Abstract
MicroRNAs (miRNAs) are a class of small non-coding RNAs that regulate gene expression at the post-transcriptional level and play critical roles in regulating physiological function, and are becoming worldwide research hot spot in brain development and diseases. However, the exact value of miRNAs in brain physiological and pathological processes remain to be fully elucidated, which is vital for the application of miRNAs as diagnostic, prognostic, and therapeutic biomarkers for brain diseases. MicroRNA-7 (miR-7), as a highly expressed miRNA molecule in the mammalian brain, is well documented to play a critical role in development of various diseases. Importantly, accumulating evidence has shown that miR-7 is involved in a range of developmental and pathological processes of brain. Expressively, miR-7, encoded by three genes located different chromosomes, is dominantly expressed in neurons with sensory or neurosecretory. Moreover, the expression of miR-7 is regulated at three levels including gene transcription, process of primary and precursor sequence and formation of mature sequence. Physiologically, miR-7 principally governs the physiological development of Pituitary gland, Optic nervous system and Cerebral cortex. Pathologically, miR-7 can regulate multiple genes thereby manipulating the process of various brain diseases including neurodegenerative diseases, neuroinflammation, and mental disorders and so on. These emerging studies have shown that miR-7, a representative member of miRNA family, might be a novel intrinsic regulatory molecule involved in the physiological and pathological process of brain. Therefore, in-depth studies on the role of miR-7 in brain physiology and pathology undoubtedly not only provide a light on the roles of miRNAs in brain development and diseases, but also are much helpful for ultimate development of therapeutic strategies against brain diseases. In this review, we provide an overview of current scientific knowledge regarding the expression and function of miR-7 in development and disease of brain and raise many issues involved in the relationship between miR-7 and brain physiological and pathological processes.
Keywords: MiR-7, Expression regulation, Physiological function, Brain diseases
Introduction
MicroRNA-7 (miR-7), as a unique member of the miRNAs family, has a high degree of conserved mature sequences with a length of 21–23 nt in different species (Fig. 1). In humans, three genes, including miR-7-1, miR-7-2 and miR-7-3 located on different chromosomes, respectively encode the corresponding precursor sequence which is processed and sheared into the same mature miR-7 sequence [1]. Similarly, in mice, miR-7a-1, miR-7a-2 and miR-7b genes are also located on three different chromosomes and encode the corresponding precursor sequence which is eventually spliced into mature miR-7. Currently, the relevant studies on miR-7 molecule mainly focus on oncology. For example, Kabir et al. reported that miR-7 regulated the growth and invasion of sorafenib-resistant cells in human hepatocellular carcinoma through targeting TYRO3 [2]; Zhao et al. found that miR-7 prevented gastric cancer cell proliferation and tumorigenesis via repressing Nuclear Factor NF-Kappa-B P65 Subunit (P65) and Fos Proto-Oncogene (FOS, AP-1 Transcription Factor Subunit) expression [3]. Rodríguez-Antolín et al. showed that miR-7 methylation was a biomarker to predict poor survival in early-stage non-small cell lung cancer patients [4]. Our previous studies also showed that miR-7 overexpression could reduce the proliferation and metastatic capacity of human lung cancer cells in vitro and in vivo [5–8]. Moreover, the downregulation of miR-7 was closely related to the site mutation of the promoter region, which was correlated with poor prognosis of human lung cancer patients [9]. These foundings indicate that miR-7 might be used as an important potential target molecule for tumor diagnosis and treatment. Significantly, many recent studies have shown that miR-7 is highly enriched in brain tissue and is closely related to physiological and pathological process of brain [10–17], suggesting that it plays an important novel role in brain tissue development and disease occurrence, hence, it is may be a new promising therapeutic target for brain diseases.
Fig. 1.
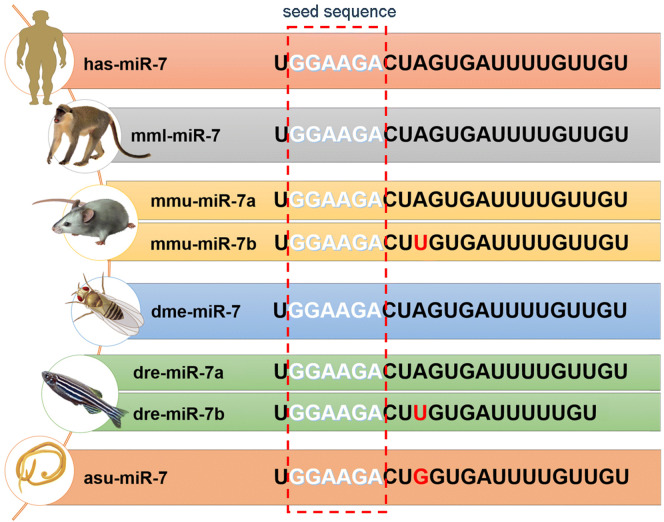
Homologous sequences of mature miR-7 in different species. The mature sequence of miR-7 has a perfectly conserved type in many species, including Homo sapiens miR-7 (has-miR-7), Macaca mulatta miR-7 (mml-miR-7), Mus musculus miR-7 (mmu-miR-7), Drosophila melanogaster miR-7 (dme-miR-7), Danio rerio miR-7 (dre-miR-7), Asacris suum miR-7 (asu-miR-7). A change in one of the bases of miR-7b (highlighted in red) exists in several species. The red virtual box is the seed sequence of mature miR-7
MiR-7 expression in brain
MiR-7 distribution in brain
Many studies have shown that miR-7 and its precursors are expressed in the mammal lens [18], Nucleus accumbens (NACC) and suprachiasmatic nucleus (associated with the reward behavior and circadian rhythm of animals) [19] and neocortical and hippocampal regions [11]. However, the comprehensive data further show that the expression level of miR-7 and its precursor are higher in Pituitary and Hypothalamus [11, 20–22], while lower in Substantia nigra, Striatum, Cerebral cortex and Cerebellum [23]. Researchers speculate that this phenomenon may be related to miR-7-3 (the precursor of mature miR-7), which remains with the Pituitary specific factor 1 (PIT1) gene intron sequence [24]. These data suggest that miR-7 may play distinct roles in different brain regions.
The expression pattern of miR-7 in human brain tissue is also found in Mice [16], Rats [18], Cattle [20], Zebrafish [11], Amphioxus [25] and other species [26, 27]. For example, in Mice, miR-7 is also expressed in neurons with sensory or neurosecretory functions in the Hypothalamus [11, 16, 28]. Furthermore, in Amphioxus, miR-7 is expressed not only in the central nervous system, but also in the most anterior end of the pharyngeal endoderm at the later stage of neuronal development (20–22 h after fertilization) [25]. Moreover, other studies have shown that, in Zebrafish and Medaka, miR-7 has a conserved and highly restricted expression in the Medial forebrain at differentiation stages [29]; Besides, miR-7 is also restricted to be expressed in the developing forebrain with the prohormone vasotocin (vasopressin/oxytocin)-neurophysin from Annelids worms and Zebrafish. Importantly, the cell types characterized with dual sensory-neurosecretory properties in the forebrain are the starting point for the evolution of neurosecretory brain centers in Bilateria [11]. These data suggest that miR-7 expression is closely related with the sensory or/and neurosecretory neurons.
The regulatory mechanisms of miR-7 expression
The expression level of mature miR-7 is affected by a variety of factors, including the direct or indirect regulation on the precursor sequence and mature miR-7. At the level of gene transcription: studies have shown that transcription factors Homeobox D10 (HOXD10) [30] and c-Myc [31] can bind to different parts of the pri-miR-7-1 promoter core sequence respectively to regulate the level of mature miR-7 (the region of c-Myc binding sequence is − 539 to − 534; The binding regions of HOXD10 are − 1028 to − 1019 and − 968 to − 958). In addition, miR-7-1 is the intron of the Heterogeneous Nuclear Ribonucleoprotein K (HNRNPK) gene, and miR-7-3 is the intron of PIT1 gene [16]. As an intron miRNAs molecule, the expression level of miR-7 is also affected by the self-expression regulation mechanism of the host genes. For example, the expression of HNRNPK and PIT1 increases or decreases by the host under the action of certain physiological development and diseases, the expression levels of pri-miR-7-1 and pri-miR-7-3 will also be changed correspondingly, and then the level of mature miR-7 is affected inevitably [16].
In terms of the expression level of primary sequence and precursor sequence: Choudhury et al. found that the level of mature miR-7 enriched in specific brain regions was not consistent with the corresponding primary sequence expression [16], suggesting that there was a relevant regulatory mechanism during the process of the primary sequence of miR-7 (pri-miR-7) to the precursor sequence of miR-7 (pre-miR-7). Similarly, Kumar et al. found that Oleic Acid (OA) could prevent RNA recognition base sequence protein, namely Musashi homologous body 2/human HuR protein, from binding pri-miR-7-1 conservative terminal ring parts [32]. Thus, this process interferes with the formation of pre-miR-7-1 (precursor sequence of miR-7-1), resulting in the inconsistency between the expression of pri-miR-7 sequence and the expression of mature miR-7 sequence.
In addition, multiple regulatory mechanisms also exist at the mature level of miR-7. Studies have found that long non-coding RNA-circR-7 contains about 70 binding sites of miR-7 mature sequence and can effectively interfere with the level of mature miR-7 in a variety of tissue cells [13, 33]. Meanwhile, another long non-coding RNA-Cyrano (linc-oip5, 1700020I14Rik) also has a conserved binding site to mature miR-7 sequence, which can directly bind and reduce the level of mature miR-7, and indirectly regulate the expression of miR-7 target molecules [10, 34]. Interestingly, recent studies also showed that Cerebellar Degeneration Related protein 1 antisense transcription (CDR1as, a complementary antisense sequence of miR-7), as a circular RNA highly expressed in brain neurons cell bodies, neurons axons and retina, could maintain miR-7 expression stability and positively regulate miR-7 level to ensure its regulation effect in neuronal cell activity [10, 12]. In addition, other members of the miRNA family, such as miR-671 also can negatively regulate the level of CDR1as and indirectly affect miR-7 expression [12, 35, 36]. These studies indicate that the regulatory mechanism of mature miR-7 expression is relatively complex (Fig. 2).
Fig. 2.
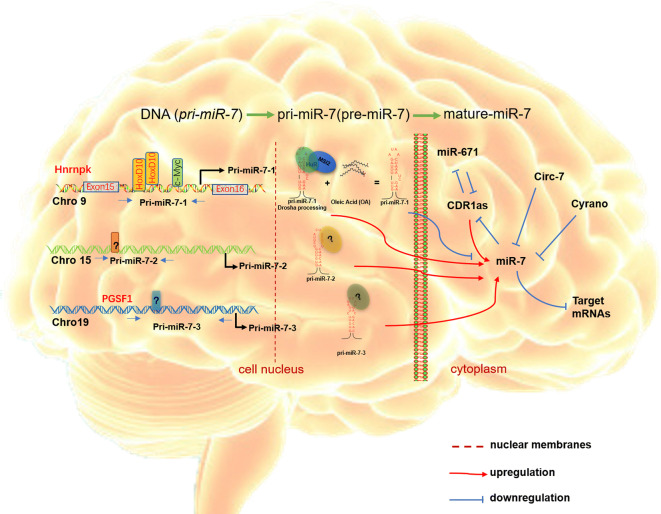
Schematic diagram of the regulatory mechanism of miR-7 expression. MiR-7 is transcribed from three different genomic loci on chromosomes 9, 15, 19 into primary miR-7 transcripts (pri-miR-7-1, pre-miR-7-2, pri-miR-7-3 respectively), which are processed into hairpin precursor molecules pre-miR-7, and then further into the same mature miR-7 sequences. There are always some molecules which affect miR-7 transcription involved in this process. Mature miR-7 sequence can target mRNAs to repress their expression, on the contrary, other antisense sequences interfere with miR-7 level
The role of miR-7 in physiological function of brain
Pituitary gland
Just as we have mentioned above, compared with other regions of the brain, the expression level of miR-7 is higher in the Pituitary gland, which is the most important endocrine gland in the body to regulate hormone secretion, indicating that miR-7 plays an important regulatory role in the process of pituitary hormone secretion. For example, Yuan et al. showed that miR-7 could directly bind to Prostaglandin F2 receptor negative regulator (PTGFRN), and then inhibit the expression of Prostaglandin F2 Receptor (PTGFR), thereby affecting the uterine contraction, ovulation, embryo implantation and other vital reproductive processes [20]. Recently, Ahmed et al. further reported that the effect of miR-7 expression on the Hypothalamus-Pituitary gonad axis. Their studies have shown that, in male and female mice, miR-7a-2 gene deletion, following lower level of miR-7, leads to a low level of gonadotropin and sex steroid hormone, small testicular or ovarian, impaired sperm production, and lack of ovulation, resulting in infertility respectively. But when miR-7-2 overexpression in the Pituitary gland, a raised miR-7 level can inhibit Golgi glycoprotein 1 (GLG1) expression and downstream bone morphogenetic protein 4 (BMP4) signaling pathway, and then reduce the level of PTGFRN and secretion of both Follicle-stimulating hormone (FSH) and Luteinizing Hormone (LH) [37]. These results suggest that the miR-7a-2/miR-7 axis regulates the secretion of FSH and LH by regulating the pituitary prostaglandin production and BMP4 signaling pathway, ultimately affects sexual maturity and reproductive function.
Optic nervous system
MiR-7 also has an important regulating role in the visual system [18, 38–41]. Medullary neurons, as the largest visual processing center of the Drosophila brain, are derived from a sheet of neuroepithelial cells. During larval development, a wave of differentiation sweeps across the neuroepithelium, converting neuroepithelial cells into neuroblasts that sequentially express transcription factors specifying different neuronal cell fates. The switch from neuroepithelial cells to neuroblasts is controlled by a complex gene regulatory network. Caygill et al. found that during this transformation process, the expression level of miR-7 in neuroepithelial cells gradually increased, and the continuously up-regulated miR-7 promoted the stable transformation of neuroepithelial cells into neuroblasts by targeting Notch effector [39]. It is worth noting that miR-7 plays a key buffer and regulatory role in this process to ensure that a precise and rigorous transformation process which can be also maintained even under conditions of environmental stress, echoing the role of miR-7 in the eye imaginal disc [39]. Subsequent studies have shown that miR-7 expression is activated when progenitor cells begin to differentiate into photoreceptor cells, the process that is dependent on the EGF receptor (EGFR) signal which triggers ERK-mediated degradation of transcription factor Yan (a retinal cell differentiation inhibitor). In progenitor cells, Yan can prevent the transcription of miR-7, meanwhile miR-7 can bind with Yan mRNA 3′UTR sequences to inhibit Yan protein expression in the photoreceptor cells. These data have shown that Yan interacts with miR-7 to form a strict feedback effect model, in which Yan is highly expressed in the progenitor cells, while the miR-7 level is higher in the photoreceptor cells; when the EGFR signal shortly triggers Yan degradation in progenitor cells, companied with gradually increased miR-7 level to ensure accurate differentiation of photoreceptor cells [42], indicating that the spatiotemporal expression patterns of Yan and miR-7 are difference. In addition, human Paired box 6 (PAX6) is an important mediator of ocular development and formation. Needhamsen et al. further showed that miR-7 could inhibit the expression of PAX6 protein by directly binding with the 3′UTR of PAX6 [43]. These above studies have proved that miR-7 plays a key regulatory role in the development and formation of visual nerve cells through a sophisticated feedback regulatory network. However, whether miR-7 is involved in the functional regulation of visual neurons remains to be clarified.
Cerebral cortex
Studies have found that, similar to photoreceptor cells, the expression level of miR-7 increases during the differentiation of cerebral cortical nerve cells or the development of Cortex after birth. However, compared with early differentiation of embryonic stem cells into nerve cells in mice, in the early development of cortex cell differentiation, miR-7 level is lower. However, in the seventh day of differentiation, 60–80% of the cells express higher level of miR-7 and neural precursor cells markers (CD57 and SOX1), suggested that miR-7 is closely related to the development of cerebral cortex cells [44]. Subsequent studies have showed downregulated expression of miR-7 in the mouse embryo cortical cells by miRNA-Sponge technology results in brain defect in mice. The mechanism is that when miR-7 is reduced, its target genes related to the p53 pathway, such as the upregulation of adenylate kinase 1 (AK1) and Cyclin dependent kinase inhibitor 1a (Cdkn1a), affects the normal transmission of p53 pathway and leads to the decreased production of cortical progenitor cells during brain development [45]. In addition, Sarangdhar et al. also found that the reduction of miR-7 level during embryonic development resulted in the change of brain morphology of mouse embryos, which was related to the increased expression of Cyrano, a lncRNA that is conservative expression in fertilized egg cells but closely related to brain development. Conversely, lowering the level of Cyrano can increase miR-7 level to improve the morphological abnormalities of fetal brain [34]. Similar studies have also shown that the loss of GLI Family Zinc Finger 3 (GLI3) in mouse embryonic cortex can increase the proportion of neural progenitor cells and newborn neurons in the brain, which resulting in the brain enlargement after birth and the abnormal folding structure of cortical midline. However, altered expression of miR-7 can restore the abnormal brain morphology in mice, as well as the production and migration of neurons. This mechanism is related to the up-regulation of the target molecule PAX6 expression after miR-7 change [46]. However, the relationship between PAX6 and GLI3 remains unclear. In conclusion, it has been proved that miR-7 is a stabilizer of these complex chain feedback and circulatory regulatory network, which is critical for stabilizing gene expression and determining cell fate. However, the complex molecular mechanism of its regulation needs to be further explored.
Other brain functions
The enriched expression of miR-7 in brain regions indicates that it may have other potentially unknown important roles in the brain. Such as Li et al. have shown that miR-7 is involved in the development and functional maintenance of various receptors in the body [41]. Recent studies have shown that miR-7 can enhance the formation of neurons in the subventricular region by inhibiting the Nod-like Receptor Protein 3 (NLRP3)/Caspase-1 pathway in human stem cells, and has a nice repair effect on neurons [47]. Other studies have also shown that miR-7 can be involved in synaptic plasticity in the Hippocampus through targeting the expression of Selenoprotein P (Sepp) [48] and Nuclear Receptor Subfamily 4 Group A Member 3 (NR4A3) [49, 50]. In addition, in Drosophila, mutations in miR-7 sequence do not cause a difference in appearance, but lead to abnormalities in sensory structures in stressful conditions [42, 44]. These researches indicate the potential role of miR-7 in physiological process of brain, which remains to be fully illuminated (Fig. 3).
Fig. 3.
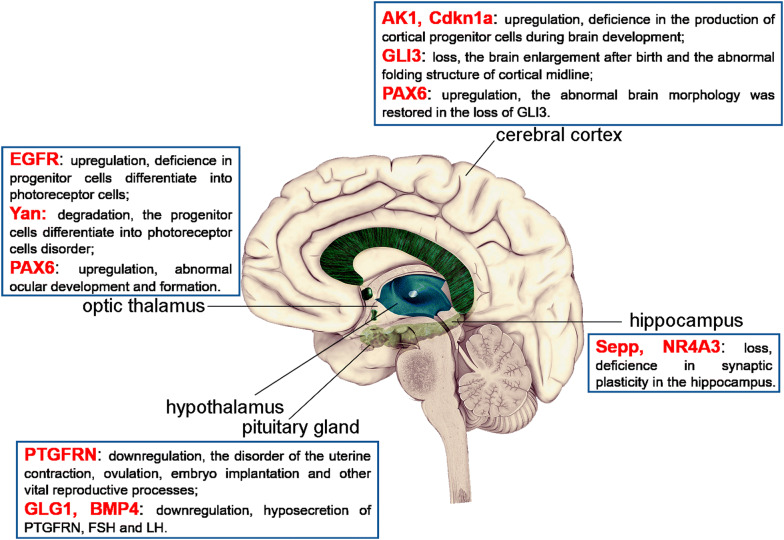
The targets of miR-7 and their function in physiological function of brain. The biological function of miR-7 target molecules in the Pituitary gland, Hypothalamus, Optic thalamus, Cerebral cortex and Hippocampus, respectively. PTGFRN prostaglandin F2 receptor negative regulator, GLG1 golgi glycoprotein 1, BMP4 bone morphogenetic protein 4, FSH follicle-stimulating hormone, EGFR EGF receptor, PAX6 paired box 6, AK1 adenylate kinase 1, Cdkn1a cyclin dependent kinase inhibitor 1a, GLI3 GLI family zinc finger 3, Sepp selenoprotein P, NR4A3 Nuclear Receptor Subfamily 4 Group A Member 3
The role of miR-7 in brain diseases
Accumulating evidence has shown that abnormal expression of miR-7 is involved in the development of various brain diseases, indicating miR-7 has an important role in pathological processes of brain (Fig. 4).
Fig. 4.
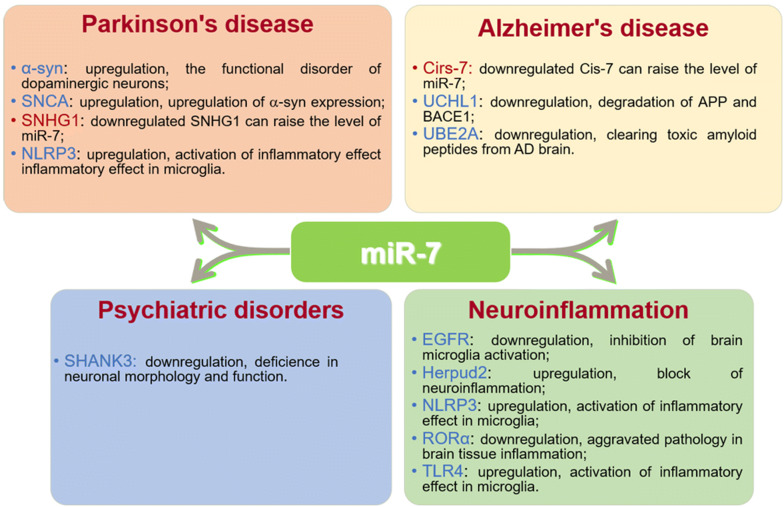
The targets of miR-7 and their function in brain diseases. The effects of miR-7 target molecules in the brain diseases including Parkinson’s disease, Alzheimer’s disease, Psychiatric disorders and Neuroinflammation respectively. The genes highlighted in red are the antisense interfering gene of miR-7. PD Parkinson’s disease, AD Alzheimer’s disease, α-syn α-synuclein, SNCA synuclein alpha, SNHG1 small nucleolar RNA host gene 1, NLRP3 nod-like receptor protein 3, Aβ beta-amyloid proteins, BACE1 beta secretase 1, APP amyloid beta precursor protein, UCHL1 ubiquitin carboxyl‐terminal hydrolase L1, UBE2A ubiquitin conjugating enzyme E2 A, SHANK3 SH3 and multiple ankyrin repeat 3 domains
Neurodegenerative diseases
Parkinson’s disease
The incidence of Parkinson’s disease (PD) accounts for 1% in people over 65 years old. The main characteristics of PD is the gradual loss of dopaminergic neurons in the Substantia nigra, in which abnormal expression of α-synuclein (α-syn) gathered in the form of fiber aggregates in the Substantia nigra and then mediates neurotoxicity to damage the function of dopaminergic neurons [51]. Studies have shown that the level of miR-7 in brain tissues of PD patients and PD animal models decreased significantly [52]. Currently, the mechanism of miR-7 is mainly focused on its regulatory effect on α-syn through multiple ways. Some studies have shown that the lack of miR-7 in brain tissues of PD patients is closely related to the accumulation of α-syn and the loss of dopaminergic neurons in the Substantia nigra, as well as the decreased dopamine secretion in the Striatum [53–55]. Choi et al. reported that miR-7 could accelerate the clearance of α-syn and its polymers, thereby promoting differentiation of ReNcell VM cells (human neural precursor cells) [55]. Junn et al. further found that miR-7 could inhibit α-syn protein level by directly acting on the 3′UTR of α-syn mRNA, thus protecting neurons from oxidative stress and proteasomal damage in MTPT-induced in vitro PD neurotoxin cell model and mouse model [23]. Meanwhile, other studies have shown that miR-7 can bind to the Synuclein Alpha (SNCA) 3′UTR of the Amyloid Precursor and then modulate the expression of α-syn [54]. In addition, miR-7 has been shown to promote the degradation of extracellular synthetic fibers of α-syn [55].
Microglia mediated inflammation reaction is involved in pathogenesis of PD [56, 57]. Studies have shown that miR-7 might regulate the activation of microglia, thereby controlling pathogenesis of PD. For example, in PD patients and dopaminergic SH SY5Y cells treated with MPPT, the expression of Small Nucleolar RNA Host Gene 1 (SNHG1) is increased, while miR-7 expression is decreased. Interestingly, downregulated SNHG1 in vitro can raise the level of miR-7 and inhibit LPS-induced BV2 microglial cells activation and inflammatory effect, and then prevent a potential loss of dopaminergic neurons in the Substantia nigra. Mechanistically, miR-7, as a regulator of SNHG1/NLRP3 axis, has an important regulating role in the inflammatory effect of microglia [58]. Besides, miR-7 can also inhibit the NF-κB signaling pathways to protect neurons from MTPT-induced cytotoxicity (dopaminergic SH SY5Y cells, human neural progenitor cells ReNcell VM cells and primary neurons in mice) [59]. In addition, miR-7 can restore anti-apoptotic protein molecule BCL2 level by inhibiting mTOR signaling pathway, thus protecting the neuron cells damage induced by MTPT in PD model [60]. These studies suggest that miR-7 has a great potential role in the pathogenesis of PD though regulating multiple cells and molecules.
Alzheimer’s disease
Recently, miR-7 has also been found to play an important regulatory role in the development of Alzheimer’s disease (AD). For example, Puthiyedth et al. analyzed the differentially expressed genes in different brain regions, including the Entorhinal Cortex, Hippocampus, Middle temporal gyrus, Posterior cingulate cortex, Superior frontal gyrus and visual cortex brain regions, from 161 clinical brain tissue samples (74 non-demented controls, 87 AD). They found that miR-7-1, a precursor of miR-7, was significantly upregulated in the all regions of brain tissue from AD patients, suggesting that miR-7 was closely related to the occurrence of AD [61]. The abnormal accumulation of beta-amyloid proteins (Aβ) in the brain is a key feature of AD progression. Beta Secretase 1 (BACE1) is a rate-limiting enzyme that forms Aβ and Amyloid beta Precursor Protein (APP) which is a precursor that forms Aβ. Shi et al. showed that Cirs-7, a regulator of BACE1 and APP expression, was significantly down-regulated in the brain tissue of AD patients [62]. Further studies have shown that low CDR1as level can lead to the increased expression of miR-7 which downregulates the activity of ubiquitin conjugating enzyme E2 A (UBE2A), thereby resulting in impaired clearing of toxic amyloid peptides from brain in AD, suggesting that miR-7 may be a new target for AD treatment [36, 63].
Psychiatric disorders
The abnormal miR-7 expression has been also linked with a neurological disorder called Schizophrenia which affects about 1 percent of adults worldwide and has the greatest impact on quality of life compared to other neurological disorders. However, the exact etiology of Schizophrenia is still unknown. Studies have shown that miR-7 expression is up-regulated in the plasma and the frontal lobe of brain in patients with Schizophrenia; Moreover, the effect of miR-7 on Schizophrenia might be linked to directly target SH3 and Multiple Ankyrin Repeat 3 Domains (SHANK3) which is closely related to synaptic plasticity and memory [64–66]. Furthermore, the abnormal level of miR-7 in Schizophrenia is associated with its primary sequence pri-miR-7-3 [67]. These findings indicate that miR-7 might be involved in the development of Schizophrenia, which still need to be further illustrated.
Neuroinflammation
It is well known that inflammation is a common pathological basis for various neurological diseases. Recently, a large number of studies have shown that miR-7 is involved in the occurrence of neuroinflammation [58, 68]. Such as, Cao’s study found that long noncoding RNA SNHG1 promoted neuroinflammation in the pathogenesis of PD via modulating miR-7/NLRP3 pathway [58]; Dong et al. also proved that miR-7 could target the 3′UTR of Herpud2 which encoded Endoplasmic Reticulum (ER) stress protein-HERP2, indicating miR-7-targeted ER stress acted as a molecular brake on neuroinflammation [68]. Furthermore, Zhang et al. showed that miR-7 could inhibit EGFR/STAT3 pathway and TLR4 expression to block brain microglia activation, cytokine production and slow the secondary damage of brain tissue in a Cerebral Hemorrhage model [69]. Separately, in our recent study, we found that miR-7 could govern the pathology of brain tissue inflammation through controlling the inflammatory reaction of neuronal cells in brain tissue inflammation model (BTI) [70]. Therefore, these current findings might highlight not only the important role of miR-7 but also the relationship among neurons and other cells such as microglia in pathogenesis of neuroinflammation.
Other brain diseases
MiR-7 also plays an important regulatory role in other brain diseases. For instance, Nelson et al. found that there was abnormal expression of miR-7 in brain tissue from the patients with Lewy body dementia (DLB), which is characterized by fluctuating cognitive dysfunction, visual hallucinations and Parkinson’s disease syndrome [71]. Moreover, other studies have shown that miR-7 is significantly increased in the peripheral blood of patients with Cerebral artery malformation and is involved in the pathological development of Cerebral artery malformation through targeting VEGF pathway [72]. Recently, Lee et al. also found that the serum expression level of miR-7-5p was significantly increased in Bipolar II disorder patients [73]. Besides, miR-7 expression in peripheral blood from patients with Acute ischemic stroke is higher than healthy controls [74]. Furthermore, inhibition of miR-7 level, in the process of ischemic postconditioning, can prevent mitochondria damage and restore the ATP activity, thereby improving neuron function after focal ischemia, suggesting miR-7 may be a potential therapeutic target in Acute ischemic stroke [74].
Conclusions
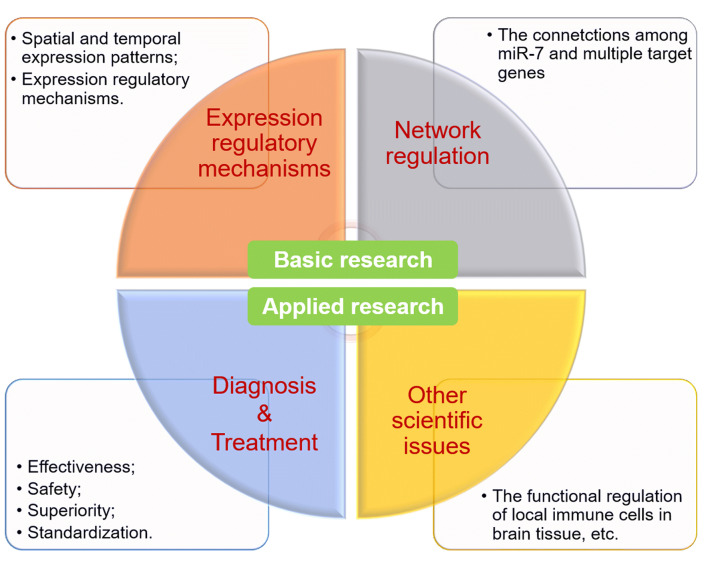
Up to now, the important progression on biological role of miR-7 in brain development and diseases has been reached. Among all of miRNA family members, miR-7 is dominantly expressed in brain tissue and plays important roles in the development of brain tissue and the progression of brain diseases (Table 1), suggesting that miR-7 may be a promising novel intrinsic regulator for brain development and disease occurrence. Alongside, many important scientific issues, at least three major aspects, still need to be further illustrated in the future (Fig. 5). Firstly, what are the spatial and temporal expression patterns of miR-7 and their regulatory mechanisms in the physiological and pathological processes of brain? Especially, the expression patterns of miR-7 in different types of cells in the process of brain development and disease occurrence are still largely unknown. Secondly, the current knowledge thus far on the underlying interactions among miR-7 and its target molecules in brain development and diseases are still limited. Interestingly, in our most recent work, we found that miR-7 synergizes with, but not antagonizes, its target RORα to control the pathology of BTI [70], indicating the complexity of network among miRNAs and their targets in biological process. Therefore, what are the exact connections among miR-7 and its multiple targets in distinct types of cells in different brain diseases? Thirdly, the potential value of miR-7 in the diagnosis and therapeutic strategies of brain diseases need to be further clarify. For instance, the expression level of miR-7 is variable in different brain diseases. We propose it reflects the complexity of the role of miR-7, especially in distinct regions of brain, in different brain diseases. Therefore, the combination of miR-7 expression and other factors, including clinical characters, of different diseases might be much valuable for the application of miR-7 as a bio-maker in diagnosis of brain diseases.
Table 1.
The effects of miR-7 target in physiological and pathological processes of brain
| Targets | Brain regions | Expression | Effects | Refs. |
|---|---|---|---|---|
| PTGFRN | Pituitary gland | Downregulation | lower level of PTGFR and LH release; the decrease of uterine contraction, ovulation, embryo implantation and other vital reproductive processes | [20] |
| GLG1, BMP4 | Pituitary gland | Downregulation | The lower secretion of PTGFRN, FSH and LH | [37] |
| Notch effector | Optic nerve epithelial cells | Downregulation | The more stable transformation of neuroepithelial cells to neuroblastocytes | [39] |
| Yan | Visual progenitor cell | Downregulation | The inhibition of miR-7 transcription, ensure that photoreceptors are successfully and accurately differentiated | [42] |
| PAX6 | Eye and brain | Downregulation | The maintenance of ocular development and formation | [43] |
| Ak1, Cdkn1a | Embryo cortical | Upregulation | Decreased production of cortical progenitor cells during brain development | [45] |
| GLI3 | Embryonic cortex | Loss | The increased proportion of neural progenitor cells and newborn neurons in the brain, which resulting in the brain enlargement after birth and the abnormal folding structure of cortical midline | [46] |
| Sepp, NR4A3 | Hippocampus | Loss | Deficience in the synaptic plasticity in the hippocampus | [48, 49] |
| α-syn | Substantia nigra | Upregulation | Increased neurons from oxidative stress and proteasomal damage, | [23] |
| SNCA | Substantia nigra | Upregulation | The increased expression of α-syn | [54] |
| mTOR | Substantia nigra | Downregulation | Protecting the neuron cells damage | [60] |
| SHANK3 | Cerebral cortex | Downregulation | Deficience in synaptic plasticity and relation with memory and learning | [66] |
| Herpud2 | Brain tissue | Upregulation | Brake on neuroinflammation | [68] |
| TLR4 | Microglia | Downregulation | Block of brain microglia activation, cytokine production and slow the secondary damage of brain tissue | [69] |
| RORα | Neuron | Downregulation | aggravated pathology of brain tissue inflammation | [70] |
PTGFRN Prostaglandin F2 receptor negative regulator; PTGFR prostaglandin F2 receptor; LH luteinizing hormone; GLG1 golgi glycoprotein 1; BMP4 bone morphogenetic protein 4; FSH follicle-stimulating hormone; PAX6 paired box 6; AK1 adenylate kinase 1; Cdkn1a cyclin dependent kinase inhibitor 1a; GLI3 GLI family zinc finger 3; Sepp selenoprotein P; NR4A3 Nuclear Receptor Subfamily 4 Group A Member 3; α-syn α-synuclein; SNCA synuclein alpha; SHANK3 SH3 and multiple ankyrin repeat 3 domains
Fig. 5.
A sketch of scientific issues of miR-7 in the physiological and pathological processes of brain. Currently, there are still many unclear scientific issues, belongs to basic and applied research fields, on the role miR-7 in the development and diseases of brain
In all, the in-depth studies on the expression patterns of miR-7, the networks among miR-7 and its targets, and the role of miR-7 in different types of cells, as well as the prognostic and therapeutic value of miR-7 in brain diseases, will undoubtedly throw a new light on the knowledge on biological roles of miRNAs in physiological and pathological process of brain and ultimately benefit clinical outcome of patients with brain diseases.
Acknowledgements
Not applicable.
Abbreviations
- miR-7
MicroRNA-7
- P65
Nuclear Factor NF-Kappa-B P65 Subunit
- FOS
Fos proto-oncogene
- NACC
Nucleus accumbens
- PIT1
Pituitary specific factor 1
- HOXD10
Homeobox D10
- HNRNPK
Heterogeneous nuclear ribonucleoprotein K
- pri-miR-7
Primary sequence of miR-7
- pre-miR-7
Precursor sequence of miR-7
- MSI2
Musashi homologous body 2
- HuR
Hu antigen R
- pre-miR-7-1
Precursor sequence of miR-7-1
- CDR1as
Cerebellar degeneration related protein 1 antisense transcription
- PTGFRN
Prostaglandin F2 receptor negative regulator
- PTGFR
Prostaglandin F2 receptor
- LH
Luteinizing hormone
- GLG1
Golgi glycoprotein 1
- BMP4
Bone morphogenetic protein 4
- FSH
Follicle-stimulating hormone
- EGFR
EGF receptor
- PAX6
Paired box 6
- AK1
Adenylate kinase 1
- Cdkn1a
Cyclin dependent kinase inhibitor 1a
- GLI3
GLI family zinc finger 3
- Sepp
Selenoprotein P
- NR4A3
Nuclear Receptor Subfamily 4 Group A Member 3
- α-syn
α-synuclein
- SNCA
Synuclein alpha
- SNHG1
Small nucleolar RNA host gene 1
- NLRP3
Nod-like receptor protein 3
- PD
Parkinson’s disease
- AD
Alzheimer’s disease
- Aβ
Beta-amyloid proteins
- BACE1
Beta secretase 1
- APP
Amyloid beta precursor protein
- UCHL1
Ubiquitin carboxyl‐terminal hydrolase L1
- UBE2A
Ubiquitin conjugating enzyme E2 A
- SHANK3
SH3 and multiple ankyrin repeat 3 domains
- ER
Endoplasmic reticulum
- BTI
Brain tissue inflammation model
- DLB
Lewy Body Dementia
Authors’ contributions
This manuscript was written by JZ, YZ, DY, MG and CC Modifications were suggested by GL and LX. critically edited the final manuscript. All authors read and approved the final manuscript.
Funding
This manuscript was supported by National Natural Science foundation of China (31760258, 81960051, 31960509, 81560058), Program for New Century Excellent Talents in University, Ministry of Education of China (NCET-12-0661), Program for High level innovative talents in Guizhou Province (QKH-RC-2016-4031), Program for Excellent Young Talents of Zunyi Medical University (15ZY-001), Program for Science and Technology Joint Fund Project in Zunyi Science and Technology Bureau and Zunyi Medical University (ZSKH-SZ-2016-38) and Project of Guizhou Provincial Department of Science and Technology (QKH-JC-2018-1428).
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Guiyou Liang, Email: guiyou515@163.com.
Lin Xu, Email: xulinzhouya@163.com.
References
- 1.Zhao J, Tao Y, Zhou Y, Qin N, Chen C, Tian D, et al. MicroRNA-7: a promising new target in cancer therapy. Cancer Cell Int. 2015;15:103. doi: 10.1186/s12935-015-0259-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kabir TD, Ganda C, Brown RM, Beveridge DJ, Richardson KL, Chaturvedi V, et al. A microRNA-7/growth arrest specific 6/TYRO3 axis regulates the growth and invasiveness of sorafenib-resistant cells in human hepatocellular carcinoma. Hepatology. 2018;67(1):216–231. doi: 10.1002/hep.29478. [DOI] [PubMed] [Google Scholar]
- 3.Zhao XD, Lu YY, Guo H, Xie HH, He LJ, Shen GF, et al. MicroRNA-7/NF-kappaB signaling regulatory feedback circuit regulates gastric carcinogenesis. J Cell Biol. 2015;210(4):613–627. doi: 10.1083/jcb.201501073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodríguez-Antolín C, Felguera-Selas L, Pernía O, et al. MiR-7 methylation as a biomarker to predict poor survival in early-stage non-small cell lung cancer patients. Cell Biosci. 2019;9:63. doi: 10.1186/s13578-019-0326-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y, Wang C, Zhou Y, Liao Z, Zhu S, Hu Y, et al. TLR9 signaling repressed tumor suppressor miR-7 expression through up-regulation of HuR in human lung cancer cells. Cancer Cell Int. 2013;13(1):90. doi: 10.1186/1475-2867-13-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lei L, Chen C, Zhao J, Wang H, Guo M, Zhou Y, et al. Targeted expression of miR-7 operated by TTF-1 promoter inhibited the growth of human lung cancer through the NDUFA4 pathway. Mol Ther Nucleic Acids. 2017;6:183–197. doi: 10.1016/j.omtn.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen C, Yue D, Lei L, Wang H, Lu J, Zhou Y, et al. Promoter-operating targeted expression of gene therapy in cancer: current stage and prospect. Mol Ther Nucleic Acids. 2018;11:508–514. doi: 10.1016/j.omtn.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu L, Wen Z, Zhou Y, Liu Z, Li Q, Fei G, et al. MicroRNA-7-regulated TLR9 signaling-enhanced growth and metastatic potential of human lung cancer cells by altering the phosphoinositide-3-kinase, regulatory subunit 3/Akt pathway. Mol Biol Cell. 2013;24(1):42–55. doi: 10.1091/mbc.E12-07-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao J, Wang K, Liao Z, Li Y, Yang H, Chen C, et al. Promoter mutation of tumor suppressor microRNA-7 is associated with poor prognosis of lung cancer. Mol Clin Oncol. 2015;3(6):1329–1336. doi: 10.3892/mco.2015.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kleaveland B, Shi CY, Stefano J, Bartel DP. A network of noncoding regulatory RNAs acts in the mammalian brain. Cell. 2018;174(2):350–362. doi: 10.1016/j.cell.2018.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tessmar-Raible K, Raible F, Christodoulou F, Guy K, Rembold M, Hausen H, et al. Conserved sensory-neurosecretory cell types in annelid and fish forebrain: insights into hypothalamus evolution. Cell. 2007;129(7):1389–1400. doi: 10.1016/j.cell.2007.04.041. [DOI] [PubMed] [Google Scholar]
- 12.Piwecka M, Glazar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017 doi: 10.1126/science.aam8526. [DOI] [PubMed] [Google Scholar]
- 13.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 14.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 15.Zhao J, Chen C, Guo M, Tao Y, Cui P, Zhou Y, et al. MicroRNA-7 deficiency ameliorates the pathologies of acute lung injury through elevating KLF4. Front Immunol. 2016;7:389. doi: 10.3389/fimmu.2016.00389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choudhury NR, Alves FL, de Andrés-Aguayo L, Graf T, Cáceres JF, Rappsilber J, et al. Tissue-specific control of brain-enriched miR-7 biogenesis. Genes Dev. 2013;27(1):24–38. doi: 10.1101/gad.199190.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smigielski L, Jagannath V, Rössler W, Walitza S, Grünblatt E. Epigenetic mechanisms in schizophrenia and other psychotic disorders: a systematic review of empirical human findings. Mol Psychiatry. 2020 doi: 10.1038/s41380-019-0601-3. [DOI] [PubMed] [Google Scholar]
- 18.Frederikse PH, Donnelly R, Partyka LM. MiRNA and Dicer in the mammalian lens: expression of brain-specific miRNAs in the lens. Histochem Cell Biol. 2006;126(1):1–8. doi: 10.1007/s00418-005-0139-0. [DOI] [PubMed] [Google Scholar]
- 19.Yaw AM, Duong TV, Nguyen D, Hoffmann HM. Circadian rhythms in the mouse reproductive axis during the estrous cycle and pregnancy. J Neurosci Res. 2020 doi: 10.1002/jnr.24606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan B, Sun GJ, Zhang GL, Wu J, Xu C, Dai LS, et al. Identification of target genes for adenohypophysis-prefer miR-7 and miR-375 in cattle. Genet Mol Res. 2015;14(3):9753–9763. doi: 10.4238/2015. [DOI] [PubMed] [Google Scholar]
- 21.Sanek NA, Young SW. Investigating the in vivo expression patterns of miR-7 microRNA family members in the adult mouse brain. MicroRNA. 2012;1(1):11–18. doi: 10.2174/2211536611201010011. [DOI] [PubMed] [Google Scholar]
- 22.Amar L, Benoit C, Beaumont G, Vacher CM, Crepin D, Taouis M, et al. MicroRNA expression profiling of hypothalamic arcuate and paraventricular nuclei from single rats using Illumina sequencing technology. J Neurosci Methods. 2012;209(1):134–143. doi: 10.1016/j.jneumeth.2012.05.033. [DOI] [PubMed] [Google Scholar]
- 23.Junn E, Lee KW, Byeong SJ, Chan TW, Im JY, Mouradian MM. Repression of α-synuclein expression and toxicity by microRNA-7. Proc Natl Acad Sci USA. 2009;106(31):13052–13057. doi: 10.1073/pnas.0906277106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, et al. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310(5755):1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 25.Candiani S, Moronti L, De Pietri Tonelli D, Garbarino G, Pestarino M. A study of neural-related microRNAs in the developing amphioxus. Evodevo. 2011;2:15. doi: 10.1186/2041-9139-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen J, Zhang S, Tong J, Teng X, Zhang Z, Li S, et al. Whole transcriptome-based miRNA-mRNA network analysis revealed the mechanism of inflammation-immunosuppressive damage caused by cadmium in common carp spleens. Sci Total Environ. 2020;717:137081. doi: 10.1016/j.scitotenv.2020.137081. [DOI] [PubMed] [Google Scholar]
- 27.Li X, Li H, Zhang D, Xu G, Zhang J, Cui S. MiR-7 mediates the signaling pathway of NE affecting FSH and LH synthesis in pig pituitary. J Endocrinol. 2020;244(3):459–471. doi: 10.1530/JOE-19-0331. [DOI] [PubMed] [Google Scholar]
- 28.Lee H, Palkovits M, Young WS. MiR-7b, a microRNA up-regulated in the hypothalamus after chronic hyperosmolar stimulation, inhibits Fos translation. Proc Natl Acad Sci USA. 2006;103(42):15669–15674. doi: 10.1073/pnas.0605781103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ason B, Darnell DK, Wittbrodt B, Berezikov E, Kloosterman WP, Wittbrodt J, et al. Differences in vertebrate microRNA expression. Proc Natl Acad Sci USA. 2006;103(39):14385–14389. doi: 10.1073/pnas.0603529103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reddy SD, Ohshiro K, Rayala SK, Kumar R. MicroRNA-7, a homeobox D10 target, inhibits p21-activated kinase 1 and regulates its functions. Cancer Res. 2008;68(20):8195–8200. doi: 10.1158/0008-5472.CAN-08-2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chou YT, Lin HH, Lien YC, Wang YH, Hong CF, Kao YR, et al. EGFR promotes lung tumorigenesis by activating miR-7 through a Ras/ERK/Myc pathway that targets the Ets2 transcriptional repressor ERF. Cancer Res. 2010;70(21):8822–8831. doi: 10.1158/0008-5472.CAN-10-0638. [DOI] [PubMed] [Google Scholar]
- 32.Kumar S, Downie Ruiz Velasco A, Michlewski G. Oleic acid induces MiR-7 processing through remodeling of Pri-MiR-7/Protein complex. J Mol Biol. 2017;429(11):1638–1649. doi: 10.1016/j.jmb.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73(18):5609–5612. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- 34.Sarangdhar MA, Chaubey D, Srikakulam N, Pillai B. Parentally inherited long non-coding RNA Cyrano is involved in zebrafish neurodevelopment. Nucleic Acids Res. 2018;46(18):9726–9735. doi: 10.1093/nar/gky628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, Liu Y, Liu H, Tang WH. Exosomes: biogenesis, biologic function and clinical potential. Cell Biosci. 2019;9:19. doi: 10.1186/s13578-019-0282-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akhter R. Circular RNA and Alzheimer’s Disease. Adv Exp Med Biol. 2018;1087:239–243. doi: 10.1007/978-981-13-1426-1_19. [DOI] [PubMed] [Google Scholar]
- 37.Ahmed K, LaPierre MP, Gasser E, Denzler R, Yang Y, Rulicke T, et al. Loss of microRNA-7a2 induces hypogonadotropic hypogonadism and infertility. J Clin Invest. 2017;127(3):1061–1074. doi: 10.1172/JCI90031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yongblah K, Alford SC, Ryan BC, Chow RL, Howard PL. Protecting Pax6 3′UTR from MicroRNA-7 partially restores PAX6 in islets from an Aniridia Mouse Model. Mol Ther Nucleic Acids. 2018;13:144–153. doi: 10.1016/j.omtn.2018.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caygill EE, Brand AH. MiR-7 buffers differentiation in the developing drosophila visual system. Cell Rep. 2017;20(6):1255–1261. doi: 10.1016/j.celrep.2017.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arora A, McKay GJ, Simpson DAC. Prediction and verification of miRNA expression in human and rat retinas. Invest Ophthalmol Vis Sci. 2007;48(9):3962–3967. doi: 10.1167/iovs.06-1221. [DOI] [PubMed] [Google Scholar]
- 41.Li X, Cassidy JJ, Reinke CA, Fischboeck S, Carthew RW. A microRNA imparts robustness against environmental fluctuation during development. Cell. 2009;137(2):273–282. doi: 10.1016/j.cell.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li X, Carthew RW. A microRNA mediates EGF receptor signaling and promotes photoreceptor differentiation in the Drosophila eye. Cell. 2005;123(7):1267–1277. doi: 10.1016/j.cell.2005.10.040. [DOI] [PubMed] [Google Scholar]
- 43.Needhamsen M, White RB, Giles KM, Dunlop SA, Thomas MG. Regulation of human PAX6 expression by miR-7. Evol Bioinform Online. 2014;10:107–113. doi: 10.4137/EBO.S13739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen H, Shalom-Feuerstein R, Riley J, Zhang SD, Tucci P, Agostini M, et al. MiR-7 and miR-214 are specifically expressed during neuroblastoma differentiation, cortical development and embryonic stem cells differentiation, and control neurite outgrowth in vitro. Biochem Biophys Res Commun. 2010;394(4):921–927. doi: 10.1016/j.bbrc.2010.03.076. [DOI] [PubMed] [Google Scholar]
- 45.Pollock A, Bian S, Zhang C, Chen Z, Sun T. Growth of the developing cerebral cortex is controlled by microRNA-7 through the p53 pathway. Cell Rep. 2014;7(4):1184–1196. doi: 10.1016/j.celrep.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang L, Mubarak T, Chen Y, Lee T, Pollock A, Sun T. Counter-balance between Gli3 and miR-7 is required for proper morphogenesis and size control of the mouse brain. Front Cell Neurosci. 2018;12:259. doi: 10.3389/fncel.2018.00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fan Z, Lu M, Qiao C, Zhou Y, Ding JH, Hu G. MicroRNA-7 enhances subventricular zone neurogenesis by inhibiting NLRP3/caspase-1 axis in adult neural stem cells. Mol Neurobiol. 2016;53(10):7057–7069. doi: 10.1007/s12035-015-9620-5. [DOI] [PubMed] [Google Scholar]
- 48.Dewing AST, Rueli RH, Robles MJ, Nguyen-Wu ED, Zeyda T, Berry MJ, et al. Expression and regulation of mouse selenoprotein P transcript variants differing in non-coding RNA. RNA Biol. 2012;9(11):1361–1369. doi: 10.4161/rna.22290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stevanato L, Sinden JD. The effects of microRNAs on human neural stem cell differentiation in two- and three-dimensional cultures. Stem Cell Res Ther. 2014;5(2):49. doi: 10.1186/scrt437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pönniö T, Conneely OM. Nor-1 regulates hippocampal axon guidance, pyramidal cell survival, and seizure susceptibility. Mol Cell Biol. 2004;24(20):9070–9078. doi: 10.1128/MCB.24.20.9070-9078.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mouradian MM. MicroRNAs in Parkinson’s disease. Neurobiol Dis. 2012;46(2):279–284. doi: 10.1016/j.nbd.2011.12.046. [DOI] [PubMed] [Google Scholar]
- 52.Horst CH, Schlemmer F, de Aguiar Montenegro N, Domingues ACM, Ferreira GG, da Silva Ribeiro CY, et al. Signature of aberrantly expressed microRNAs in the striatum of rotenone-induced Parkinsonian Rats. Neurochem Res. 2018;43(11):2132–2140. doi: 10.1007/s11064-018-2638-0. [DOI] [PubMed] [Google Scholar]
- 53.Titze-de-Almeida R, Titze-de-Almeida SS. MiR-7 replacement therapy in Parkinson’s disease. Curr Gene Ther. 2018;18(3):143–153. doi: 10.2174/1566523218666180430121323. [DOI] [PubMed] [Google Scholar]
- 54.McMillan KJ, Murray TK, Bengoa-Vergniory N, Cordero-Llana O, Cooper J, Buckley A, et al. Loss of MicroRNA-7 regulation leads to alpha-synuclein accumulation and dopaminergic neuronal loss in vivo. Mol Ther. 2017;25(10):2404–2414. doi: 10.1016/j.ymthe.2017.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Choi DC, Yoo M, Kabaria S, Junn E. MicroRNA-7 facilitates the degradation of alpha-synuclein and its aggregates by promoting autophagy. Neurosci Lett. 2018;678:118–123. doi: 10.1016/j.neulet.2018.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yun SP, Kam T-I, Panicker N, Kim S, Oh Y, Park J-S, et al. Block of A1 astrocyte conversion by microglia is neuroprotective in models of Parkinson’s disease. Nat Med. 2018;24(7):931–938. doi: 10.1038/s41591-018-0051-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ho MS. Microglia in Parkinson’s Disease. Adv Exp Med Biol. 2019;1175:335–353. doi: 10.1007/978-981-13-9913-8_13. [DOI] [PubMed] [Google Scholar]
- 58.Cao B, Wang T, Qu Q, Kang T, Yang Q. Long noncoding RNA SNHG1 promotes neuroinflammation in Parkinson’s disease via regulating miR-7/NLRP3 pathway. Neuroscience. 2018;388:118–127. doi: 10.1016/j.neuroscience.2018.07.019. [DOI] [PubMed] [Google Scholar]
- 59.Choi DC, Chae YJ, Kabaria S, Chaudhuri AD, Jain MR, Li H, et al. MicroRNA-7 protects against 1-methyl-4-phenylpyridinium-induced cell death by targeting RelA. J Neurosci. 2014;34(38):12725–12737. doi: 10.1523/JNEUROSCI.0985-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fragkouli A, Doxakis E. MiR-7 and miR-153 protect neurons against MPP(+)-induced cell death via upregulation of mTOR pathway. Front Cell Neurosci. 2014;8:182. doi: 10.3389/fncel.2014.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Puthiyedth N, Riveros C, Berretta R, Moscato P. Identification of differentially expressed genes through integrated study of Alzheimer’s disease affected brain regions. PLoS ONE. 2016;11(4):e0152342. doi: 10.1371/journal.pone.0152342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shi Z, Chen T, Yao Q, Zheng L, Zhang Z, Wang J, et al. The circular RNA ciRS-7 promotes APP and BACE1 degradation in an NF-kappaB-dependent manner. FEBS J. 2017;284(7):1096–1109. doi: 10.1111/febs.14045. [DOI] [PubMed] [Google Scholar]
- 63.Madadi S, Schwarzenbach H, Saidijam M, Mahjub R, Soleimani M. Potential microRNA-related targets in clearance pathways of amyloid-β: novel therapeutic approach for the treatment of Alzheimer’s disease. Cell Biosci. 2019;9:91. doi: 10.1186/s13578-019-0354-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Perkins DO, Jeffries CD, Jarskog LF, Thomson JM, Woods K, Newman MA, et al. MicroRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol. 2007;8(2):R27. doi: 10.1186/gb-2007-8-2-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Choi SY, Pang K, Kim JY, Ryu JR, Kang H, Liu Z, et al. Post-transcriptional regulation of SHANK3 expression by microRNAs related to multiple neuropsychiatric disorders. Mol Brain. 2015;8(1):74. doi: 10.1186/s13041-015-0165-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang J, Sun XY, Zhang LY. MicroRNA-7/Shank3 axis involved in schizophrenia pathogenesis. J Clin Neurosci. 2015;22(8):1254–1257. doi: 10.1016/j.jocn.2015.01.031. [DOI] [PubMed] [Google Scholar]
- 67.Zhao H, Xu J, Pang L, Zhang Y, Fan H, Liu L, et al. Genome-wide DNA methylome reveals the dysfunction of intronic microRNAs in major psychosis. BMC Med Genomics. 2015;8:62. doi: 10.1186/s12920-015-0139-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dong YF, Chen ZZ, Zhao Z, Yang DD, Yan H, Ji J, et al. Potential role of microRNA-7 in the anti-neuroinflammation effects of nicorandil in astrocytes induced by oxygen-glucose deprivation. J Neuroinflamm. 2016;13(1):60. doi: 10.1186/s12974-016-0527-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang XD, Fan QY, Qiu Z, Chen S. MiR-7 alleviates secondary inflammatory response of microglia caused by cerebral hemorrhage through inhibiting TLR4 expression. Eur Rev Med Pharmacol Sci. 2018;22(17):5597–5604. doi: 10.26355/eurrev_201809_15824. [DOI] [PubMed] [Google Scholar]
- 70.Yue D, Zhao J, Chen H, Guo M, Chen C, Zhou Y, et al. MicroRNA-7, synergizes with RORalpha, negatively controls the pathology of brain tissue inflammation. J Neuroinflamm. 2020;17(1):28. doi: 10.1186/s12974-020-1710-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nelson PT, Wang WX, Janse SA, Thompson KL. MicroRNA expression patterns in human anterior cingulate and motor cortex: a study of dementia with Lewy bodies cases and controls. Brain Res. 2018;1678:374–383. doi: 10.1016/j.brainres.2017.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen Y, Li Z, Shi Y, Huang G, Chen L, Tan H, et al. Deep sequencing of small RNAs in blood of patients with brain arteriovenous malformations. World Neurosurg. 2018;115:e570–e579. doi: 10.1016/j.wneu.2018.04.097. [DOI] [PubMed] [Google Scholar]
- 73.Lee S, Lu R, Wang L, Chang C, Lu T, Wang T, et al. Serum miRNA as a possible biomarker in the diagnosis of bipolar II disorder. Sci Rep. 2020;10(1):1131. doi: 10.1038/s41598-020-58195-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yao GY, Zhu Q, Xia J, Chen FJ, Huang M, Liu J, et al. Ischemic postconditioning confers cerebroprotection by stabilizing VDACs after brain ischemia. Cell Death Dis. 2018;9(10):1033. doi: 10.1038/s41419-018-1089-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.