Abstract
Purpose:
Response assessment of radiotherapy for the treatment of intrahepatic cholangiocarcinoma (IHCC) across longitudinal images is challenging due to anatomical changes. Advanced deformable image registration (DIR) techniques are required to correlate corresponding tissues across time. In this study, the accuracy of five commercially available DIR algorithms in four treatment planning systems (TPS) is investigated for the registration of planning images with post-treatment follow-up images for response assessment or re-treatment purposes.
Methods:
Twenty-nine IHCC patients treated with hypofractionated radiotherapy and with pre-treatment and post-treatment contrast-enhanced CT images were analyzed. Liver segmentations were semi-automatically generated on all CTs and the post-treatment CT was then registered to the pre-treatment CT using five commercially available algorithms (Demon’s, B-splines, salient feature-based, anatomically-constrained and finite element-based) in four TPSs. This was followed by an in-depth analysis of ten DIR strategies (plus global and liver-focused rigid registration) in one of the TPSs. Eight of the strategies were variants of the anatomically-constrained DIR while the two were based on a finite element-based biomechanical registration. The anatomically constrained techniques were combinations of: (1) initializations with the two rigid registrations; (2) two similarity metrics – correlation coefficient (CC) and mutual information (MI); and (3) with and without a controlling ROI (liver). The finite element-based techniques were initialized by the two rigid registrations. The accuracy of each registration was evaluated using target registration error (TRE) based on identified vessel bifurcations. The results were statistically analyzed with a one-way ANOVA and pairwise comparison tests. Stratified analysis was conducted on the inter-TPS data (plus the liver-focused rigid registration) using treatment volume changes, slice thickness, time between scans and abnormal lab values as stratifying factors.
Results:
The complex deformation observed following treatment resulted in average TRE exceeding the image voxel size for all techniques. For the inter-TPS comparison, the Demon’s algorithm had the lowest TRE, which was significantly superior to all the other algorithms. The respective mean (standard deviation) TRE (in mm) for the Demons’s, B-splines, salient feature-based, anatomically-constrained and finite element-based algorithms were 4.6 (2.0), 7.4 (2.7), 7.2 (2.6), 6.3 (2.3) and 7.5 (4.0). In the follow-up comparison of the anatomically-constrained DIR, the strategy with liver-focused rigid registration initialization, CC as similarity metric and liver as a controlling ROI had the lowest mean TRE – 6.0 (2.0). The maximum TRE for all techniques exceeded 10 mm. Selection of DIR strategy was found to be a statistically significant factor for registration accuracy. Tumor volume change had a significant effect on TRE for finite element- based registration and B-splines DIR. Time between scans had a substantial effect on TRE for all registrations but was only significant for liver-focused rigid, finite element-based and salient feature-based DIRs.
Conclusions:
This study demonstrates the limitations of commercially available DIR techniques in treatment planning systems for alignment of longitudinal images of liver cancer presenting complex anatomical changes including local hypertrophy and fibrosis/necrosis. DIR in this setting should be used with caution and careful evaluation.
Keywords: Deformable image registration, Biomechanical Registration, Cholangiocarcinoma, Liver Cancer, Treatment Planning Systems
INTRODUCTION
Intrahepatic cholangiocarcinoma (a bile-duct cancer, IHCC) is a rare form of liver cancer that affects about 500 patients in the US every year1. Its incidence however, has seen an increase in the past decade2,3. The disease is considered incurable unless complete resection is possible; however, only 15% of the tumors are operable4. In such cases, chemotherapy and radiation therapy (RT), particularly stereotactic body radiotherapy (SBRT) and hypofractionated RT, are used to extend survival and improve quality of life5. Functional imaging has been investigated to better understand the normal tissue changes in the liver after RT6. These images are acquired serially over several months during and after the completion of RT. It is important to accurately align the planning CT scan with the delivered dose to the follow-up images to understand the dose-response relationship in order to optimize the delivery of radiation to reduce liver damage. In addition, multiple courses of radiation therapy may be required if additional tumors develop in the liver7. Therefore, it is also important to understand the cumulative dose to the normal liver, most often done by mapping the previously administered dose to the subsequent treatment plans. Unfortunately, dose accumulation in a deformable organ such as the liver can be quite challenging. Simple rigid registration does not achieve a sufficient level of accuracy when volumetric changes and large patient motion occurs8. Studies have demonstrated the large volumetric response of the liver six weeks post-radiation therapy, including hypertrophy and fibrosis9,10.
One of the first studies to compare deformable image registration (DIR) techniques for pre- and post-RT liver cancer was conducted by Fukumitsu et al.11. The study compared commercially available DIR techniques in two treatment planning systems, MIM Maestro (MIM Software Inc., Cleveland, OH, USA) and Velocity (Velocity Medical Solutions, Atlanta, GA, USA). Mean fiducial registration errors in both TPSs were found to be about 1 cm (standard deviation 1 cm) for CT with slice thickness of 0.5 cm. A parallel study compared rigid and DIR for registering pre- and post-RT liver CTs using MIM Maestro12. Fiducial registration errors of 1.1 (0.7) cm and 0.7 (0.8) cm were reported (slice thickness 0.5 cm). In cases tumor resection, where large sections of the liver are not removed, similar issues of volumetric changes arise when deciding upon subsequent treatment plans. Hence, DIR of pre and post-surgery liver CTs is a related problem. Gunay et al. proposed a DIR for this application that reported a mean target registration error of 0.6 (0.5) cm for slice thicknesses between 0.1 and 0.2 cm13.
The goal of this work is to compute the accuracy of DIR algorithms available in four commercial treatment planning systems (TPS) and assess their clinical usability for IHCC treatment. We also analyze various treatment-related factors that may affect the accuracy of particular registration methods. This is followed by an in-depth analysis with the DIR parameters in one of the TPSs where five different techniques including intensity-based, biomechanical model-based, and region of interest-driven DIRs are compared.
METHODS
Patients
Twenty-nine patients with IHCC treated with RT at The University of Texas MD Anderson Cancer Center between 2001 and 2016 were retrospectively evaluated under an institutional review board approved study. For each case, a contrast-enhanced CT scan was obtained prior to radiation therapy (RT) and after the completion of RT (mean follow-up time 12 weeks, range 4–31 weeks). The image resolution for the pre-treatment CT images was between 0.66 and 0.98 mm in-plane with a slice thickness of 2.5 mm (n = 25 cases) or 5.0 mm (n = 4 cases). The tumors on both the pre- and post-RT CT images were contoured manually by a non-radiologist scientist (DE) and reviewed and approved by a radiation oncologist (EJK). Demographic (age and gender) data and three liver-related laboratory traits (aspartate aminotransferase [AST], alanine aminotransferase [ALT] and cancer antigen CA19–9) were extracted from the electronic health records database. These characteristics are summarized in Table 1.
Table 1:
Baseline characteristics of the twenty-nine patients with intrahepatic cholangiocarcinoma included in this study. Abnormal cases refer to lab values outside clinically defined normal ranges.
| Gender (#) | |
| Male | 14 |
| Female | 15 |
| Age (years) | |
| Mean (Standard Deviation) | 62.10 (9.95) |
| Range | 32 – 79 |
| Seniors (>65 years) | 13 |
| AST (international units/L) | |
| Mean (Standard Deviation) | 49.14 (35.72) |
| Range | 20 – 213 |
| Abnormal cases | 16 |
| ALT (international units/L) | |
| Mean (Standard Deviation) | 42.00 (37.28) |
| Range | 12 – 177 |
| Abnormal cases | 9 |
| Cancer Antigen 19–9 (units/ml) | |
| Mean (Standard Deviation) | 240.08 (1040.32) |
| Range | 1.00 – 4716.00 |
| Abnormal cases | 12 |
AST (Aspartate Aminotransferase); ALT (Alanine Aminotransferase)
Liver Segmentation
A pre-trained fully convolutional neural network, the DeepLab v3 plus network14, was adapted for rapid segmentation of the liver. A skip-layer architecture, similar to that found in the ‘U-net’ design15, was implemented to improve efficiency and consistency of the liver segmentation. Details of this segmentation validation can be found in Anderson et al16. Once segmented, the contours were evaluated by a non-radiologist scientist (GC) experienced in liver image analysis and edited when necessary. The pre- and post-treatment volumes for the liver and the tumor along with the time between scans were calculated and are presented in Table 2.
Table 2:
Treatment-related characteristics of the twenty-nine patients with intrahepatic cholangiocarcinoma included in this study
| Follow-up time between pre- and post-treatment scans (days) | |
| Mean (Standard Deviation) | 115 (37) |
| Range | 32–217 |
| Pre-treatment tumor volume (cm3) | |
| Mean (Standard Deviation) | 140.21 (112.99) |
| Range | 20.53 – 481.97 |
| Post-treatment tumor volume (cm3) | |
| Mean (Standard Deviation) | 108.63 (99.12) |
| Range | 13.37 – 396.59 |
| Post-treatment tumor volume change | |
| Actual change - Mean (Std. Dev.) (cm3) | −31.58 (49.59) |
| Actual change - Range (cm3) | −176.36 – 43.63 |
| Percentage change - Mean (Std. Dev.) | −19.16 (32.34) |
| Percentage change - Range | −68.82 – 78.59 |
| Large change (>30%) (#) | 10 |
| Volume increased (#) | 7 |
| Volume decreased (#) | 22 |
| Pre-treatment liver volume (cm3) | |
| Mean (Standard Deviation) | 1746.67 (360.34) |
| Range | 1256.81 – 2713.68 |
| Post-treatment liver volume (cm3) | |
| Mean (Standard Deviation) | 1708.50 (365.40) |
| Range | 1207.04 – 2696.61 |
| Post-treatment liver volume change | |
| Actual change - Mean (Std. Dev.) (cm3) | −38.17 (151.86) |
| Actual change - Range (cm3) | −329.01 – 298.19 |
| Percentage change - Mean (Std. Dev.) | −1.87 (9.03) |
| Percentage change - Range | −17.78 – 22.83 |
| Large change (>10%) (#) | 8 |
| Volume increased (#) | 12 |
| Volume decreased (#) | 17 |
Deformable Image Registration Methods
The post-treatment image was registered to the pre-treatment image using five DIR techniques available in four TPSs. The TPSs used in this study were: Velocity (v3.0, Velocity Medical Solutions, Atlanta, GA, USA), Pinnacle (v9.10, Philips, Madison, Wisconsin, USA), Eclipse (v13.6, Varian Medical Systems, Palo Alto, CA, USA) and RayStation (v7.99, RaySearch Laboratories, Stockholm, Sweden). Velocity uses an intensity-based B-spline multipass algorithm17 with mutual information (Matte’s formulation) as the similarity metric. Pinnacle uses the salient feature-based registration (SFBR) algorithm for DIR18,19. This is a landmark-based registration where distinctive features are automatically extracted from each image based on a similarity metric (cross correlation, normalized mutual information or local correlation). Eclipse implements an accelerated version20 of the Demon’s algorithm21. The options for similarity are mutual information, cross correlation and pattern intensity. RayStation has two DIR algorithms: Anatomically constrained deformation algorithm (ANACONDA)22 and a finite element-based biomechanical DIR – Morfeus23. ANACONDA employs a hybrid approach that combines intensity data and contours of anatomical regions. The optimization problem accounts for image similarity (correlation coefficient or mutual information), regularization of the deformation and alignment of optional structures of interest. Morfeus is a finite element model (FEM) based DIR technique where boundary conditions are determined on the surface of the structure and then used in a finite element analysis solver to estimate the deformation of the internal structures based on linear elastic material properties.
Comparison between Treatment Planning Systems
All the DIRs across systems were initialized by a rigid registration available within the corresponding TPS. The rigid registrations were monitored and manually edited if necessary by non-radiologist scientists (MMM for Velocity, AS for the other systems). For Pinnacle and Eclipse, a focus region was determined by a bounding box computed from the liver contours plus a one-centimeter margin. For Velocity, though the liver contours were not used directly, a bounding box containing the liver was manually created as a focus region. ANACONDA used the liver contours as a controlling ROI. For Morfeus, a triangular mesh was generated using the contours of the liver, which was then used for FEM-based DIR. The similarity metrics used in the various algorithms were mutual information (Velocity and Eclipse), cross-correlation (Pinnacle) and correlation coefficient (RayStation - ANACONDA).
Comparison within a Treatment Planning System
Due to Raystation’s greater flexibility in adjusting for parameters, a follow-up intra-TPS comparison was conducted using different DIR strategies. Eight of the strategies were variants of ANACONDA. In this study, ANACONDA was performed with two different rigid registration initializations: global and liver-focused; two different intensity similarity measures: correlation coefficient (CC) and mutual information (MI); and with and without using the liver as a controlling structure. The other parameters for ANACONDA along with the values used were: controlling-ROI weight (0.5), Gaussian smoothing standard deviation – initial (20 mm), final (3.3 mm) and initial/final grid regularization weight (400). Two strategies involved the implementation of Morfeus in RayStation – using the two rigid registration initializations. For Morfeus, a triangular mesh with edge length of 6 mm and smoothing radius of 1 mm was generated from the liver contours. The elastic properties of the liver included a Young’s modulus of 1000 Pa and a Poisson’s ratio of 0.48. The two rigid registrations also served as data points for comparisons.
Parameter Optimization
Continuing with the detailed analysis of ANACONDA within RayStation, we performed an optimization for the parameters mentioned above. For ANACONDA, the following combination of parameters were tested:
Rigid registrations: global and liver-focused
Similarity measure: correlation coefficient and mutual information
ROI weight: 0.4, 0.6 and 0.9
Initial Gaussian filter standard deviation: 16 mm, 20 mm and 24 mm
Initial/final grid regularization weight: 320, 400 and 480
The final Gaussian filter standard deviation was dependent on the grid regularization weight: 2 mm for 320, 3 mm for 400 and 4 mm for 480. Hence, 108 sets of parameters were tested for registration accuracy of ANACONDA.
Performance Evaluation
In all three of the above comparisons (comparison between TPSs, comparison within a TPS and parameter optimization), target registration error (TRE) was used for evaluating registration accuracy24. Five anatomical landmarks (vessel bifurcations) were identified in both pre- and post-treatment images. The TRE is the mean distance between corresponding points on the reference image and the deformed target image. The bifurcations were marked by an experienced medical physicist (KKB). This marking was completely independent of the liver contouring and statistical analysis described below.
An example of a pre-RT and post-RT image is shown in Figure 1. Contours for the liver and the tumor are shown. Two of the points used for the TRE evaluation are also marked.
Figure 1:
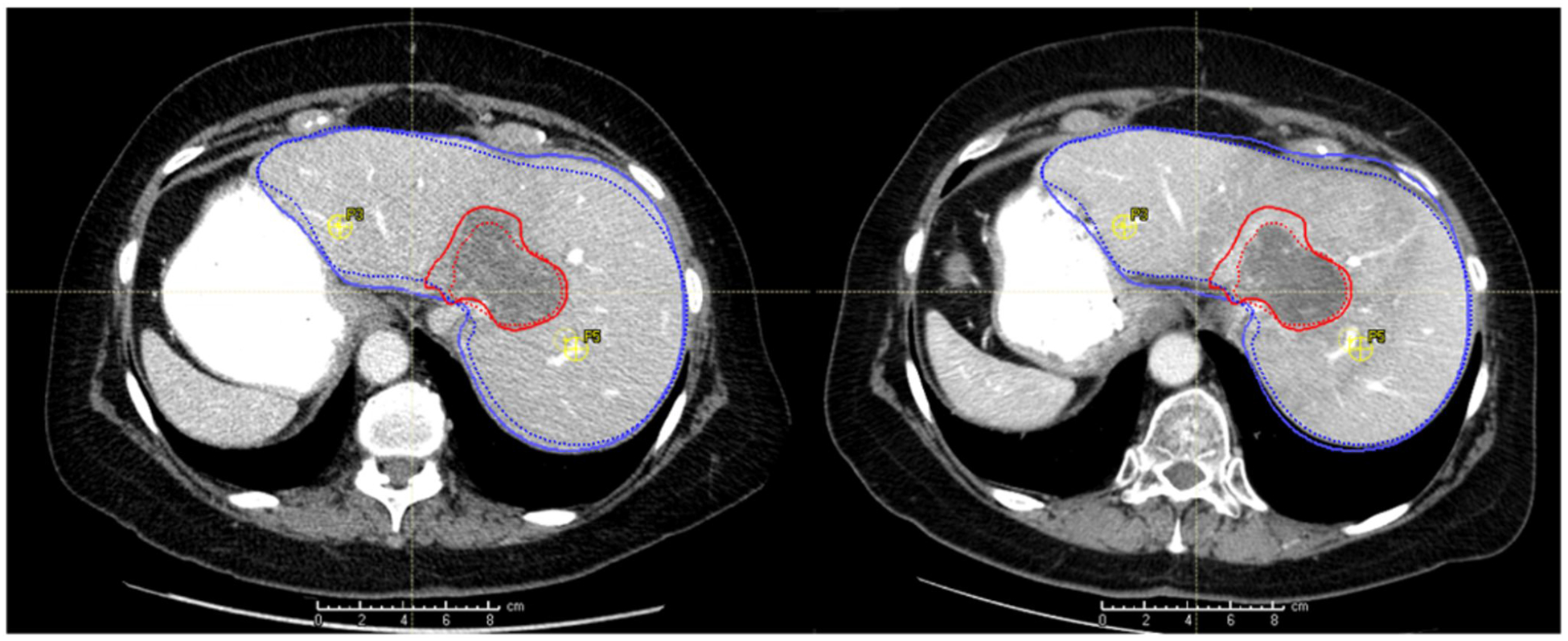
Pre- and post-treatment images for a patient. The liver and tumor are contoured in blue and red, respectively. Contours for the pre-treatment image are in solid lines while those for the post-treatment image are in dotted lines. Two of the points (vessel bifurcations) for the TRE calculation are also marked in yellow.
Statistical Analysis
For both the inter-TPS and intra-TPS comparisons, the TREs for the registration methods were analyzed for statistical differences with a Kruskal-Wallis ANOVA (analysis of variance). This was followed by pairwise ANOVAs to test for significant differences between any pair of registration methods. For the intra-TPS comparison, the analysis was carried out in two parts based on the type of rigid registration used. For the parameter optimization, a multi-factor ANOVA was used to test the statistical significance of each parameter within the registration process.
Stratified statistical analyses were conducted on the inter-TPS data (plus the liver-focused rigid registration in RayStation for comparison) by grouping the patients by tumor and liver volume changes, time duration between scans, CT slice thickness and lab test abnormalities. Large tumor volume change was defined as >30% change from pre-treatment volume while the corresponding threshold for large liver volume change was 10%. It is hypothesized that large changes in tumor and liver volume may lead to lower registration accuracy. This is something to be particularly aware of in Morfeus, where no boundary condition is placed on the tumor. A thick CT slice criterion was applied for patients imaged with a slice thickness of 5 mm. While it is expected that thicker slices will lead to higher TREs, the effect may be challenging to quantify in the present study because only four such patients were analyzed. A large time duration between scans was defined to be greater than the mean duration of 115 days. It is hypothesized that the larger the time duration, the more complex changes in the liver will occur. This may not be captured in total liver volume change as hypertrophy and necrosis/fibrosis may offset for a neutral overall change in liver volume, but still lead to complexities in deformation. Similarly, it is hypothesized that abnormal laboratory test results may indicate complexities in deformation. For any pair of stratifying trait (e.g. abnormal ALT, large liver volume change) and registration method, a one-way Kruskal-Wallis ANOVA was performed with the stratifying trait status as the factor. For all statistical analyses, the threshold for statistical significance was 0.05.
RESULTS
The baseline and treatment-related characteristics of the cohort of patients with IHCC are described in Table 1 and 2 respectively. Age was calculated in completed years at the start of RT.
Figure 2 shows a representative slice for the five registration algorithms within the four TPSs using a translucent fusion of the pre and post-treatment images. A box and whisker plot for the accuracy of these algorithms is shown in Figure 3 and a corresponding summary is provided in Table 3. The accelerated Demon’s algorithm in Eclipse had the best average performance (mean TRE 4.6±2.0 mm). The performances of DIRs in Pinnacle SFBR, Velocity B-spline and RayStation Morfeus were very similar (mean TREs between 7.2 and 7.7 mm) while Raystation ANACONDA was comparatively superior (mean TRE 6.0 mm±2.0 mm). The Kruskal-Walis ANOVA showed registration algorithm to be a statistically significant factor with p<0.01. The pairwise ANOVAs showed the Eclipse DIR to be significantly superior to all four methods (p<0.01) while the other methods did not differ significantly from each other.
Figure 2:
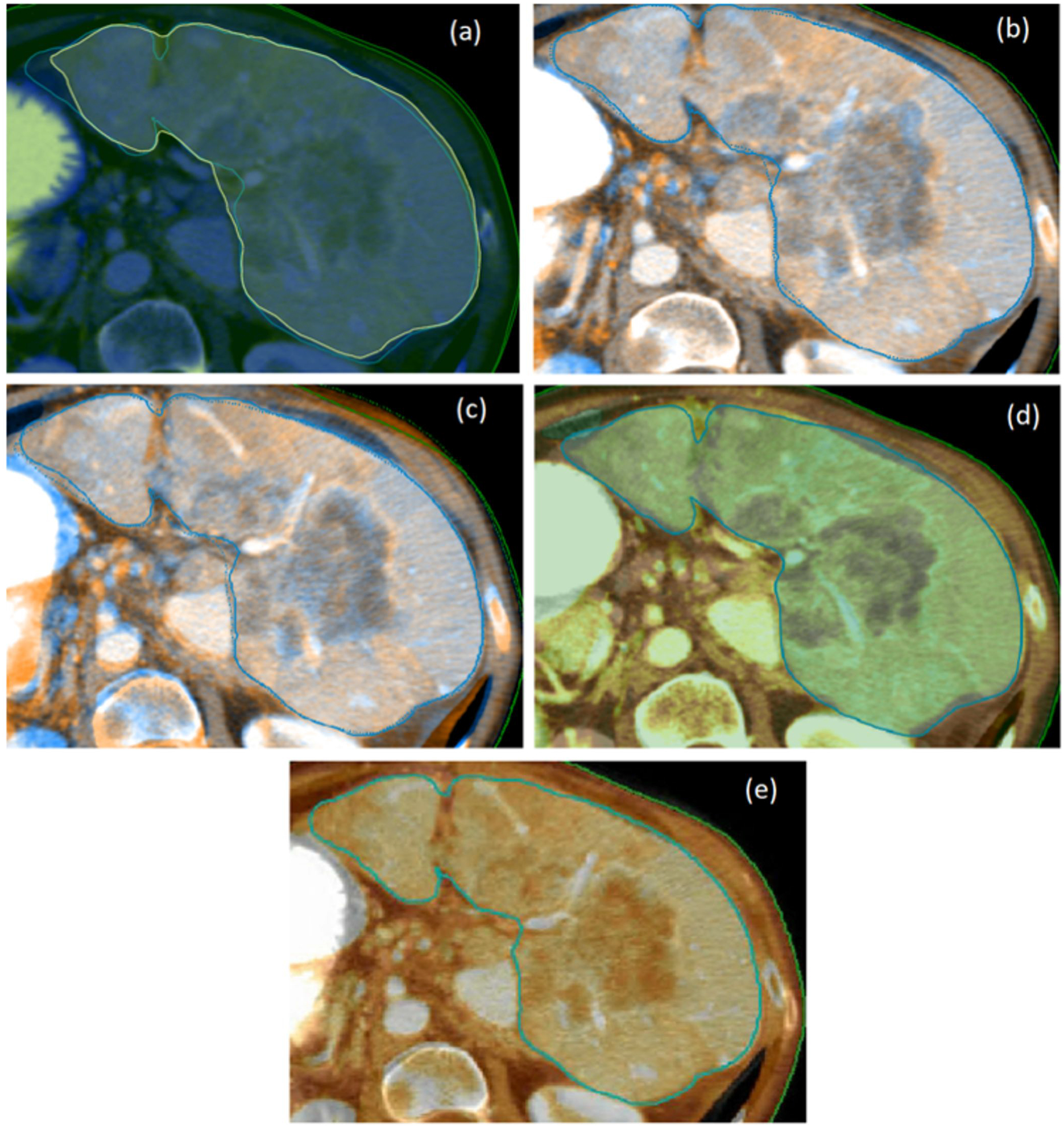
Demonstration of the liver registrations of in four TPSs: (a) Eclipse – Demon’s (b) RayStation - ANACONDA (c) RayStation - Morfeus (d) Velocity – B-spline (e) Pinnacle - SFBR. The pre- and post-treatment CT images are shown in different color maps, respectively. The liver contoured on the pre-RT image is marked in solid blue in all the images. The deformed post-treatment contours are shown for RayStation (dotted) and Eclipse (yellow). For Pinnacle and Velocity there was no option of viewing the deformed post-treatment contours in the fused DIR rendering.
Figure 3:
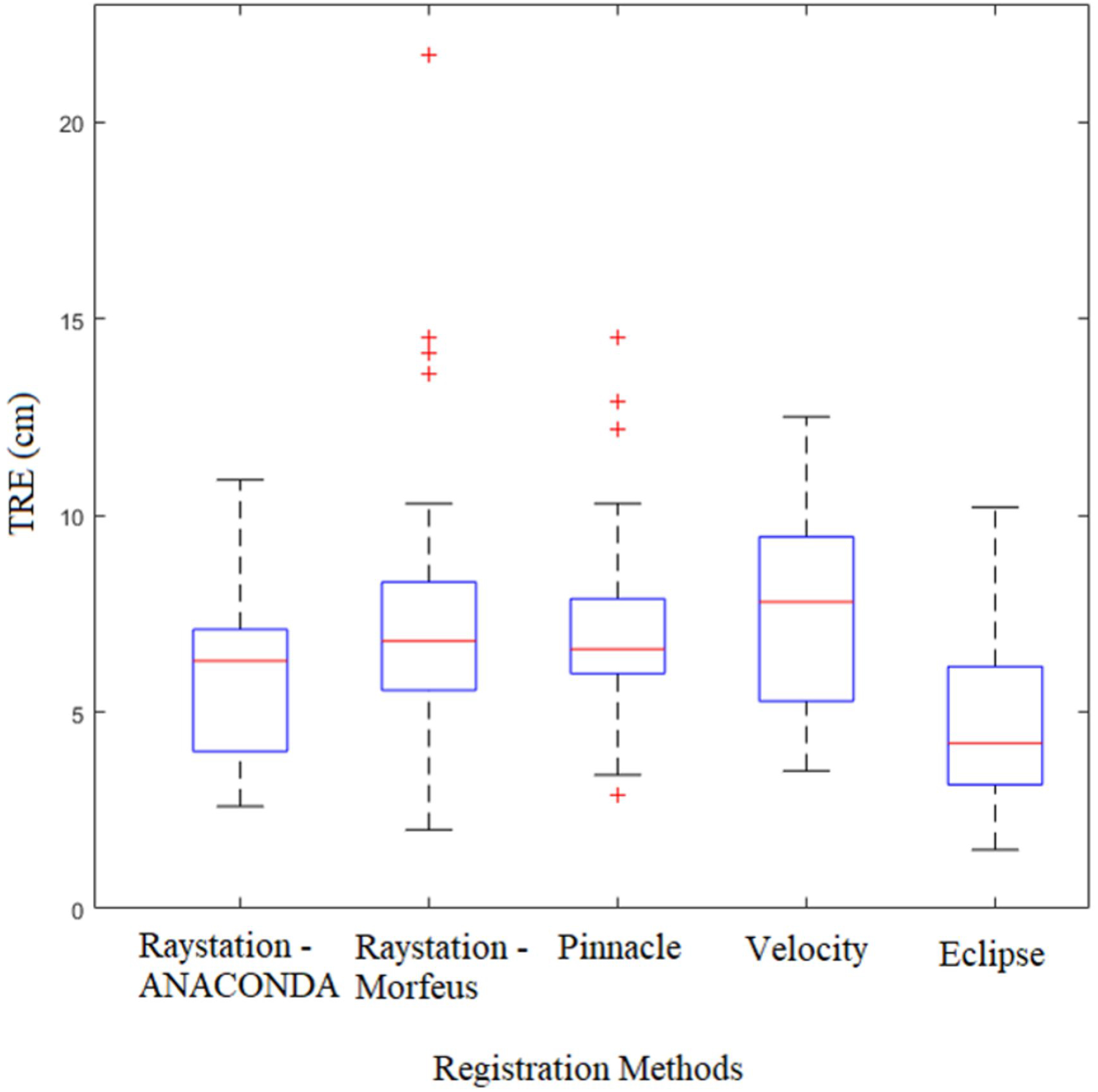
Box and whisker plot for the performance of the five registration algorithms within the four treatment planning systems (summarized in Table 3). For each registration method, the central red line denotes the median. The blue box is the interquartile range (IQR). The whisker length is defined as 1.5 times the IQR beyond its extremities. The actual whisker is drawn up to the furthest data point that is still within the whisker length. Data points that are beyond the whisker length are considered outliers and marked individually.
Table 3:
Summary of registration accuracy (TREs) for the DIRs computed in the four TPSs
| RayStation - ANACONDA | RayStation - Morfeus | Pinnacle - SFBR | Velocity - B-splines | Eclipse - Demons | |
|---|---|---|---|---|---|
| Mean TRE (mm) | 6.0 | 7.7 | 7.2 | 7.4 | 4.6 |
| Standard Deviation (mm) | 2.0 | 3.9 | 2.6 | 2.6 | 2.0 |
| Median TRE (mm) | 6.3 | 6.8 | 6.6 | 7.8 | 4.2 |
| Minimum (mm) | 2.6 | 2.0 | 2.9 | 3.5 | 1.5 |
| Maximum (mm) | 10.9 | 21.7 | 14.5 | 12.5 | 10.2 |
A summary for the intra-TPS comparison is provided in Table 4. As expected, the rigid registrations had the lowest performance, though there was a substantial improvement from the global to the liver-focused registration (mean TREs 9.1±3.5 mm and 8.4±3.1 mm respectively). Consequently, there were improvements in ANACONDA when using the liver-focused rigid registrations. The improvements were greater for the strategies that did not use liver contours as controlling ROIs (mean differences of 1.2 mm and 0.8 mm with CC and MI, respectively) as opposed to the strategies that used the liver as a controlling ROI (0.3 mm with both CC and MI). Among all DIR techniques, the ANACODA initialized by liver-focused rigid registration with CC as similarity metric and liver as the controlling ROI had the best overall performance (mean TRE 6.0±2.0 mm), while the ANACODA initialized by global rigid registration with CC as similarity metric and no controlling ROI performed the worst (mean TRE 8.6±3.5 mm). The choice of rigid registration had negligible impact on Morfeus (limited to round-off differences). Morfeus had a substantially higher standard deviation than the other DIR techniques.
Table 4:
Summary of registration accuracy (TREs) for registrations performed in RayStation
| Mean | Std. Dev. | Median | Minimum | Maximum | |||
|---|---|---|---|---|---|---|---|
| RIGID | |||||||
| Global | 9.1 | 3.5 | 8.4 | 3.2 | 16.2 | ||
| Liver-focused | 8.4 | 3.1 | 7.6 | 3.6 | 17.3 | ||
| ANACONDA | |||||||
| Initialization | Similarity metric | Control ROI | |||||
| Global Rigid Registration | CC | None | 8.6 | 3.5 | 8.1 | 3.4 | 19.9 |
| Liver | 6.3 | 2.3 | 6.4 | 2.6 | 11.5 | ||
| MI | None | 7.1 | 2.7 | 6.2 | 4.0 | 13.6 | |
| Liver | 6.4 | 2.4 | 6.6 | 3.1 | 12.2 | ||
| Liver-focused Rigid Registration | CC | None | 7.4 | 2.9 | 6.8 | 3.3 | 16.1 |
| Liver | 6.0 | 2.2 | 6.3 | 2.6 | 10.9 | ||
| MI | None | 6.3 | 2.4 | 5.4 | 2.6 | 12.6 | |
| Liver | 6.1 | 2.7 | 5.3 | 1.8 | 15.0 | ||
| MORFEUS | |||||||
| Initialization | |||||||
| Global Rigid Registration | 7.7 | 3.9 | 6.8 | 2.0 | 21.7 | ||
| Liver-focused Rigid Registration | 7.8 | 3.9 | 6.8 | 2.1 | 21.6 | ||
CC (Correlation Coefficient); MI (Mutual Information)
The Kruskal-Wallis ANOVA comparing all six registration strategies with global rigid registration showed registration technique to be statistically significant (p = 0.005). The subsequent pairwise one-way ANOVAs demonstrated that the rigid registration performed significantly worse than all DIR techniques besides ANACONDA with CC (p-values between 0.001 and 0.033). The strategies with the lowest mean TREs – ANACONDA with liver as controlling ROI and CC/MI as similarity metric – performed significantly better than ANACONDA with CC and no controlling ROI (p = 0.009 and 0.011, respectively). No other pairs of DIR registration techniques were statistically different in terms of registration accuracy.
The corresponding analysis with the liver-focused rigid registration yielded similar results but less pairs with significant differences. Registration technique was still statistically significant (p = 0.008). However, the only pairs significantly different involved rigid registration and (i) ANACONDA with CC and liver as controlling ROI (p = 0.004), (ii) ANACONDA with MI and no controlling ROI (p = 0.006), and (iii) ANACONDA with MI and liver as controlling ROI (p = 0.003).
For the parameter optimization of ANACONDA the mean TREs of all 108 parameter sets were between 5.8 and 7.0 mm. The multifactor ANOVA found the choices of rigid registration (global and liver-focused) and similarity metric (CC and MI) to be statistically significant factors (p< 0.01) for ANACONDA registration accuracy. The peak performance (5.8 mm) was attained for the parameters: MI as similarity metric, ROI weight of 0.9, initial Gaussian filter standard deviation of 24 mm and grid regularization weight of 400.
The summary of the stratified analysis by lab tests and treatment-related features is shown in Table 5. High tumor volume change was associated with significantly poorer performance of Raystation Morfeus and Velocity B-spline (p=0.048 and 0.004). This is illustrated in Figure 4(a) that plots TREs for each registration method as a function of the tumor volume change. A quadratic least-square trend line is also shown. While Morfeus shows a distinctive parabolic shape with higher TREs away from the vertex, Velocity B-spline shows a monotonic decrease.
Table 5:
TREs for the stratified analysis comparing the effect of treatment, imaging and lab-related factors on registration accuracy. For ANACONDA and Morfeus, the best performing strategies in Table 4 are presented here. Significant differences are italicized and the corresponding p-values are reported in the last row.
| Registrations→ Stratifying trait↓ | Rigid | Raystation - ANACONDA | Raystation - Morfeus | Pinnacle - SFBR | Velocity - B-spline | Eclipse - Demons | |
|---|---|---|---|---|---|---|---|
| Tumor volume change | >30% | 9.8 | 6.5 | 10.4 | 7.93 | 9.3 | 5.2 |
| Liver volume change | >10% | 9.9 | 6.0 | 7.5 | 7.9 | 7.4 | 4.2 |
| Time between scans | >115 d | 10.1 | 7.2 | 10.1 | 8.6 | 8.5 | 5.5 |
| CT Slice thickness | 2.5 mm | 8.3 | 5.8 | 7.4 | 7.1 | 7.4 | 4.3 |
| CA 19–9 | Normal | 8.9 | 6.2 | 8.5 | 7.8 | 6.8 | 4.8 |
| ALT | Normal | 8.5 | 5.8 | 8.1 | 7.0 | 7.3 | 4.9 |
| AST | Normal | 8.9 | 5.7 | 8.2 | 7.0 | 7.2 | 4.7 |
| Significant p-values | a0.014 | b0.036 | d0.049 | e0.004 | |||
AST (Aspartate Aminotransferase); ALT (Alanine Aminotransferase); CA 19–9 (Cancer Antigen 19–9)
Figure 4:
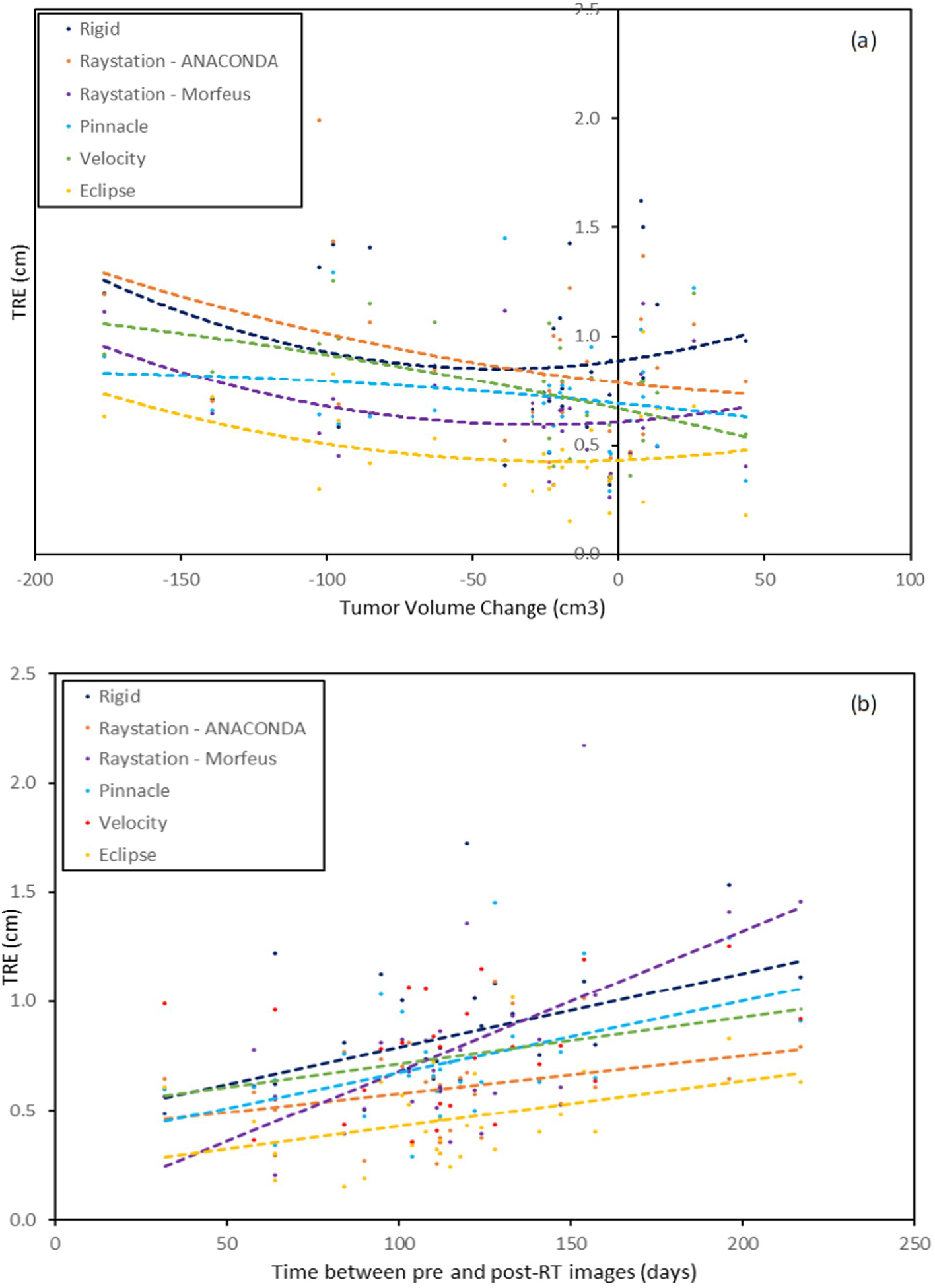
The TRE plotted as a function of the (a) liver volume change and (b) time duration between images for liver-focused rigid registration and the five DIRs across four TPSs. The quadratic least-square fits for each registration method is also shown.
A large time duration between scans was associated with higher TREs for all registration methods. Statistical significance was achieved for rigid registration, Raystation Morfeus and Pinnacle SFBR (p-values 0.014, 0.012 and 0.048, respectively). The other registration methods also recorded substantial differences. Figure 4(b) shows the correlation between the time duration between scans and the TRE for each registration method. Linear least-square trend lines are shown alongside. While most of the registrations show small increases in TREs with time between scans, the increase is particularly striking for Morfeus and to a lesser extent for rigid registration and Pinnacle. None of the other factors (lab abnormalities, liver volume change and slice thickness) led to significant differences in any of registration methods. All these associations are further discussed below.
DISCUSSION
The analysis presented above is supported by the mechanisms of the algorithms used. Focusing on the liver at the rigid registration stage substantially improved the performance of the ANCONDA strategies that did not involve the liver at the DIR stage. The inclusion of the liver for rigid registration for these strategies levelled the field as compared to the ANACONDA strategies that used the liver as a controlling ROI and led to fewer statistically significant pairs. A large tumor volume change resulted in significantly poor performance of Morfeus as the tumor boundary conditions were not modeled. In fact, the overall performance of Morfeus was affected by a major outlier (TRE >20 mm). This patient had by far had the highest percentage change in liver volume (~79%). Morfeus was also unaffected by the choice of rigid registration since its implementation in RayStation performs an internal realignment.
This study highlights the limitations associated with currently available DIR algorithms to handle complex deformations that include volumetric changes. The TPSs used in this study encompass a wide variety of DIR algorithms developed over a period of time from b-splines in the early 2000s to recent ones such as ANACONDA (2015). All DIR algorithms and strategies tested achieved relatively poor accuracy with mean TREs exceeding voxel thickness and maximum TREs greater than 1 cm. Among these, the best performance was obtained in Eclipse (mean TRE 4.6 mm), but even this was nearly double the average slice thickness (2.8 mm). The low performance can partly be attributed to the limitations associated with the flexibility of the registration algorithms on TPSs. Three of the four TPSs (other than RayStation) have limited scope in altering registration parameters, but even different parameterizations of an algorithm (ANACONDA) failed to improve the performance significantly. While more recent state-of-the art DIR algorithms such as GIFTed Demon’s, and DIS-CO,25,26 and advanced boundary conditions in biomechanical models8 may be able to provide some improvements, the full integration of new algorithms into existing TPSs is a lengthy process.
In addition limitations associated with TPSs, the challenge of this task is the major factor for suboptimal performance. The liver is a large organ and undergoes large anatomical changes after radiation therapy. Such challenges have provided similar conclusions in previous studies that encompass a variety of applications27–30. One of the limitations of our study – the low number of points used for TRE calculations, was a result of these challenges. It took an experienced medical physicist about twenty minutes to mark the five points in each image. Adding additional points would not only be time consuming but increase the uncertainty of the TREs as the chances of incorrect markings would increase for non-obvious markers. Due to the focus of this paper on DIR of the liver, structures outside the liver were not considered. However, in applications such as dose accumulation, alignment of surrounding structures (especially bone) may also be important. An optimization scheme that performs DIR within the liver and simultaneous rigid registration of the surrounding bone structures can be one way of addressing this.
CONCLUSIONS
In this study, a comparison of a range of commercially available image registration algorithms was performed on 29 planning and follow-up image pairs from patients with intrahepatic cholangiocarcinoma. Due to the complexity of deformations within the liver, simpler techniques such as rigid and intensity-based registrations had severe limitations. Advanced techniques with greater flexibility can provide an acceptable alternative in some cases. However, significant errors often exist, especially when large liver and tumor volume changes are noted. As repeat treatment for patients continues to be developed, requiring the mapping of dose, and functional imaging advances are made, requiring correlation with delivered dose, improvements in DIR algorithm in this area are needed.
Acknowledgements
This research was supported in part by the National Cancer Institute of the National Institutes of Health under award number 5R01CA221971-02 (PI: Brock) and the Helen Black Image Guided Cancer Therapy Research (IGCTR) Program at UT MD Anderson Cancer Center. The authors would like to thank Jared Ohrt and the members of the MORFEUS laboratory at UT MD Anderson Cancer Center for continuous feedback during the course of this project. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of Interest Statement
Kristy K. Brock reports a conflict of interest with RaySearch Laboratories, Stockholm, Sweden.
References
- 1.Ghouri YA, Mian I, Blechacz B. Cancer review: Cholangiocarcinoma. J Carcinog. 2015;14:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patel T Worldwide trends in mortality from biliary tract malignancies. BMC cancer. 2002;2:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patel N, Benipal B. Incidence of Cholangiocarcinoma in the USA from 2001 to 2015: A US Cancer Statistics Analysis of 50 States. Cureus. 11(1):e3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buettner S, van Vugt JL, JN IJ, Groot Koerkamp B. Intrahepatic cholangiocarcinoma: current perspectives. Onco Targets Ther. 2017;10:1131–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glimelius B, Departments of Oncology UoU, Hoffman K, et al. Chemotherapy improves survival and quality of life in advanced pancreatic and biliary cancer. Annals of Oncology. 1996;7(6):593–600. [DOI] [PubMed] [Google Scholar]
- 6.Thorwarth D Functional imaging for radiotherapy treatment planning: current status and future directions—a review. Br J Radiol. 2015;88(1051). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Treatments for recurrent liver cancer. 2019; https://www.cancer.ca:443/en/cancer-information/cancer-type/liver/treatment/recurrent/?region=on, 2019.
- 8.Polan DF, Feng M, Lawrence TS, Ten Haken RK, Brock KK. Implementing Radiation Dose-Volume Liver Response in Biomechanical Deformable Image Registration. International journal of radiation oncology, biology, physics. 2017;99(4):1004–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim J, Jung Y. Radiation-induced liver disease: current understanding and future perspectives. Exp Mol Med. 2017;49(7):e359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teo JY, Allen JC Jr., Ng DC, et al. A systematic review of contralateral liver lobe hypertrophy after unilobar selective internal radiation therapy with Y90. HPB: the official journal of the International Hepato Pancreato Biliary Association. 2016;18(1):7–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukumitsu N, Nitta K, Terunuma T, et al. Registration error of the liver CT using deformable image registration of MIM Maestro and Velocity AI. BMC medical imaging. 2017;17(1):30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fukumitsu N, Terunuma T, Okumura T, et al. Comparison of rigid and deformable image registration accuracy of the liver during long-term transition after proton beam therapy. Imaging in Medicine. 2017;9(6):149–154. [Google Scholar]
- 13.Gunay G, Luu MH, Moelker A, Walsum Tv, Klein S. Semiautomated registration of pre ‐ and intraoperative CT for image ‐ guided percutaneous liver tumor ablation interventions - Gunay - 2017 - Medical Physics - Wiley Online Library. Medica Physics. 2019;44(7):3718–3725. [DOI] [PubMed] [Google Scholar]
- 14.Chen L-C, Zhu Y, Papandreou G, Schroff F, Adam H. Encoder-Decoder with Atrous Separable Convolution for Semantic Image Segmentation. 2018.
- 15.Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. Paper presented at: Medical Image Computing and Computer-Assisted Intervention (MICCAI)2015. [Google Scholar]
- 16.Anderson BM, Lin E, Cardenas CE, Koay EJ, Brock KK. Automated Contouring of Contrast and Non-Contrast CT Liver Images with Fully Convolutional Neural Networks Paper presented at: ASTRO Annual Conference2018; San Antonio. [Google Scholar]
- 17.Kadoya N, Fujita Y, Katsuta Y, et al. Evaluation of various deformable image registration algorithms for thoracic images. Journal of Radiation Research. 2014;55(1):175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardcastle N, Elmpt Wv, Ruysscher DD, Bzdusek K, Tomé WA. Accuracy of deformable image registration for contour propagation in adaptive lung radiotherapy. Radiation Oncology. 2013;8(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allaire S, Pekar V, Hope AJ, Breen SL, Jaffray DA. Automatic Contour Propagation in Head & Neck IGRT Based on 3D Salient Interest Points. International Journal of Radiation Oncology. 2008;72(1):S87. [Google Scholar]
- 20.Wang H, Dong L, O’Daniel J, et al. Validation of an accelerated ‘demons’ algorithm for deformable image registration in radiation therapy. Physics in medicine and biology. 2005;50(12):2887–2905. [DOI] [PubMed] [Google Scholar]
- 21.Thirion JP. Image matching as a diffusion process: an analogy with Maxwell’s demons. Medical image analysis. 1998;2(3):243–260. [DOI] [PubMed] [Google Scholar]
- 22.Weistrand O, Svensson S. The ANACONDA algorithm for deformable image registration in radiotherapy. Medical physics. 2015;42(1):40–53. [DOI] [PubMed] [Google Scholar]
- 23.Brock KK, Sharpe MB, Dawson LA, Kim SM, Jaffray DA. Accuracy of finite element model-based multi-organ deformable image registration. Medical physics. 2005;32(6):1647–1659. [DOI] [PubMed] [Google Scholar]
- 24.Van Herk M Image Registration Using Chamfer Matching In: Bankman IN, ed. Handbook of Medical Image Processing and Analysis (Second Edition). Academic Press; 2009:591–603. [Google Scholar]
- 25.Papież BW, Kingdom) UoOU, Franklin JM, et al. GIFTed Demons: deformable image registration with local structure-preserving regularization using supervoxels for liver applications. Journal of Medical Imaging. 2019;5(2):024001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rühaak J, Polzin T, Heldmann S, et al. Estimation of Large Motion in Lung CT by Integrating Regularized Keypoint Correspondences into Dense Deformable Registration - IEEE Journals & Magazine. IEEE Transactions on Medical Imaging. of Publication: 05 April 2017;36(8):1746–1757. [DOI] [PubMed] [Google Scholar]
- 27.Zhang A, Li J, Qiu H, Wang W, Guo Y. Comparison of rigid and deformable registration through the…: Medicine. Medicine. 2017;96(50):e9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carrell TWG, Modarai B, Brown JRI, Penney GP. Feasibility and Limitations of an Automated 2D-3D Rigid Image Registration System for Complex Endovascular Aortic Procedures:. Journal of Endovascular Therapy. 2010. [DOI] [PubMed] [Google Scholar]
- 29.Brock KK, Mutic S, McNutt TR, Marc HL, Kessler L. Use of image registration and fusion algorithms and techniques in radiotherapy: Report of the AAPM Radiation Therapy Committee Task Group No. 132 - Brock - 2017 - Medical Physics - Wiley Online Library. Medical physics. 2017(44):7. [DOI] [PubMed] [Google Scholar]
- 30.Laaksonen L, Claridge E, Fält P, Hauta-Kasaric M, Uusitalo H, Lensu L. Comparison of image registration methods for composing spectral retinal images. Biomedical Signal Processing and Control. 2017;36:234–245. [Google Scholar]


