Abstract
Proteins of modern terrestrial organisms are composed of nearly 20 amino acids; however, the amino acid sets of primitive organisms may have contained fewer than 20 amino acids. Furthermore, the full set of 20 amino acids is not required by some proteins to encode their function. Indeed, simplified variants of Escherichia coli (E. coli) orotate phosphoribosyltransferase (OPRTase) constructed by Akanuma et al. and composed of a limited amino acid set exhibit significant catalytic activity for the growth of E. coli. However, its structural details are currently unclear. Here, we predict the structures of simplified variants of OPRTase using molecular dynamics (MD) simulations and evaluate the accuracy of the MD simulations for simplified proteins. The three-dimensional structure of the wild-type was largely maintained in the simplified variants, but differences in the catalyst loop and C-terminal helix were observed. These results are considered sufficient to elucidate the differences in catalytic activity between the wild-type and simplified OPRTase variants. Thus, using MD simulations to make structural predictions appears to be a useful strategy when investigating non-wild-type proteins composed of reduced amino acid sets.
Introduction
Proteins are key functional molecules in contemporary organisms on planet Earth. They are involved in many biological processes, catalysis of metabolic reactions, signal transduction, and providing structure to cells.1−3 The information required for protein synthesis is coded on nucleic acids. In addition, proteins play a significant role in the replication of nucleic acid polymers, that is, DNA and RNA.4 Although each organism possesses different features, they are derived from the last universal common ancestor (LUCA).5,6 The size of the amino acid alphabet of proteins in the LUCA must have been dependent on the genetic code.
Protein function is associated with tertiary structures that depend on amino acid sequences.7 Modern organisms use a set of 20 amino acids to synthesize proteins. However, much fewer than 20 amino acids are considered essential for maintaining stable protein structures and catalytic functions.8,9 Indeed, previous studies have revealed that the full set of 20 amino acids is not vital for the production of functional proteins.10−23 Primitive proteins are hypothesized to have been composed of limited sets of amino acids. [GADV]-proteins, which are one of the primitive proteins, are thought to be constructed of only glycine, alanine, aspartic acid, and valine.10−12 In previous studies, [GADV]-proteins composed of 20 residues exhibited the ability to form secondary structures, and multimers of [GADV]-proteins have been suggested to have catalytic moieties.13,14 Other simplified amino acid sets have been tested to construct simplified proteins.15−22 Simple helical proteins of approximately 100 residues can be constructed from sets of 7–9 amino acids.15,16 A simplified four-helix protein bundle with a rigid structure can be constructed from Ala, Gln, Glu, Gly, Leu, Lys, and Ser.15 Furthermore, a simplified variant of AroQ chorismate mutase, constructed from a nine-amino acid set (Asn, Asp, Arg, Glu, Ile, Leu, Lys, Met, and Phe), is fully functional in vivo, although the residues at the active site are substituted.16 Moreover, this catalytic activity can be improved 10-fold by two mutations (Ile30Thr and Leu61Val).17 Functional de novo α-helical proteins with heme-binding, peroxidase, esterase, or lipase activity can be developed with sets of fewer than 15 different amino acids (from Asn, Asp, Arg, Gln, Glu, Gly, His, Leu, Lys, Met, Phe, Ser, Trp, Tyr, and Val).18 In addition, small β-sheet proteins have been constructed from various combinations of restricted amino acid sets, and the simplified variants are stable and functional.19 More complex proteins can also be reconstructed from limited sets of amino acids.20−22 A protein composed of three α-helices and three β-strands can be generated from seven amino acids (Ala, Glu, Ile, Leu, Lys, Thr, and Val).20 Moreover, ancestral nucleoside diphosphate kinase of archaea (Arc1), which is composed of 139 residues, can be reconstructed from 10 to 16 amino acids.21 Although the wild-type of Arc1 is hexameric, a simplified protein with 10 amino acids (Ala, Asn, Asp, Arg, Glu, Gly, His, Leu, Pro, and Val) is dimeric; a simplified variant composed of 13 amino acids (Ala, Asn, Asp, Arg, Glu, Gly, His, Leu, Lys, Pro, Ser, Tyr, and Val) is also functional. Previously, Akanuma et al.22 succeeded in reconstructing the 213-residue Escherichia coli (E. coli) orotate phosphoribosyltransferase (OPRTase) with a 13-amino acid alphabet (Ala, Asp, Arg, Glu, Gly, Leu, Lys, Met, Phe, Pro, Ser, Tyr, and Val). OPRTase catalyzes Mg2+-dependent orotidine 5′-monophosphate formation from orotate and α-d-ribosyldiphosphate 5-phosphate (PRPP).23 This function is involved in a de novo biosynthesis pathway of pyrimidine nucleotides. Akanuma et al.22 evaluated OPRTase activity using E. coli strains with a simplified OPRTase in a minimum medium without uracil. Despite the estimated catalytic activity of the simplified variant constructed by these authors being reduced relative to that of the wild-type, it had sufficient catalytic activity for uracil auxotrophic growth of the E. coli strain. However, the three-dimensional (3D) structures of the simplified variants were not resolved on the atomic level, and a detailed study of active site properties for the variants was not conducted. In addition, analyses of stability and secondary structure formation were not performed. In the present study, we therefore attempted to predict the structures of simplified variants using molecular dynamics (MD) simulations.
MD simulations are used to predict or refine protein structures. In previous studies, the structural features of natural mutant proteins have been investigated by MD simulations.24−26 In our study, we attempted to analyze the structural features of artificially simplified variants of OPRTase using MD simulations and investigate whether MD simulations are useful for evaluating proteins composed of artificially simplified amino acid sets. To assess the effects of the number of substituted residues on the structural changes and to confirm the results obtained from simplified variants, simulations were performed for the wild-type, a moderately simplified variant, and a completely simplified variant. Because OPRTase forms a homodimer, simulations of monomeric and dimeric structures for the wild-type and variants were performed.
Results and Discussion
The completely simplified OPRTase (Simp-2), previously constructed by Akanuma et al.,22 was used in this study.23 Simp-2 was constructed by introducing 23 rounds of mutagenesis. The structure of the middle (moderately simplified) variant, which was used for comparison purposes, was simplified through 12 rounds of mutagenesis, that is, between the wild-type and Simp-2. In addition, simulations for two monomer structures were performed to verify that the calculated structures converged to similar structures. Amino acid sequences of all targets for MD simulations are presented in Figure 1. In the middle variant and Simp-2, the amino acid residues of the catalyst loop are mutated (Asn99Asp, Lys103Arg, His105Tyr, and Glu107Asp). On the other hand, the residues related to the interaction with the cofactor PRPP are unmutated (Lys26, Tyr72, Lys73, Arg99, Lys100, Lys103, Asp124, Asp125, Thr128, Ala129, Gly130, Thr131, and Ala132). Moreover, Lys26, Phe35, and Arg156, which interact with the substrate, are not substituted.
Figure 1.
Amino acid sequences of the wild-type and two simplified variants of OPRTase. Substituted amino acid residues are indicated in red. Wild: wild-type; middle: moderately simplified variant; Simp-2: completely simplified variant.
Root-Mean-Square Deviations
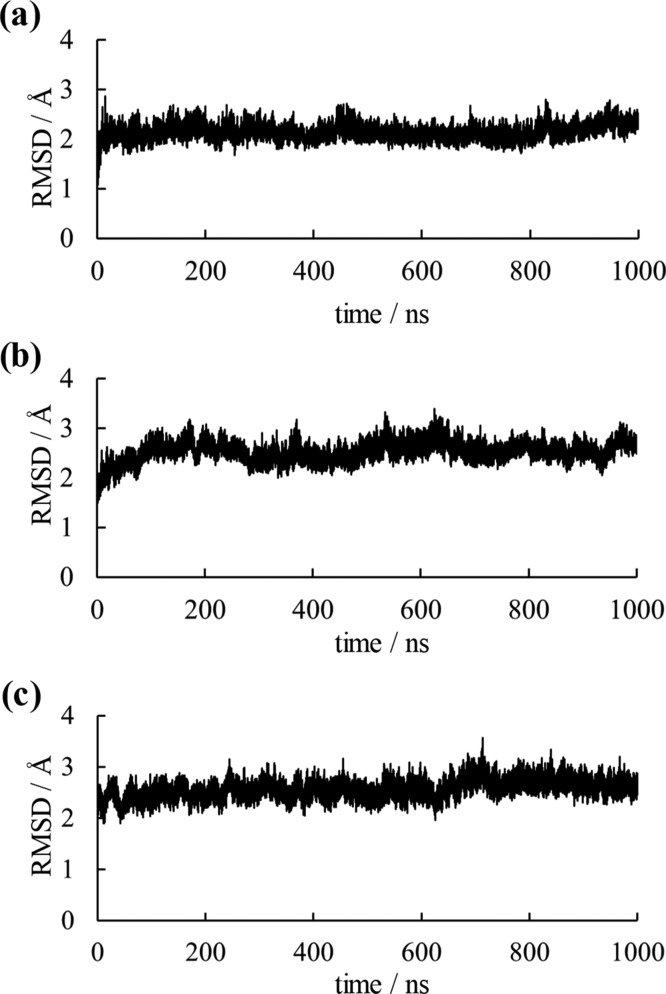
To evaluate simulation convergences, root-mean-square deviations (RMSDs) for main-chain atoms are presented in Figures 2 and S1. Where MD simulations were not effective for structural predictions of artificially simplified proteins, RMSD values would be large with structural collapse. As shown in Figure 2, the RMSD values of dimeric structures converged for all simulations. For monomeric structures, although fluctuations of the RMSD values were observed during simulations, these values did not increase continuously and were almost constant by the end of the simulations. Because fluctuations in RMSD values were observed even in wild-type simulations, these fluctuations were considered to have resulted from the solvent exposure of the protein–protein interaction surface caused by the monomerization of OPRTase. Therefore, MD simulations are suggested to be applicable to the structural investigation of simplified proteins.
Figure 2.
RMSD plots for the main-chain atom of OPRTase dimers. RMSD plots of (a) the wild-type, (b) the middle variant, and (c) Simp-2 for dimers are shown.
Structural Domains
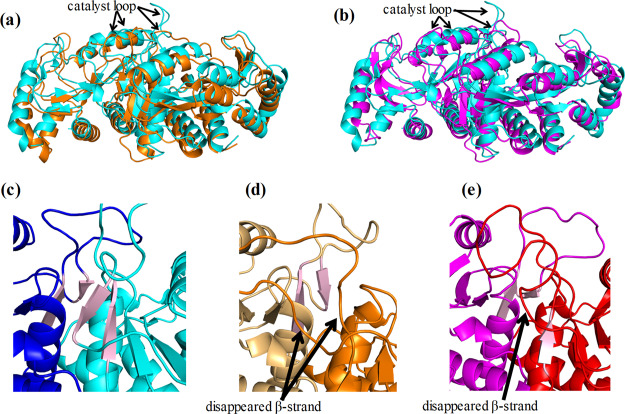
To investigate the effects of the simplification of amino acid sequences on protein structures, 3D structures were compared. To provide an example of the converged structures, the structures at the end of the simulations of monomers and dimers are presented in Figures S2 and 3, respectively. The structural motifs were almost maintained in all calculated structures. For monomers, although the RMSD values throughout the simulations were larger than those for dimers (Figure S1), the overall structures of the monomers were conserved similar to those of the dimers (Figure S2). The RMSD values between monomers 1 and 2 were 3.69, 3.37, and 3.63 Å in the wild-type, the middle variant, and Simp-2, respectively. The differences between loops were relatively substantial for the monomers. In dimers, however, the structures of each simplified variant retained nearly the same structure as the wild-type; this explains why Simp-2 exhibits sufficient activity for the growth of E. coli in a minimum medium without uracil. Nevertheless, two structural changes were observed in the simplified variants: structural changes were found at the positions of the C-terminal helices and secondary structures around the catalyst loops. Secondary structure formation in the last 100 ns simulations is shown in Table 1. In this table, H and S represent helix and beta-strand structures, respectively. Although the helix formation of the C-terminus remained in both chains of the middle variant, it unfolded at Tyr207, Arg208, and Asp209 in chain B of Simp-2. Furthermore, structural changes were observed in the catalyst loops of the middle variant and Simp-2. The β-strand formations on the C-terminal side of the catalyst loops disappeared in chain A of the middle variant. In Simp-2, the residues of β-strand S4 on the C-terminal side of the catalyst loop were changed in chain A. Although S5 in the wild-type was composed of the residues 111–113, S5 in Simp-2 was composed of the residues 109–111. In the final structures, the β-strand formation on the N-terminal side of the catalyst loop in the middle variant and Simp-2 disappeared (Figure 3c–e). The secondary structures of the calculated structures for monomers were similar to those for the dimers (Table S1). These structural changes may be related to the attenuation of the catalytic activity of OPRTase.
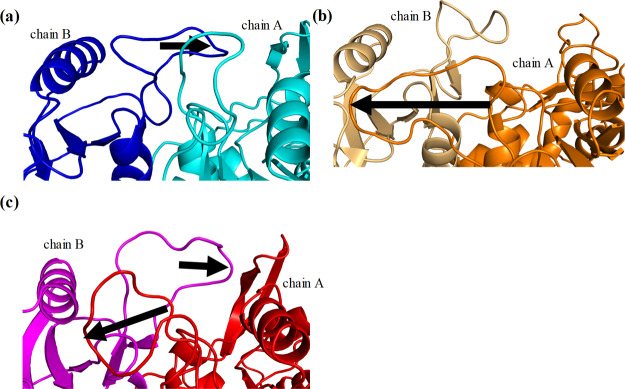
Figure 3.
Comparison between the calculated dimer structures of the wild-type and those of two simplified variants. The structural superposition of the calculated structures for the (a) wild-type–middle variant and (b) wild-type–Simp-2 are shown. The structures of the wild-type, the middle variant, and Simp-2 are indicated in cyan, orange, and purple, respectively. The β-strand formations around the catalyst loops of (c) the wild-type, (d) the middle variant, and (e) Simp-2 in the final structures of simulations are also illustrated. The β-strands S4 and S5 are shown in pink.
Table 1. Secondary Structure Formation in >50% of Trajectories for the Last 100 ns of Simulations for the Dimersa.
| wild A | wild B | middle A | middle B | Simp-2 A | Simp-2 B | |
|---|---|---|---|---|---|---|
| H1 | 3–14 | 3–14 | 3–14 | 3–14 | 3–14 | 3–14 |
| S1 | 18–21, 23–24 | 18–21, 23–24 | 18–21, 23–24 | 18–21, 23–24 | 18–21, 23–24 | 18–21, 23 |
| S2 | 30–35 | 30–35 | 30–35 | 30–35 | 30–34 | 31–35 |
| H2 | 37–39 | 37–39 | 37–39 | 37–39 | 37–39 | 37–39 |
| H3 | 43–59 | 43–60 | 42–59 | 43–60 | 43–59 | 43–60 |
| S3 | 66–68 | 66–69 | 66–68 | 66–69 | 66–68 | 66–69 |
| H4 | 75–89 | 75–89 | 76–88 | 74–89 | 75–88 | 75–89 |
| S4 | 95–98 | 95–98 | 95–96 | 95–97 | 95 | 95–97 |
| S5 | 111–113 | 111–113 | not detected | 112 | 109–111 | 112–113 |
| S6 | 119–123 | 119–123 | 119–123 | 119–123 | 119–124 | 119–123 |
| H5 | 132–142 | 134–142 | 132–142 | 129–134, 137–142 | 133–142 | 134–142 |
| S7 | 146–147, 149–155 | 146–147, 149–153 | 146–147, 149–154 | 146–147, 149–153, 155 | 146–152 | 146–147, 149–155 |
| H6 | 166–173 | 168–174 | 166–174 | 169–173 | 166–173 | 167–174 |
| S8 | 177–183 | 179–182 | 177–182 | 178–181, 183 | 177–180 | 177–183 |
| H7 | 184–191 | 184–191 | 184–191 | 184–192 | 184–192 | 184–193 |
| H8 | 195–211 | 195–210 | 195–210 | 195–210 | 198–209 | 195–206, 210–211 |
H: Helix (α-helix and 3–10 helix); S: para and anti-β-sheet.
C-Terminal Helices in Simplified Variants
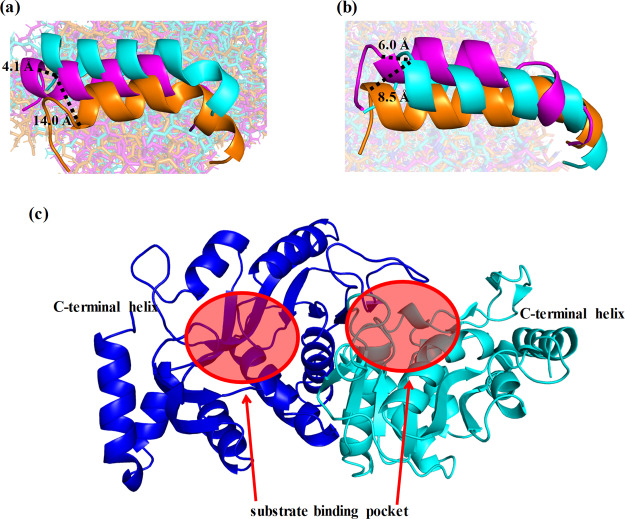
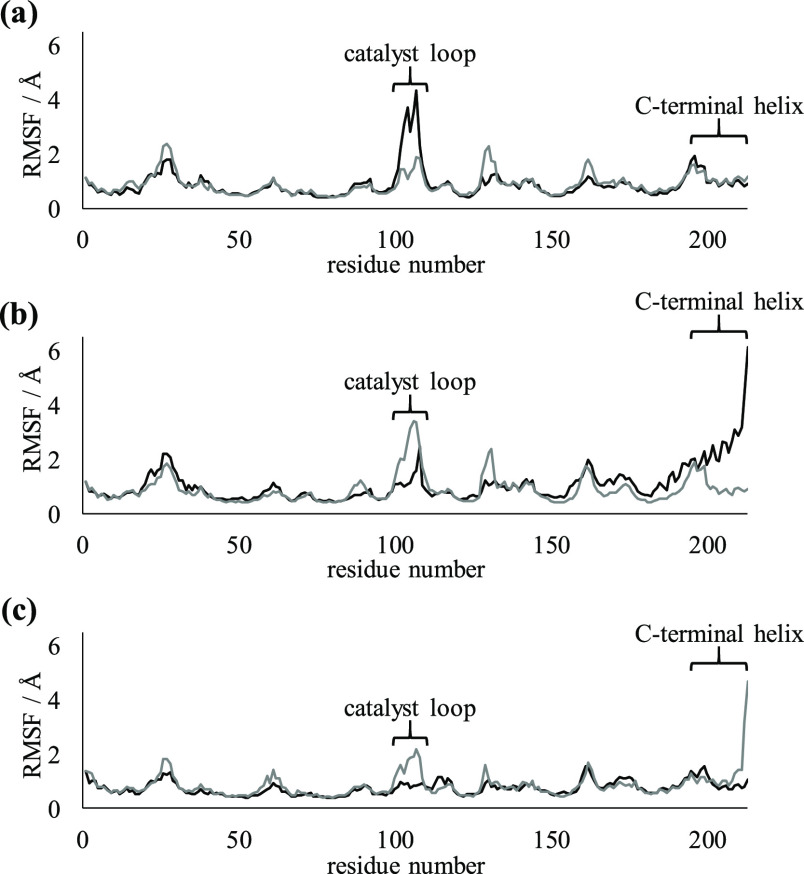
To investigate the effects of simplification on shifts in C-terminal helices in dimers, the locations of the C-terminal helices were evaluated (Figure 4). The C-terminal helix of chain A in the middle variant was largely shifted into the vicinity of H7, in comparison with that of the wild-type. Because no mutations are introduced in the C-terminal helix in the middle variant, the shift of this helix was assumed to be caused by an increase in flexibility because of the simplification of other regions. The root-mean-square fluctuations (RMSFs) of the C-terminal helices for chain A in the middle variant and chain B in Simp-2 were higher than that of the wild-type (Figure 5). The RMSF value of the C-terminal helix in monomer 1 was also higher than that in monomer 2, even in the wild-type (Figure S3). The C-terminal helices are exposed to solvent molecules and not involved in interactions with another subunit. The increase in the RMSF values in the C-terminal helices are not related to monomerization. Because C-terminal helices were adjacent to the residues composing substrate-binding pockets, these helices may be stabilized by ligand binding. However, the unfolding of the C-terminal helix was observed only in Simp-2. The simplification is considered to affect the flexibility of that helix. In addition, because the C-terminal helix-deleted mutant of OPRTase exhibited less activity than the wild-type in a previous study of Thermus thermophilus, the structural flexibilities of C-terminal helices could affect the catalytic activity of OPRTase, even in E. coli.27,28 Hence, we suggest that the simplification of amino acid sequences in OPRTase has a negative effect on substrate binding through the increased flexibility of C-terminal helices.
Figure 4.
Differences in the locations and structures of C-terminal helices of dimers. C-terminal helices in (a) chain A and (b) chain B were compared among the wild-type (cyan), middle variant (orange), and Simp-2 (purple). The dotted lines indicate the distance among the Cα atoms of C-terminal residues in the helices. (c) Positions of substrate-binding pockets and C-terminal helices are shown.
Figure 5.
Comparison of RMSFs between chain A and chain B in dimers. The RMSF plots of dimers in (a) the wild-type, (b) the middle variant, and (c) Simp-2. Plots for chain A and chain B are indicated by black lines and gray lines, respectively.
Catalyst Loops of Simplified Variants
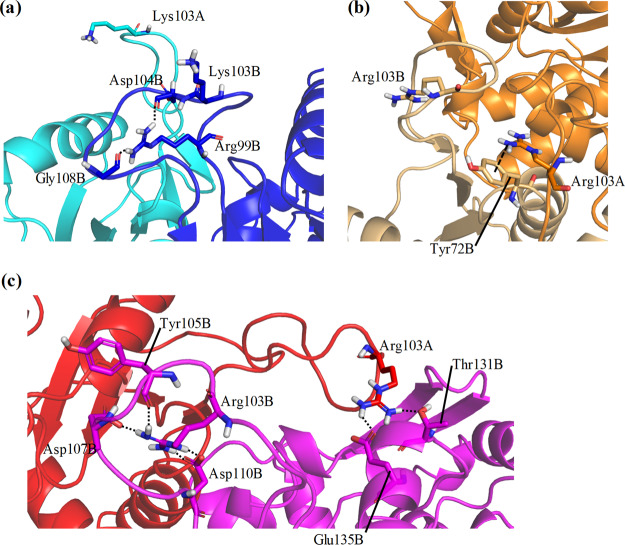
To understand the mechanism underlying the alteration in flexibilities and shifts of catalyst loops, their specific structures and hydrogen-bond formation were analyzed. The catalyst loops are important for the catalyst activity of OPRTase; in particular, Lys103Ala or Lys103Gln mutations considerably reduce the catalyst activity.29 The RMSFs of catalyst loops in one chain of dimeric structures were larger than those in other regions (Figure 5). In monomeric structures, the flexibilities of catalyst loops were high in the wild-type and in the middle variant (Figure S3). This phenomenon was observed in only one calculated structure in Simp-2 monomers. Because the catalyst loops are located on the interface of the OPRTase dimer, they are considered to be unstable in monomeric structures. In addition, differences in the flexibilities of catalyst loops were observed between the wild-type and the two simplified variants in dimeric structures. The RMSF values of catalyst loops in the middle variant were slightly lower than those of the wild-type, and the values in Simp-2 were even lower. In E. coli OPRTase, the catalyst loop of one subunit is flexible, whereas another interacts with the adjacent subunit.30 This difference is considered to be important for substrate recognition. For further details on how simplification affected the catalyst loops, we assessed the structures in the vicinity of these loops (Figure 6). In the wild-type, the catalyst loop of chain B, for which the RMSF values were lower than those of the loop in chain A, was attracted to chain A. However, the catalyst loop of chain A was not attracted to chain B. In the middle variant, the catalyst loop of chain B was not attracted to chain A, but that of chain A was attracted to chain B. Although the detailed reaction mechanism is under debate, the interaction between catalyst loops and another subunit is postulated to be important for PRPP binding and catalytic activity.30−32 The RMSF value of the attracted catalyst loop in chain A was lower than that in chain B. In Simp-2, each catalyst loop was attracted to the other chain. The distance between chain B and the catalyst loop of chain A was relatively small compared with that between chain A and the catalyst loop of chain B. The RMSF value of the largely attracted catalyst loop was smaller than that of the catalyst loop of another chain. This structural asymmetry is thought to cause differences in the RMSFs of catalyst loops. Furthermore, interactions between the side chain of Lys103 (wild-type) or Arg103 (middle variant and Simp-2) and the catalyst loop were important to the latter’s flexibility (Figure 7). The hydrogen bond shown in Figure 7 was formed in more than 70% of the trajectory frames in the last 100 ns of simulations. In Simp-2, for which the RMSF values of the catalyst loops were lower than those of the wild-type and the middle variant, Arg103 of chain B (Arg103B) formed hydrogen bonds with Asp107B, Asp110B, and Tyr105B (Figure 7C). In addition, Arg103A of Simp-2 formed hydrogen bonds with Thr131B and Glu135B. In the middle variant, Arg103B did not frequently form hydrogen bonds with any residues. Although the frequencies of hydrogen-bond formations for Arg103A in the middle variant were lower than those of Simp-2, the cation−π interaction between Arg103A and Tyr72B appeared to stabilize the position of the catalyst loop (Figure 7B). Because mutations of Lys103 have been reported to decrease the catalytic activity of OPRTase and this lysine residue interacts with PRPP,31,32 these interactions in the middle variant and Simp-2 are thought to affect catalytic activity. However, Lys103 in chain A and B of the wild-type formed few hydrogen bonds with any residues. Instead, Arg99B formed two hydrogen bonds with Asp104B and Gly108B (Figure 7A). The average distances between hydrogen of Arg99B and hydrogen-bond forming oxygen of Asp104B and Gly108B were 1.98 Å [standard deviation (SD) 0.289 Å] and 2.33 Å (SD 0.695 Å), respectively. In Simp-2, the distances between the oxygen atoms in the side chain of Asp110B and hydrogen of Arg103B were 2.30 Å (SD 0.363 Å) and 2.26 Å (SD 0.369 Å), respectively, indicating that the interaction between these atoms may be relatively weak in comparison to the interaction between hydrogen of Arg99B and oxygen of Asp104B. However, Arg103B formed four hydrogen bonds in Simp-2, and the average distances between hydrogen of Arg104B and oxygen of Asp107B and Tyr105B were 1.91 Å (SD 0.247 Å) and 2.03 Å (SD 0.321 Å), respectively. Overall, the distances between the hydrogen-bond forming atoms in the catalyst loop are not considered to be different. In the wild-type, the distance between hydrogen of Lys103B and nitrogen atoms of His105 or oxygen atoms of Glu107 was approximately 10 Å. In addition, the distances between hydrogen of Lys103A and oxygen of Thr131B were >20 Å. Therefore, the distances among the hydrogen-bond forming atoms of catalyst loops were similar for the wild-type and Simp-2. The number of hydrogen bonds is expected to effectively alter the structures and flexibilities of catalyst loops. These results suggest that structural stabilization through interactions involved in the side chain of Arg/Lys residues is an important factor in changing the flexibility of the catalyst loop. Furthermore, changes in catalyst loop flexibilities are expected to affect substrate recognition.
Figure 6.
Structural changes in the catalyst loops through attraction to another subunit. Structures of catalyst loops of (a) the wild-type, (b) the middle variant, and (c) Simp-2 are shown. Arrows indicate that the catalyst loops are close enough to interact with another subunit.
Figure 7.
Hydrogen-bond formation in catalyst loops. The catalyst loops of (a) the wild-type (b) the middle variant, and (c) Simp-2 in dimers are shown. Black dotted lines indicate the hydrogen bonds, and the dashed line indicates a possible cation−π interaction. The rates of occurrence of hydrogen bonds shown here were >70% of trajectories for the last 100 ns of simulations.
Conclusions
We investigated the effects of simplification of amino acid sequences on the 3D structure of OPRTase of E. coli using MD simulations. All simulations converged, and the dimer structures of the simplified variants closely resembled that of the wild-type. The 3D structures of OPRTase did not collapse because of the simplification of the amino acid sequences. Experimental data suggest that Simp-2 has adequate catalytic activity,22 and the calculated 3D structures were consistent with the experimental data. However, structural differences in the C-terminal helices and catalyst loops among the wild-type and simplified variants were observed. In particular, simplification caused changes to hydrogen-bond formation in the catalyst loops. The altered structural features observed for the simplified OPRTase variants are likely to be the cause of their decreased catalytic activities. Indeed, where the catalytic activities of Simp-2 have been experimentally observed, they are reportedly lower than those of the wild-type.22 Further simulations including complexes of simplified OPRTase and substrates will fully clarify the effects of simplification on these catalytic reactions. The results of the present study, however, indicate that MD simulations can reproduce the 3D structures of artificially simplified proteins. Such computational methods can be expected to aid and develop the study of primitive proteins.
Computational Methods
The initial structure of the wild-type was constructed from an experimental structure registered in the protein data bank (PDB). The crystal structure of the OPRTase dimer (PDB ID: 1ORO) was obtained from PDB.30 Initially, all water and sulfuric acid molecules were deleted from this structure. Because residues of the catalyst loop in chain B are disordered, chain A was used to simulate monomers. To construct the initial structure of the dimer, disordered residues in chain B were generated using the geometries of the catalyst loop in chain A. The system was then solvated using TIP3P water33 and neutralized by adding sodium ions. All calculations were performed under periodic boundary conditions. The particle mesh Ewald method was used to calculate electrostatic interactions.34 The cutoff distance for the calculations of the nonbonding interactions was set at 10 Å. The time step of MD simulations was 2 fs. SHAKE algorithm was applied to constrain the lengths of bonds containing hydrogen atoms.35 Energy minimization of water molecules and ions was performed for 1000 cycles, and then energy minimization of the entire system was performed for 2500 cycles. After these minimizations, temperature-increasing MD simulations were performed for 20 ps, with the temperature increased from 0 to 300 K. Equilibrating MD simulations were subsequently performed under a constant temperature and pressure. The simulation times of equilibrating MD simulations for monomers and dimers were 3000 and 1000 ns, respectively, which converged the RMSDs. AMBER16 was used to perform all calculations.36 AMBER ff14SB force field was used for the amino acid parameters,37 and RMSDs (for the main-chain atom) and RMSFs (for the Cα atom using the last 100 ns MD trajectories) were calculated using the cpptraj module of AmberTools16. Hydrogen bonds were analyzed using the last 100 ns MD trajectories. Secondary structure formations were detected by DSSP. The structural features of natural mutant proteins have previously been efficiently investigated using typical MD simulations from initial structures that were constructed and based on wild-type structures.24−26 Therefore, the initial structures of Simp-2 and the middle variant were based on the calculated structure of the wild-type after MD simulations. For the two simplified variants, MD simulations were performed under the same conditions as those for the wild-type.
Acknowledgments
This work was supported by Grants-in-Aid for Scientific Research (17K08257) from the Japan Society for the Promotion of Science.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.0c01012.
RMSD plots; final structures; secondary structure formations; and RMSF plots of monomers for the wild-type, the middle variant, and Simp-2 (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Agarwal P. K. A biophysical perspective on enzyme catalysis. Biochemistry 2019, 58, 438–449. 10.1021/acs.biochem.8b01004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss G.Biochemistry of dignal transduction and regulation; Wiley-VCH, 2003; pp. 7–51. [Google Scholar]
- Wickstead B.; Gull K. The evolution of the cytoskeleton. J. Cell Biol. 2011, 194, 513–525. 10.1083/jcb.201102065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane A. N.; Fan T. W.-M. Regulation of mammalian nucleotide metabolism and biosynthesis. Nucleic Acids Res. 2015, 43, 2466–2485. 10.1093/nar/gkv047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penny D.; Poole A. The nature of the last universal common ancestor. Curr. Opin. Genet. Dev. 1999, 9, 672–677. 10.1016/s0959-437x(99)00020-9. [DOI] [PubMed] [Google Scholar]
- Camprubí E.; de Leeuw J. W.; House C. H.; Raulin F.; Russell M. J.; Spang A.; Tirumalai M. R.; Westall F. The emergence of life. Space Sci. Rev. 2016, 205, 285–348.28057962 [Google Scholar]
- Chou P. Y.; Fasman G. D. Prediction of protein conformation. Biochemistry 1974, 13, 222–245. 10.1021/bi00699a002. [DOI] [PubMed] [Google Scholar]
- Sander C.; Schulz G. E. Degeneracy of the information contained in amino acid sequences: evidence from overlaid genes. J. Mol. Evol. 1979, 13, 245–252. 10.1007/bf01739483. [DOI] [PubMed] [Google Scholar]
- Tanaka J.; Doi N.; Takashima H.; Yanagawa H. Comparative characterization of random-sequence proteins consisting of 5, 12, and 20 kinds of amino acids. Protein Sci. 2010, 19, 786–795. 10.1002/pro.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikehara K. Origins of gene, genetic code, protein and life: comprehensive view of life systems from a GNC-SNS primitive genetic code hypothesis. J. Biosci. 2002, 27, 165–186. 10.1007/bf02703773. [DOI] [PubMed] [Google Scholar]
- Oba T.; Fukushima J.; Maruyama M.; Iwamoto R.; Ikehara K. Catalytic activities of [GADV]-peptides. Formation and establishment of [GADV]-protein world for the emergence of life. Orig. Life Evol. Biosph. 2005, 35, 447–460. 10.1007/s11084-005-3519-5. [DOI] [PubMed] [Google Scholar]
- Ikehara K. Evolutionary steps in the emergence of life deduced from the bottom-up approach and GADV hypothesis (Top-Down Approach). Life 2016, 6, E6 10.3390/life6010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda A.; Fukuyoshi S. Predicting three-dimensional conformations of peptides constructed of only glycine, alanine, aspartic acid, and valine. Orig. Life Evol. Biosph. 2015, 45, 183–193. 10.1007/s11084-015-9418-5. [DOI] [PubMed] [Google Scholar]
- Oda A.; Nakayoshi T.; Kato K.; Fukuyoshi S.; Kurimoto E. Three dimensional structures of putative, primitive proteins to investigate the origin of homochirality. Sci. Rep. 2019, 9, 11594. 10.1038/s41598-019-48134-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafmeister C. E.; LaPorte S. L.; Miercke L. J. W.; Stroud R. M. A designed four helix bundle protein with native-like structure. Nat. Struct. Mol. Biol. 1997, 4, 1039–1046. 10.1038/nsb1297-1039. [DOI] [PubMed] [Google Scholar]
- Walter K. U.; Vamvaca K.; Hilvert D. An active enzyme constructed from a 9-amino acid alphabet. J. Biol. Chem. 2005, 280, 37742–37746. 10.1074/jbc.m507210200. [DOI] [PubMed] [Google Scholar]
- Müller M. M.; Allison J. R.; Hongdilokkul N.; Gaillon L.; Kast P.; van Gunsteren W. F.; Marlière P.; Hilvert D. Directed evolution of a model primordial enzyme provides insights into the development of the genetic code. PLoS Genet. 2013, 9, e1003187 10.1371/journal.pgen.1003187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S. C.; Bradley L. H.; Jinadasa S. P.; Hecht M. H. Cofactor binding and enzymatic activity in an unevolved superfamily of de novo designed 4-helix bundle proteins. Protein Sci. 2009, 18, 1388–1400. 10.1002/pro.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka J.; Yanagawa H.; Doi N. Comparison of the frequency of functional SH3 domains with different limited sets of amino acids using mRNA display. PLoS One 2011, 6, e18034 10.1371/journal.pone.0018034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumawid M. T.; Takahashi T.; Yamazaki T.; Ashigai H.; Mihara H. Selection and structural analysis of de novo proteins from an α3β3 genetic library. Protein Sci. 2009, 18, 384–398. 10.1002/pro.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibue R.; Sasamoto T.; Shimada M.; Zhang B.; Yamagishi A.; Akanuma S. Comprehensive reduction of amino acid set in a protein suggests the importance of prebiotic amino acids for stable proteins. Sci. Rep. 2018, 8, 1227. 10.1038/s41598-018-19561-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akanuma S.; Kigawa T.; Yokoyama S. Combinatorial mutagenesis to restrict amino acid usage in an enzyme to a reduced set. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 13549–13553. 10.1073/pnas.222243999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musick W. D. L.; Nyhan W. L. Structural features of the phosphoribosyltransferases and their relationship to the human deficiency disorders of purine and pyrimidine metabolism. Crit. Rev. Biochem. 1981, 11, 1–34. 10.3109/10409238109108698. [DOI] [PubMed] [Google Scholar]
- Watanabe Y.; Fukuyoshi S.; Kato K.; Hiratsuka M.; Yamaotsu N.; Hirono S.; Gouda H.; Oda A. Investigation of substrate recognition for cytochrome P450 1A2 mediated by water molecules using docking and molecular dynamics simulations. J. Mol. Graph. Model. 2017, 74, 326–336. 10.1016/j.jmgm.2017.04.006. [DOI] [PubMed] [Google Scholar]
- Bouard C.; Terreux R.; Tissier A.; Jacqueroud L.; Vigneron A.; Ansieau S.; Puisieux A.; Payen L. Destabilization of the TWIST1/E12 complex dimerization following the R154P point-mutation of TWIST1: an in silico approach. BMC Struct. Biol. 2017, 17, 6. 10.1186/s12900-017-0076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narang S. S.; Shuaib S.; Goyal D.; Goyal B. Assessing the effect of D59P mutation in the DE loop region in amyloid aggregation propensity of β2-microglobulin: A molecular dynamics simulation study. J. Cell. Biochem. 2018, 119, 782–792. 10.1002/jcb.26241. [DOI] [PubMed] [Google Scholar]
- Hamana H.; Shinozawa T. Effects of C-terminal deletion on the activity and thermostability of orotate phosphoribosyltransferase from Thermus thermophilus. J. Biochem. 1999, 125, 109–114. 10.1093/oxfordjournals.jbchem.a022246. [DOI] [PubMed] [Google Scholar]
- Surekha K.; Prabhu D.; Richard M.; Nachiappan M.; Biswal J.; Jeyakanthan J. Investigation of vital pathogenic target orotate phosphoribosyltransferases (OPRTase) from Thermus thermophilus HB8: Phylogenetic and molecular modeling approach. Gene 2016, 583, 102–111. 10.1016/j.gene.2016.02.006. [DOI] [PubMed] [Google Scholar]
- Ozturk D. H.; Dorfman R. H.; Scapin G.; Sacchettini J. C.; Grubmeyer C. Locations and functional roles of conserved lysine residues in Salmonella typhimurium orotate phosphoribosyltransferase. Biochemistry 1995, 34, 10755–10763. 10.1021/bi00034a007. [DOI] [PubMed] [Google Scholar]
- Henriksen A.; Aghajari N.; Jensen K. F.; Gajhede M. A flexible loop at the dimer interface is a part of the active site of the adjacent monomer of Escherichia coli orotate phosphoribosyltransferase. Biochemistry 1996, 35, 3803–3809. 10.1021/bi952226y. [DOI] [PubMed] [Google Scholar]
- Grubmeyer C.; Segura E.; Dorfman R. Active site lysines in orotate phosphoribosyltransferase. J. Biol. Chem. 1993, 268, 20299–304. [PubMed] [Google Scholar]
- Roca M.; Navas-Yuste S.; Zinovjev K.; López-Estepa M.; Gómez S.; Fernández F. J.; Vega M. C.; Tuñón I. Elucidating the catalytic reaction mechanism of orotate phosphoribosyltransferase by means of X-ray crystallography and computational simulations. ACS Catal. 2020, 10, 1871–1885. 10.1021/acscatal.9b05294. [DOI] [Google Scholar]
- Joung I. S.; Cheatham T. E. III. Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J. Phys. Chem. B 2008, 112, 9020–9041. 10.1021/jp8001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darden T.; York D.; Pedersen L. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. 10.1063/1.464397. [DOI] [Google Scholar]
- Ryckaert J.-P.; Ciccotti G.; Berendsen H. J. C. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977, 23, 327–341. 10.1016/0021-9991(77)90098-5. [DOI] [Google Scholar]
- Case D. A.; Babin V.; Berryman J. T.; Betz R. M.; Cai Q.; Cerutti D. S.; Cheatham T. E. III; Darden T. A.; Duke R. E.; Gohlke H.; Goetz A. W.; Gusarov S.; Homeyer H.; Janowski P.; Kaus J.; Kolossvary I.; Kovalenko A.; Lee T. S.; LeGrand S.; Luchko T.; Luo R.; Madej B.; Merz K. M.; Paesani F.; Roe D. R.; Roitberg A.; Sagui C.; Salomon-Ferrer R.; Seabra G.; Simmerling C. L.; Smith W.; Swails J.; Walker R. C.; Wang J.; Wolf R. M.; Wu X.; Kollman P. A.; AMBER16; University of California: San Francisco, 2012. [Google Scholar]
- Maier J. A.; Martinez C.; Kasavajhala K.; Wickstrom L.; Hauser K. E.; Simmerling C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.