDAC patients at two geographically disparate tertiary care institutions (studying long-term versus short-term survivors) to gain insights on the host-related influences that might guide this unusual long-term survival. Furthermore, we have performed human fecal microbial transplants from patients with PDAC, survivors of PDAC and healthy controls to assess the capacity of the gut microbiome to shape the tumor microbiome, modulate the immune system and, ultimately, affect tumor growth.
RESULTS.
Tumor microbial diversity is associated with better outcomes in resected PDAC patients
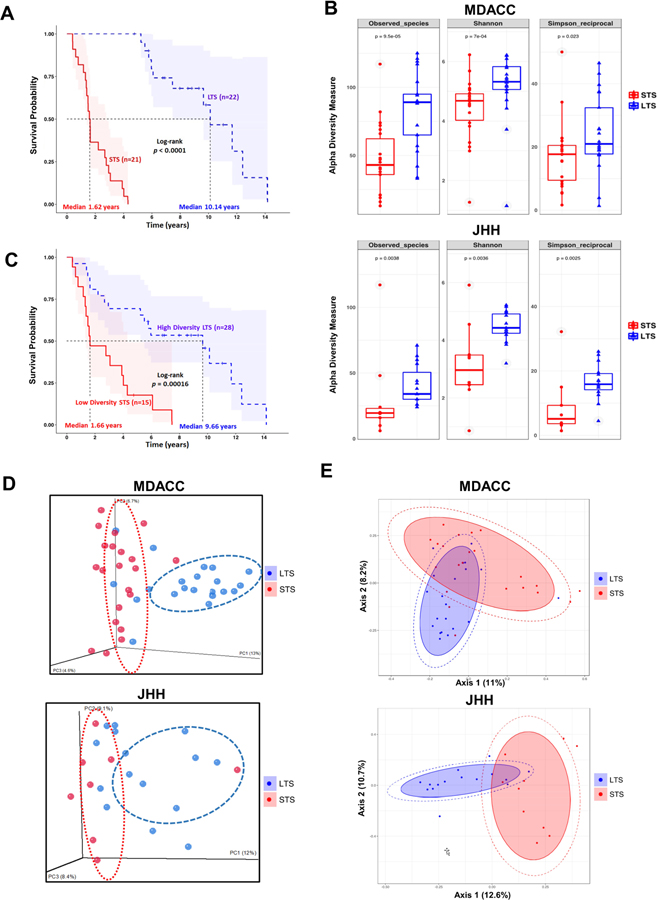
To explore the role of the human tumor microbiome composition in mediating clinical outcomes of PDAC patients, we used a discovery cohort to compare surgically resected patients who survived more than 5 years post-surgery, or long-term survivors (LTS, median survival 10.1 years), to stage-matched short-term survivors who survived less than 5 years post-surgery (STS, median survival 1.6 years) from UT MD Anderson Cancer Center (MDACC) in Houston, Texas (Figure 1A and Table 1). Patients in LTS and STS groups were matched with respect to age, gender, stage and prior therapies, including antibiotics use, neadjuvant or adjuvant treatments (Table 1). We then used a validation cohort with similar survival characteristics from Johns Hopkins Hospital (JHH) in Baltimore, Maryland. Bacterial DNA was extracted from 68 surgically resected PDAC tumor (36 LTS and 32 STS) and taxonomic profiling via 16S rRNA gene sequencing was performed. We first measured the tumor microbial diversity using different methodologies (Observed Taxonomic Units, Shannon and Simpson Indices) and found that alpha-diversity of the tumor microbiome, defined as the number of species present within each tumor sample (Kurilshikov et al., 2017), was significantly higher in the LTS patients compared to STS on both the MDACC discovery cohort (p < 0.0005, p < 0.0005 and p < 0.05, for each alpha-diversity indice, respectively) and the JHH validation cohort (p < 0.005, p < 0.005 and p < 0.005, for each alpha-diversity indice, respectively) (Figure 1B). Based on these results, we then tested the relationship berween PDAC tumor microbial diversity and overall survival (OS) in the MDACC cohort by stratifying the patients in two groups based on median diversity obtained by Shannon index. As expected, we found that patients with high alpha diversity had significantly prolonged overall survival (median survival: 9.66 years) than those with low alpha diversity (median survival: 1.66 years) using univariate Cox proportional hazard models (Figure 1C). The relationship between tumoral microbial diversity and survival actually allowed for the stratification and redistribution of PDAC patients according to alpha diversity value (high or low) (Figure 1C). Importantly, we assessed for potential contributors to microbial diversity, including clinico-pathological features, body mass index, sex, smoking, neoadjuvant/adjuvant therapies as well as antibiotics use, and were not able to find any significant association (Figure S1). Recent studies have proposed that a high microbial diversity in the gut microbiome is associated with favorable outcomes to treatment (Gopalakrishnan et al., 2018). On the contrary, an imbalance in the gut microbiome or dysbiosis, is associated with poor responses to these therapies, and associated with chronic diseases and cancer development (Ferretti et al., 2017; Human Microbiome Project, 2012b; Kundu et al., 2017; Lloyd-Price et al., 2017; Shoemark and Allen, 2015). Our findings indicate that the tumor alpha diversity could serve as a predictor of survival outcome in resected PDAC patients, suggesting the potential relevance of the microbiome composition in mediating pancreatic cancer progression.
Figure 1.
Tumor microbial diversity influences the outcome of PDAC patients.
(A) Kaplan-Meier plot of MDACC cohort PDAC patients.
(B) Alpha diversity box plot (Observed species, Shannon and Simpson reciprocal) in MDACC and JHH cohorts of PDAC patients.
(C) Kaplan-Meier plot of MDACC cohort PDAC patients defined by alpha diversity.
(D) Principal coordinate analysis (PCoA) using Unweighted-UniFrad of beta diversity.
(E) Principal coordinate analysis (PCoA) using Bray-Curtis metric distances of beta diversity.
Table 1.
Clinico-pathological Characteristics of MDACC Cohort PDAC Patients
| Patients Characteristics | STS (n=22) |
LTS (n=21) |
P value |
|---|---|---|---|
| Overall survival (yrs) | 1.62 | 10.14 | <0.0001 |
| Surgery Date | 1999–2014 | 2000–2010 | |
| Gender | |||
| Female | 9 | 11 | 0.45 |
| Male | 13 | 10 | |
| Age (yrs) | |||
| Median | 62.05 | 62.71 | 0.69 |
| Range | 46–74 | 44–73 | |
| Race | |||
| Caucasian | 19 | 20 | 0.48 |
| Asian | 0 | 1 | |
| African American | 1 | 0 | |
| Hispanic | 2 | 0 | |
| Stage | |||
| IB | 2 | 1 | 0.56 |
| IIA | 7 | 10 | |
| IIB | 13 | 10 | |
| Neoadjuvant Therapy (Chemo-radiation) | |||
| Yes | 14 | 17 | 0.31 |
| No | 8 | 4 | |
| Adjuvant Therapy | |||
| Yes | 14 | 11 | 0.45 |
| No | 8 | 10 | |
| Antibiotics Use (pre-surgery) | |||
| Yes | 14 | 11 | 0.45 |
| No | 8 | 10 | |
| Biliary Obstruction | |||
| Yes | 12 | 10 | 0.64 |
| No | 10 | 11 |
To extend our understanding of the role of microbiome diversity and its association with survival, we aimed to detect whether phylogenetic relationships exist between the bacterial communities enriched in the PDAC milieu of STS and LTS. We used beta-diversity to generate a principal coordinate analysis (PCoA) using Unweighted-UniFrac distances (Lozupone et al., 2011) and using Bray-Curtis metric distances (McMurdie and Holmes, 2013). A clear clustering between operational taxonomic units (OTUs) from LTS and STS was revealed by both methods in both independent cohorts (Figure 1D and 1E), suggesting that the tumor microbial communities exhibit phylogenetic closeness within each group (p < 0.05).
Tumor microbiome communities are significantly different between LTS and STS
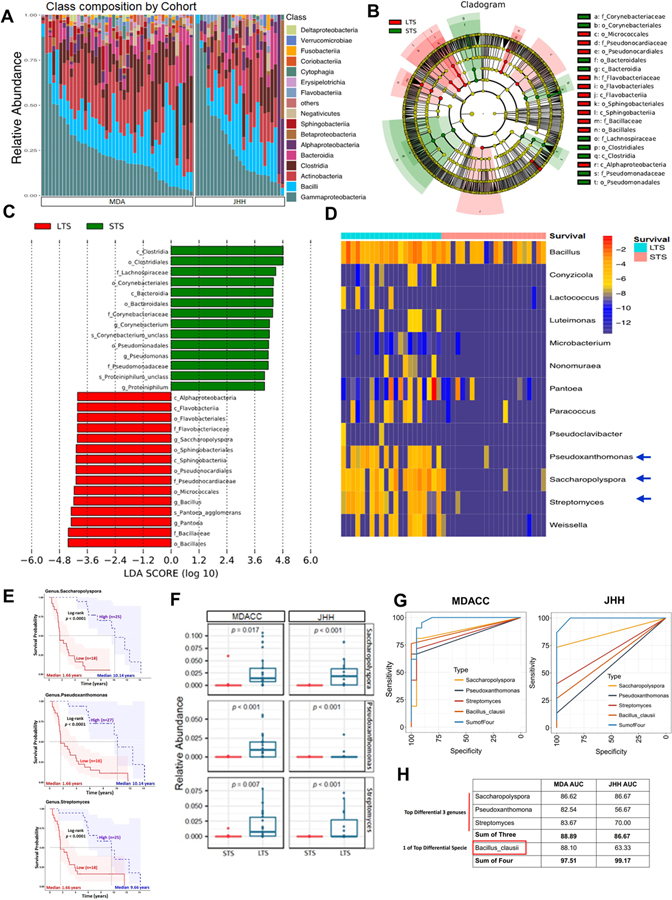
Considering the relationship between PDAC intra-tumoral bacterial diversity and overall survival, we next sought to determine if there were differences in the tumor microbiome composition between PDAC LTS and STS. To this end, we first assessed the general landscape of the tumor microbiome on all patients from both cohorts which revealed presence of similar communities (Figure 2A). We then compared enrichment of OTUs in LTS versus STS, which revealed enrichment for specific bacterial communities in each group at the various taxonomic level (Figure S2A–S2E). When we evaluated for the influence of neadjuvant/adjuvant therapies and antibiotics use in MDACC cohort, we did not detect significant differences in the taxonomic composition at the order level (Figure S2F–S2H). To further investigate these findings, we focused on our discovery cohort and conducted high dimensional class comparisons using linear discriminant analysis of effect size (LEfSe) (Segata et al., 2011) which detected marked differences in the predominance of bacterial communities between LTS and STS (Figure 2B and 2C): The LTS tumors exhibited a predominance of Alphaprotebacteria Sphingobacteria, and Flavobacteria at the Class level. In contrast, the PDAC STS cases were dominated by Clostridia and Bacteroidea at the Class level (Figure 2B and 2C). We then interrogated if the tumor microbiome can be segregated by comparison heatmap based in the OTU abundance at the genus level using patients survival as a variable (Figure 2D). The genus features were selected using logistic regression combined with LASSO (See STAR methods). We visualized the differential taxonomic communities differential segregation according to the survival of the PDAC patients. The LTS patients showed an enrichment on Proteobacteria (Pseudoxanthomonas) and Actinobacteria (Saccharopolyspora and Streptomyces), while no predominant genus was detected in the STS tumors. We then stratified patients into high versus low categories based on their median relative abundance of these three taxa (Pseudoxanthomonas, Saccharopolyspora and Streptomyces). Significantly better outcomes were predicted for PDAC patients with higher abundance of Saccharopolyspora (HR= 13.47, 95% CI 4.672–38.83), Pseudoxanthomonas (HR=5.885, 95% CI 2.37–14.61) and Streptomyces (HR= 4.572, 95% CI 2.033–10.28) (Figure 2E and Table S1A). Considering that all above comparisons were done in the discovery cohort, we then proceeded to validate the relative abundance of top 3 hits in the JHH cohort and found that Pseudoxanthomona, Saccharopolsypora and Streptomyces were also significantly more abundant in LTS vs STS from this cohort (Figure 2F). We then used these three genus with higher abundance in LTS to run Area Under Curve (AUC)-Receiver Operator Characteristic (ROC) analysis. We found that the combination of these Top 3 taxa (Pseudoxanthomonas, Saccharopolyspora and Streptomyces) resulted in an AUC of 88.89 in the discovery cohort and 86.67 in the validation cohort (Figure 2G).
Figure 2.
Tumor microbiome communities are significantly different between LTS and STS.
(A) Bar plots of the class taxonomic levels in MDA and JHH cohorts of PDAC patients. Relative abundance is plotted for each tumor.
(B) Taxonomic Cladogram from LEfSe, depicting taxonomic association from between microbiome communities from LTS and STS PDAC patients. Each node represents a specific taxonomic type. Yellow nodes denote the taxonomic features that are not significantly differentiated between LTS and STS. Red nodes denote the taxonomic types with more abundance in LTS than in STS, while the green nodes represent the taxonomic types more abundant in STS.
(C) LDA score computed from features differentially abundant between LTS and STS. The criteria for feature selection is Log LDA Score > 4.
(D) Heatmap of selected most differentially abundant features at the genus level. Highlighting three taxa enriched in LTS. The blue color represents less abundant, lighter yellow color represents intermediate abundance and red represents the most abundant. Highlighting three taxa enriched in LTS.
(E) Kaplan-Meier estimates for survival probability based on the abundance levels of microbes enriched at Genus level in LTS. Right plot, Saccharopolyspora, middle plot, Pseudoxanthomonas and left plot, Streptomyces (p < 0.0001).
(F) Plots of differentially abundant genus significantly enriched in both MDA and JHH LTS patients. FDR adjusted p-values from negative binomial test p-value.
(G) ROC analysis of Taxa relative abundance as predictive of LTS status. The top 3 differential bacteria (Genus) identified and Baccilus Clausii (One of top species) were tested individually and in aggregate in the MDA Discovery Cohort (Left panel) were then validated in the JHH Validation Cohort (Right panel).
(H) Table depicting AUC of bacteria tested in Fig2G for both MDA and JHH cohorts.
We then attempted to assess for differences in mean relative abundance of species between LTS and STS that could potentially increase predictive value of long-term survivorship and found two species significantly enriched in LTS vs STS in the discovery cohort: Bacillus clausii (1.78% vs 0%, FDR adj P-value: 0.001) and Saccharopolyspora rectivirgula (1.91% vs 0.26%, FDR adj P-value: 0.001) (Table S1B). Since Bacillus clausii belongs to a genus different from the three described, we tested its addition to the signature to predict LTS. While the individual AUC were 88.10 and 63.3 in the discovery cohort and validation cohorts, respectively, when we added Bacilus Clausii to the three genus signature, the AUC in the discovery cohort increased to 97.51 and in validation cohort to 99.17 (Figure 2G and 2H). These data suggest that the presence and abundance of these three taxa communities Saccharopolyspora, Pseudoxanthomonas and Streptomyces, added with the presence of Bacillus Clausii, could influence and predict long-term survivorship in PDAC patients.
To definitively confirm the presence of intratumoral bacteria in PDAC cases, we conducted several additional experiments. We first performed ribosomal RNA (rRNA) fluorescence in situ hybridization (FISH) in a subset of archival FFPE PDAC samples, using a specific probe that targets bacterial 16S rRNA. The FISH analysis of bacterial 16S rRNA confirmed the presence of bacterial DNA in all PDAC samples analyzed (Figure S3A). Additionally, in the same set of FFPE samples, we detected the presence of bacterial lipopolysaccharide (LPS) by immunohistochemistry using an antibacterial lipopolysaccharide antibody, as previously performed (Figure S3B) (Geller et al., 2017). Consistently, this approach confirmed the presence of intratumoral bacterial in all PDAC samples evaluated (Figure S3B). Furthermore, to verify the presence of bacteria in frozen samples, we analyzed 9 frozen PDAC tissue samples matched to FFPE specimens that were previously analyzed. The 16S rDNA PCR demonstrated the presence of bacterial DNA in the 9 PDAC frozen samples analyzed (Figure S3C). Additionally, we sought for the presence of Saccharopolyspora rectivirgula, one of the top species enriched in LTS vs STS, using primers designed for this species and isolated bacteria as positive control and we detected positive bands on 6/6 LTS PDAC frozen samples analyzed (Figure S3D).
Additionally, to confirm whether the overall tumor bacterial composition was similar between frozen and FFPE samples, and to exclude the possibility that most of bacteria previously found was an artifact of FFPE fixation, we performed 16S rRNA gene sequencing of frozen tissue and compared it to the corresponding FFPE samples (Figure S3E). The data showed similar taxonomic composition between FFPE and frozen PDAC samples, and notably, no statistically significant differences were found between their taxonomic compositions of both types of samples (Figure S3E and Table S2). Additionally, we used frozen PDAC samples to obtain and isolate bacteria from the tissue (Figure S3F). These colonies were selected and DNA was extracted for subsequent PCR amplification of the 16S rDNA (Figure S3G).
Overall, the results obtained from all of these approaches confirmed the presence of bacteria in the PDAC samples including some of the species enriched in LTS.
The tumor microbiome shapes immune responses promoting T cell activation
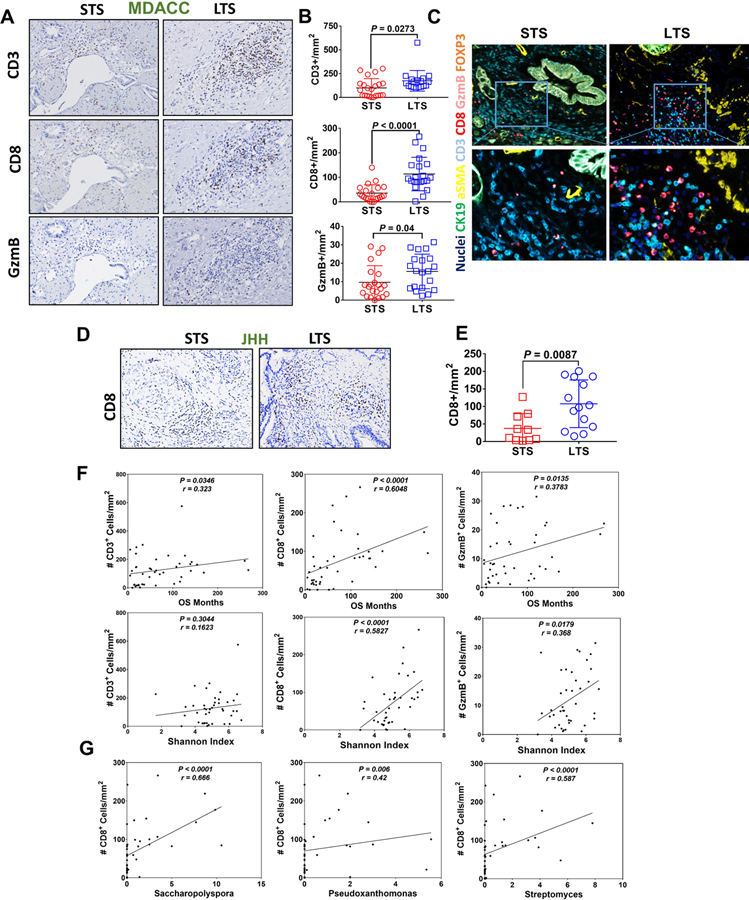
The gut microbiota plays a pivotal role in shaping the immune system (Atarashi et al., 2013; Mazmanian et al., 2008; McAllister et al., 2014). Recent studies have described that the gut microbiota composition can improve responses to immunotherapy by modulating the immune system (Gopalakrishnan et al., 2018; Matson et al., 2018; Riquelme et al., 2018; Routy et al., 2018; Vetizou et al., 2015). We hypothesized that tumoral bacteria has the ability to shape the immune tumor microenvironment which can influence the natural history of the cancer. We used singleplex immuhistochemistry, as well as multiplex immunofluorescence staining to delineate the tumor immune infiltrates in the discovery cohort (MDACC). We found greater densities of CD3+ and CD8+ T cells in the LTS compared with the STS patients (p = 0.0273 and p < 0.0001, respectively) (Figure 3A–3C). We also detected significantly higher number of Granzyme B+ (GzmB) cells in the LTS (Figure 3A and 3B, lower panel, p = 0.04), while no significant differences were detected in regulatory T cells (CD3+ FOXP3+), macrophages (CD68), or MDSC (CD66b) (data not shown). Consistently, we found greater densities of CD8+ T cells in the LTS compared with the STS patients (p = 0.008) in the validation cohort (Figure 3D and 3E). The Spearman rank-order correlation demonstrated a significant positive correlation between CD3+, CD8+ and GzmB+ tissue densities and the overall survival of PDAC patients (p = 0.03, p < 0.001 and p = 0.01, respectively) (Figure 3F). Interestingly, we found a strong significant correlation between CD8+ and GzmB+ tissue densities with microbiome diversity (Figure 3F, lower panel), suggesting that the tumor microbiome diversity may influence both the extent of immune infiltration and the degree of activation of CD8+ T cells. Finally, when we correlated the CD8+ T cell tissue densities with the top-three enriched genus in LTS patients, Saccharopolyspora, Pseudoxanthomonas and Streptomyces, we found a positive Spearman correlation between the two variables (p < 0.0001, p = 0.006 and p < 0.0001, respectivelly) (Figure 3G). These findings suggest that the tumor microbiome diversity and the presence of these three genus in the tumor may contribute to the anti-tumor immune response by favoring recruitment and activation of CD8+ T cells.
Figure. 3.
Commensal microbiome from LTS PDAC patients induces a strong immune infiltration and antitumoral immune response.
(A) Immunohistochemical (IHC) staining of CD3, CD8 and Granzyme B from tumors of STS and LTS PDAC patients (representative picture).
(B) Quantification of IHC of CD3, CD8 and Granzyme B on STS and LTS PDAC patients.
(C) Representive pictures of multiplex immunofluorescence staining (Multiplex IF) with Opal kit.
(D) Immunohistochemical staining of CD8 from tumors of STS and LTS PDAC patients from validation cohorts (JHH) (representative picture).
(E) Quantification of IHC of CD8 on STS and LTS PDAC patients from validation cohorts (JHH).
(F) Spearman correlation between CD3+, CD8+ and GzmB+ tissue densities and the overall survival (upper panel) and alpha diversity by Shannon Index (lower panel) of all PDAC patients.
(G) Spearman correlation between CD8+ tissue densities and Saccharopolyspora, Pseudoxanthomonas and Streptomyces (p < 0.0001, p = 0.006 and p < 0.0001, respectivelly) in PDAC patients.
Microbiome communities from LTS and STS are associated with different metabolic pathways.
It has been demonstrated that microbiota imbalances can induce systemic metabolic alterations (Devaraj et al., 2013; Nieuwdorp et al., 2014). Conversely, metabolic dysfunction can also induce microbiome imbalances (Cani, 2017). Based on this data, we next assessed if the intra-tumoral microbiome is associated with host metabolic pathways. We used Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) (Langille et al., 2013), a technique which uses evolutionary modeling, to predict metagenomes from 16S data and reference genome databases (Kanehisa and Goto, 2000). Predicted metagenomes were then used as inputs for metabolic reconstruction, using level 2 KEGG Pathways and/or KEGG modules between LTS and STS groups (MDACC cohort; FDR adjusted p < 0.05), were mean-centered and visualized as a heatmap by Linear Discriminant Analysis (LDA) to assess enrichment and depletion between the two groups. PICRUSt analysis identified 26 core functional modules present across all PDAC samples with a coverage of >90% and p < 0.05. We detected enrichment of differential pathways between LTS and STS groups. Predicted functional categories involved in important cellular functions were associated with diverse metabolic and energetic processes (Figure S4A and S4B). The LTS cases exhibited enrichment in the pathways related to metabolism of amino acids, xenobiotics, lipids, terpenoids and polyketides, besides other cellular functions. Significantly better outcomes were predicted for PDAC patients demonstrating enrichment in xenobiotics biodegradation and lipids metabolism pathways (HR=5.198, 95% CI 1.07–25.06 and HR=4.528, 95% CI 1.54–13.305, respectivelly) (Figure S4C). In contrast, the STS cases demonstrated enrichment in synthesis and processing of proteins, processing of genetic information, energetic and nucleotide metabolism, replication and repair. Some of the pathways shared by both groups, and probably not directly related to the differential outcomes of patients, were related to enzymes, cancer, excretory and circulatory system. The PICRUSt taxonomic functional relationships suggest that the composition of the intra-tumoral microbiome determines a differential enrichment of metabolic functional pathways between LTS and STS cases, which may influence patient survival.
Gut microbiota can influence tumor microbiota and tumor growth
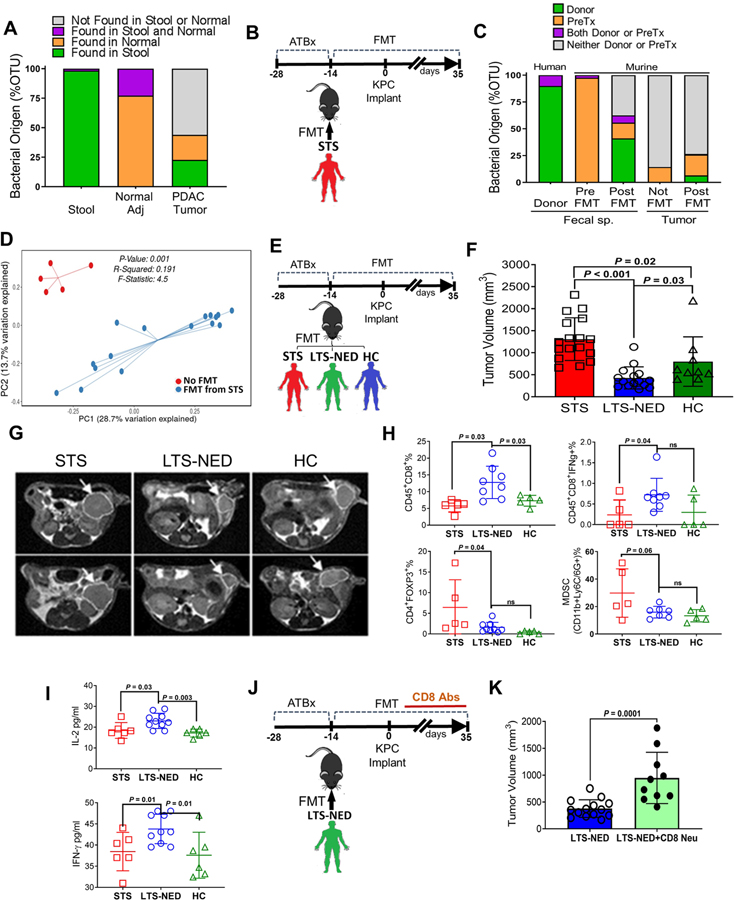
We asked if the gut microbiome can modulate the intratumoral microbiome and to this end we collected stools, PDAC tumor specimens and non-tumoral adjacent normal tissue from 3 patients who underwent Whipple surgery. We compared the taxonomic composition of matched samples and found that the human gut microbiome represents approximately 25% of the human tumor microbiome, while it is absent from the normal adjacent tissue (Figure 4A). Since the bacterial composition found in normal adjacent tissue differs from that found in the tumor, it suggests that the tumor microbiome might be unique. This data suggests that the gut microbiota has the capacity to specifically colonize pancreatic tumors. To determine if we can actively modify the tumor microbiome by changing the gut microbiome, we performed fecal microbial transplantation (FMT) from patients with advanced PDAC (“STS”) into mice previously treated with antibiotics (ATBx). Human fecal material was transferred by oral gavage three times a week and weekly after two weeks. Mice were then challenged with orthotopic implantation of syngeneic cancer lines derived from genetically engineered Pdx1-Cre, LSL-KrasG12D/+, LSL-Trp53R172H/+ (“KPC”) mice. We examined the OTUs abundance in the human STS donor samples, murine fecal samples pre-FMT (Basal condition), post-FMT, and murine tumors at the end point of 5 weeks post-tumor implantation. Interrogation of bacterial origin showed that a large number of bacteria of human donor origin was found as part of the murine gut microbiome (~40%) of recipient mice post-FMT (Figure 4B and 4C). Interestingly, we were able to detect human donor bacteria in the murine tumor microbiome post-FMT while it remained absent from mice who did not get FMT (Figure 4B and 4C). However, the bacteria coming from donors represented a small percent of the tumor microbiome (<5%), while the remaining 20% represented the basal murine gut microbiome. Since over 70% of the tumor microbiome was not representative of the gut microbiome (Figure 4C, grey bars), we then wondered if FMT could modulate or shift the overall intratumoral bacterial composition, in addition to direct translocation. We used beta-diversity to generate a principal coordinate analysis (PCoA) and a clear differential clustering was detected between operational taxonomic units (OTUs) on tumors from mice who received FMT versus mice that did not receive FMT (p < 0.001) (Figure 4D). Additionally, we looked at the taxonomic composition of tumors and found significant changes in individual bacterial populations after FMT (Figure S5A). Interestingly, one of the bacterial classes that increased in tumors from mice who received FMT from STS donors was Clostridiales, which was enriched in the original human STS tumor specimens. This data suggests that the gut microbiome can modulate the tumor microbiome, in minor part by direct translocation into the tumors, but more significantly, by altering the microbial landscape.
Figure. 4.
Gut microbiota from PDAC patients can influence tumor microbiota and tumor growth
(A) Taxonomic classification of bacterial 16S sequence detected in human samples by origin
(B) Experimental design of Fecal Microbiota Transplantation (FMT) from metastatic PDAC donors in C57BL/6 wild-type mice treated with antibiotics (ATBx).
(C) Taxonomic classification of bacterial 16S sequence detected in human donor stools and FMT recipient mice (stools/tumors) by origin.
(D) Principal coordinate analysis (PCA) using Unweighted-UniFraC of beta diversity, showing closeness between mice that received FMT from PDAC STS patients and distance from those that did not receive FMT.
(E) Experimental design of Fecal Microbiota Transplantation (FMT) from advanced PDAC (STS), PDAC LTS with no evidence of disease (LTS-NED) and Healthy Control (HC) donors in C57BL/6 wild-type mice treated with antibiotics (ATBx).
(F) Tumor volume from mice orthotopically implanted with KPC pancreatic cancer cell lines along with transplantation with stools from STS, LTS-NED and HC donors.
(G) Magnetic resonance imaging (MRI) scans of KPC-implanted mice transplanted with stools from STS, LTS-NED and HC donors (representative images).
(H) Flow cytometry analysis of CD45+CD8+, CD45+CD8+IFN-γ+, Treg (CD45+CD4+FOXP3+) and MDSC (CD45+CD11b+Ly-6G/Ly-6C+) cells from KPC-implanted mice and transplanted with stools from STS, LTS-NED and HC donors.
(I) Serum level of IL-2 and IFN-γ in KPC-implanted mice and transplanted with stools from STS, LTS-NED and HC donors.
(J) Experimental design of Fecal Microbiota Transplantation (FMT) from LTS-NED who received CD8 neutralizing antibodies vs isotype control.
(K) Tumor volume from KPC-implanted mice and transplanted with stools from LTS-NED who received CD8 neutralizing antibodies vs isotype control.
In order to expand our findings to assess how tumor growth may get affected by differential modulation of the tumor microbiome in response to recipient fecal transplants, we obtained stool specimens from the following 3 groups of patients: PDAC patients with advanced disease who would likely experience short term surivaval (“STS”), patients who had PDAC resected more than 5 years previous to collection who would be classified as LTS with no evidence of disease (“LTS-NED”), and healthy controls (“HC”). We employed a similar FMT design as described above (Figure 4E). Five weeks after tumor implantation we assessed gut/tumor microbiome and tumor growth. Gut microbial beta-diversity distinguished the three types of recipient mice (Figure S5B) and tumor microbial beta-diversity differentially clustered mice groups according to the type of FMT they received (Figure S5C–S5G).
We observed a significant reduction in tumor growth in mice that received FMT from LTS-NED donors compared with the mice transplanted with stools from STS donors (p < 0.001) or HC donors (p = 0.02) (Figure 4F and 4G). These results suggest that gut/tumor bacteria from patients who had PDAC and survived long term may have a protective effect against tumors. We also found that tumors from mice who got FMT from STS patients were larger than those from mice who got HC FMT, suggesting that PDAC-associated gut/tumor bacteria may exert a tumor-promoting effect.
To confirm that the anti-tumoral effect exerted by LTS-NED was definitively induced by change in bacterial content, we treated mice transplanted with stools from LTS-NED with antibiotics post-FMT and compared with mice that didn’t receive antibiotics post-FMT (Figure S5H). We found that short term antibiotics on mice that received FMT from LTS-NED donors induced larger tumors than untreated mice by modifying the gut microbiome (Figure S5I–S5J). When we analyzed the tumor microbiome of the two groups, we were indeed able to see differential clustering for beta-diversity between the two groups (Figure S5K). This data indicates that bacteria ablation can decrease the anti-tumoral efficacy induced by LTS-NED FMT, which validates the central role of bacteria.
Next we assessed if the gut microbiome can influence the pancreatic tumor immune infiltrates. Flow cytometry analysis demonstrated that tumors from mice that received FMT from LTS-NED had significantly higher numbers of CD8+ T cells, as well as activated T cells (CD8+/IFNg+ T cells) versus those with stools transferred from STS or HC donors, whereas those who received STS FMT had increased CD4+FOXP3+ and myeloid-derived suppressor cells (MDSC) infiltration (Figure 4H). Additionally, mice receiving FMT from LTS-NED patients had higher serum level of IFN-γ and IL-2 (p < 0.05) compared to mice receiving STS FMT (Figure 4I). To evaluate the role of T cells in mediating the observed phenotype, we depleted CD8+ T cells using neutralizing antibodies in mice transplanted with LTS-NED stools and subsequently challenged with orthotopic tumors (Figure 4J). We found that CD8+ T cells depletion blocked the anti-tumoral effect induced by LTS-NED FMT (Figure 4K), suggesting that the beneficial effect of LTS-NED-associated gut/tumor bacteria is mediated by CD8+T cells. Together, these data strongly suggest that the gut microbiome can colonize pancreatic tumors, modify its overall tumoral bacterial composition, and modulate immune function to ultimately affect the natural history and survival.
DISCUSSION
The microbiota can exert regulatory effects in other sites beyond the gut. Our study represents the first report to explore the influence of the tumor microbiome on clinical outcomes. We have performed a comprehensive analysis of the PDAC intratumoral microbiome in two independent cohorts of long and short term survival patients from different institutions. It is important to note that one of these cohorts (JHH) had already been examined for genome wide differences in the mutational landscape that could be contributing to favorable survival and none were identified (Dal Molin et al., 2015). Overall, we detected substantial abundance of microbiome in PDAC tumors from all patients, as previously reported (Geller et al., 2017). We found that PDAC patients with the uncommon phenotype of LTS had significantly higher tumor bacteria diversity that the patients with more typical shorter survival. Further, the LTS and STS cohorts each had a distinctive tumor microbiome signature with specific bacterial genus that were predictive of survival in a multi-variate analysis. Notably, we demonstrated for the first time that the microbiota reconstitution by FMT with stool from HC, STS or LTS-NED patients in tumor-bearing mice mirrors the recruitment, or lack thereof, of immune cells to the tumor milieu seen in the respective cohorts and influences tumor growth, supporting a causal role for the gut microbiome in shaping tumor immune-responses and PDAC progression.
Recent studies have shown that the gut microbiome composition can improve the outcome of cancer immunotherapy (anti-CTLA4 and anti-PD-L1) by influencing the immune system (Gopalakrishnan et al., 2018; Matson et al., 2018; Routy et al., 2018). Our finding suggests that, independent of therapy, the PDAC tumor microbiome diversity and composition can influence immune infiltration which ultimately influences PDAC survival. Importantly, we found a signature of 3 tumor bacterial taxa: Sachharopolyspora, Pseudoxanthomonas and Streptomyces significantly enriched in LTS patients. Presence of Bacillus clausii, one of top species enriched in LTS, combined with the three genus signature, were highly predictive of long term survivorship in the MDA discovery cohort and validated in the JHH cohort. In the future, tumor microbiome sequencing could be used to stratify patients for adjuvant trials, including microbiome interventions.
Although the greater microbial diversity could have an immunoregulatory impact, its role in the antitumor response is not entirely clear. Components of the Saccharopolyspora family, specifically Saccharopolyspora rectivirgula, have been described as having a role in inflammatory lung diseases, such as hypersensitivity pneumonitis that are associated with IFN-γ overproduction (Kim et al., 2010). The presence of Saccharopolyspora spp could contribute towards generating a proinflammatory microenvironment mediated by cytokines and chemokines that recruit inflammatory cells and IFN-γ secretion. However, its role in PDAC remains to be explored. Future investigations will determine if similar mechanism can be used by the tumor microbiota to modulate the immune system by improving or impairing the immune response against the tumor.
Most of the bacterial communities found in the tumoral milieu are present commonly in the gut microbiome (Human Microbiome Project, 2012b; Lloyd-Price et al., 2017), suggesting that potentially bacterial translocation from the gut to the pancreas might be occuring. We demostrate for the first time in human PDAC patients that the gut microbiota has the capacity to colonize pancreatic tumors and that this colonization can modify the overall microbiome of the tumor. Additionally, our preclinical data shows that the modification of the gut/tumor microbiome with human flora from LTS-NED patients is able to induce an antitumor response and activation of the immune system in tumor-bearing mice, which is not observed in FMT from STS PDAC patients. Furthermore, we have found that immunosuppressive populations were also modulated with stools from survivors or healthy patients by decreasing tumor infiltration by Tregs. This could be a potential mechanism for certain bacteria to promote immune activation. Microbiome-dependent CD8 T cell activation may play a key role. Further studies are needed with in human PDAC patients to determine how FMT can induce changes in the microbiome tumor and inmune activation. Additionally, the potential evaluation of cross-reactivity between T cells that recognize tumor neoantigens and microbial antigen (mimicry) present in the tumor should be conducted in future studies as they may be useful in understanding mechanisms by which bacteria can exert immuno-activating effects but also may be useful in the design on novel therapeutic strategies.
In conclusion, we found that the tumor microbiome diversity has a powerful impact in determining the survival of PDAC patients. The tumor microbiome unique to LTS may contribute towards shaping a favorable tumor microenvironment, characterized by the recruitment and activation of CD8 T cells to the tumor milieu, and it might also be useful as a predictor of patients outcomes. Besides the microbiome-based prognostic tool, the results of FMT represent an immense therapeutic opportunity to manipulate the microbiome to improve the life expectancy of PDAC patients in whom few therapeutic options exist.
STAR METHODS
CONTACT FOR REAGENTS AND RESOURCE SHARING
Further information and request for resources and reagents should be directed to and will be fulfilled by the contact, Florencia McAllister (fmcallister@mdanderson.org).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human Tumor Specimens.
We initially used a discovery cohort of patients with long-term survivors PDAC (>5-years overall survival, median survival 10.14 years, called LTS, n=22), compared with stage-matched PDAC patients who survive less than 5 years (median survival 1.62 years, called STS, n=21) from the University of Texas MD Anderson Cancer Center (MDACC), Houston, TX. Detailed clinical and pathologic information on the patients are presented in Table 1. A second validation cohort with very long-term survivors of similar characteristics of survival (>10 years overall survival, n=15) were compared with stage-matched regular PDAC patients who survive less than 5 years (n=10), from Johns Hopkins Hospital (JHH), Baltimore, MD. Archived formalin-fixed paraffin-embedded (FFPE) tumor specimens obtained from these patients who underwent surgical resection with curative intent were collected from MDACC and JHH. The staging of disease was reviewed and updated to comply with the 7th Edition of the American Joint Committee on Cancer classification. The study protocols were approved by the Institutional Review Board at MDACC and JHH.
Human Stool Samples.
Stool collection from PDAC STS, PDAC LTS-NED patients and healthy control (HC) donors were collected on OMNIgene GUT kit. Fresh stools for fecal microbiota transplantation (FMT) studies were collected and frozen at −80°C prior to FMT.
Murine studies
Antibiotic treatments and Fecal microbiota transplantation (FMT)
All animal experiments were carried out in compliance and approved by the Animal Care and Use Committee at The UT MD Anderson Cancer Center. Female C57BL/6 aged 6 weeks were purchased from Taconic Biosciences, USA. Mice were treated for two weeks with an antibiotic solution (ATBx) containing streptomycin (5 mg/ml), and clindamycin (0.1 mg/ml) added to the sterile drinking water of mice ad libitum. Solutions and bottles were changed 2 times a week. After two weeks, ATBx treatment was stopped and the mice were recolonized by FMT, receiving stool from patients with PDAC STS, LTS-NED or HC donors. Mice received stools a total of 3 times/week by oral gavage using animal feeding needles before undergoing orthotopic tumor implantation and once/week after the tumor implantation until the end point. For CD8+ T cells depletion experiments, mice were treated for two weeks, biweekly with 150ug of antibodies against mouse CD8a (by intraperitoneal injections). For late bacterial ablation experiments, mice transplanted with stools from LTS-NED were treated with antibiotics post-FMT in the last two weeks of the experiment.
Orthotopic tumor implantation
C57BL/6 mice were anesthetized with 2% isoflurane in oxygen. A lateral incision was made on the abdominal wall of each mouse and tumors were implanted orthotopically in the pancreas with 2 × 104 KPC pancreatic adenocarcinoma cell lines. Tumor growth was monitored by Magnetic Resonance Imaging (MRI) of mice body at 4 weeks after the tumor implantation. At the endpoint, tumors, fecal and blood specimens were harvested and processed for further analysis.
METHOD DETAIL
DNA extraction and bacterial 16S rRNA sequencing
16S rRNA gene sequencing methods were adapted from the methods developed for the Earth Microbiome Project (X) and NIH-Human Microbiome Project (Human Microbiome Project, 2012b) (Caporaso et al., 2012; Human Microbiome Project, 2012a, b). Briefly, three sections of 10 um of FFPE of PDAC tissue were aseptically collected and placed in 1.5ml Eppendorf tube. Normal pancreatic tissue and paraffin without tissue were used as controls. Bacterial genomic DNA was extracted using Qiagen QIAamp DNA FFPE. The 16S rDNA V4 region was amplified by PCR and sequenced in the MiSeq platform (Illumina) using the 2x250 bp paired-end protocol yielding pair-end reads that overlap almost completely. The primers used for amplification contain adapters for MiSeq sequencing and single-index barcodes so that the PCR products may be pooled and sequenced directly (Caporaso et al., 2012), targeting at least 10,000 reads per sample. 16S (variable region 4 [v4]) rRNA gene pipeline data incorporated phylogenetic and alignment based approaches to maximize data resolution. The read pairs were demultiplexed based on unique molecular barcodes added via PCR during library generation, then merged using USEARCH v7.0.1090 (Edgar, 2010).
Microbiome Pipeline Analysis steps
Raw paired-end 16S rRNA reads (V4 region) were merged into consensus fragments by FLASH (Magoc and Salzberg, 2011) and subsequently filtered for quality (targeted error rate < 0.5%) and length (minimum 200bp) using Trimmomatic (Bolger et al., 2014) [and QIIME (Caporaso et al., 2010a; Kuczynski et al., 2011). Spurious hits to the PhiX control genome were identified using BLASTN and removed. Passing sequences were trimmed of primers, evaluated for chimeras with UCLUST (de novo mode) (Edgar et al., 2011), and screened for human-associated contaminants using Bowtie2 (Langmead and Salzberg, 2012). Chloroplast and mitochondrial contaminants were detected and filtered using the RDP classifier (Wang et al., 2007) with a confidence threshold of 50%. High-quality passing 16S rRNA sequences were assigned to a high-resolution taxonomic lineage using Resphera Insight (Daquigan et al., 2017; Drewes et al., 2017) and SILVA Database v128 (Quast et al., 2013). Bacterial contaminant removal was performed using four paraffin-only samples (no tissue) and additional literature. Contaminant list can be found in Table S2C. Resulting contaminant-free 16S rRNA profiles were subsampled to 2,000 sequences per sample for downstream comparative analysis. Alpha- and beta-diversity analysis and principal coordinates analysis utilized QIIME and R. Differential abundance analysis of alpha diversity features of interest evaluated differences using the nonparametric difference test. Differential abundance analysis of taxonomic abundances evaluated differences using the negative binomial test (DESeq) (Anders and Huber, 2010). The false discovery rate (FDR) was used to correct for multiple hypothesis testing (Benjamini et al., 2001). Generalized linear modeling adjusting for cohort membership and survival status was performed using R. LEfSe was used for linear discriminant analysis (Segata et al., 2011). High-quality non-contaminant 16S rRNA sequences were analyzed for functional gene content using PICRUSt (Langille et al., 2013) which provides proportional contributions of KEGG categories for each sample (Kanehisa and Goto, 2000). Differentially abundant functional categories (KEGG Level 2, FDR adj.P < 0.05 MDA cohort) were utilized for visualization as a heatmap. Relative abundance values were mean centered by functional category and colored according to enrichment or depletion between LTS and STS groups (Magoc and Salzberg, 2011). Statistical annotations are added to denote significant correlations with metadata, enabling quick assessment of many variables. Receiver Operator Characteristic (ROC) and Area Under Curve (AUC) analysis were performed using R, evaluating variable thresholds for relative abundance of taxa individually and in aggregate for classification of LTS vs STS status. Sensitivity was calculated as 100*(number of true positive LTS / total LTS). Specificity was calculated as 100*(number of true negative STS / total STS).
Chromogenic Immunohistochemistry (IHC)
4μm sections of FFPE tumor tissue were mounted on Superfrost Plus Microscope Slides (FisherScientific) and prepared for IHC and Multiplex IF. The slides were deparaffinized in xylene and rehydrated in graded ethanol. Antigen retrieval was performed in citrate buffer (citrate pH 6.0) using microwave heating (EZ Retriever by BioGenex). Chromogen-based IHC analysis was performed by using anti-human antibodies against the following: CD3 (T-cell lymphocytes), CD8 (CD8 T cells), Granzyme B (GzmB), FOXP3 (regulatory T cell), CD68 (Macrophages), CD66b (Granulocytes/MDSC). Signal Stain DAB Substrate KIT (Cell signal, Danvers, MA) was used for IHC detection and hematoxylin counterstaining. The densities of cells expressing CD3, CD8 and GzmB were measured by quantifying positively stained cells in five random square areas (1 mm2 each) in the tumor. The average total number of cells positive for each marker in the five square areas was expressed in density per mm2.
Multiplex Immunofluorescence Staining (Multiplex IF).
Staining was performed manually using the same primary antibodies used for IHC analysis against markers: Cytokeratin AE1/AE3 (Epithelial cell marker), Smooth Muscle Actin (Alpha-smooth muscle isoform of actin), CD3, CD8, Granzyme B and FOXP3. Antibody detection was performed with Opal Polymer HRP Ms + Rb immunohistochemistry detection reagent (PelkinElmer, Boston, MA).
Staining was performed consecutively by using the same protocol as described for IHC, and the detection for each marker was completed before application of the next antibody. The Opal Polymer HRP Ms + Rb detection reagent was used for the primary antibody detection and Opal 7-Color Manual IHC, with 6 reactive fluorophores Opal 520, Opal 540, Opal 570, Opal 620, Opal 650, Opal and 690 plus 4′,6-diamidino-2-phenylindole (DAPI) nuclear counterstain, were added according to the manufacturer’s instructions. Uniplex IF and Negative controls were stained with the same protocols. At the conclusion of the staining, slides were imaged using Vectra 3.0 spectral imaging system (PerkinElmer) according to previously published instructions.
Lipopolysaccharide (LPS) staining and Ribosomal RNA (rRNA) Fluorescence In Situ Hybridization (FISH).
Slides were stained for bacteria with the automated slide stainer BOND RXm (Leica) using the Bond polymer refine detection kit, according to manufacturer’s instructions. Heat induced epitope retrieval (HIER) at pH6 was done by a 20 min heating step with the epitope retrieval solution 1 (BOND). Gram negative were stained with Lipopolysaccharide Core (1:1000 dilution).
FISH was executed using Vysis IntelliFISH Universal FFPE Tissue Pretreatment and Wash Reagents Kit (Abbott Molecular Inc, IL). 5μm sections of FFPE tumor tissue were hybridized to a probe that recognizes the 16S rRNA genes of all bacteria (green) (Salzman et al., 2010) and counterstained with DAPI to visualize nuclei (blue), and tissues were visualized using a Nikon Eclipse Ti microscope.
Flow Cytometry
To characterize different subpopulations of immune cells, murine orthotopic PDAC tumors were removed and single cell suspensions were obtained by digestion with Collagenase P. Cells were stained with anti-Mouse antibodies against: CD45, CD4, FoxP3, Ly-6G, Ly-6C, IFN-γ and CD8a. Sample acquisition was carried out on LSRFortessa X-20 Analyzer Flow Cytometer (BD Biosciences, Franklin Lakes, NJ). Analysis was performed with FlowJo version 10.
Tissue culture based and 16S rDNA PCR
Frozen PDAC tissue samples were gently digested with collagenase P in sterile conditions to obtain a single cell suspension. Once the cell fraction was obtained, it was pelleted and the supernatants were plated on Columbia agar and maintained under aerobic and anaerobic conditions overnight at 37°C. Bacterial colonies were selected and DNA was extracted for subsequent PCR amplification of the 16S rDNA using the primers 515F–806R targeting the V4 region of the 16S rRNA.
For 16S rDNA PCR, bacterial DNA was extracted from frozen PDAC tissue samples maintaining sterile conditions using DNA QIAamp DNA Mini Kit (QIAGEN). 16S rDNA PCR was executed using the primers 515F–806R targeting the V4 region of the 16S rRNA.
QUANTIFICATION AND STATISTICAL ANALYSES
The patients’ demographic and clinical information were compared using chi-squared test and Fisher’s exact test were used to evaluate the association between two categorical variables. Wilcoxon’s rank sum test was used to compare the distributions of continuous variables between two different groups (microbiota composition positive vs. negative). Overall survival (OS) was defined as the time from diagnosis to death from any cause. Patients who did not experience death were censored at date of last follow-up. Kaplan-Meier curves were estimated for the survival distributions. The Log-rank test was used to test the difference in survival distributions between subgroups. Univariate Cox proportional hazard models were used to determine the effects of microbiota composition on OS (Heller, 2001). Hazard ratios and 95% confidence intervals were provided. All tests were two-sided. P-values less than 0.05 were considered statistically significant. All analyses were conducted using SAS 9.4 and S-Plus 8.0 software. Raw 16S rRNA sequences were processed using QIIME (Caporaso et al., 2010b). The minimal sequencing depth was 817, mean: 22178 and maximal: 89566. Sequences were aligned with reference to Silva v128 (Quast et al., 2013). Alpha- and beta-diversity analysis, survival analysis, principal coordinates analysis, ecological network analysis and Logistic regression combined with LASSO method utilized R 3.4.3. For LASSO logistic regression, we ran 10-fold cross validations with logistic regression for 100 times (starting with different seeds), then we aggregated all the deviances from 100 validation results with respect to each tuning parameter of lambda. The one with minimal average deviance was set as the best lambda value. Then we fit the LASSO logistic regression again with this best lambda value to get a stable set of selected features. Linear discriminant analysis (LDA) effect size (LEfSe) was performed under bioconda environment (Segata et al., 2011) to determine the genomic features most likely to explain differences between biological classes (STS and LTS of MDACC cohorts). All p-values were adjusted for multiple comparisons with the FDR algorithm (Benjamini et al., 2001). For the discovery, validation and prioritization of candidate taxa, the procedure was the following; all genera/species from the discovery cohort (MDACC) with an FDR adjusted p-value < 0.05 between LTS and STS were included. Those genera/species that remained significantly associated with LTS and STS in the validation cohort (JHH) were selected as candidates. Finally, the prioritization of the final candidates list required the candidate genera/species remain significantly associated with LTS/STS status (P<0.01) independent of cohort using a generalized linear model. IHC, Flow cytometry, Mouse Chemokine Assays and tumor size data were analyzed and expressed as the mean ± standard deviation using GraphPad Prism 7 (GraphPad Software, Inc., San Diego, CA).
DATA AND SOFTWARE AVAILABLE
The raw data is deposited and available on NCBI BioProject Accession Number: PRJNA542615. All analyses were conducted using SAS 9.4 and S-Plus 8.0 software. Raw 16S rRNA sequences were processed using QIIME (Caporaso et al., 2010b). Logistic regression combined with LASSO method utilized R 3.4.3. Linear discriminant analysis (LDA) effect size (LEfSe) was performed under bioconda environment (Segata et al., 2011). All p-values are adjusted for multiple comparisons with the FDR algorithm (Benjamini et al., 2001).
Supplementary Material
(A-E) Bar plots displaying taxonomic composition in both cohorts at Phylum level (A), Class level (B), Order level (C), Family level (D), and Genus level (E). Relative abundance is plotted for each tumor (0–100%).
(F-H) Bar plots comparing taxonomic composition (Order level) in MDACC cohort based on following potentially influential factors: Neoadjuvant therapies (F), Adjuvants therapies (G) and Antibiotics usage prior to surgery (+3 days) (H).
(A) Fluorescence in situ hybridization (FISH) in human FFPE PDAC samples to detect bacterial 16S rRNA sequences (green). Cell nuclei stained with 4′,6-diamidino-2-phenylindole (DAPI). (B) Lipopolysaccharide (LPS) staining in FFPE PDAC samples by immunohistochemistry using an antibacterial-LPS antibody. (C) 16S rDNA PCR executed using the primers 515F–806R target the V4 region of the 16S rRNA, shows the presence of bacterial DNA in 9 PDAC frozen samples. (D) PCR of Saccharopolyspora rectivirgula on PDAC LTS frozen samples. (E) Bar plots of taxonomic composition from FFPE and frozen PDAC samples at Genus level. (F) Culture based assay from frozen PDAC samples. (G) 16S rDNA PCR from colonies selected on agar plate.
(A) Genera and (B) Species ranked by Mann-Whitney FDR adjusted P-value between LTS and STS in Discovery and Validation Cohorts.
(A). Taxa in FFPE and matching Frozen PDAC samples. (B) Taxa in FFPE and matching Frozen PDAC samples. (C) Taxa filtered out as contaminants based on 4 negative controls and additional literature prior to downstream statistical analysis.
(A) Heat map of PICRUSt analysis identifing 26 core functional modules present across all PDAC samples with a coverage of >90% and p < 0.05. (B) LDA score computed from enrichment metabolic pathways between LTS and STS. (C) Kaplan-Meier estimates for survival probability based on the top two enrich metabolic pathways in LTS. Upper plot, Xenobiotics Biodegradation and Metabolism (p < 0.00001) and lower plot, Lipids Metabolism (p < 0.00001).
(A) Stacked Bar plots showing taxonomic composition (Order level) on tumors from mice who did not receive FMT (no FMT) vs those who received PDAC FMT. (B) Beta diversity through LDA comparing gut microbiome from: HC, LTS-NED and STS. (C-G) Beta-diversity through PCA/Soransen methods comparing tumor microbiome from the different groups. (H) Experimental design of the bacterial ablation by antibiotics after FMT. (I) Tumor volume from mice orthotopically implanted with KPC pancreatic cancer cell lines and FMT with stool from LTS-NED donors and antibiotic treated after FMT.(J) Taxonomic classification of bacterial 16S sequence detected in stools from human LTS donors and FMT recipient mice (pre/post antibiotics) by origin. (K) Beta diversity by PCA, showing closeness between mice that received FMT from LTS-NED and distance from those that received FMT and were treated with antibiotics post-FMT.
(A-F) Alpha diversity box plots (Observed species and Shannon) in MDACC cohorts PDAC patients. Analysis of following factors: Body Mass Index (BMI) (A), sex (B), Smoking status (C), Neoadjuvant therapies (D), Adjuvants therapies (E) and Antibiotics (F).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Polyclonal Rabbit Anti-Human CD3 | DAKO | Cat#AO452 |
| Mouse anti-human CD8 | Thermo Fisher Scientific | Cat#MS-457-S |
| Mouse anti-human Granzyme B | Leica Biosystems | Cat#PA0291 |
| Mouse anti-human FOXP3 | BioLegend | Cat#320102 |
| Mouse anti-human CD68 | DAKO | Cat#M0876 |
| Mouse anti-human CD66b | BioLegend | Cat#392902 |
| Monoclonal Mouse Anti-Human Cytokeratin AE1/AE3 | DAKO | Cat#M351529-2 |
| Rabbit Smooth Muscle Actin Polyclonal Antibody | Thermo Fisher Scientific | Cat#PA5-16697 |
| Lipopolysaccharide Core, mAb WN1 222-5 | HycultBiotech | Cat#HM6011 |
| Rat Anti-Mouse CD45 PerCP-Cy™5.5 | BD Pharmingen™ | Cat#550994 |
| Rat Anti-Mouse CD4 PE-CF594 | BD Horizon™ | Cat#562285 |
| Rat Anti-Mouse Foxp3 V450 | BD Horizon™ | Cat#561293 |
| Rat Anti-Mouse Ly-6G PE | BD Pharmingen™ | Cat#551461 |
| Rat Anti-Mouse IFN-γ PE | BD Pharmingen™ | Cat#554412 |
| CD8a Monoclonal Antibody FITC | eBioscience™ | Cat#11-0087-41 |
| InVivoMAb anti-mouse CD8α | BioXcell | Cat#BE0061 |
| Bacterial and Virus Strains | ||
| Saccharopolyspora rectivirgula | ATCC® | Cat#33515TM |
| Biological Samples | ||
| Human Pancreatic cancer specimens (Discovery cohort) | University of Texas MD Anderson Cancer Center Tissue Biobank | https://www.mdanderson.org |
| Human Pancreatic cancer specimens (Validation cohort) | Johns Hopkins Hospital | https://www.hopkinsmedicine.org/the_johns_hopkins_hospital/index.html |
| Human Stool specimens | University of Texas MD Anderson Cancer Center | https://www.mdanderson.org |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Cleocin Phosphate (clyndamycin injection, USP 300mg/2ml | Pfizer | Cat#NDC0009-0870-26 |
| Streptomycin for Injection, USP | X-GEN Pharmaceuticals, Inc | Cat#NDC 39822-0706-2 |
| Critical Commercial Assays | ||
| OMNIgene GUT kit | DNA Genotek | Cat#OMR-200 |
| QIAamp DNA FFPEQIAamp DNA FFPE Tissue Kit | Qiagen | Cat#56404 |
| Opal Polymer HRP Ms Plus Rb, 1X, 50 mL | PerkinElmer | Cat#ARH1001EA |
| Bio-Plex Pro Mouse Chemokine Panel, 31-plex | BIO-RAD | Cat#12009159 |
| Deposited Data | ||
| NCBI BioProject | This paper | Accession Number:PRJNA542615 |
| Experimental Models: Cell Lines | ||
| Murine pancreatic ductal adenocarcinoma cell lines | This paper | KPC,Derived from genetically engineered Pdx1-Cre, LSL-KrasG12D/+, LSLTrp53R172H/+ (“KPC”) mice |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6NTac | Taconic Bioscience | B6-F |
| Oligonucleotides | ||
| 16S 15F (Parada)-806R (Apprill), forward-barcoded: FWD:GTGYCAGCMGCCGCGGTAA; REV:GGACTACNVGGGTWTCTAAT | Integrated DNA technologies, IDT | 515F-806R target the V4 region |
| Saccharo5 F AAGAGCTCGTAGGCGGTTTG Saccharo5 R GCATTTCACCGCTACACCAG | This paper | N/A |
| Software and Algorithms | ||
| QIIME | Caporaso et al., 2010a | http://qiime.org/ |
| FlowJo version 10 | Tree Star Inc | https://www.flowjo.com/solutions/flowjo |
| SAS 9.4 | SAS, Cary, NC | www.sas.com/en_us/software/sas9.html |
| S-Plus 8.0 | TIBCO Software Inc | www.tibco.com |
| Logistic regression combined with LASSO method | Tibshirani et al., 1996 | https://www.jstor.org/stable/2346178 |
| LEfSe | Segata et al., 2011 | https://bioconda.github.io/recipes/lefse/README.html |
| FDR algorithm | Benjamini et al., 2001 | N/A |
| R 3.4.3 | This paper | https://cran.r-project.org/bin/windows/base/ |
| GraphPad Prism 7 | GraphPad Software | www.graphpad.com |
Highlights.
PDAC long term survivors display high tumor microbial diversity and immunoactivation.
A PDAC tumoral microbiome signature predicts PDAC long-term survival.
The gut microbiome modulates the PDAC tumor microbiome landscape.
Fecal microbial transplants can modulate tumors immunosuppression and growth.
Acknowledgments:
We thank Peter Davies for his scientific vision and feedback during the study conduction. Funding support: FM has been funded by American Gastroenterological Association Research Foundation, PanCAN/AACR Career Development Award Number 14–20-25-MCAL, MD Anderson Philanthropic Funds, Emerson Collective Award, K12 Paul Calabresi Clinical Scholarship Award (K12CA088084–16A1). AM and FM were funded by MD Anderson Philanthropic Funds and Stand Up To Cancer-Lustgarten Foundation Pancreatic Cancer Interception Translational Cancer Research Grant (Grant Number: SU2C-AACR-DT25–17) administered by the American Association for Cancer Research. ER acknowledges Becas Chile. PQ was supported by National Cancer Institute grant R25E (CA056452) and MD Anderson’s Cancer Center Support Grant (CA016672).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests: J. Wargo is an inventor on a US patent application (PCT/US17/53.717) submitted by the UT MDACC, and reports compensation for speaker’s bureau and honoraria from Imedex, Dava Oncology, Omniprex, Illumina, Gilead, MedImmune and Bristol-Myers Squibb (BMS). J. Wargo serves as a consultant/advisory board member for Roche/Genentech, Novartis, AstraZeneca, GlaxoSmithKline (GSK), BMS, Merck, Biothera Pharmaceuticals and Microbiome DX, and receives research support from GSK, Roche/Genentech, BMS, and Novartis. A.M. and S.M.H. receive royalties from Hangzhou Guangkeande (Cosmos) Biotechnology Company LTD for a license managed by the MD Anderson Conflict of Interest Committee. FM, ER and YZ are filing a patent with findings presented in this manuscript.
REFERENCES
- Anders S, and Huber W(2010). Differential expression analysis for sequence count data. Genome Biol 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, et al. (2013). Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500, 232–236. [DOI] [PubMed] [Google Scholar]
- Balachandran VP, Luksza M, Zhao JN, Makarov V, Moral JA, Remark R, Herbst B, Askan G, Bhanot U, Senbabaoglu Y, et al. (2017). Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature 551, 512–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Drai D, Elmer G, Kafkafi N, and Golani I(2001). Controlling the false discovery rate in behavior genetics research. Behav Brain Res 125, 279–284. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B(2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cani PD (2017). Gut cell metabolism shapes the microbiome. Science 357, 548–549 [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, and Knight R (2010a). PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26, 266–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. (2010b). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7, 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6, 1621–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Molin M, Zhang M, de Wilde RF, Ottenhof NA, Rezaee N, Wolfgang CL, Blackford A, Vogelstein B, Kinzler KW, Papadopoulos N, et al. (2015). Very Long-term Survival Following Resection for Pancreatic Cancer Is Not Explained by Commonly Mutated Genes: Results of Whole-Exome Sequencing Analysis. Clin Cancer Res 21, 1944–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daquigan N, Seekatz AM, Greathouse KL, Young VB, and White JR (2017). High-resolution profiling of the gut microbiome reveals the extent of Clostridium difficile burden. NPJ Biofilms Microbiomes 3, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis CE, Lin CC, Mariotto AB, Siegel RL, Stein KD, Kramer JL, Alteri R, Robbins AS, and Jemal A (2014). Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin 64, 252–271. [DOI] [PubMed] [Google Scholar]
- Devaraj S, Hemarajata P, and Versalovic J (2013). The human gut microbiome and body metabolism: implications for obesity and diabetes. Clin Chem 59, 617–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewes JL, White JR, Dejea CM, Fathi P, Iyadorai T, Vadivelu J, Roslani AC, Wick EC, Mongodin EF, Loke MF, et al. (2017). High-resolution bacterial 16S rRNA gene profile meta-analysis and biofilm status reveal common colorectal cancer consortia. NPJ Biofilms Microbiomes 3, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. [DOI] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, and Knight R (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti P, Farina S, Cristofolini M, Girolomoni G, Tett A, and Segata N (2017). Experimental metagenomics and ribosomal profiling of the human skin microbiome. Exp Dermatol 26, 211–219. [DOI] [PubMed] [Google Scholar]
- Garrido-Laguna I, and Hidalgo M (2015). Pancreatic cancer: from state-of-the-art treatments to promising novel therapies. Nat Rev Clin Oncol 12, 319–334. [DOI] [PubMed] [Google Scholar]
- Geller LT, Barzily-Rokni M, Danino T, Jonas OH, Shental N, Nejman D, Gavert N, Zwang Y, Cooper ZA, Shee K, et al. (2017). Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 357, 1156–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman K, Wei SC, et al. (2018). Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359, 97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller G (2001). The Cox proportional hazards model with a partly linear relative risk function. Lifetime Data Anal 7, 255–277. [DOI] [PubMed] [Google Scholar]
- Hidalgo M (2010). Pancreatic cancer. N Engl J Med 362, 1605–1617. [DOI] [PubMed] [Google Scholar]
- Human Microbiome Project C (2012a) A framework for human microbiome research. Nature 486, 215–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project C (2012b). Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamisawa T, Wood LD, Itoi T, and Takaori K (2016). Pancreatic cancer. Lancet 388, 73–85. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, and Goto S (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YI, Park JE, Brand DD, Fitzpatrick EA, and Yi AK (2010). Protein kinase D1 is essential for the proinflammatory response induced by hypersensitivity pneumonitis-causing thermophilic actinomycetes Saccharopolyspora rectivirgula. J Immunol 184, 3145–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuczynski J, Stombaugh J, Walters WA, Gonzalez A, Caporaso JG, and Knight R (2011). Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr Protoc Bioinformatics Chapter 10, Unit 10 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu P, Blacher E, Elinav E, and Pettersson S (2017). Our Gut Microbiome: The Evolving Inner Self. Cell 171, 1481–1493. [DOI] [PubMed] [Google Scholar]
- Kurilshikov A, Wijmenga C, Fu J, and Zhernakova A (2017). Host Genetics and Gut Microbiome: Challenges and Perspectives. Trends Immunol 38, 633–647. [DOI] [PubMed] [Google Scholar]
- Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31, 814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, et al. (2015). PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 372, 2509–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Price J, Mahurkar A, Rahnavard G, Crabtree J, Orvis J, Hall AB, Brady A, Creasy HH, McCracken C, Giglio MG, et al. (2017). Strains, functions and dynamics in the expanded Human Microbiome Project. Nature 550, 61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Lladser ME, Knights D, Stombaugh J, and Knight R (2011). UniFrac: an effective distance metric for microbial community comparison. ISME J 5, 169–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoc T, and Salzberg SL (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makohon-Moore AP, Zhang M, Reiter JG, Bozic I, Allen B, Kundu D, Chatterjee K, Wong F, Jiao Y, Kohutek ZA, et al. (2017). Limited heterogeneity of known driver gene mutations among the metastases of individual patients with pancreatic cancer. Nat Genet 49, 358–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manji GA, Olive KP, Saenger YM, and Oberstein P (2017). Current and Emerging Therapies in Metastatic Pancreatic Cancer. Clin Cancer Res 23, 1670–1678. [DOI] [PubMed] [Google Scholar]
- Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, Luke JJ, and Gajewski TF (2018). The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 359, 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Round JL, and Kasper DL (2008). A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453, 620–625. [DOI] [PubMed] [Google Scholar]
- McAllister F, Housseau F, and Sears CL (2014). Microbiota and immune responses in colon cancer: more to learn. Cancer J 20, 232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie PJ, and Holmes S (2013). phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. Plos One 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, Stein KD, Alteri R, and Jemal A (2016). Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin 66, 271–289. [DOI] [PubMed] [Google Scholar]
- Nieuwdorp M, Gilijamse PW, Pai N, and Kaplan LM (2014). Role of the microbiome in energy regulation and metabolism. Gastroenterology 146, 1525–1533. [DOI] [PubMed] [Google Scholar]
- Ock CY, Hwang JE, Keam B, Kim SB, Shim JJ, Jang HJ, Park S, Sohn BH, Cha M, Ajani JA, et al. (2017). Genomic landscape associated with potential response to anti-CTLA-4 treatment in cancers. Nat Commun 8, 1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushalkar S, Hundeyin M, Daley D, Zambirinis CP, Kurz E, Mishra A, Mohan N, Aykut B, Usyk M, Torres LE, et al. (2018). The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer Discov 8, 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, and Glockner FO (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41, D590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riquelme E, Maitra A, and McAllister F (2018). Immunotherapy for Pancreatic Cancer: More Than Just a Gut Feeling. Cancer Discov 8, 386–388. [DOI] [PubMed] [Google Scholar]
- Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillere R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, et al. (2018). Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97. [DOI] [PubMed] [Google Scholar]
- Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjoberg J, Amir E, Teggatz P, Barman M, Hayward M, Eastwood D, et al. (2010). Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol 11, 76–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, and Huttenhower C (2011). Metagenomic biomarker discovery and explanation. Genome Biol 12, R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemark DK, and Allen SJ (2015). The microbiome and disease: reviewing the links between the oral microbiome, aging, and Alzheimer’s disease. J Alzheimers Dis 43, 725–738. [DOI] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, and Jemal A (2018). Cancer statistics, 2018. CA Cancer J Clin 68, 7–30. [DOI] [PubMed] [Google Scholar]
- Vetizou M, Pitt JM, Daillere R, Lepage P, Waldschmitt N, Flament C, Rusakiewicz S, Routy B, Roberti MP, Duong CP, et al. (2015). Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 350, 1079–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr., and Kinzler KW (2013). Cancer genome landscapes. Science 339, 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, and Cole JR (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73, 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A-E) Bar plots displaying taxonomic composition in both cohorts at Phylum level (A), Class level (B), Order level (C), Family level (D), and Genus level (E). Relative abundance is plotted for each tumor (0–100%).
(F-H) Bar plots comparing taxonomic composition (Order level) in MDACC cohort based on following potentially influential factors: Neoadjuvant therapies (F), Adjuvants therapies (G) and Antibiotics usage prior to surgery (+3 days) (H).
(A) Fluorescence in situ hybridization (FISH) in human FFPE PDAC samples to detect bacterial 16S rRNA sequences (green). Cell nuclei stained with 4′,6-diamidino-2-phenylindole (DAPI). (B) Lipopolysaccharide (LPS) staining in FFPE PDAC samples by immunohistochemistry using an antibacterial-LPS antibody. (C) 16S rDNA PCR executed using the primers 515F–806R target the V4 region of the 16S rRNA, shows the presence of bacterial DNA in 9 PDAC frozen samples. (D) PCR of Saccharopolyspora rectivirgula on PDAC LTS frozen samples. (E) Bar plots of taxonomic composition from FFPE and frozen PDAC samples at Genus level. (F) Culture based assay from frozen PDAC samples. (G) 16S rDNA PCR from colonies selected on agar plate.
(A) Genera and (B) Species ranked by Mann-Whitney FDR adjusted P-value between LTS and STS in Discovery and Validation Cohorts.
(A). Taxa in FFPE and matching Frozen PDAC samples. (B) Taxa in FFPE and matching Frozen PDAC samples. (C) Taxa filtered out as contaminants based on 4 negative controls and additional literature prior to downstream statistical analysis.
(A) Heat map of PICRUSt analysis identifing 26 core functional modules present across all PDAC samples with a coverage of >90% and p < 0.05. (B) LDA score computed from enrichment metabolic pathways between LTS and STS. (C) Kaplan-Meier estimates for survival probability based on the top two enrich metabolic pathways in LTS. Upper plot, Xenobiotics Biodegradation and Metabolism (p < 0.00001) and lower plot, Lipids Metabolism (p < 0.00001).
(A) Stacked Bar plots showing taxonomic composition (Order level) on tumors from mice who did not receive FMT (no FMT) vs those who received PDAC FMT. (B) Beta diversity through LDA comparing gut microbiome from: HC, LTS-NED and STS. (C-G) Beta-diversity through PCA/Soransen methods comparing tumor microbiome from the different groups. (H) Experimental design of the bacterial ablation by antibiotics after FMT. (I) Tumor volume from mice orthotopically implanted with KPC pancreatic cancer cell lines and FMT with stool from LTS-NED donors and antibiotic treated after FMT.(J) Taxonomic classification of bacterial 16S sequence detected in stools from human LTS donors and FMT recipient mice (pre/post antibiotics) by origin. (K) Beta diversity by PCA, showing closeness between mice that received FMT from LTS-NED and distance from those that received FMT and were treated with antibiotics post-FMT.
(A-F) Alpha diversity box plots (Observed species and Shannon) in MDACC cohorts PDAC patients. Analysis of following factors: Body Mass Index (BMI) (A), sex (B), Smoking status (C), Neoadjuvant therapies (D), Adjuvants therapies (E) and Antibiotics (F).