Summary
One of the most common brain tumors in children and adults is the glioma or astrocytoma. There are few effective therapies for these cancers, and patients with malignant glioma fare poorly, even after aggressive surgery, chemotherapy and radiation. Over the past decade, it is now appreciated that these tumors are comprised of numerous distinct neoplastic and non-neoplastic cell populations, which could each influence overall tumor biology and response to therapy. Among these non-cancerous cell types, monocytes (microglia and macrophages) predominate. In this review, we discuss the complex interactions involving microglia and macrophages relevant to glioma formation, progression, and response to therapy.
Introduction
The most common brain tumor arising in children and adults is the glioma (or astrocytoma). These tumors are classified by the World Health Organization according to histologic features, running the spectrum from low-grade (I and II) to high-grade (III and IV) gliomas. With the advent of molecular diagnostics, the nosology of adult glioma has changed [Louis et al., 2016], now incorporating the presence or absence of specific genetic (e.g., IDH1 and IDH2 mutations) and genomic (e.g., chromosome 1p/19q co-deletions) alterations. Similarly, pediatric diffuse gliomas are molecularly defined by mutations in histone H3 genes (H3-K27M-mutant diffuse midline gliomas).
Glioma tissue is not only composed of cancer cells, but is also infiltrated by non-transformed cells, predominantly resident microglia from the brain and circulating blood monocytes (macrophages), which comprise 30–50% of the cellular content of these tumors [Simmons et al., 2011]. Microglia are the brain resident immune cells, first described by Pio del Rio Hortega in 1919 [Sierra et al., 2016], and later reported by Wilder Penfield to infiltrate glioma tissue as cells with an amoeboid morphology, similar to microglia found in multiple sclerosis or Alzheimer disease tissue [Penfield, 1925].
Microglia develop from primitive yolk sac myeloid precursors, and enter the brain during embryogenesis [Ginhoux et al., 2010] to mature into distinct populations of central nervous system (CNS) monocytes. Additionally, there may be other ontologies for brain microglia that reflect different waves of yolk sac hematopoiesis [De et al., 2018]. In contrast to resident microglia, macrophages most typically enter the brain in the context of pathology, either from the blood (bone marrow-derived macrophages) or through direct vascular channels that connect the skull bone marrow to the brain [Herisson et al., 2018].
Over the last decade, it has become clear that glioma associated microglia and infiltrated macrophages from the peripheral system (GAM) are not merely bystander or “reactive” immune system-like cells, but interact with numerous other cell types to actively influence brain tumor biology. However, there is still no consensus on the clinical significance of GAM infiltration relevant to patient outcome. In one study, there was a positive association between the numbers of CD68-, CD163- and CD206-positive GAM in the vital tumor core and prolonged overall survival of patients with IDH1R132H-non-mutant GBM [Zeiner et al., 2018]. Similarly, others found that microglia density, as determined by Iba1 labelling, correlated with improved outcomes for patients with gliomas, whereas CD204 expression in GAM was associated with poorer survival [Sorensen et al., 2018]. In contrast, the expression of microglia/macrophage-related genes in adult and pediatric malignant gliomas of the mesenchymal subtype revealed a negative correlation between GAM accumulation and patient survival in adults, but not children [Engler et al., 2012]. While GAM enrichment is higher in the mesenchymal subtype of high-grade glioma relative to proneural or classical gliomas [Kaffes et al., 2019], the mechanisms underlying GAM-mediated patient survival remain to be determined.
GAM regulate glioma formation and progression
Numerous studies have revealed that GAM are critical for both low-grade and high-grade gliomagenesis and continued tumor growth. In experimental malignant (high-grade) glioma models, microglia depletion reduces glioma growth [Hambardzumyan et al., 2016] (Figure 1). While the mechanisms underlying this pro-tumoral effect likely vary from tumor to tumor, numerous potential etiologies have been identified. In these studies, TGF-ß, stress-inducible Protein (STI)-1, IL-6, IL-1ß and EGF are factors released from GAM, which each can promote tumor growth [Hambardzumyan et al., 2016]. Moreover, microglia, but not bone marrow-derived macrophages, induce platelet-derived growth factor receptor expression in a subset of tumor cells of mouse and human low and high-grade glioma. This receptor expression stimulates the migratory capacity of glioma cells, which accelerates tumor progression [Wallmann et al., 2018]. Similarly, GAM are the predominant source of the secreted form of osteopontin/SPP1, which suppresses glioma growth in some contexts [Szulzewsky et al., 2018] and reduces glioma cell apoptosis in other experimental settings [P Chen et al., 2019].
Figure 1. GAM increase glioma growth through the release of mitogens and invasion promoting factors.
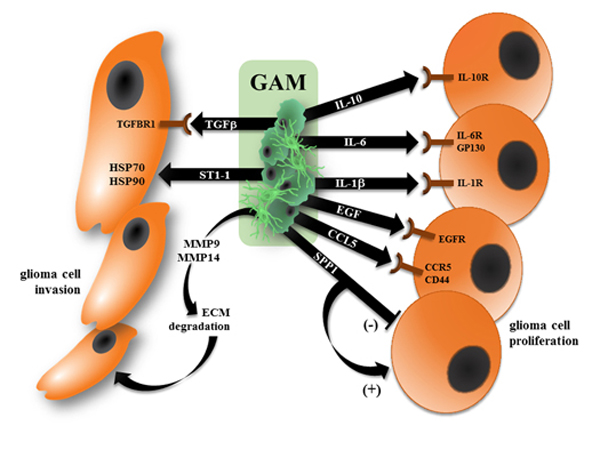
Brain microglia and infiltrating peripheral macrophages (GAM) become reprogrammed to produce growth factors that increase glioma cell proliferation, attenuate glioma cell apoptosis, and promote tumor cell migration. In this fashion, GAM produce factors that enhance (IL-10, IL-6, IL-1, EGF, and CCL5) through binding to their cognate receptors (right). In addition, SPP1 has been reported to both increase (“+”) and inhibit (“−“) glioma cell growth. Moreover, GAM elaborate other factors that increase extracellular matrix (ECM) degradation and directly promote glioma cell invasion and motility (left).
In contrast to their malignant counterparts, low-grade gliomas predominate in children and harbor a paucity of genetic mutations. These tumors arise either sporadically, caused frequently by genomic alterations involving the BRAF kinase gene (KIAA1549:BRAF fusion) [Pfister et al., 2008] [J Yu et al., 2009], or in the setting of the Neurofibromatosis type 1 (NF1) cancer predisposition syndrome [Listernick et al., 1989]. Importantly, Nf1 loss or BRAF fusion is usually not sufficient for glioma formation in rodents, unless coupled with supportive growth factors from the tumor microenvironment. In this regard, Nf1 loss [Larribere et al., 2015] or KIAA1549:BRAF expression [Jacob et al., 2011] in numerous cell types induces cellular senescence, which requires stromal signals to overcome and lead to neoplasia.
To explore the role of these permissive stromal signals, studies using Nf1 genetically engineered mouse strains that develop optic glioma, the signature brain tumor seen in children with NF1, have uncovered essential roles for microglia in glioma formation and maintenance. First, optic glioma formation is delayed in mice with reduced expression of the key receptor involved in directed microglia migration (CX3CR1) [Pong et al., 2013a]. Second, optic glioma growth in vivo is attenuated following treatment with either minocycline (crude microglia inhibitor) [Daginakatte and Gutmann, 2007] [Pan et al., 2017b] or JNK inhibitors (targeting the signalling pathway hyperactivated in Nf1-mutant microglia) [Daginakatte et al., 2008]. Third, RNA sequencing of murine Nf1 optic glioma-associated microglia demonstrated that these GAM secrete CCL5, which is a potent growth factor for glioma cells [Solga et al., 2015]. Treatment of Nf1 optic glioma-bearing mice with a neutralizing CCL5 antibody dramatically attenuated tumor growth in vivo. Consistent with the idea that high-grade gliomas gain some measure of stromal independence, malignant gliomas harboring NF1 mutations express CCL5, establishing an autocrine loop for mesenchymal glioblastoma survival [Pan et al., 2017a]. The requirement for microglia and microglia-produced Ccl5 is further underscored by studies in which murine Nf1 optic glioma stem cells are implanted into mice. While wild-type mice support the formation of glioma-like lesions following transplantation [Y H Chen et al., 2015], no tumors form in mice lacking the two chemokine receptors that direct monocyte migration (Cx3cr1 and Ccr2) or in mice deficient in Ccl5 expression [Pan et al., 2018] [Guo et al., 2019]. While less well studied, KIAA1549:BRAF-expressing low-grade gliomas are also dependent on monocytes in the tumor microenvironment, such that gliomas fail to form following the transplantation of murine KIAA1549:BRAF-expressing neural stem cells in Ccr2-deficient mice [R Chen et al., 2019].
Glioma cells attract/recruit GAM
There are a variety of glioma-derived factors that can function as chemoattractants for GAM, including CCL2, CX3CL1, SDF-1, CSF-1, GM-CSF, LOX and potentially EGF [Roesch et al., 2018]. This directed monocyte recruitment occurs through the establishment of chemokine gradients that attract monocytes to the evolving tumor bed (Figure 2). Studies employing Ccr2 and Cx3cr1 reporter mice have revealed that two of the most important chemoattractants are Ccl2 and Cx3cl1, which traditionally have been thought to drive directional migration of macrophages and microglia, respectively. In this regard, Ccl2 is produced by low-grade glioma stem cells expressing the KIAA1549:BRAF genomic alteration, leading to monocyte attraction [R Chen et al., 2019], whereas NF1 mutation in both low-grade and high-grade gliomas attracts microglia through Cx3cl1 [Z Chen et al., 2017] [Guo et al., 2019]. Similarly, in experimental glioblastoma models, Ccl2 is produced by the tumor cells, which attract macrophages [Platten et al., 2003], such that Ccl2/Ccr2 inhibition prolongs mouse survival [Z Chen et al., 2017] [Hutter et al., 2019]. While there are few studies that specifically focused on chemokine networks responsible for monocyte attraction, other molecules are also likely involved in recruiting monocytes to the tumor [Dijksterhuis et al., 2015] [Brandenburg et al., 2016].
Figure 2. Glioma cells recruit and reprogram GAM through the elaboration of chemokines and other soluble factors.
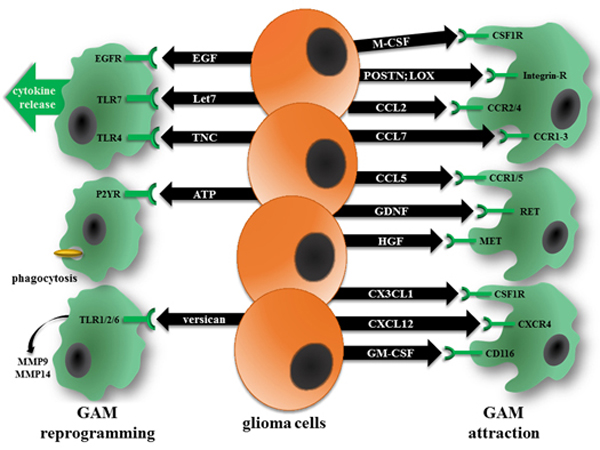
Chemokines produced by glioma cells actively recruit resident microglia from the brain, as well as macrophages from the blood, through binding to their cognate receptors on GAM (right). In addition, glioma cells produce proteins that increase cytokine release (EGF, Let7, and Tenascin-C; TNC), phagocytosis (ATP), and matrix metalloprotease (MMP9, MMP14) expression (versican) (left).
Moreover, emerging evidence supports the view that microglia and macrophages colonize different regions of malignant gliomas, such that macrophages appear to be recruited early during tumorigenesis and occupy perivascular regions [Z Chen et al., 2017]. However, conflicting data exist regarding the major monocyte population in these tumors, with some studies reporting a microglia predominance [Hutter et al., 2019] and others demonstrating that infiltrating macrophages represent the majority of the GAM population [Z Chen et al., 2017] [K Yu et al., 2019]. These differences could reflect specific experimental mouse model systems (RCAS model versus GL261 or T387 cell lines) used in each of these studies, suggesting that variations in GAM populations may be differentially dictated by the molecular properties of the glioma.
Support for potential mutation-specific effects on GAM recruitment comes from two studies: First, murine high-grade gliomas harboring a mutant IDH1 allele exhibited reduced macrophage and microglia infiltration, which correlated with lower levels of chemokine expression (e.g., CCL2, CXCL2) [Amankulor et al., 2017]. Moreover, the IDH1/2-mediated accumulation of the oncometabolite 2-hydroxyglutarate reduced the expression of pro-inflammatory chemokines [Han et al., 2019]. Second, differences in microglia recruitment were observed in mice genetically engineered to develop low-grade gliomas with distinct patient-derived germline NF1 gene mutations and cooperating genetic alterations (e.g., heterozygous Pten loss) through the elaboration of chemokines [Guo et al., 2019]. These findings suggest that variations in the GAM composition of gliomas could be dictated by types of immune chemoattractant molecules produced by cancer cells with different mutations.
GAM acquire unique phenotypes in the setting of glioma
Once GAM are recruited to the glioma milieu, they adopt new cellular and molecular identities that reflect the enactment of epigenetic or transcriptional programs that create monocyte populations critical for glioma homeostasis and progression. To identify these GAM transcriptional programs, two mouse glioma models, inoculated GL261 cells and the RCAS transgenic system, have been used to compare the expression profiles of glioma-associated microglia/macrophages and naive control cells. Not surprisingly, these profiles do not correspond to the M1/M2 classification scheme often used to describe macrophages/microglia in homeostatic or pro-inflammatory settings. Two genes were increased in GAM, Gpnmb and Spp1, which have also been described in other specialized microglia populations (e.g., disease-associated microglia [DAM] and proliferative region-associated microglia [PAM]), and were associated with poor prognosis in human GBM [Szulzewsky et al., 2015]. Work in many laboratories, using different microglia isolation methods and RNA analysis platforms have identified other GAM-specific markers; however, no marker has been discovered to date that uniquely distinguishes GAM from other microglia populations.
While microglia are the professional phagocytes of the CNS, in the setting of glioma, they do not attack or phagocytose glioma cells. This may reflect increased expression of the anti-phagocytic (“don’t eat me”) surface protein CD47 that binds to its cognate receptor SIRPα on phagocytic cells to inhibit its phagocytic activity. In this regard, anti-CD47 antibody treatment induces microglial tumor phagocytosis in a glioblastoma xenograft model and reduced tumor expansion [Hutter et al., 2019]. A similar effect of a humanized anti-CD47 antibody was found on five aggressive and etiologically distinct pediatric brain tumors: group 3 medulloblastoma (primary and metastatic), atypical teratoid rhabdoid tumor, primitive neuroectodermal tumor, pediatric glioblastoma, and diffuse intrinsic pontine glioma [Gholamin et al., 2017]. Moreover, in glioma patients, CD47 expression inversely correlated with histopathologic grading (low-grade versus high-grade glioma), and high levels of CD47 were associated with lower overall survival rates [F Li et al., 2018].
How microglia become reprogrammed in the setting of glioma remains to be fully elucidated. One family of receptors expressed on microglia may play an instructive role in the establishment of these new functional states. Toll-like receptors (TLR) were initially recognized as pathogen sensors, but it is now appreciated that they can also be activated by endogenous ligands. TLR2 expression is increased in GAM [Hu et al., 2015], such that mouse GL261 glioma cells implanted into the brains of Tlr2 knockout mice have smaller tumors and enhanced survival. This TLR2-dependency is partially mediated by increased production of a membrane-residing protease (MMP14) essential for activating MMP2 and promoting malignant glioma invasion [Vinnakota et al., 2013]. In addition to MMP14, TLR2 signaling also triggers the release of MMP9 from microglia, which serves as an additional factor for degrading the extracellular matrix to promote glioma invasion and growth [Hu et al., 2014]. TLR2 signaling is also involved in the interaction of GAM with T cells. As such, TLR2-mediated growth glioma is reduced using TLR2 blocking antibodies [Hu et al., 2015]. Beyond its role in glioma growth, TLR2 activation may also function to facilitate glioma immune evasion through reduced MHC class II molecule expression and reduced CD4+ T cells activation [J Qian et al., 2018].
In addition to TLR2, TLR4 is also important for regulating microglial IL-6 secretion, a mitogen for glioma stem cells. Analysis of human glioma tissue confirmed that GAM are the major source of IL-6 in gliomas [a Dzaye et al., 2016]. One of the endogenous ligands for TLR4 is the cell adhesion molecule, tenascin C. In tumors with genetically reduced tenascin C expression, microglia acquire a more amoeboid morphology and increased expression of MHCII molecules [Xia et al., 2016], similar to that observed with TLR2 silencing.
Finally, monocytes likely receive instructive cues from other cell types. One of these cell types is the T lymphocyte. Using a low-grade glioma stem cell transplantation system, glioma-like lesions did not form in mice with impaired T cell function [Pan et al., 2018]. These T cells, found in small abundance in both human and mouse NF1-mutant tumors, secrete paracrine factors important for inducing microglia to express CCL5 and drive glioma growth. Understanding how these T cells are recruited to the tumor, how they are activated, and the mechanisms that underlie their induction of microglia represent opportunities to dissect other immunologic determinants that underlie glioma formation and maintenance.
Experimental model systems to study glioma-microglia interactions
Current available platforms to examine the interactions between monocytes and glioma cells involve co-culture systems and organotypic systems, as well as small and large animal models. In addition to several genetically engineered mouse models (for review see [Gargiulo, 2018], large animal platforms involving pigs and dogs have been utilized. Spontaneously occurring low-and high-grade gliomas have been observed in dogs, but a systematic study of GAM in these tumors has not been performed [Bentley et al., 2017]. Similarly, swine engineered with a patient germline NF1 gene mutation develop low-grade optic gliomas, as observed in their human counterparts; however, little is known about the role of GAM populations in these tumors [Isakson et al., 2018]. Moreover, human glioma cell lines can be injected into the brains of immunosuppressed pigs as xenograft models [Selek et al., 2014] [Khoshnevis et al., 2017], but the use of immunocompromised animals limits a full analysis of GAM contributions due to secondary defects in microglia function [Pan et al., 2018].
In vitro culture assays have been employed for many years, typically involving established glioma cell lines (e.g., mouse GL261 or human U87 high-grade glioma cells) and freshly isolated microglia or the BV2 microglia-like cell line [S Muller et al., 2017] [Gu et al., 2017]. These platforms in vitro are well suited to determine defined functions of defined cell types, such as quantifying chemoattraction by tumor cells or microglia-induced glioma growth and migratory properties, but have only limited value in determining the interactions of these cell types in the natural context of a glioma tissue.
An intermediate model between in vivo and cell culture is represented by the organotypic brain slice model, in which rodent glioma cells are introduced into rodent brain slices. This allows for direct visualization of microglia/tumor interactions using fluorescent protein-expressing tumor cells and transgenic mice with microglia-restricted expression of different fluorescent proteins [Bayerl et al., 2016] [Ghoochani et al., 2016] [Resende et al., 2016]. Further refinements of these platforms enable the analysis of microglia and tumor motility using induced models of glioma, rather than using established cell lines [Juliano et al., 2018].
A newer experimental platform is the zebrafish larval brain. The optically transparent zebrafish larva allows for real-time visualizations of the interactions between implanted mammalian (even human) glioma cells and genetically labelled GAM [Astell and Sieger, 2017] [Yan et al., 2019]. Using the Irf8−/− zebrafish mutant, which lacks microglia, the contribution of microglia to the tumor growth has been explored [Hamilton et al., 2016]. While the current xenotransplantation models have employed established human glioblastoma cell lines, future modifications may incorporate reporter fish in which gliomas are induced by specific cancer-causing genetic alterations [Chia et al., 2018] [Jung et al., 2013], thus providing a more physiologic context in which to examine the glioma ecosystem. This model may have good potential as an important tool for drug screening and the development of future immunotherapeutic approaches that target microglia.
While these vertebrate models have been very instructive, they do not fully recapitulate their human counterparts. In addition to structural and lifespan differences between rodents and people, the human brain contains more white matter relative to the mouse brain. For this reason, the generation of human cerebral organoids from induced pluripotent stem cells (iPSCs) has recently been developed as a proxy for the human brain [X Qian et al., 2019], allowing for the implantation of patient-derived glioma stem cells [Linkous et al., 2019] [Hubert et al., 2016]. Alternatively, brain tumors can be generated by genomic editing. For example, CRISPR/CAS9 insertion of an oncogenic RAS allele into the TP53 locus in human iPSCs was used to generate high-grade gliomas following transplantation into immunodeficient animals [Ogawa et al., 2018]. These humanized systems facilitate a close examination of the interactions between glioma cells and the non-neoplastic cells in the human brain.
Outlook
Unfortunately to date, the clinical application of these basic science advances has been modest. In this regard, numerous strategies aimed at silencing GAM function has not translated well to human clinical trials. For example, the tetracycline analog, minocycline, which blocks microglia activation and reduces glioma expansion in experimental glioma mouse models of both high-grade [Markovic et al., 2011] and low-grade [Daginakatte and Gutmann, 2007; Toonen et al., 2017] glioma, has led to three clinical trials with no clear clinical benefit (NCT01580969, NCT02272270, NCT02770378). Similarly, PLX3397-induced depletion of microglia attenuates malignant glioma growth in mice [Pyonteck et al., 2013]; however, no efficacy was observed in human clinical trials [NCT01349036], despite good patient tolerability and adequate blood-brain-barrier penetration [Butowski et al., 2016]. Finally, antibody targeting of microglial TLRs reduces high-grade glioma growth in a mouse brain tumor slice model [Hu et al., 2015] and exhibits good safety and tolerability profiles in healthy subjects [Reilly et al., 2013], but has not progressed further in clinical trials for glioma.
While targeting microglia and macrophages represents a novel and potentially efficacious therapeutic approach, it is possible that adaptive changes in the tumor ecosystem will occur, creating a treatment-induced whack-a-mole scenario. In this regard, microglia can be reprogrammed by radiation [Monje et al., 2002] [Allen et al., 2014] [Monje et al., 2003] and/or chemotherapy [Gibson et al., 2019] to create new functional states, with different abilities to promote tumor growth. Similarly, hypoxia in necrotic areas of high-grade gliomas harbor high levels of extracellular ATP, which could act on microglia expressing purinergic receptors to increase cell motility, phagocytic activity and cytokine release [Kettenmann et al., 2011]. Moreover, it is possible that immunotherapies, like chimeric antigen receptor T cell therapy, will similarly alter microglia dynamics to establish new functional states. For this reason, it becomes exceeding important to understand the complex relationships established between GAM and the other cell types in the tumor in order to develop treatments that disrupt these interactions that support glioma maintenance (Figure 3).
Figure 3. Microglia are reprogrammed by numerous cell types and conditions in the context of glioma.
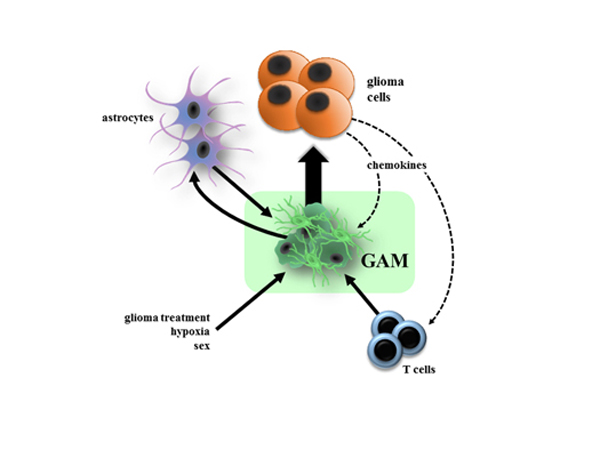
As highly adaptive cells, GAM gene expression and function can be changed by glioma treatment (chemotherapy, radiation therapy), environmental stresses (hypoxia), interactions with other non-neoplastic cell types (e.g., astrocytes), and signals from the glioma cells themselves. Each of these modifications could alter the properties of GAM in ways that either promote or inhibit continued tumor growth or invasion.
Additionally, it is also vital to consider other factors that could influence microglia function, including brain location and patient sex. To this end, the incidence of malignant glioma is higher in males [Ostrom et al., 2018], but females generally have better overall survival [Gittleman et al., 2018]. While these sexually dimorphic differences may reflect direct effects on cancer cells, it is equally possible that they act at the level of GAM. Emerging evidence from numerous laboratories have revealed differences in brain microglia gene expression and function in normal male and female mice [Guneykaya et al., 2018; Villa et al., 2018] [Thion et al., 2018], as well as between male and female mice in the setting of a diverse number of different brain diseases, including murine Nf1 optic glioma microglia-induced neuronal damage and vision loss [Toonen et al., 2017]
Finally, the lack of robust GAM-specific markers has limited our ability to define the distinct contributions of microglia and macrophages to glioma biology. In most studies, CD45 expression by FACS is used to distinguish microglia (CD45low) from peripheral macrophages (CD45high) [Badie and Schartner, 2000], leading to conflicting conclusions regarding the distribution of microglia and macrophages within gliomas. Moreover, microglia increase CD45 expression in the context of glioma [A Muller et al., 2015], whereas macrophages often express markers traditionally associated with microglia following integration into the cancer ecosystem, including downregulation of CD45 and upregulation of CX3CR1 expression [Pong et al., 2013b] [Z Chen et al., 2017]. These adaptations make it difficult to confidently ascribe unique functions to distinct monocyte populations [S Muller et al., 2017]. However, with the availability of more stably expressed markers and the use of spatial proteomics and single cell RNA sequencing strategies [Q Li et al., 2019] [Keren-Shaul et al., 2017] [Masuda et al., 2019], it should become possible to define the individual contributions of these monocyte populations to overall glioma biology [Haage et al., 2019] [Z Chen et al., 2017] [Bowman et al., 2016] [S Muller et al., 2017]. Understanding how distinct subpopulations of microglia/macrophages contribute to glioma pathobiology may also help to resolve conflicting reports in the literature regarding monocyte content and overall patient survival with glioma. Importantly, defining these different GAM species is necessary in order to design effective therapeutic approaches that selectively impair the monocyte populations most critical for tumor maintenance and progression.
Acknowledgements
We appreciate the support from a Berlin Institute of Health/Einstein fellowship grant to D.H.G. and H.K., an Alexander von Humboldt Foundation grant to D.H.G. and the Helmholtz-Gemeinschaft, Zukunftsthema “Immunology and Inflammation” to H.K. (ZT-0027).
References
- a Dzaye OD, Hu F, Derkow K, Haage V, Euskirchen P, Harms C, Lehnardt S, Synowitz M, Wolf SA, and Kettenmann H (2016), Glioma Stem Cells but Not Bulk Glioma Cells Upregulate IL-6 Secretion in Microglia/Brain Macrophages via Toll-like Receptor 4 Signaling, Journal of neuropathology and experimental neurology, 75(5), 429–440, doi: 10.1093/jnen/nlw016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen AR, Eilertson K, Chakraborti A, Sharma S, Baure J, Habdank-Kolaczkowski J, Allen B, Rosi S, Raber J, and Fike JR (2014), Radiation exposure prior to traumatic brain injury induces responses that differ as a function of animal age, Int J Radiat Biol, 90(3), 214–223, doi: 10.3109/09553002.2014.859761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amankulor NM, et al. (2017), Mutant IDH1 regulates the tumor-associated immune system in gliomas, Genes Dev, 31(8), 774–786, doi: 10.1101/gad.294991.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astell KR, and Sieger D (2017), Investigating microglia-brain tumor cell interactions in vivo in the larval zebrafish brain, Methods Cell Biol, 138, 593–626, doi: 10.1016/bs.mcb.2016.10.001. [DOI] [PubMed] [Google Scholar]
- Badie B, and Schartner JM (2000), Flow cytometric characterization of tumor-associated macrophages in experimental gliomas, Neurosurgery, 46(4), 957–961; discussion 961–952. [DOI] [PubMed] [Google Scholar]
- Bayerl SH, Niesner R, Cseresnyes Z, Radbruch H, Pohlan J, Brandenburg S, Czabanka MA, and Vajkoczy P (2016), Time lapse in vivo microscopy reveals distinct dynamics of microglia-tumor environment interactions-a new role for the tumor perivascular space as highway for trafficking microglia, Glia, 64(7), 1210–1226, doi: 10.1002/glia.22994. [DOI] [PubMed] [Google Scholar]
- Bentley RT, Ahmed AU, Yanke AB, Cohen-Gadol AA, and Dey M (2017), Dogs are man’s best friend: in sickness and in health, Neuro-oncology, 19(3), 312–322, doi: 10.1093/neuonc/now109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman RL, et al. (2016), Macrophage Ontogeny Underlies Differences in Tumor-Specific Education in Brain Malignancies, Cell Rep, 17(9), 2445–2459, doi: 10.1016/j.celrep.2016.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandenburg S, et al. (2016), Resident microglia rather than peripheral macrophages promote vascularization in brain tumors and are source of alternative pro-angiogenic factors, Acta neuropathologica, 131(3), 365–378, doi: 10.1007/s00401-015-1529-6. [DOI] [PubMed] [Google Scholar]
- Butowski N, et al. (2016), Orally administered colony stimulating factor 1 receptor inhibitor PLX3397 in recurrent glioblastoma: an Ivy Foundation Early Phase Clinical Trials Consortium phase II study, Neuro-oncology, 18(4), 557–564, doi: 10.1093/neuonc/nov245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, et al. (2019), Symbiotic Macrophage-Glioma Cell Interactions Reveal Synthetic Lethality in PTEN-Null Glioma, Cancer Cell, 35(6), 868–884 e866, doi: 10.1016/j.ccell.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Keoni C, Waker CA, Lober RM, Gutmann DH, and Chen YH (2019), KIAA1549-BRAF Expression Establishes a Permissive Tumor Microenvironment Through NFkappaB-Mediated CCL2 Production, Neoplasia, 21(1), 52–60, doi: 10.1016/j.neo.2018.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, McGowan LD, Cimino PJ, Dahiya S, Leonard JR, Lee DY, and Gutmann DH (2015), Mouse low-grade gliomas contain cancer stem cells with unique molecular and functional properties, Cell Rep, 10(11), 1899–1912, doi: 10.1016/j.celrep.2015.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, et al. (2017), Cellular and Molecular Identity of Tumor-Associated Macrophages in Glioblastoma, Cancer research, 77(9), 2266–2278, doi: 10.1158/0008-5472.CAN-16-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia K, Mazzolini J, Mione M, and Sieger D (2018), Tumor initiating cells induce Cxcr4-mediated infiltration of pro-tumoral macrophages into the brain, Elife, 7, doi: 10.7554/eLife.31918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daginakatte GC, and Gutmann DH (2007), Neurofibromatosis-1 (Nf1) heterozygous brain microglia elaborate paracrine factors that promote Nf1-deficient astrocyte and glioma growth, Hum Mol Genet, 16(9), 1098–1112, doi: 10.1093/hmg/ddm059. [DOI] [PubMed] [Google Scholar]
- Daginakatte GC, Gianino SM, Zhao NW, Parsadanian AS, and Gutmann DH (2008), Increased c-Jun-NH2-kinase signaling in neurofibromatosis-1 heterozygous microglia drives microglia activation and promotes optic glioma proliferation, Cancer research, 68(24), 10358–10366, doi: 10.1158/0008-5472.CAN-08-2506. [DOI] [PubMed] [Google Scholar]
- De S, Van Deren D, Peden E, Hockin M, Boulet A, Titen S, and Capecchi MR (2018), Two distinct ontogenies confer heterogeneity to mouse brain microglia, Development, 145(13), doi: 10.1242/dev.152306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijksterhuis JP, Arthofer E, Marinescu VD, Nelander S, Uhlen M, Ponten F, Mulder J, and Schulte G (2015), High levels of WNT-5A in human glioma correlate with increased presence of tumor-associated microglia/monocytes, Experimental cell research, 339(2), 280–288, doi: 10.1016/j.yexcr.2015.10.022. [DOI] [PubMed] [Google Scholar]
- Engler JR, Robinson AE, Smirnov I, Hodgson JG, Berger MS, Gupta N, James CD, Molinaro A, and Phillips JJ (2012), Increased microglia/macrophage gene expression in a subset of adult and pediatric astrocytomas, PloS one, 7(8), e43339, doi: 10.1371/journal.pone.0043339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargiulo G (2018), Next-Generation in vivo Modeling of Human Cancers, Front Oncol, 8, 429, doi: 10.3389/fonc.2018.00429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gholamin S, et al. (2017), Disrupting the CD47-SIRPalpha anti-phagocytic axis by a humanized anti-CD47 antibody is an efficacious treatment for malignant pediatric brain tumors, Sci Transl Med, 9(381), doi: 10.1126/scitranslmed.aaf2968. [DOI] [PubMed] [Google Scholar]
- Ghoochani A, Yakubov E, Sehm T, Fan Z, Hock S, Buchfelder M, Eyupoglu IY, and Savaskan NE (2016), A versatile ex vivo technique for assaying tumor angiogenesis and microglia in the brain, Oncotarget, 7(2), 1838–1853, doi: 10.18632/oncotarget.6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson EM, et al. (2019), Methotrexate Chemotherapy Induces Persistent Tri-glial Dysregulation that Underlies Chemotherapy-Related Cognitive Impairment, Cell, 176(1–2), 43–55 e13, doi: 10.1016/j.cell.2018.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F, et al. (2010), Fate mapping analysis reveals that adult microglia derive from primitive macrophages, Science, 330(6005), 841–845, doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittleman H, Boscia A, Ostrom QT, Truitt G, Fritz Y, Kruchko C, and Barnholtz-Sloan JS (2018), Survivorship in adults with malignant brain and other central nervous system tumor from 2000–2014, Neuro-oncology, 20(suppl_7), vii6-vii16, doi: 10.1093/neuonc/noy090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu R, Zhang X, Zhang G, Tao T, Yu H, Liu L, Dou Y, Li A, and Qin J (2017), Probing the Bi-directional Interaction Between Microglia and Gliomas in a Tumor Microenvironment on a Microdevice, Neurochem Res, 42(5), 1478–1487, doi: 10.1007/s11064-017-2204-1. [DOI] [PubMed] [Google Scholar]
- Guneykaya D, et al. (2018), Transcriptional and Translational Differences of Microglia from Male and Female Brains, Cell Rep, 24(10), 2773–2783 e2776, doi: 10.1016/j.celrep.2018.08.001. [DOI] [PubMed] [Google Scholar]
- Guo X, Pan Y, and Gutmann DH (2019), Genetic and genomic alterations differentially dictate low-grade glioma growth through cancer stem cell-specific chemokine recruitment of T cells and microglia, Neuro-oncology, doi: 10.1093/neuonc/noz080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haage V, et al. (2019), Comprehensive gene expression meta-analysis identifies signature genes that distinguish microglia from peripheral monocytes/macrophages in health and glioma, Acta neuropathologica communications, 7(1), 20, doi: 10.1186/s40478-019-0665-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hambardzumyan D, Gutmann DH, and Kettenmann H (2016), The role of microglia and macrophages in glioma maintenance and progression, Nature neuroscience, 19(1), 20–27, doi: 10.1038/nn.4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton L, Astell KR, Velikova G, and Sieger D (2016), A Zebrafish Live Imaging Model Reveals Differential Responses of Microglia Toward Glioblastoma Cells In Vivo, Zebrafish, 13(6), 523–534, doi: 10.1089/zeb.2016.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han CJ, Zheng JY, Sun L, Yang HC, Cao ZQ, Zhang XH, Zheng LT, and Zhen XC (2019), The oncometabolite 2-hydroxyglutarate inhibits microglial activation via the AMPK/mTOR/NF-kappaB pathway, Acta Pharmacol Sin, doi: 10.1038/s41401-019-0225-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herisson F, et al. (2018), Direct vascular channels connect skull bone marrow and the brain surface enabling myeloid cell migration, Nature neuroscience, 21(9), 1209–1217, doi: 10.1038/s41593-018-0213-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu F, Ku MC, Markovic D, a Dzaye OD, Lehnardt S, Synowitz M, Wolf SA, and Kettenmann H (2014), Glioma-associated microglial MMP9 expression is upregulated by TLR2 signaling and sensitive to minocycline, International journal of cancer, 135(11), 2569–2578, doi: 10.1002/ijc.28908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu F, et al. (2015), Glioma-derived versican promotes tumor expansion via glioma-associated microglial/macrophages Toll-like receptor 2 signaling, Neuro-oncology, 17(2), 200–210, doi: 10.1093/neuonc/nou324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert CG, Rivera M, Spangler LC, Wu Q, Mack SC, Prager BC, Couce M, McLendon RE, Sloan AE, and Rich JN (2016), A Three-Dimensional Organoid Culture System Derived from Human Glioblastomas Recapitulates the Hypoxic Gradients and Cancer Stem Cell Heterogeneity of Tumors Found In Vivo, Cancer research, 76(8), 2465–2477, doi: 10.1158/0008-5472.CAN-15-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter G, et al. (2019), Microglia are effector cells of CD47-SIRPalpha antiphagocytic axis disruption against glioblastoma, Proceedings of the National Academy of Sciences of the United States of America, 116(3), 997–1006, doi: 10.1073/pnas.1721434116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob K, et al. (2011), Genetic aberrations leading to MAPK pathway activation mediate oncogene-induced senescence in sporadic pilocytic astrocytomas, Clin Cancer Res, 17(14), 4650–4660, doi: 10.1158/1078-0432.CCR-11-0127. [DOI] [PubMed] [Google Scholar]
- Juliano J, et al. (2018), Comparative dynamics of microglial and glioma cell motility at the infiltrative margin of brain tumours, J R Soc Interface, 15(139), doi: 10.1098/rsif.2017.0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung IH, Leem GL, Jung DE, Kim MH, Kim EY, Kim SH, Park HC, and Park SW (2013), Glioma is formed by active Akt1 alone and promoted by active Rac1 in transgenic zebrafish, Neuro-oncology, 15(3), 290–304, doi: 10.1093/neuonc/nos387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren-Shaul H, et al. (2017), A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease, Cell, 169(7), 1276–1290 e1217, doi: 10.1016/j.cell.2017.05.018. [DOI] [PubMed] [Google Scholar]
- Kettenmann H, Hanisch UK, Noda M, and Verkhratsky A (2011), Physiology of microglia, Physiol Rev, 91(2), 461–553, doi: 10.1152/physrev.00011.2010. [DOI] [PubMed] [Google Scholar]
- Khoshnevis M, et al. (2017), Development of induced glioblastoma by implantation of a human xenograft in Yucatan minipig as a large animal model, Journal of neuroscience methods, 282, 61–68, doi: 10.1016/j.jneumeth.2017.03.007. [DOI] [PubMed] [Google Scholar]
- Larribere L, et al. (2015), NF1 loss induces senescence during human melanocyte differentiation in an iPSC-based model, Pigment Cell Melanoma Res, 28(4), 407–416, doi: 10.1111/pcmr.12369. [DOI] [PubMed] [Google Scholar]
- Li F, et al. (2018), Blocking the CD47-SIRPalpha axis by delivery of anti-CD47 antibody induces antitumor effects in glioma and glioma stem cells, Oncoimmunology, 7(2), e1391973, doi: 10.1080/2162402X.2017.1391973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, et al. (2019), Developmental Heterogeneity of Microglia and Brain Myeloid Cells Revealed by Deep Single-Cell RNA Sequencing, Neuron, 101(2), 207–223 e210, doi: 10.1016/j.neuron.2018.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkous A, et al. (2019), Modeling Patient-Derived Glioblastoma with Cerebral Organoids, Cell Rep, 26(12), 3203–3211 e3205, doi: 10.1016/j.celrep.2019.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listernick R, Charrow J, Greenwald MJ, and Esterly NB (1989), Optic gliomas in children with neurofibromatosis type 1, J Pediatr, 114(5), 788–792. [DOI] [PubMed] [Google Scholar]
- Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, and Ellison DW (2016), The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary, Acta neuropathologica, 131(6), 803–820, doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- Markovic DS, Vinnakota K, van Rooijen N, Kiwit J, Synowitz M, Glass R, and Kettenmann H (2011), Minocycline reduces glioma expansion and invasion by attenuating microglial MT1-MMP expression, Brain, behavior, and immunity, 25(4), 624–628, doi: 10.1016/j.bbi.2011.01.015. [DOI] [PubMed] [Google Scholar]
- Masuda T, et al. (2019), Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution, Nature, 566(7744), 388–392, doi: 10.1038/s41586-019-0924-x. [DOI] [PubMed] [Google Scholar]
- Monje ML, Toda H, and Palmer TD (2003), Inflammatory blockade restores adult hippocampal neurogenesis, Science, 302(5651), 1760–1765, doi: 10.1126/science.1088417. [DOI] [PubMed] [Google Scholar]
- Monje ML, Mizumatsu S, Fike JR, and Palmer TD (2002), Irradiation induces neural precursor-cell dysfunction, Nat Med, 8(9), 955–962, doi: 10.1038/nm749. [DOI] [PubMed] [Google Scholar]
- Muller A, Brandenburg S, Turkowski K, Muller S, and Vajkoczy P (2015), Resident microglia, and not peripheral macrophages, are the main source of brain tumor mononuclear cells, International journal of cancer, 137(2), 278–288, doi: 10.1002/ijc.29379. [DOI] [PubMed] [Google Scholar]
- Muller S, et al. (2017), Single-cell profiling of human gliomas reveals macrophage ontogeny as a basis for regional differences in macrophage activation in the tumor microenvironment, Genome Biol, 18(1), 234, doi: 10.1186/s13059-017-1362-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa J, Pao GM, Shokhirev MN, and Verma IM (2018), Glioblastoma Model Using Human Cerebral Organoids, Cell Rep, 23(4), 1220–1229, doi: 10.1016/j.celrep.2018.03.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrom QT, Gittleman H, Truitt G, Boscia A, Kruchko C, and Barnholtz-Sloan JS (2018), CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2011–2015, Neuro-oncology, 20(suppl_4), iv1-iv86, doi: 10.1093/neuonc/noy131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Smithson LJ, Ma Y, Hambardzumyan D, and Gutmann DH (2017a), Ccl5 establishes an autocrine high-grade glioma growth regulatory circuit critical for mesenchymal glioblastoma survival, Oncotarget, 8(20), 32977–32989, doi: 10.18632/oncotarget.16516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Bush EC, Toonen JA, Ma Y, Solga AC, Sims PA, and Gutmann DH (2017b), Whole tumor RNA-sequencing and deconvolution reveal a clinically-prognostic PTEN/PI3K-regulated glioma transcriptional signature, Oncotarget, 8(32), 52474–52487, doi: 10.18632/oncotarget.17193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, et al. (2018), Athymic mice reveal a requirement for T-cell-microglia interactions in establishing a microenvironment supportive of Nf1 low-grade glioma growth, Genes Dev, 32(7–8), 491–496, doi: 10.1101/gad.310797.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield W (1925), Microglia and the process of phagocytosis in glioma, Amercan Journal of Pathology, 1, 77–90. [PMC free article] [PubMed] [Google Scholar]
- Pfister S, et al. (2008), BRAF gene duplication constitutes a mechanism of MAPK pathway activation in low-grade astrocytomas, J Clin Invest, 118(5), 1739–1749, doi: 10.1172/JCI33656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platten M, Kretz A, Naumann U, Aulwurm S, Egashira K, Isenmann S, and Weller M (2003), Monocyte chemoattractant protein-1 increases microglial infiltration and aggressiveness of gliomas, Ann Neurol, 54(3), 388–392, doi: 10.1002/ana.10679. [DOI] [PubMed] [Google Scholar]
- Pong WW, Higer SB, Gianino SM, Emnett RJ, and Gutmann DH (2013. a), Reduced microglial CX3CR1 expression delays neurofibromatosis-1 glioma formation, Ann Neurol, 73(2), 303–308, doi: 10.1002/ana.23813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pong WW, et al. (2013b), F11R is a novel monocyte prognostic biomarker for malignant glioma, PloS one, 8(10), e77571, doi: 10.1371/journal.pone.0077571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyonteck SM, et al. (2013), CSF-1R inhibition alters macrophage polarization and blocks glioma progression, Nat Med, 19(10), 1264–1272, doi: 10.1038/nm.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J, et al. (2018), TLR2 Promotes Glioma Immune Evasion by Downregulating MHC Class II Molecules in Microglia, Cancer Immunol Res, 6(10), 1220–1233, doi: 10.1158/2326-6066.CIR-18-0020. [DOI] [PubMed] [Google Scholar]
- Qian X, Song H, and Ming GL (2019), Brain organoids: advances, applications and challenges, Development, 146(8), doi: 10.1242/dev.166074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly M, et al. (2013), Randomized, double-blind, placebo-controlled, dose-escalating phase I, healthy subjects study of intravenous OPN-305, a humanized anti-TLR2 antibody, Clin Pharmacol Ther, 94(5), 593–600, doi: 10.1038/clpt.2013.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resende FF, Bai X, Del Bel EA, Kirchhoff F, Scheller A, and Titze-de-Almeida R (2016), Evaluation of TgH(CX3CR1-EGFP) mice implanted with mCherry-GL261 cells as an in vivo model for morphometrical analysis of glioma-microglia interaction, BMC Cancer, 16, 72, doi: 10.1186/s12885-016-2118-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch S, Rapp C, Dettling S, and Herold-Mende C (2018), When Immune Cells Turn Bad-Tumor-Associated Microglia/Macrophages in Glioma, International journal of molecular sciences, 19(2), doi: 10.3390/ijms19020436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selek L, et al. (2014), Imaging and histological characterization of a human brain xenograft in pig: the first induced glioma model in a large animal, Journal of neuroscience methods, 221, 159–165, doi: 10.1016/j.jneumeth.2013.10.002. [DOI] [PubMed] [Google Scholar]
- Sierra A, de Castro F, Del Rio-Hortega J, Rafael Iglesias-Rozas J, Garrosa M, and Kettenmann H (2016), The “Big-Bang” for modern glial biology: Translation and comments on Pio del Rio-Hortega 1919 series of papers on microglia, Glia, 64(11), 1801–1840, doi: 10.1002/glia.23046. [DOI] [PubMed] [Google Scholar]
- Simmons GW, Pong WW, Emnett RJ, White CR, Gianino SM, Rodriguez FJ, and Gutmann DH (2011), Neurofibromatosis-1 heterozygosity increases microglia in a spatially and temporally restricted pattern relevant to mouse optic glioma formation and growth, Journal of neuropathology and experimental neurology, 70(1), 51–62, doi: 10.1097/NEN.0b013e3182032d37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solga AC, et al. (2015), RNA Sequencing of Tumor-Associated Microglia Reveals Ccl5 as a Stromal Chemokine Critical for Neurofibromatosis-1 Glioma Growth, Neoplasia, 17(10), 776–788, doi: 10.1016/j.neo.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen MD, Dahlrot RH, Boldt HB, Hansen S, and Kristensen BW (2018), Tumour-associated microglia/macrophages predict poor prognosis in high-grade gliomas and correlate with an aggressive tumour subtype, Neuropathology and applied neurobiology, 44(2), 185–206, doi: 10.1111/nan.12428. [DOI] [PubMed] [Google Scholar]
- Szulzewsky F, Schwendinger N, Guneykaya D, Cimino PJ, Hambardzumyan D, Synowitz M, Holland EC, and Kettenmann H (2018), Loss of host-derived osteopontin creates a glioblastoma-promoting microenvironment, Neuro-oncology, 20(3), 355–366, doi: 10.1093/neuonc/nox165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulzewsky F, et al. (2015), Glioma-associated microglia/macrophages display an expression profile different from M1 and M2 polarization and highly express Gpnmb and Spp1, PloS one, 10(2), e0116644, doi: 10.1371/journal.pone.0116644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thion MS, et al. (2018), Microbiome Influences Prenatal and Adult Microglia in a Sex-Specific Manner, Cell, 172(3), 500–516 e516, doi: 10.1016/j.cell.2017.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toonen JA, Solga AC, Ma Y, and Gutmann DH (2017), Estrogen activation of microglia underlies the sexually dimorphic differences in Nf1 optic glioma-induced retinal pathology, J Exp Med, 214(1), 17–25, doi: 10.1084/jem.20160447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa A, et al. (2018), Sex-Specific Features of Microglia from Adult Mice, Cell Rep, 23(12), 3501–3511, doi: 10.1016/j.celrep.2018.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinnakota K, et al. (2013), Toll-like receptor 2 mediates microglia/brain macrophage MT1-MMP expression and glioma expansion, Neuro-oncology, 15(11), 1457–1468, doi: 10.1093/neuonc/not115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallmann T, et al. (2018), Microglia Induce PDGFRB Expression in Glioma Cells to Enhance Their Migratory Capacity, iScience, 9, 71–83, doi: 10.1016/j.isci.2018.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S, Lal B, Tung B, Wang S, Goodwin CR, and Laterra J (2016), Tumor microenvironment tenascin-C promotes glioblastoma invasion and negatively regulates tumor proliferation, Neuro-oncology, 18(4), 507–517, doi: 10.1093/neuonc/nov171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C, et al. (2019), Visualizing Engrafted Human Cancer and Therapy Responses in Immunodeficient Zebrafish, Cell, 177(7), 1903–1914 e1914, doi: 10.1016/j.cell.2019.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Deshmukh H, Gutmann RJ, Emnett RJ, Rodriguez FJ, Watson MA, Nagarajan R, and Gutmann DH (2009), Alterations of BRAF and HIPK2 loci predominate in sporadic pilocytic astrocytoma, Neurology, 73(19), 1526–1531, doi: 10.1212/WNL.0b013e3181c0664a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, et al. (2019), A nonmyeloablative chimeric mouse model accurately defines microglia and macrophage contribution in glioma, Neuropathology and applied neurobiology, 45(2), 119–140, doi: 10.1111/nan.12489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiner PS, et al. (2018), Distribution and prognostic impact of microglia/macrophage subpopulations in gliomas, Brain pathology, doi: 10.1111/bpa.12690. [DOI] [PMC free article] [PubMed] [Google Scholar]


