Abstract
ZC4H2 encodes a C4H2 type zinc-finger nuclear factor, the mutation of which has been associated with disorders with various clinical phenotypes in human, including developmental delay, intellectual disability and dystonia. ZC4H2 has been suggested to regulate spinal cord patterning in zebrafish as a co-factor for RNF220, an ubiquitin E3 ligase involved in Gli signaling. Here we showed that ZC4H2 and RNF220 knockout animals phenocopy each other in spinal patterning in both mouse and zebrafish, with mispatterned progenitor and neuronal domains in the ventral spinal cord. We showed evidence that ZC4H2 is required for the stability of RNF220 and also proper Gli ubiquitination and signaling in vivo. Our data provides new insights into the possible etiology of the neurodevelopmental impairments observed in ZC4H2-associated syndromes.
Keywords: ZC4H2, RNF220, spinal cord, patterning, Gli signaling
Introduction
ZC4H2 is located on the X chromosome, which encodes a C4H2 type zinc-finger nuclear factor. Mutations in ZC4H2 have been reported to be associated with various disorders, including arthrogryposis multiplex congenita (AMC), X-linked intellectual disability, Wieacker–Wolff syndrome, and Miles–Carpenter syndrome (Hirata et al., 2013; May et al., 2015; Kondo et al., 2018). The clinical phenotypes associated with ZC4H2 deficiency are variable, including AMC, intellectual disability, epilepsy, spasticity, dystonia, developmental delay, etc. (Hirata et al., 2013; May et al., 2015). Since ZC4H2 is located on the X-chromosome, the disorder is usually severe or even lethal in males. However, females with heterozygous ZC4H2 mutations can also be severely affected with AMC and neurodevelopmental impairments, although most female carriers are affected to a lesser degree with mild or no symptoms (Zanzottera et al., 2017; Okubo et al., 2018). The different manifestation in ZC4H2 heterozygous females has been suggested to be related to varying X inactivation.
Mechanically, ZC4H2 has been suggested to be involved in neural development and dendritic spine density (Hirata et al., 2013; May et al., 2015). ZC4H2 knockout in zebrafish affects the specification of GABAergic V2 interneurons in spinal cord (May et al., 2015), implying a role of ZC4H2 during spinal cord patterning process. Our previous study also showed that ZC4H2 is involved in neural induction by modulating BMP/Smad signaling in Xenopus (Ma et al., 2017). In a recent report, Kim et al. (2018) suggested that ZC4H2 works as a co-factor for RNF220, an ubiquitin E3 ligase, to regulate the stability of several transcription factors involved in ventral spinal cord patterning. However, the exact roles and mechanism of ZC4H2 during mouse neural development remain unexplored.
We recently showed that RNF220 works as an ubiquitin E3 ligase for Gli, the transcriptional effector of Shh pathway (Ma et al., 2019). Shh activity forms a decreasing gradient along the ventral-dorsal axis, which regulates the expression of a group of Class I and Class II transcription factors at distinct dorsal-ventral levels, by which the ventral spinal cord is divided into five domains where V0, V1, V2, motor neuron (MN) and V3 neurons are generated respectively (Jessell, 2000; Dessaud et al., 2008). RNF220 mediated non-proteolytic ubiquitination of Glis promotes their nuclear exportation to refine the Shh/Gli signaling gradient to regulate local neural cell fates. Loss of RNF220 leads to expansion of both the intermediate V0 and ventral V3 neurons, at the expenses of MN, V1 and V2 neurons in between (Ma et al., 2019). Here we confirmed that ZC4H2 and RNF220 knockout animals phenocopy each other in spinal cord patterning in both mouse and zebrafish. We showed evidence that ZC4H2 is required for the stability of RNF220 and also proper Gli ubiquitination and signaling in vivo. Our data provides new insights into the possible etiology of the neurodevelopmental impairments observed in ZC4H2 associated syndromes.
Results
Enriched expression of ZC4H2 in the developing nervous system and construction of ZC4H2 knockout mice
Using whole-mount in situ hybridization, we first examined the expression pattern of ZC4H2 during mouse embryonic development. As in zebrafish and Xenopus (Hirata et al., 2013; May et al., 2015; Ma et al., 2017), ZC4H2 transcripts were mainly detected throughout the developing central neural system, including regions from telencephalon to spinal cord at E8.5, E9.5 and E10.5 mouse embryos (Figure 1A–E). At E10.5, ZC4H2 transcripts were also detected in the developing limb buds (Figure 1D and E). Paraffin section of the stained mouse embryos showed that ZC4H2 mainly expressed at ventral domain of developing spinal cord (Figure 1C).
Figure 1.
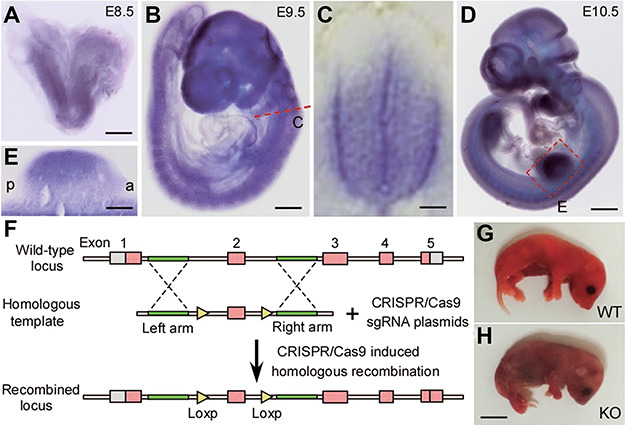
Developmental expression of ZC4H2 and construction of ZC4H2 knockout mice. (A–E) Whole-mount in situ hybridization showing that ZC4H2 is expressed in the developing neural system at E8.5 (A), E9.5 (B and C), and E10.5 (D and E). (C) Section of E9.5 embryos at spinal cord shows that ZC4H2 is expressed in the ventral zone of spinal cord. (E) In E10.5 embryos, ZC4H2 expression is also detected in the limb buds. Scale bar, 400 μm in A, 300 μm in B, 50 μm in C, 700 μm in D, and 150 μm in E. (F) Diagram of the targeting construct and expected recombination events. Exon 2 of ZC4H2 is floxed by two Loxp sites. (G and H) Photographs showing control (G, WT) and ZC4H2−/− (H, KO) pups on the day of birth. The ZC4H2−/− pups die after birth. Scale bar, 5 mm (H, applies to G).
To investigate the function of ZC4H2 in mouse development, we created a conditional allele of ZC4H2 in which exon 2 with its surrounding intron sequence was flanked by loxP sites using CRISPR/Cas9 system (Figure 1F). ZC4H2fl/fl or ZC4H2fl/Y mouse was crossed with Vasa-Cre line to get the ZC4H2fl/wt;Vasa-Cre or ZC4H2+/− mouse. Although the resulting female ZC4H2+/− mice were born at the expected Mendelian ratio, ~50% of the ZC4H2+/− mice showed postnatal developmental delay and failed to survive to adult stage (data not shown). The ZC4H2fl/wt;Vasa-Cre or ZC4H2+/− mouse was mated with ZC4H2fl/Y mouse, and the resulting embryos were genotyped using polymerase chain reaction (PCR) and processed for phenotypic analysis. The resulting ZC4H2−/− female and ZC4H2−/Y male embryos were found neonatal lethal (Figure 1G and H). There are no obvious defects in the skeleton pattern in ZC4H2 knockout mouse limbs (data not shown).
Progenitor domains and post-mitotic neurons are altered in the ventral spinal cord of ZC4H2 knockout mice
We examined the genes expressed in different progenitor domains of spinal cord in ZC4H2 knockout mice at E10.5 to determine whether the deletion of ZC4H2 affects the patterning of spinal ventricular zones (Figure 2A–H). The Vp3 domain is located dorsal to the floor plate and expresses Nkx2.2 (Briscoe et al., 2000; Jessell, 2000). Nkx2.2+ Vp3 domain expanded dorsally in ZC4H2 knockout mice as compared with controls (Figure 2A and B). In contrast, the pMN domain expressing Olig2 was decreased in ZC4H2 knockout mice at this stage (Figure 2A–D). The Vp2 domain was delineated as an Nkx6.1+ domain located dorsal to the Nkx6.1+/Olig2+ pMN domain (Briscoe et al., 2000; Vallstedt et al., 2001), which was almost completely lost in ZC4H2 KO mice at E10.5 (Figure 2C and D). In contrast, the medial Dbx1+ Vp0 domain expands ventrally, leaving the Vp1 domain (the gap region between the Dbx1+ Vp0 and Nkx6.1+ Vp2 domains) greatly reduced. Dbx2 is expressed in the Vp0 and Vp1 domains in the control mice, but it expanded ventrally to the ventral end of the ventricular zone in ZC4H2 knockout mice (Figure 2G and H).
Figure 2.
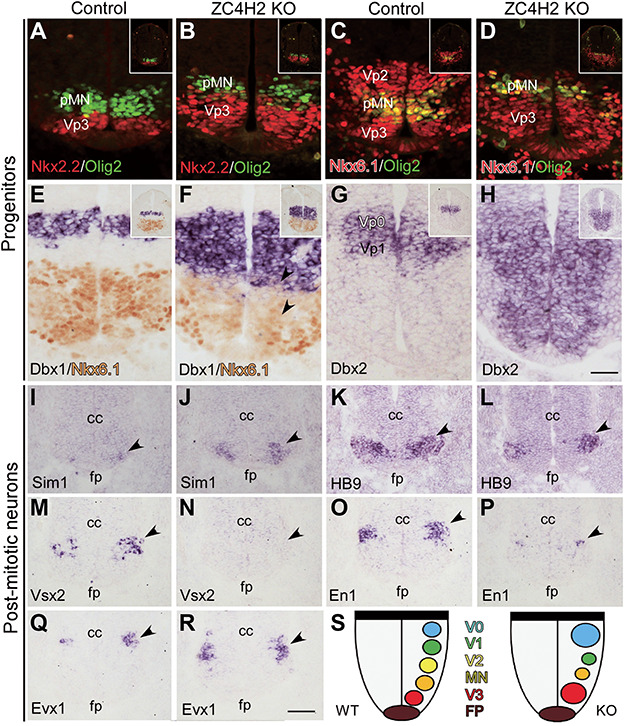
Progenitor and post-mitotic neuron domains are altered in the ventral spinal cord of the ZC4H2 knockout (KO) mice at E10.5. (A and B) The Nkx2.2+ (red) VP3 domain is expanded dorsally, while the Olig2+ (green) pMN domain is reduced in the ZC4H2 KO mice relative to controls. (C and D) Double labelling of Dbx1 and Nkx6.1 (red) and Olig2 (green) shows a dorsal shift and reduction of the Olig2+ pMN domain and the Vp2 domain was decreased in ZC4H2 KO spinal tube. (E and F) In situ immunostaining of Dbx1 and Nkx6.1 shows that the Dbx1+ Vp0 domain is increased and expanded ventrally to the dorsal boundary of the Nkx6.1+ domain in the ZC4H2 KO mice, whereas there is a gap region (i.e. Vp1 domain) between the Nkx6.1+ and Dbx1+ Vp0 domains in controls. (G and H) Dbx2 expression is expanded ventrally to the floor plate in ZC4H2 KO mice. Inserts in A–H are low magnification pictures of the whole spinal tube. Scale bar, 50 μm (H, applies to A–G). (I and J) The number of Sim1+ V3 neurons is dramatically increased in the ZC4H2 KO mice (J) compared with controls (I). Triangles point to their expressions in the ventral tube. (K and L) The number of Hb9+ MN neurons is reduced in the ZC4H2 KO mice (L) compared with controls (K). (M and N) Expression of Vsx2 is not detected in the ZC4H2 KO mice (N) but observed in control spinal cord (M). (O and P) The number of En1+ V1 neurons is greatly reduced in the ZC4H2 KO mice (P) compared with controls (O). (Q and R) The number of Evx1+ V0 neurons is increased in the ZC4H2 KO mice (R) compared with controls (Q). Triangles in I–R point to their expressions in the ventral tube. cc, central canal; fp, floor plate. Scale bar, 100 μm (R, applies to I–Q). (S) A cartoon showing the relative changes of the different neuronal territories in the ZC4H2 KO ventral spinal cord. Note that there is no difference in the changes of the progenitor and neuronal domains between female ZC4H2−/− and male ZC4H2−/Y embryos (data not shown). The data shown here were all from ZC4H2−/− female embryos.
To further confirm the phenotype in ZC4H2 knockout mice, we next examined the post-mitotic cells generated from individual progenitor domains in ZC4H2 knockout mice at E10.5 and later stages. There were more Sim1+ V3 neurons, which are generated from Vp3 domain (Briscoe et al., 2000; Jessell, 2000) in ZC4H2 knockout mice compared with that in control mice at E10.5 (Figure 2I and J). Consistent with the reduction of Olig2+ pMN domain, the Hb9+ motor neurons were decreased in ZC4H2 knockout mice at this stage (Figure 2K and L). More dorsally, the Vsx2+ V2a neurons generated from the Vp2 domain were absent in ZC4H2 knockout mice, whereas a group of Vsx2+ neurons was observed in E10.5 control spinal tube (Figure 2M and N). En1 is expressed in post-mitotic neurons produced from the Vp1 domain, and the number of En1+ cells was significantly reduced in ZC4H2 knockout mice relative to controls (Figure 2O and P). In contrast, there were more Evx1+ neurons which are generated from the Vp0 domain in ZC4H2 knockout mice at E10.5 (Figure 2Q and R). The relative changes of the different neuronal domains in the ZC4H2 knockout ventral spinal cord are summarized in Figure 2S. The alterations of post-mitotic spinal neurons were also found in the spinal cord of ZC4H2 knockout mice at E12.5 (data not shown).
Taken together, both progenitor domains and their progenies in the ventral spinal cord are altered in the absence of ZC4H2, similar to that observed in RNF220 knockout mice (Ma et al., 2019).
Spinal cord was mispatterned in some ZC4H2 mouse heterozygotes
ZC4H2 gene is located on the X chromosome, which is subjected to random inactivation during development. It has been reported that some females with heterozygous mutations in ZC4H2 can also be affected with various degreed defects (Zanzottera et al., 2017; Okubo et al., 2018). We found that about half of the ZC4H2 heterozygous female mice failed to grow to adult stage, suggesting that they are likely affected. Indeed, similar patterning defects were observed in some ZC4H2 heterozygotes, with increased Vp3 domain and reduced Vp2 domain, similar to that in ZC4H2 KO mice (Figure 3A–D). Besides, on some sections of ZC4H2 heterozygotes, it was also interesting to find that there were some Olig2+ neurons scattered ventrally (data not show), and their location was similar to that in control mice. Thus, the defective patterning of progenitor domain is also present in some female ZC4H2 heterozygotes. The varying manifestation in the ZC4H2 heterozygous female mice is likely due to random X inactivation.
Figure 3.
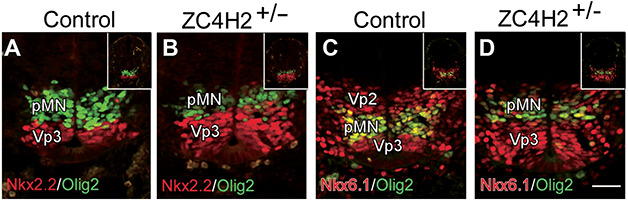
Changes of the progenitor domains in the ZC4H2 heterozygote (ZC4H2+/−) spinal cord at E10.5. (A and B) Double immunostaining shows that the Nkx2.2+ (red) Vp3 domain is expanded, while the Olig2+ (green) pMN domain is reduced along the D–V axis of the ventricular zone in ZC4H2+/− mice relative to controls. (C and D) Double immunostaining for Nkx6.1 (red) and Olig2 (green) shows a dorsal shift of the Nkx6.1+/Olig2+ pMN domain and a reduction of the Vp2 domain in the ZC4H2+/− spinal cord. Inserts in A–D are low magnification pictures of the whole spinal tube. Scale bar, 50 μm (D, applies to A–C).
rnf220a mutants phenocopy zc4h2 null embryos in zebrafish
In zebrafish, Zc4h2 has been shown to be required for the specification of V2 interneurons in the spinal cord (May et al., 2015). Here, we constructed an rnf220a mutant zebrafish line using CRISPR/Cas9 system (Figure 4A). To address a potential spinal cord mispatterning defect in the rnf220a null embryos, we stained wild-type and mutant fish embryos at 24 h with antisense probes for the markers of the related neural progenitors and their differentiated neurons (Figure 4B–Q). We found that, similar to the zc4h2 mutant fish, dbx2 expression was clearly expanded with a corresponding loss of nkx6.1 expression in both hindbrain and spinal cord (Figure 4B–I) (May et al., 2015). And also, vsx1 and vsx2, which label V2a and V2b interneurons, respectively, were both significantly decreased in spinal cord (Figure 4J–Q).
Figure 4.
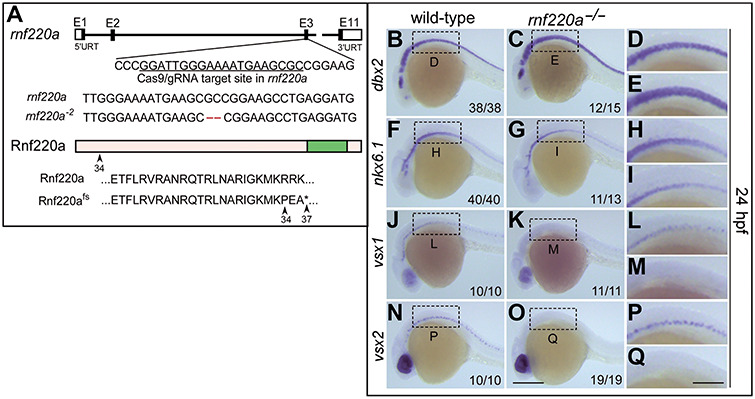
Neural progenitors and post-mitotic neurons are altered in rnf220a knockout zebrafish embryos. (A) Schematic representation of CRISPR/Cas9-mediated genome modification. The black box represents the exon and the white box represents the non-coding region. The CRISPR/Cas9 target site is located in the third exon of rnf220a genomic locus, marked with a black underline. Lastly, a 2-bp deletion (labelled in red) induced by CRISPR/Cas9 results in a frame shift. Both wild-type Rnf220a and Rnf220fs proteins are shown. The wild-type protein is truncated to 37 amino acids by this mutation. fs, frame shit. (B–I) Genes marking the progenitor domains of ventral spinal cord were examined by whole-mount in situ hybridization in rnf220a homozygous mutant (rnf220a−/−) embryos at 24 hpf. dbx2 gene expression domain is expanded ventrally (B–E) at the expense of the reduced nkx6.1 expression domain (F–I). (J–Q) Whole-mount in situ hybridization analysis showing the expression of the post-mitotic neuronal markers in rnf220a mutants. Neurons labelled with either vsx1 (J–M) or vsx2 (N–Q) are reduced in rnf220a mutants. The statistical data are annotated in the bottom right corner of each panel. Scale bar, 200 μm (O, applies to B, C, F, G, J, K, N, and O) and 80 μm (Q, applies to D, E, H, I, L, M, and P).
Thus, ZC4H2 and RNF220 mutants phenocopy each other in both mice and zebrafish as far as spinal patterning is concerned (May et al., 2015; Kim et al., 2018; Ma et al., 2019), implying their functional interaction.
ZC4H2 interacts and stabilizes RNF220 in vitro and in vivo
Using yeast two hybridization assays, we identified ZC4H2 as a potential interactor for RNF220 (data not shown), as also reported by Kim et al. (2018). Their interaction was confirmed by coimmunoprecipitation assay in HEK293 cells (Figure 5A and B). Given that RNF220 is an ubiquitin E3 ligase that often regulates the stability of its targets (Kong et al., 2010), we then examined if the ZC4H2 protein stability was regulated by RNF220. Unexpectedly, neither the ZC4H2 protein level nor its polyubiquitination level was affected by RNF220 coexpression in HEK293 cells (data not shown). However, we found that RNF220 protein was stabilized by ZC4H2 overexpression (Figure 5C). We also used a series of cycloheximide base protein chase assays to test the protein stability of RNF220 in presence of ZC4H2. The results showed that ZC4H2 overexpression indeed prolonged the half-life of RNF220 protein (Figure 5D and E). Furthermore, RNF220 polyubiquitination modification was reduced when ZC4H2 was coexpressed (Figure 5F). To test whether ZC4H2 regulates RNF220 protein level in vivo, we isolated embryonic neural stem cells from E15.5 wild-type and ZC4H2 knockout mouse embryos and compared the RNF220 protein levels by western blotting. The results showed that RNF220 protein decreased significantly in ZC4H2 knockout neural stem cells (Figure 5G). Using immunofluorescence staining assays, we also examined the spinal cord RNF220 expression in ZC4H2 knockout embryos. RNF220 protein was indeed hardly detected in the ZC4H2 knockout spinal cord (Figure 5H). Note that the ZC4H2 protein level in RNF220 knockout spinal cord is comparable to control (Figure 5H). Collectively, these data suggested that ZC4H2 stabilizes RNF220 both in vitro and in vivo.
Figure 5.
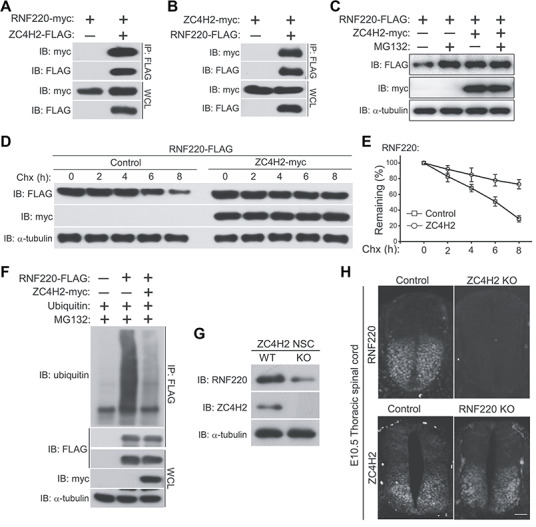
ZC4H2 interacts with and stabilizes RNF220 in vitro and in vivo. (A) RNF220 is coimmunoprecipitated with ZC4H2. (B) ZC4H2 is coimmunoprecipitated with RNF220. HEK293 cells were transiently transfected with different combinations of RNF220 and ZC4H2 expression vectors. Cell lysates were incubated with anti-FLAG beads, washed, and subsequently analyzed by western blotting. (C) RNF220 protein is stabilized by ZC4H2 overexpression. FLAG-tagged RNF220 and myc-tagged ZC4H2 plasmids were transfected into HEK293 cells as indicated. After 40 h, cells were treated with 15 μM MG132 or not before harvest and then cell lysates were analyzed by western blotting. (D and E) Effect of ZC4H2 on the stablity of RNF220. HEK293 cells were transiently transfected with the indicated plasmids. At 48 h post-transfection, cycloheximide (Chx) was added to all samples, and the cells were harvested at the time points indicated. Levels of RNF220 were determined by western blotting with anti-FLAG antibody. In all cases, α-tubulin was used as a loading control. The relative levels of RNF220 were quantified densitometrically and normalized against α-tubulin and the statistics are shown in E. (F) In vitro ubiquitination assay showing that polyubiquitination level of RNF220 is reduced by ZC4H2 coexpression in HEK293 cells. (G) Western blotting assay showing the protein level of RNF220 in wild-type (WT) or ZC4H2 knockout (KO) neural stem cells. (H) Immunofluoscence staining showing the RNF220 or ZC4H2 protein expression in wild-type (Control) or ZC4H2/RNF220 knockout (KO) mouse spinal cords at E10.5. Scale bar, 75 μm. IB, immunoblot; IP, immunoprecipitation, WCL, whole-cell lysate.
ZC4H2 knockout alters Shh/Gli signaling in vivo
We previously reported that RNF220 modulates Gli gradient through targeting Glis for K63-linked polyubiquitination and nuclear exportation (Ma et al., 2019). We tested whether ZC4H2 is also involved in Shh/Gli signaling through stabilizing RNF220 protein. The expression of Gli1, Ptch1 and Hhip1, three established Shh target genes, was up-regulated in ZC4H2 knockout neural stem cells (Figure 6A), implying that ZC4H2 modulates Shh signaling like RNF220 (Ma et al., 2019). We also examined the effect of ZC4H2 knockout on the protein level and K63-linked polyubiquitination level of endogenous Glis in neural stem cells or MEF cells. ZC4H2 knockout does not affect the protein level of total endogenous Glis (Figure 6C and data not shown); however, the distribution of Glis (including Gli1, Gli2, full length and repressor forms of Gli3) in the nuclear and soluble cytoplasmic fractions clearly increased, while that in the cytoskeleton fraction reduced (Figure 6B). Furthermore, the levels of K63-linked polyubiquitinated Glis dramatically reduced in both neural stem cells and MEF cells (Figure 6C and data not shown), as observed in the RNF220−/− cells (Ma et al., 2019). Together, the above data suggest that ZC4H2 regulates Shh/Gli signaling in vivo through RNF220.
Figure 6.
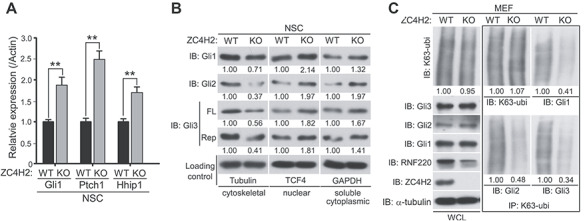
ZC4H2 regulates Gli signaling, subcellular distribution, and ubiquitination in vivo. (A) Real-time PCR results showing the relative Gli1, Ptch1, and Hhip1 expression in wild-type (WT) or ZC4H2 knockout (KO) neural stem cells. ** P < 0.01 (Student’s t-test). (B) Western blotting assays showing the subcellular distribution of endogenous Glis in ZC4H2 WT or KO neural stem cells. (C) The levels of K63-ubiquitinated Glis in ZC4H2 WT or KO MEF cells. The cell lysates were immunoprecipitated with K63-ubiquitin chain-specific antibody or control IgG before blotted using Gli antibodies. The relative densitometrical statistics are shown below.
Discussion
In human, ZC4H2 has been reported to be associated with X-linked neurodevelopment disorders while its mechanism of action remains unclear (Hirata et al., 2013; May et al., 2015). Here we showed that ZC4H2 knockout mice are neonatal lethal with ventral spinal cord patterning defects. The ZC4H2 mouse heterozygotes also showed various neural developmental defects, making it a model to study the development and mechanisms of related human disease. Mechanically, we showed evidence that ZC4H2 is required for the stability of RNF220 and also proper Gli ubiquitination and subcellular localization in vivo. Using both mouse and zebrafish models, we confirmed that ZC4H2 and RNF220 knockout animals phenocopy each other in neural patterning. Our work established that ZC4H2 is an RNF220 stabilizer and is involved in the regulation of Shh/Gli signaling and ventral spinal cord patterning.
Kim et al. (2018) suggested that RNF220/ZC4H2 complex is directly involved in the neural patterning transcriptional network by targeting Dbx1, Dbx2, Nkx2.2 etc., with no clear effect on the upstream signaling network. We have provided compelling evidence in vitro and in vivo that RNF220 and ZC4H2 have a direct role on Shh/Gli signaling (Figure 6; Ma et al., 2019) and play a more upstream role accounting for the observed patterning defects in the knockout animals. Their roles in the control of the stability of the patterning transcription factors is likely involved in the refinement of the neural progenitor domains.
The study by Kim et al. (2018) and our study have established that RNF220 is an important E3 ubiquitin ligase during neural patterning, regulating the localization and stability of its target proteins (Ma et al., 2019). We showed here that ZC4H2 stabilizes RNF220 in vitro and in vivo and reduces its polyubiquitination level, suggesting that RNF220 itself is subjected to regulation by ubiquitination. The mechanisms, including the structural basis of such regulation, remain to be investigated. Also, whether ZC4H2 is required physically for the full activity of RNF220 is not known, although RNF220 protein purified from cells overexpressing exogenous RNF220 is active by itself in in vitro ubiquitination assays (Ma et al., 2019).
Materials and methods
Mouse, staging and genotyping
All mice were maintained and handled according to guidelines approved by the Animal Care and Use Committee of the Kunming Institute of Zoology, Chinese Academy of Sciences. All mice were maintained on a C57BL/6 background. The conditional ZC4H2 knockout allele, ZC4H2fl was generated by insertion of two LoxP sites into introns flanking exon 2 through CRISPR/Cas9 mediated genome editing technology. To obtain ZC4H2 knockout embryos, Vasa-Cre mice were used to generate germ cell ZC4H2 conditional knockout female mice (ZC4H2fl/wt;Vasa-Cre) firstly and then the female mice were used to mate with the male ZC4H2 floxed mice (ZC4H2fl/Y).
The stage of mouse embryos was determined by taking the morning when the copulation plug was seen as embryonic day 0.5 (E0.5). All genotypes described were confirmed by PCR. ZC4H2 alleles were genotyped using genome DNAs prepared from either tail tips or yolk sac. PCR primers were: forward, 5′-GACTAGGAAGACTTTTCCTGG-3′ and reverse, 5′-TCCCAAGATATGTGGCACATG-3′. PCR amplified DNA was analyzed on a 2.5% TBE agarose gel.
Zebrafish
Wild-type (AB) zebrafish were raised at 28.5 C. Embryos were collected by spontaneous spawning and staged as described (Westerfield, 1995). Embryos were fixed in 4% paraformaldehyde (PFA) overnight at 4°C. Whole-mount in situ hybridization was performed as previously described (Thisse and Thisse, 2008). The antisense RNAs were labelled using digoxigenin-11-UTP (Roche Diagnostics). CRISPR/Cas9 mediated genome editing technology was used to construct rnf220a mutants (Shao et al., 2017). RNF220 homozygous mutant were identified by allele specific PCR with the following primers: common forward, 5′-TTAATCCTTCAACTGTTCCCT-3′; rnf220a wild-type reverse, 5′-GGATTGGGAAAATGAAGCGCC-3′; rnf220a mutant reverse, 5′-GGATTGGGAAAATGAAGCCTG-3′ (Cheng et al., 2018).
Cell culture, transfection, immunoprecipitation, immunoblotting, and ubiquitination assays
HEK293 cell culture, plasmids transfection, immunoprecipitation, western blotting and in vivo ubiquitination assays were all carried out as previously reported (Ma et al., 2014).
Immunohistochemistry, mouse in situ hybridization, and analysis
Mouse whole-mount/section in situ hybridization assays, immunohistochemical staining, and related analysis were carried out as previously described (Rosen and Beddington, 1993; Song et al., 2011). Antibodies and probes were used as described in previous report (Ma et al., 2019). A 2-kbp-long ZC4H2 probe covering ZC4H2 CDS plus most of its 3′UTR was used in whole-mount in situ hybridization assays.
Statistical analysis
GraphPad and Origin 8 software were used for statistical analysis. Comparisons were performed using the two tails Student’s t-test. P-values of <0.05 or 0.01 were considered statistically significant or very significant, respectively. All experiments were carried out at least three times and samples were analyzed in at least triplicate.
Funding
This study was supported by the National Natural Science Foundation of China (31871483 and 31671521 to B.M., 31500847 to P.M., 31771134 to N.S., 81571332 and 91232724 to Y.D., and 31671509 to D.S.), the National Key R&D Program of China (2017YFA0104002 to Y.-Q.D.), Shanghai Municipal Science and Technology Major Project (2018SHZDZX01) and ZJLab.
Conflict of interest: none declared.
References
- Briscoe J., Pierani A., Jessell T.M., et al. (2000). A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell 101, 435–445. [DOI] [PubMed] [Google Scholar]
- Cheng X.N., Shao M., and Shi D.L. (2018). Mutation of Frizzled8a delays neural retinal cell differentiation and results in microphthalmia in zebrafish. Int. J. Dev. Biol. 62, 285–291. [DOI] [PubMed] [Google Scholar]
- Dessaud E., McMahon A.P., and Briscoe J. (2008). Pattern formation in the vertebrate neural tube: a sonic Hedgehog morphogen-regulated transcriptional network. Development 135, 2489–2503. [DOI] [PubMed] [Google Scholar]
- Hirata H., Nanda I., van Riesen A., et al. (2013). ZC4H2 mutations are associated with arthrogryposis multiplex congenita and intellectual disability through impairment of central and peripheral synaptic plasticity. Am. J. Hum. Genet. 92, 681–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessell T.M. (2000). Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat. Rev. Genet. 1, 20–29. [DOI] [PubMed] [Google Scholar]
- Kim J., Choi T.I., Park S., et al. (2018). Rnf220 cooperates with Zc4h2 to specify spinal progenitor domains. Development 145, pii: dev165340. [DOI] [PubMed] [Google Scholar]
- Kondo D., Noguchi A., Takahashi I., et al. (2018). A novel ZC4H2 gene mutation, K209N, in Japanese siblings with arthrogryposis multiplex congenita and intellectual disability: characterization of the K209N mutation and clinical findings. Brain Dev. 40, 760–767. [DOI] [PubMed] [Google Scholar]
- Kong Q., Zeng W., Wu J., et al. (2010). RNF220, an E3 ubiquitin ligase that targets Sin3B for ubiquitination. Biochem. Biophys. Res. Comm. 393, 708–713. [DOI] [PubMed] [Google Scholar]
- Ma P., Ren B., Yang X., et al. (2017). ZC4H2 stabilizes Smads to enhance BMP signalling, which is involved in neural development in Xenopus. Open Biol. 7, 170122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma P., Song N., Li Y., et al. (2019). Fine-tuning of Shh/Gli signaling gradient by non-proteolytic ubiquitination during neural patterning. Cell Rep. 28, 541–553. [DOI] [PubMed] [Google Scholar]
- Ma P., Yang X., Kong Q., et al. (2014). The ubiquitin ligase RNF220 enhances canonical Wnt signaling through USP7-mediated deubiquitination of β-catenin. Mol. Cell. Biol. 34, 4355–4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May M., Hwang K.S., Miles J., et al. (2015). ZC4H2, an XLID gene, is required for the generation of a specific subset of CNS interneurons. Hum. Mol. Genet. 24, 4848–4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okubo Y., Endo W., Inui T., et al. (2018). A severe female case of arthrogryposis multiplex congenita with brain atrophy, spastic quadriplegia and intellectual disability caused by ZC4H2 mutation. Brain Dev. 40, 334–338. [DOI] [PubMed] [Google Scholar]
- Rosen B., and Beddington R.S. (1993). Whole-mount in situ hybridization in the mouse embryo: gene expression in three dimensions. Trends Genet. 9, 162–167. [DOI] [PubMed] [Google Scholar]
- Shao M., Wang M., Liu Y.Y., et al. (2017). Vegetally localised Vrtn functions as a novel repressor to modulate bmp2b transcription during dorsoventral patterning in zebrafish. Development 144, 3361–3374. [DOI] [PubMed] [Google Scholar]
- Song N.N., Xiu J.B., Huang Y., et al. (2011). Adult raphe-specific deletion of Lmx1b leads to central serotonin deficiency. PLoS One 6, e15998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C., and Thisse B. (2008). High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69. [DOI] [PubMed] [Google Scholar]
- Vallstedt A., Pattyn A., Pierani A., et al. (2001). Different levels of repressor activity assign redundant and specific roles to Nkx6 genes in motor neuron and interneuron specification. Neuron 31, 743–755. [DOI] [PubMed] [Google Scholar]
- Westerfield M. (1995). The Zebrafish Book. Guide for the Laboratory Use of Zebrafish (Danio rerio). Eugene: University of Oregon Press. [Google Scholar]
- Zanzottera C., Milani D., Alfei E., et al. (2017). ZC4H2 deletions can cause severe phenotype in female carriers. Am. J. Med. Genet. A 173, 1358–1363. [DOI] [PubMed] [Google Scholar]


