Tissue-resident stem cells are important for maintaining proper organ and tissue function throughout the lifetime of mammals. Although some types of stem cells constantly proliferate and give rise to committed progeny, such as intestine and skin, others reside mainly in a quiescent (noncycling) state, such as skeletal muscle. How stem cells maintain their quiescence while contributing to homeostatic tissue turnover is not well understood and is an active topic of investigation because of the potential of stem cell biology in regenerative medicine and healthy aging (1). On page 734 of this issue, de Morree et al. (2) characterize the underlying mechanisms that control muscle stem cell (MuSC) behavior in mice. Unexpectedly, they show that multiple species of RNAs coordinately confer precise regulation of quiescence and proliferation in MuSCs under homeostatic conditions.
The paired-box homeodomain transcription factors PAX3 and PAX7 are critical regulators of skeletal muscle specification and function in a range of animal species, including mouse (3). Although PAX7 is expressed in all adult mouse MuSCs, PAX3 is only expressed in a subset of skeletal muscles, such as the diaphragm (4). Previously, the same laboratory showed that this variation of PAX3 protein expression is regulated by the microRNA miR206, which mediates translational repression of Pax3 messenger RNA (mRNA). The extent of miR206 regulation is dependent on the generation of Pax3 mRNA isoforms of different lengths. mRNA length is controlled through polyadenylation, the addition of a polyadenosine tail to the 39 untranslated region (UTR) of an mRNA at polyadenylation signal sequences. Longer Pax3 mRNAs are generated through the use of a distal polyadenylation site and are subject to miR206-mediated translational repression, whereas shorter isoforms are produced when proximal polyadenylation sites are used, which produces an mRNA that does not contain miR206 targeting sequences and are therefore not susceptible to miR206 (5).
To elucidate the underlying mechanisms that regulate differential polyadenylation of Pax3 mRNAs, de Morree et al. compared the expression of known alternative polyadenylation factors in MuSCs from hindlimb (which contain longer Pax3 mRNAs and lower PAX3 protein expression) and diaphragm (shorter Pax3 mRNAs and higher PAX3 protein expression). They found that the expression of U1 small nuclear RNA (snRNA), the RNA scaffold of U1 small nuclear ribonucleoprotein that prevents nascent mRNA from premature transcription termination and polyadenylation (6, 7), was up-regulated in hindlimb compared to diaphragm MuSCs. The authors elegantly demonstrated that higher expression of U1 snRNA favors the use of the distal polyadenylation site in the 39 UTR of the Pax3 mRNA, leading to generation of longer Pax3 mRNAs and lower PAX3 protein expression owing to miR206-mediated repression (see the figure).
Regulation of stem cell behavior by multiple RNA species.
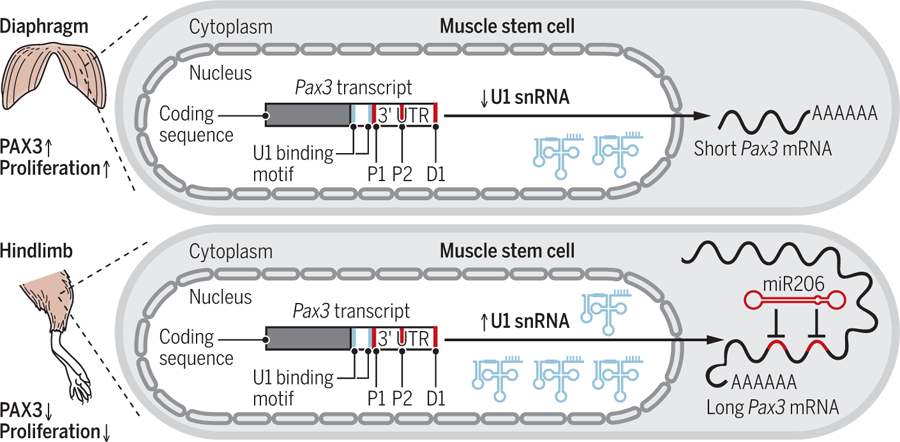
Diaphragm muscle stem cells express high amounts of paired box protein 3 (PAX3), causing increased rates of proliferation. This is mediated by low expression of U1 small nuclear RNA (snRNA), which promotes use of the proximal polyadenylation sites (P1 and P2) in the untranslated region (UTR) and thus short messenger RNAs (mRNAs) that are not targeted by microRNA-206 (miR206). In hindlimb muscle stem cells, high amounts of U1 snRNA favor the distal polyadenylation site (D1) and the generation of long mRNAs with miR206 binding sites.
Functionally, de Morree et al. showed that this U1 snRNA–mediated control of Pax3 transcript length and PAX3 protein expression regulates the propensity of quiescent MuSCs to become proliferative by exiting quiescence and entering the cell cycle in muscles without external injury. They found that a higher proportion of MuSCs from diaphragm, which expresses low amounts of U1 snRNA and high amounts of PAX3 protein, are actively cycling compared to those from hindlimb with high U1 snRNA and low PAX3 expression. Using conditional Pax3 deletion in mice, the authors further showed that under homeostatic conditions, PAX3 controls long-term contribution of MuSCs to myofibers, multinucleated single muscle cells that constitute the basic structural and functional units of skeletal muscle tissue. When Pax3 was deleted in muscle groups that normally contain high PAX3-expressing MuSCs, 9 months later the mice were found to have fewer MuSC-derived myofibers compared to wild-type animals. Moreover, myofibers from affected muscles, including diaphragm and triceps, were smaller in size in the Pax3-depleted mice, which also showed compromised muscle-associated function such as shortened running distance and reduced grip strength.
The study of de Morree et al. helps to answer the long-standing question of the functional importance of the observed heterogeneity of PAX3 protein expression and myofiber turnover rate among different skeletal muscle groups (4, 8, 9). Perhaps certain muscles that undergo constant contraction to sustain vital processes, such as diaphragm for respiratory control, benefit from harboring MuSCs with higher PAX3 protein expression that are more prone to proliferate and repair damaged myofibers owing to elevated rates of wear and tear.
Posttranscriptional regulation of stem cell behaviors by microRNAs on cognate mRNAs has been extensively studied in many tissue and organ systems, including skeletal muscle (10) The study of de Morree et al. demonstrates how simple nongenomic modifications can add an exquisite layer of regulatory control and specific fine-tuning of MuSC function by a third RNA species, U1 snRNA, which affects alternative polyadenylation and thus determines Pax3 mRNA targeting by miR206. Indeed, it has been shown that more than half of human genes are regulated by alternative polyadenylation, which influences many physiological or pathological processes in various cell types (11). Therefore, the intricate interplay between distinct RNA species might have a central role in precise regulation of stem cell behaviors not only in skeletal muscles but also in other tissues and organs. Furthermore, it will be interesting to examine whether similar regulatory mechanisms occur in developmental myogenesis (5, 12).
It has also been well documented that distinct skeletal muscle groups possess differential susceptibility to various types of muscular dystrophies (13). For example, in Duchenne muscular dystrophy, clinical symptoms are mainly manifested first in proximal limb muscles in the early teens, followed by respiratory complications involving diaphragm muscles later. This could be associated with differential MuSC activity and muscle turnover in distinct muscle groups mediated by RNA control beyond the genome level. Furthermore, in sarcopenia (age-related muscle loss), RNA control of stem cell fate may also play a role in regulating functional decline of skeletal muscle with age (14). Thus, it will be interesting to explore the potential functional relevance of RNA control of MuSCs in the context of development and disease settings in future studies.
REFERENCES AND NOTES
- 1.Cho IJ et al. , Stem Cell Reports 12, 1190 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Moree A et al. , Science 366, 734 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buckingham M, Relaix F, Semin. Cell Dev. Biol 44, 115 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Relaix F et al. , J. Cell Biol 172, 91 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boutet SC et al. , Cell Stem Cell 10, 327 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaida D et al. , Nature 468, 664 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berg MG et al. , Cell 150, 53 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pawlikowski B et al. , Skelet. Muscle 5, 42-015-0067-1 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keefe AC et al. , Nat. Commun 6, 7087 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shenoy A, Blelloch RH, Nat. Rev. Mol. Cell Biol 15, 565 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Giammartino DC et al. , Mol. Cell 43, 853 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goljanek-Whysall K et al. , Proc. Natl. Acad. Sci. U.S.A 108, 11936 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emery AE, Lancet 359, 687 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Snijders T, Parise G, Curr. Opin. Clin. Nutr. Metab. Care 20, 186 (2017). [DOI] [PubMed] [Google Scholar]


