Abstract
Background
The established clinical criteria for gastric cancer prognosis are insufficient due to molecular heterogeneity. Therefore, constructing a robust prognostic model is essential to predict gastric cancer patient survival.
Material/Methods
A comprehensive method, which combined weighted gene co-expression network analysis (WGCNA) with elastic-net Cox regression, was utilized to identify prognostic long non-coding RNAs (lncRNAs) from Gene Expression Omnibus database for overall survival (OS) prediction. Methods using WGCNA or elastic-net Cox regression alone were treated as “contrast” methods. The univariate and multivariate Cox regression was used to identify independent prognostic clinical factors. We performed 3-year and 5-year area under the curve (AUC) of the time-dependent receiver operating characteristic comparison of 3 different methods in gene and clinical-gene models to explore the prediction ability of the comprehensive method. The optimal model identified in the training set were validated in the validation set. Biological information analysis for the optimal model was also explored.
Results
The clinical-gene model containing 13 co-expression lncRNAs identified by the comprehensive method and 3 clinical factors including molecular subtype, recurrence status and operation type, was the found to be the optimal model in the study, with 0.832 and 0.830 for the 3-year and 5-year AUC in the training set, and 0.764 and 0.778 in the validation set, respectively. Biological information analysis suggested that lipid metabolism played an important role in the occurrence and development of gastric cancer.
Conclusions
We constructed a novel prognostic model containing 13 co-expression lncRNAs and 3 clinical factors for gastric cancer patients.
MeSH Keywords: Survival Analysis; Gene Expression Profiling; Prognosis; RNA, Long Noncoding; Stomach Neoplasms
Background
Gastric cancer (GC) is a widely known malignant cancer, identified as the third leading cause of death from cancer worldwide. In 2015, approximately 1 313 000 people were diagnosed with the condition, and 819 000 patients died worldwide with a mortality rate of nearly 50% [1]. Although development in neoadjuvant therapy and surgical techniques can improve the potential survival of GC patients, the 5-year overall survival (OS) rate of GC patients remains at an unsatisfactory level [2]. Earlier studies have researched that the time of initial diagnosis, disease stage, recurrence or distance-metastasis of GC, the infection status of Helicobacter pylori and other demographic and clinical factors that are associated with a poor prognosis [3,4]. However, established clinical criteria of prognostic strategies based on these factors, such as tumor/node/metastasis (TNM) classification, Lauren classification, and the World Health Organization (WHO) classification, were insufficient to predict the OS of patients with the tumor involving complex genetic alteration [5–7]. Therefore, constructing a robust prognostic model including genetic factors is essential to effectively predict the prognosis of GC patients.
Recent studies have focused on high-throughput sequencing technology to identify biomarkers related to the survival of GC patients. Since less than 2% of the human genome encode proteins, there is increasing evidence that has indicated that the non-coding RNA is involved in tumor occurrence and progression by regulating gene expression [8,9]. Long non-coding RNA (lncRNA), a group of non-coding RNA with a length greater than 200 nucleotides, which is a 3 times greater quantity than protein-coding RNA, has been shown to exhibit a vital role in cancer prognosis in recent years [10]. Zhu et al. [11] identified 24 lncRNAs which were related to the prognosis of GC patients by using a multivariable Cox regression model, with area under the curve (AUC) was 0.85 when combined with the American Joint Committee on Cancers (AJCC) stage. Fan et al. [12] selected 5 lncRNAs as OS biomarkers through a random survival forest algorithm, with an AUC of 0.86. Cheng et al. [13] screened 3 prognostic lncRNAs using the least absolute shrinkage and selection operator (LASSO) Cox regression and the 19-month AUC was 0.737. Peng et al. [14] identified 7 prognostic lncRNA pairs as a prognostic signature using a permutation method and LASSO Cox regression with concordance index of 0.872. Zhang et al. [15] identified 11 lncRNAs as an independent survival signature for GC patients by utilizing the co-expression of genes and LASSO Cox regression. However, most of these studies were faced with the challenge of high dimensionality and collinearity in data analysis, and they might not reflect the interconnection among genes, which might cause model over-fitting and lose meaningful molecules during analysis [16]. Therefore, it is necessary to consider the interconnection among genes and avoid model over-fitting simultaneously when predicting GC prognosis.
In recent years, various statistical methods have been introduced to reduce over-fitting in microarray data analysis [10,12,17]. Penalized regression combined with the Cox proportional hazards model, including LASSO Cox regression, ridge regulated Cox regression, and elastic-net Cox regression, can achieve greater performance of genomic survival analysis by adjusting the parameters, rather than using traditional Cox regression [18]. The LASSO Cox regression can reduce the dimensions of microarray data, but it cannot solve the collinearity problem; while the ridge Cox regression can address the multicollinearity issues but cannot execute the variable selection. The elastic-net Cox regression, which combines the advantages of both LASSO and ridge Cox regressions, has been used in a number of research studies to screen genes associated with cancer prognosis [19,20]. In addition, weighted gene co-expression network analysis (WGCNA) has been widely performed to identify highly interconnected genes and to explore the correlation between co-expression modules and clinical traits [21].
Thus, in this study, in order to construct a robust prognostic model of GC, we used a comprehensive method, which combined WGCNA with elastic-net Cox regression, to identify the OS prediction lncRNAs. Methods using WGCNA or elastic-net Cox regression alone were treated as “contrast” methods. Three- and 5-year AUC of the time-dependent receiver operating characteristic (ROC) were calculated to evaluate the prediction ability of different models and identify an optimal model as the robust prognostic model of GC patients in our study. Stratification analysis based on independent clinical factors was used to validate the independence of the optimal model. Biological information analysis such as the Gene Ontology (GO) function and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses was used to identify the biological functions of the lncRNAs involved in the optimal model, so as to provide a more comprehensive reference for future prognostic researches and the treatment of GC patients.
Material and Methods
Data resource and preprocessing
The chip data were obtained from Gene Expression Omnibus (GEO) database. Due to the chip platform requirement of the lncRNA re-annotation pipeline, the inclusion criteria were as follows: gene expression profiles of GC specimens could be accessed; a total sample of GC >50; the chip platform was GPL570 (Affymetrix Human Genome U133 Plus 2.0 Array, Santa Clara, CA, USA); basic clinical data containing OS information was available. Lastly, the data set GSE62254 was selected, which consisted of 300 GC samples, the corresponding clinical variables were obtained from the original research. Next, we randomly selected 70% of the samples without returning to be our training set and the other 30% of samples were used as the validation set. All statistical analysis was conducted by R 3.5.0 software, significance level was set as P<0.05.
In general, lncRNA expression levels are lower than those of protein-coding genes. The robust multichip average (RMA) method is an effective method used to obtain a consistent estimate value of lncRNA expression profiles [22,23]. Therefore, the raw CEL file of GSE62254 was processed with background correction, quantile normalization and log2 transformation by using the RMA method of R package “affy”. Several missing clinic values were filled by using R package “rpart”.
LncRNA re-annotation
Affymetrix HG-U133 Plus 2.0 probe set ID annotation was based on the annotation of NetAffx, RefSeq and Ensembl databases for lncRNAs [24,25]. Firstly, we mapped the chip probe set ID to the NetAffx Annotation Files (HG-U133 Plus 2.0 Annotations, CSV format, release 36, January 2017), the Refseq IDs of NetAffx Annotation Files which were labeled “NR” and “XR” were retained. Secondly, the probe sets from the latest Refseq database annotation with the gene type of “long non-coding RNA” were retained. For the next step, we retained the probe sets from both the NetAffx and Refseq database annotations. Then, in the annotation of the Ensembl database, the IDs with “3prime-overlapping-ncRNA”, “antisense”, “bidirectional_promoter_lncRNA”, “lincRNA”, “macro-lncRNA”, “misc-RNA”, “processed transcripts”, “sense-overlapping”, “sense_intronic” in the Ensembl database were retained. Finally, the probe sets which were assigned with a Refseq transcript ID or Ensembl gene ID were retained for annotation.
Prognostic model construction and comparison
Construction of weighted gene co-expression networks
The lncRNA expression profiles were used to construct weighted co-expression networks through the R package “WGCNA” [26,27]. Firstly, a co-expression similarity matrix was constructed by using Pearson’s correlation coefficients for all pairwise lncRNAs. Secondly, we transformed the co-expression similarity matrix into an adjacency matrix by choosing the soft threshold power β=7 (scale-free topology fitting index R2=0.92) for scale-free topology network construction. A topological matrix (TOM) was created using a topological overlap measure. Then, we calculated the corresponding dissimilarity of TOM (dissTOM) for further analysis. The average linkage hierarchical clustering method was used to define the network modules by node dissimilarity, and the hybrid dynamic tree cutting method was used to cut branches by setting a minimum gene group size of 30 with a cut height of 0.99 for the resulted dendrogram. Additionally, the module eigengenes (MEs) were used to represent the gene expression profiles of modules, defined as the first principal component following principal component analysis in the expression profiles of lncRNAs within a given module. The module dissimilarity correlation was calculated based on MEs to merge the modules with similar expression profiles greater than 20%. A univariate Cox regression analysis was performed to identify prognostic modules with P value <0.05. Then, the genes in prognostic modules with P value <0.01 were selected as hub genes by the univariate Cox regression analysis.
Development of prognostic models
To construct a robust prognostic model, we used 3 methods to identify prognosis candidate biomarkers: the comprehensive method which used the WGCNA algorithm and elastic-net Cox regression simultaneously, and the “contrast” methods using the WGCNA or the elastic-net Cox regression alone. The hub genes of the co-expression modules play important roles within biological processes and have generally high interconnection. Therefore, for the WGCNA method, we used hub genes of prognostic modules as candidate prognostic genes. For the elastic-net Cox regression method, all lncRNAs were included in an elastic-net Cox regression, of which lncRNAs with non-zero regression coefficient were considered as candidate genes. For the comprehensive method, we incorporated the lncRNAs of the prognostic modules which was identified by WGCNA into the elastic-net Cox regression to further screen candidate genes. The significant independent prognostic clinical variables were selected using univariate and multivariate Cox regression analysis with a threshold α=0.05. We then constructed clinical, gene, and clinical-gene models of the 3 methods by using risk score (RS) formula for GC patients. The risk score was calculated as follows [27,28]:
where expi indicates the expression of the candidate variable i, and βi is the regression coefficient of i which is calculated using ridge regulated Cox regression to ensure consistency of all models in our study [16]. The penalized Cox regression, including the elastic-net Cox regression and ridge regulated Cox regression, were performed using R package “glmnet” with 10 000 iterations and 10-fold cross-validations.
Prediction ability comparison of prognostic models
The 3-year and 5-year AUC of each model were calculated by R package “timeROC” to explore the model predictive ability. The Z-test of the AUC values were used to compare the predictive ability of the models and identify an optimal prognostic model of GC in our study. Bonferroni correction was used for multiple comparisons with threshold α=0.025.
Furthermore, all GC patients were divided into 2 groups (high-risk group and low-risk group) based on their risk score using the median value of risk score as the cutoff value. Overall survival comparison of the 2 risk groups was carried out using Kaplan-Meier (K-M) analysis and a log-rank test to identify the prognostic value of the RS index. In addition, stratification analysis was performed based on the independent prognostic clinical variables to assess the independence of the optimal model which we identified.
Biological function analysis
The top 200 mRNAs of the Spearman correlation coefficient between the optimal lncRNAs and the data corresponding with mRNA were identified as model target genes [28]. The GO function and KEGG pathway enrichment analyses of the target genes were used to explore potential biological functions involved in the optimal model. In our study, the analysis was performed through the database for annotation, visualization and integrated discovery (DAVID) 6.8 (http://david.abcc.ncifcrf.gov/).
Results
Basic characteristics of the data microarray
A total of 14 different clinical factors were utilized when conducting this study, which were age, sex, T stage, M stage, N stage, AJCC stage, WHO classification, Lauren subtype, molecular subtype, recurrence status, tumor site, adjuvant concurrent chemoradiation therapy (adjuvant CCRT), operation (OP) type, and Epstein-Barr virus (EBV) status. The basic characteristics of these clinical variables were all displayed in Table 1. Based on random sampling, 210 independent samples were classified as a training data set and 90 samples were classified as a validation data set. Using the data re-annotation method, a total of 7150 probes (containing the 5238 lncRNAs) were identified for further analysis.
Table 1.
Basic characteristics of the clinical variables in GC patients.
| Clinical variables | Total | Training dataset | Validate dataset |
|---|---|---|---|
| Sample size | 300 | 210 | 90 |
| Survival status | |||
| Dead | 152 | 102 | 50 |
| Survived | 148 | 108 | 40 |
| Median age (year) | 61.94 | 47.59 | 62.76 |
| Sex | |||
| Female | 101 | 74 | 27 |
| Male | 199 | 136 | 63 |
| T stage | |||
| 2 | 188 | 130 | 58 |
| 3 | 91 | 67 | 24 |
| 4 | 21 | 13 | 8 |
| N stage | |||
| 0 | 38 | 28 | 10 |
| 1 | 131 | 94 | 37 |
| 2 | 80 | 49 | 31 |
| 3 | 51 | 39 | 12 |
| M stage | |||
| 0 | 273 | 191 | 82 |
| 1 | 27 | 19 | 8 |
| AJCC stage | |||
| I | 30 | 21 | 9 |
| II | 97 | 70 | 27 |
| III | 96 | 65 | 31 |
| IV | 77 | 54 | 23 |
| Recurrent status | |||
| Yes | 125 | 110 | 53 |
| No | 157 | 77 | 29 |
| Unknown | 18 | 19 | 8 |
| Tumor cite | 4 | ||
| Antrum | 155 | 110 | 45 |
| Body | 107 | 77 | 30 |
| Cardia | 32 | 19 | 13 |
| Whole | 6 | 4 | 2 |
| WHO classification | |||
| W/D and M/D tubular | 114 | 89 | 25 |
| P/D tubular | 116 | 73 | 43 |
| Signet ring cell | 37 | 25 | 12 |
| Mucinous | 8 | 5 | 3 |
| Papillary | 9 | 6 | 3 |
| Other | 16 | 12 | 4 |
| Lauren type | |||
| Intestinal | 150 | 110 | 40 |
| Diffuse | 142 | 95 | 47 |
| Mixed | 8 | 5 | 3 |
| Molecular subtype | |||
| MSS/TP53− | 107 | 79 | 28 |
| MSS/TP53+ | 79 | 53 | 26 |
| MSI | 68 | 44 | 24 |
| MSS/EMT | 46 | 34 | 12 |
| EBV status | |||
| Positive | 18 | 181 | 76 |
| Negative | 257 | 13 | 5 |
| Missing | 12 | 16 | 9 |
| Adjuvant CCRT | |||
| Completed | 73 | 46 | 27 |
| Not completed | 7 | 5 | 2 |
| Not done | 220 | 129 | 61 |
| OP type | |||
| TG | 135 | 94 | 41 |
| STG | 165 | 116 | 49 |
GC – gastric cancer; AJCC – American Joint Committee on Cancer; WHO – World Health Organization; W/D – well-differentiated; M/D – moderately differentiated; P/D – poorly differentiated; MSS – microsatellite stable; TP53− – tumor protein 53 inactive; TP53+ – tumor protein 53 active; MSI – microsatellite instability; EMT – epithelial-mesenchymal transition; TG – total gastrectomy; STG – subtotal gastrectomy.
Construction and comparison of prognostic models for GC patients
Clinical model
The univariate and multivariate Cox regression analyses of the clinical information only screened 3 clinical variables related to OS which were molecular subtype, recurrence status, and OP type, shown in Table 2. Clinical model RSclinical was constructed based on these 3 independent prognostic factors, the corresponding 3-year and 5-year AUC were 0.765 and 0.780.
Table 2.
Prognostic clinical factors of OS for GC patients estimated by univariate and multivariate Cox regression.
| Clinical variables | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | P value | 95% CI | HR | P value | 95% CI | |
| Sex | 0.80 | 0.311 | 0.53–1.23 | 0.66 | 0.074 | 0.42–1.04 |
| Age | 1.01 | 0.416 | 0.99–1.03 | 1.03 | 0.014 | 1.01–1.05 |
| Molecular subtype* | 0.79 | 0.028 | 0.64–0.98 | 0.70 | 0.003 | 0.55–0.88 |
| Lauren subtype | 1.25 | 0.220 | 0.87–1.79 | 1.15 | 0.506 | 0.76–1.75 |
| WHO classification | 1.11 | 0.148 | 0.96–1.28 | 1.03 | 0.752 | 0.87–1.22 |
| T stage | 1.68 | 0.001 | 1.24–2.28 | 1.51 | 0.059 | 0.98–2.32 |
| N stage | 1.99 | 0.000 | 1.55–2.56 | 1.33 | 0.266 | 0.81–2.19 |
| M stage | 3.81 | 0.002 | 1.61–9.04 | 1.83 | 0.257 | 0.64–5.19 |
| AJCC stage | 2.20 | 0.000 | 1.7–2.86 | 1.37 | 0.307 | 0.75–2.49 |
| Tumor site | 1.48 | 0.003 | 1.14–1.91 | 1.18 | 0.352 | 0.83–1.66 |
| Recurrence status* | 1.39 | 0.013 | 1.07–1.81 | 1.57 | 0.007 | 1.13–2.17 |
| OP type* | 0.53 | 0.001 | 0.36–0.78 | 0.56 | 0.049 | 0.32–0.99 |
| EBV status | 0.58 | 0.441 | 0.14–2.35 | 0.59 | 0.471 | 0.14–2.49 |
| Adjuvant. CCRT | 1.07 | 0.576 | 0.85–1.33 | 1.00 | 1.000 | 0.78–1.28 |
Independent prognostic clinical variables with statistical significance at P<0.05 level both in univariate and multivariate Cox analyses.
OS – overall survival; GC – gastric cancer; HR – hazard ratio; CI – confidence interval; WHO – World Health Organization; AJCC – American Joint Committee on Cancer; OP – operation; EBV – Epstein-Barr virus; CCRT – concurrent chemoradiation therapy.
Gene model
Seven modules including blue, brown, green, red, turquoise, yellow, and gray modules were identified by WGCNA in our study. The modules were represented by branches of different colors, shown in Figure 1. The univariate Cox regression based on MEs of co-expression modules indicated only the red module (containing 55 lncRNAs) had a significant association with OS, and the increased expression of lncRNAs in the red module was associated with poor prognosis (hazard ratio [HR]=42.25, P=0.013, Table 3).
Figure 1.
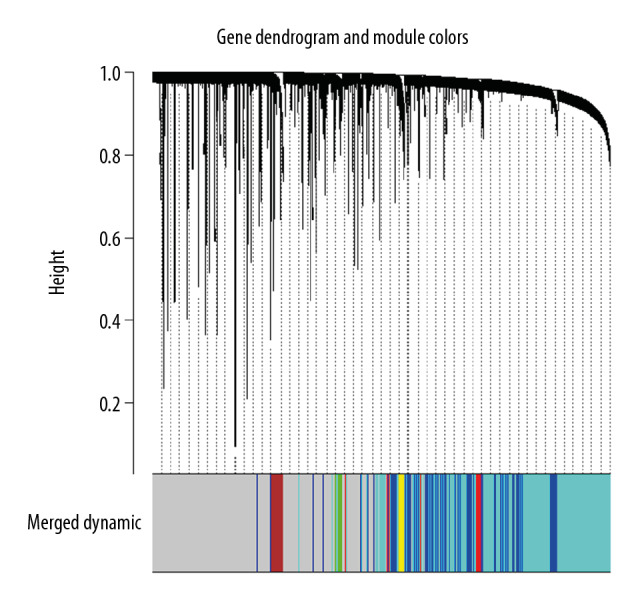
Gene clustering dendrogram and merged module colors based on a dissimilarity measure (1-TOM).
Table 3.
Prognostic modules of OS for GC patients estimated by univariate Cox regression analysis.
| Modules# | HR | P-Value | 95%CI | Gene numbers | Hub genes |
|---|---|---|---|---|---|
| Blue | 0.18 | 0.234 | 0.01–3.090 | 826 | – |
| Brown | 1.03 | 0.981 | 0.06–17.34 | 110 | – |
| Turquoise | 1.87 | 0.671 | 0.10–33.44 | 2064 | – |
| Yellow | 1.72 | 0.712 | 0.10–30.39 | 86 | – |
| Green | 3.29 | 0.392 | 0.22–50.32 | 65 | – |
| Red* | 42.25 | 0.013 | 2.20–811.4 | 55 | 11 |
| Gray | 0.05 | 0.053 | 0.00–1.040 | 2032 | – |
Modules identified by WGCNA;
prognostic module with statistical significance at P<0.05 level in univariate Cox analysis.
OS – overall survival; GC – gastric cancer; HR – hazard ratio; CI – confidence interval. WGCNA – weighted gene co-expression network analysis.
Firstly, 11 hub genes of red module were identified as candidate lncRNAs of WGCNA method to construct the gene model RSW. The 3-year and 5-year AUC of the RSW model were 0.689 and 0.682, respectively. Next, the 3 lncRNAs (LINC00930, AP000550.1, and AC009052.1) that were screened by the elastic-net Cox regression were used to construct the gene model RSe. The 3-year and 5-year AUC of RSe were 0.715 and 0.694, respectively. Then, the 13 candidate lncRNAs screened by the comprehensive method were used to construct the gene model RSc, the corresponding 3-year and 5-year AUC were 0.731 and 0.732, respectively. The candidate prognostic lncRNAs of different methods are shown in Table 4.
Table 4.
The candidate LncRNAs identified by 3 different methods.
| RSW | RSe | RSc | |||
|---|---|---|---|---|---|
| lncRNA | Coef | lncRNA | Coef | lncRNA | Coef |
| LOC644656 | 0.0375 | LINC00930 | −1.4153 | LOC644656 | 0.1710 |
| VWA8.AS1 | 0.0435 | AP000550.1 | −0.6437 | VWA8.AS1 | 0.2159 |
| LINC01085 | 0.0216 | AC009052.1 | −1.9817 | LOC101928069 | −0.4790 |
| LINC00606 | 0.0302 | LINC01206 | −0.2111 | ||
| KMT2E.AS1 | 0.0679 | LINC01085 | 0.1170 | ||
| DLG1.AS1 | 0.0186 | KMT2E.AS1 | 0.3330 | ||
| BVES.AS1 | 0.0666 | DAPK1.IT1 | 0.1238 | ||
| ADAMTSL4.AS1 | 0.0275 | AC139713.2 | 0.3901 | ||
| AC139713.2 | 0.0773 | AC023509.1 | −0.7042 | ||
| AC017091.1 | 0.1007 | AC017091.1 | 0.2683 | ||
| PXN.AS1 | 0.0679 | PXN.AS1 | 0.1654 | ||
| PTPRD.AS1 | −0.5058 | ||||
| PRKAG2.AS1 | −0.3523 | ||||
Coef was the corresponding ridge regression coefficient of the lncRNA (long noncoding RNA).
Clinical-gene model
The integrated clinical-gene models consisted of the candidate lncRNAs with 3 independent clinical factors for further analysis. The model RSW-clinical, which combined the candidate lncRNAs of RSW with 3 independent clinical factors, had 3-year and 5-year AUC of 0.816 and 0.805, respectively. The model RSe-clinical, containing 3 lncRNAs of RSe, had 3-year and 5-year AUC of 0.814 and 0.796, respectively. Furthermore, the 3-year and 5-year predictive ability of the model RSc-clinical, containing 13 lncRNAs identified by the comprehensive method, were 0.832 and 0.830, respectively.
Comparison of predictive ability
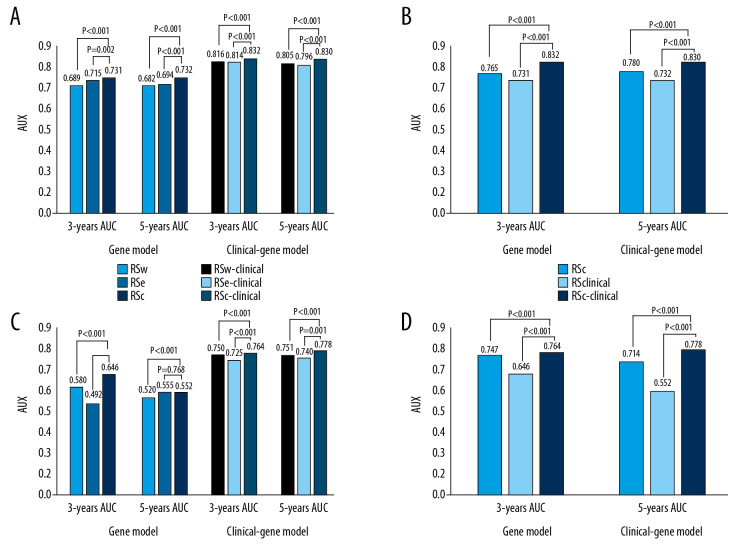
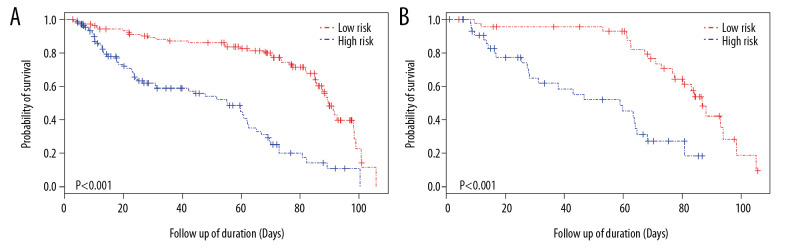
The comparison between the different models were illustrated in Figure 2. The 3-year and 5-year AUC comparisons of gene models indicated that the model RSc performed much better than models RSW and RSe with P<0.05. Considering the influence of clinical factors on model predictive ability, we compared the clinical-gene models of different methods. The model RSc-clinical exhibited a higher predictive ability than the RSW-clinical and RSe-clinical models in 3-year and 5-year survival prediction with P<0.001 (Figure 2A). Furthermore, the comparison between clinical, gene, and clinical-gene models of the comprehensive method indicated the RSc-clinical exhibited the highest predictive ability of GC patient’s OS (Figure 2B). Therefore, the optimal model RSc-clinical were identified as the robust prognostic model in the study. The median of the RSc-clinical index was used as our cutoff value to divide GC patients into 2 separate groups (high-risk group and low-risk group). The K-M survival analysis indicated that the low-risk group had a significantly better survival than the high-risk group (P<0.001, Figure 3A).
Figure 2.
The 3-year and 5-year area under the curve (AUC) comparison. (A) The 3-year and 5-year AUC comparison between 3 methods in gene and clinical-gene models in the training set. (B) The 3-year and 5-year AUC comparison between clinical, gene, and clinical-gene models of the comprehensive method in the training set. (C) The 3-year and 5-year AUC comparison between 3 methods in gene and clinical-gene models in the validation set. (D) The 3-year and 5-year AUC comparison between clinical, gene, and clinical-gene models of the comprehensive method in the validation set.
Figure 3.
Kaplan-Meier curve. (A) Kaplan-Meier curve of high-risk and low-risk groups in the training set (the median of RSc-clinical as a cutoff value for patients grouping); (B) Kaplan-Meier curve of high-risk and low-risk groups in the validation set (the median of RSc-clinical as a cutoff value for patients grouping).
Validation of predictive ability
The predictive ability of models aforementioned were validated using the remaining 30% of samples. The 3-year and 5-year AUC of gene model RSc were 0.646 and 0.552, respectively. A comparison between gene models proved that the model RSc had a relatively better performance than models RSW and RSe in 3-year OS prediction of GC patients (the 3-year AUC of model RSW was 0.580; and the 3-year AUC of model RSe was 0.492; P<0.001, Figure 2C). There was no significant difference in the 5-year survival prediction between RSc and RSe, the 5-year AUC of gene model RSe was 0.555, P=0.768. In terms of comparison between the clinical-gene models, the model RSc-clinical, with 3-year and 5-year AUCs of 0.764 and 0.778, demonstrated a better OS predictive ability than RSW-clinical and RSe-clinical (the 3-year and 5-year AUC values of RSW-clinical were 0.750 and 0.751; and the 3-year and 5-year AUC values of RSe-clinical were 0.725 and 0.740, Figure 2C). The 3-year and 5-year AUC of the clinical model in the validation set were 0.747 and 0.714, respectively. Additionally, the comparison between clinical, gene, and clinical-gene models of the comprehensive method indicated that the model RSc-clinical had the most precise predictive ability in the validation set (P<0.05, Figure 2D). Using the model RSc-clinical, the K-M survival analysis between high-risk group and low-risk group in the validation set also indicated that high risk patients had a lower survival rate than the rate among low risk patients (Figure 3B).
Stratification analysis
Stratification analysis of the optimal model RSc-clinical was performed based on molecular subtypes, recurrent status and operation type (Table 5). The molecular subtypes of GC were identified based on different patterns of disease occurrence, progression, and prognosis from several studies [29–33]. The 4 molecular subtypes, including microsatellite instability (MSI), microsatellite stable with epithelial-mesenchymal transition (MSS/EMT), MSS with active tumor protein 53 (MSS/TP53+), and MSS with inactive tumor protein 53 (MSS/TP53−), were proven to have a significant difference regarding survival rates. Of the 4 subtypes, MSI subtype led to the best outcome, followed by MSS/TP53+ and MSS/TP53−, and MSS/EMT had the worst prognosis [34]. By the stratification analysis, the model performed stably and reliably in MSS/TP53+ and MSS/TP53− subgroups than other 2 subgroups: the 3-year and 5-year AUC values of MSS/TP53+ were 0.860 and 0.816; and the 3-year and 5-year AUC values of MSS/TP53− were 0.843 and 0.800. The AUC values of the MSI and MSS/EMT subgroups displayed a large fluctuation: the 3-year and 5-year AUC of MSI were 0.677 and 0.717; the 3-year and 5-year AUC values of MSS/EMT were 0.782 and 0.881. In addition, stratification analysis in recurrent status and operation type indicated the model performed well in patients without recurrence (the 3-year and 5-year AUC were 0.812 and 0.730), and had a good prediction ability for patients with total gastrectomy (the 3-year and 5-year AUC were 0.765 and 0.804). The stratified analysis was not performed in the validation set due to the insufficient sample size of subgroups.
Table 5.
Stratification analysis of the optimal model based on 3 independent clinical factors in the training set.
| Independent clinical factors | 3-year AUC | 5-year AUC |
|---|---|---|
| Molecular type | ||
| MSS/TP53+ | 0.860 | 0.816 |
| MSS/TP53− | 0.843 | 0.800 |
| MSI | 0.677 | 0.717 |
| MSS/EMT | 0.782 | 0.881 |
| Recurrent status | ||
| Yes | 0.664 | 0.748 |
| No | 0.812 | 0.730 |
| Operation type | ||
| TG | 0.765 | 0.804 |
| STG | 0.770 | 0.684 |
AUC – area under the curve; MSS – microsatellite stable; TP53+ – tumor protein 53 active; TP53− – tumor protein 53 inactive; MSI – microsatellite instability; EMT – epithelial-mesenchymal transition; TG – total gastrectomy; STG – subtotal gastrectomy.
Biological function analysis of 13-lncRNA
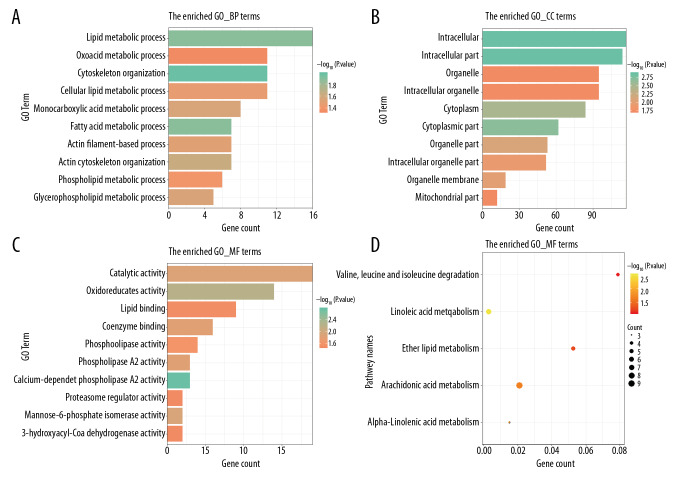
The list of 200 target genes was inputted into the DAVID database for GO and KEGG enrichment analysis and the results are shown in Figure 4. The GO analysis was performed for 3 different categories: biological process (BP); cell component (CC); and molecular function (MF). The top 10 items of BP, indicated that these lncRNAs were all associated with “cytoskeleton organization” and “fatty acid metabolic process”(Figure 4A); the top 10 CC terms shown in Figure 4B, such as “intracellular part” and “cytoplasmic part”, indicated that it might be involved in the composition of intracellular components. For the top 10 GO MF categories, items such as “oxidoreductase activity”, “phospholipase A2 activity”, and “catalytic activity” indicated that these biological activity processes potentially play a significant role in the occurrence of GC (Figure 4C). The top 5 significant KEGG pathways, which included “linoleic acid metabolism”, “alpha-linolenic acid metabolism”, “arachidonic acid metabolism”, “ether lipid metabolism”, and “valine, leucine and isoleucine degradation”, are displayed in Figure 4D. The biological pathways analysis indicated that the alterations in lipid metabolism were associated with cell proliferation of GC, and might play distinctive roles at various stages of tumor development [35].
Figure 4.
The most significantly enriched Gene Ontology (GO) annotations and pathways of 13 lncRNAs which we identified in the study. The length of bars and the size of dots represent the numbers of genes; the color of bars and dots corresponds to P value according to legend. (A) Top 10 significantly enriched biological process GO annotations. (B) Top 10 significantly enriched cellular component GO annotations. (C) Top 10 significantly enriched molecular function GO annotations. (D) Top 5 significantly enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway.
Discussion
Gastric cancer (GC) is a widely known cancer with unsatisfied survival. It is crucial to predict the survival of GC patients by constructing a robust prognostic model. However, most of the studies could not cope with the problems of high dimensionality and collinearity in data analysis, and they did not consider the interconnection among genes. Therefore, we combined the weighted co-expression gene analysis with elastic-net Cox regression based on the lncRNA expression, and we identified 13 co-expression lncRNAs as prognostic biomarkers of GC, which were LOC644656, VWA8-AS1, LOC101928069, LINC01206, LINC01085, KMT2E-AS1, DAPK1-IT1, AC139713.2, AC023509.1, AC017091.1, PXN-AS1, PTPRD-AS1, and PRKAG2-AS1.
The 3-year and 5-year AUC comparison of 3 different methods in gene and clinical-gene models indicated that the 13 co-expression lncRNAs identified by the comprehensive method were the best prognostic biomarkers in the study. The comparison between clinical, gene, and clinical-gene models of the comprehensive method identified the model RSc-clinical was the optimal prognostic model, with 0.832 and 0.830 for 3-year and 5-year AUC in the training set, respectively. Compared with previous prognostic research, although the 3-year and 5-year AUC of RSc-clinical were not the highest, the comprehensive method performed well in avoiding model over-fitting in high dimensional data and considering the association between genes, which were the limitations in previous studies. The results also indicated that both the clinical and genetic factors were indispensable in prognosis prediction.
Analysis in the validation set further validated our findings. The comparison between different methods in the clinical-gene models validated the value of the comprehensive method in prognostic prediction for GC. The 3-year and 5-year AUC of the optimal model RSc-clinical were 0.764 and 0.778 in the validation set, respectively. However, the predictive ability of model RSc was performed unstably in the 3-year and 5-year survival prediction, which might be due to insufficient sample size. Furthermore, the K-M analysis of GC patients indicated that the RSc-clinical index might be an effective prognostic factor to distinguish high-risk and low-risk patients with GC. In addition, stratification analysis indicated the model RSc-clinical performed stably in the MSS/TP53+ and MSS/TP53− subgroups. And the optimal model had a good prediction ability for patients without recurrence and patients with total gastrectomy. However, the model performed inaccurately in other subgroups. The large fluctuations in the 3-year and 5-year survival prediction of the optimal model in other subgroups might be caused by the small sample size and the extreme survival status of subgroup GC patients. A more reliable predictive model is required in future research for patients with special subtypes of GC.
One of the 13 prognostic lncRNAs PXN-AS1 has been reported to play an important role in tumor development, apoptosis, metastasis, and drug-resistance in several previous studies. Yuan et al. [36] identified PXN-AS1 as an alternative splicing factor which was modulated by Muscleblind-like-3. It was associated with focal adhesion protein, involved in transducing signals of the extracellular matrix, post-transcriptional gene regulation, and promoted cell proliferation in hepatocellular carcinoma. Zhang et al. [37] reported the mechanisms of PXN-AS1-L in non-small cell lung cancer (NSCLC). The over-expression of PXN-AS1-L increased the diversity of NSCLC cell and was significantly associated with advanced TNM stages and poor prognosis of NSCLC patients, and could be a potential prognostic biomarker and therapeutic target of NSCLC. Furthermore, lncRNA LINC01206 has been reported over-expression in lung squamous cell carcinoma or lung adenocarcinoma, and might be involved in cancer-related pathways such as apoptosis and migration of cell [38].
However, other lncRNAs which we identified in this study have not been reported currently, and the biological role of these lncRNAs in GC remains unknown. Therefore, we performed the GO and KEGG pathway enrichment analyses to briefly describe the potential molecular mechanisms of these 13 prognostic lncRNAs in GC. The results of the GO analysis indicated that these lncRNAs were associated with cytoskeleton components and intracellular components such as cytoplasm and organelle membrane, and participated in the fatty acid metabolic process and various biological activities such as oxidoreductase activity and catalytic activity. The KEGG enrichment analysis suggested that lipid metabolism plays an important role in GC cell proliferation, differentiation, and survival. The lipids are a diverse group of hydrophobic molecules which includes fats, oils, waxes, phospholipids, and steroids. Several studies have confirmed that various human cancers displayed aberrant activation of lipid metabolism, and this enabled cancer cells to proliferate, grow, and metastasize [35]. The alterations in lipid metabolism might be associate with GC progression and prognosis, and it might provide a novel diagnostic and therapeutic target for clinics. The biological functions of the 13 lncRNAs included in this study requires further investigation to provide a better understanding of the molecular mechanism in GC.
In addition, some limitations in our study need to be improved in future research. Firstly, the lncRNA re-annotation pipeline was based on the Affymetrix HG-U133 Plus 2.0 platform, which only represented a part of lncRNAs. A more comprehensive and reliable lncRNA re-annotation pipeline for all platforms is required. In addition, we only used a 30% internal sample as our validation set; large external cohorts of GC patients are required to further assess the robustness of the optimal model. Also, corresponding cell experiments and clinical trials are needed to validate our findings in future investigation. In addition, some biases might exist in selecting prognostic biomarkers based on lncRNAs profiles in this study; thus, analysis based on multi-omics data is required to better understand the molecular functions and disease etiology.
Conclusions
In summary, we used a comprehensive method which combined WGCNA with elastic-net Cox regression to identify potential biomarkers in the OS prediction of GC patients. The clinical-gene model, which contained 13 co-expression lncRNAs (LOC644656, VWA8-AS1, LOC101928069, LINC01206, LINC01085, KMT2E-AS1, DAPK1-IT1, AC139713.2, AC023509.1, AC017091.1, PXN-AS1, PTPRD-AS1, and PRKAG2-AS1) and 3 independent clinical variables (molecular subtypes, recurrent status, and operation type) were identified as a robust prognostic model for this study. The novel prognosis model might provide molecular knowledge to improve the clinical findings for the OS of GC patients. Further studies are required to validate our findings and explain the biological functions of these lncRNAs.
Acknowledgement
We sincerely thanked the GEO public database.
Footnotes
Conflict of interest
None.
Source of support: This study was supported by the Fundamental Research Funds for the Central Universities of Central South University (Grant numbers: 2019zzts743); partly supported by the grants from Natural Science Foundation of China (Grant numbers: 81101655) and the grant from National Science Foundation of Hunan Province (Grant numbers: 2011QNZT133)
References
- 1.Fitzmaurice C, Allen C, Barber RM, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: A systematic analysis for the global burden of disease study. JAMA Oncol. 2017;3:524–48. doi: 10.1001/jamaoncol.2016.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orditura M, Galizia G, Sforza V, et al. Treatment of gastric cancer. World J Gastroenterol. 2014;20:1635–49. doi: 10.3748/wjg.v20.i7.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang J, Liu X. Correlation analysis between helicobacter pylori infection status and tumor clinical pathology as well as prognosis of gastric cancer patients. Iran J Public Health. 2018;47:1529–36. [PMC free article] [PubMed] [Google Scholar]
- 4.Yusefi AR, Bagheri Lankarani K, Bastani P, et al. Risk factors for gastric cancer: A systematic review. Asian Pac J Cancer Prev. 2018;19:591–603. doi: 10.22034/APJCP.2018.19.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berlth F, Bollschweiler E, Drebber U, et al. Pathohistological classification systems in gastric cancer: Diagnostic relevance and prognostic value. World J Gastroenterol. 2014;20:5679–84. doi: 10.3748/wjg.v20.i19.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cislo M, Filip AA, Arnold Offerhaus GJ, et al. Distinct molecular subtypes of gastric cancer: from Lauren to molecular pathology. Oncotarget. 2018;9:19427–42. doi: 10.18632/oncotarget.24827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wittekind C. The development of the TNM classification of gastric cancer. Pathol Int. 2015;65:399–403. doi: 10.1111/pin.12306. [DOI] [PubMed] [Google Scholar]
- 8.Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–8. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnsson P, Lipovich L, Grander D, Morris KV. Evolutionary conservation of long non-coding RNAs; Sequence, structure, function. Biochim Biophysic Acta. 2014;1840:1063–71. doi: 10.1016/j.bbagen.2013.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gu Y, Chen T, Li G, et al. LncRNAs: Emerging biomarkers in gastric cancer. Future Oncol. 2015;11:2427–41. doi: 10.2217/fon.15.175. [DOI] [PubMed] [Google Scholar]
- 11.Zhu X, Tian X, Yu C, et al. A long non-coding RNA signature to improve prognosis prediction of gastric cancer. Mol Cancer. 2016;15:60. doi: 10.1186/s12943-016-0544-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan YZ, Liu W, Yan C, et al. Identification of a 5-lncRNA signature for the diagnosis and prognosis of gastric cancer. Tumour Biol. 2016;37:13265–77. doi: 10.1007/s13277-016-5185-9. [DOI] [PubMed] [Google Scholar]
- 13.Cheng P. A prognostic 3-long noncoding RNA signature for patients with gastric cancer. J Cell Biochem. 2018;119:9261–69. doi: 10.1002/jcb.27195. [DOI] [PubMed] [Google Scholar]
- 14.Peng PL, Zhou XY, Yi GD, et al. Identification of a novel gene pairs signature in the prognosis of gastric cancer. Cancer Med. 2018;7:344–50. doi: 10.1002/cam4.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Li H, Zhang W, et al. LASSO based CoxPH model identifies an 11 lncRNA signature for prognosis prediction in gastric cancer. Mol Med Rep. 2018;18:5579–93. doi: 10.3892/mmr.2018.9567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang R, Xiong J, Deng D, et al. An integrated model of clinical information and gene expression for prediction of survival in ovarian cancer patients. Trans Res. 2016;172:84–95.e11. doi: 10.1016/j.trsl.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 17.Lai Y, Hayashida M, Akutsu T. Survival analysis by penalized regression and matrix factorization. ScientificWorldJournal. 2013;2013 doi: 10.1155/2013/632030. 632030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiong J, Bing Z, Su Y, et al. An integrated mRNA and microRNA expression signature for glioblastoma multiforme prognosis. PLoS One. 2014;9:e98419. doi: 10.1371/journal.pone.0098419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim YH, Jeong DC, Pak K, et al. Gene network inherent in genomic big data improves the accuracy of prognostic prediction for cancer patients. Oncotarget. 2017;8:77515–26. doi: 10.18632/oncotarget.20548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simon N, Friedman J, Hastie T, Tibshirani R. Regularization paths for Cox’s proportional hazards model via coordinate descent. J Stat Softw. 2011;39:1–13. doi: 10.18637/jss.v039.i05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen PF, Wang F, Nie JY, et al. Co-expression network analysis identified CDH11 in association with progression and prognosis in gastric cancer. Onco Targets Ther. 2018;11:6425–36. doi: 10.2147/OTT.S176511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–93. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 23.Yang F, Zhang L, Huo XS, et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54:1679–89. doi: 10.1002/hep.24563. [DOI] [PubMed] [Google Scholar]
- 24.Wang S, Fan W, Wan B, et al. Characterization of long noncoding RNA and messenger RNA signatures in melanoma tumorigenesis and metastasis. PLoS One. 2017;12:e0172498. doi: 10.1371/journal.pone.0172498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X, Sun S, Pu JK, et al. Long non-coding RNA expression profiles predict clinical phenotypes in glioma. Neurobiol Dis. 2012;48:1. doi: 10.1016/j.nbd.2012.06.004. - [DOI] [PubMed] [Google Scholar]
- 26.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article17. [DOI] [PubMed] [Google Scholar]
- 28.Li H, Gong M, Zhao M, et al. LncRNAs KB-1836B5, LINC00566 and FAM27L are associated with the survival time of patients with ovarian cancer. Oncol Lett. 2018;16:3735–45. doi: 10.3892/ol.2018.9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–9. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lei Z, Tan IB, Das K, et al. Identification of molecular subtypes of gastric cancer with different responses to PI3-kinase inhibitors and 5-fluorouracil. Gastroenterology. 2013;145:554–65. doi: 10.1053/j.gastro.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 31.Tan IB, Ivanova T, Lim KH, et al. Intrinsic subtypes of gastric cancer, based on gene expression pattern, predict survival and respond differently to chemotherapy. Gastroenterology. 2011;141:476–85. 485.e1–11. doi: 10.1053/j.gastro.2011.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tay ST, Leong SH, Yu K, et al. A combined comparative genomic hybridization and expression microarray analysis of gastric cancer reveals novel molecular subtypes. Cancer Res. 2003;63:3309–16. [PubMed] [Google Scholar]
- 33.Zouridis H, Deng N, Ivanova T, et al. Methylation subtypes and large-scale epigenetic alterations in gastric cancer. Sci Transl Med. 2012;4:156ra140. doi: 10.1126/scitranslmed.3004504. [DOI] [PubMed] [Google Scholar]
- 34.Cristescu R, Lee J, Nebozhyn M, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med. 2015;21:449–56. doi: 10.1038/nm.3850. [DOI] [PubMed] [Google Scholar]
- 35.Chen M, Huang J. The expanded role of fatty acid metabolism in cancer: New aspects and targets. Prec Clin Med. 2019;2:183–91. doi: 10.1093/pcmedi/pbz017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan JH, Liu XN, Wang TT, et al. The MBNL3 splicing factor promotes hepatocellular carcinoma by increasing PXN expression through the alternative splicing of lncRNA-PXN-AS1. Nat Cell Biol. 2017;19:820–32. doi: 10.1038/ncb3538. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Z, Peng Z, Cao J, et al. Long noncoding RNA PXN-AS1-L promotes non-small cell lung cancer progression via regulating PXN. Cancer Cell Int. 2019;19:20. doi: 10.1186/s12935-019-0734-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang HY, Yang W, Zheng FS, et al. Long non-coding RNA SNHG1 regulates zinc finger E-box binding homeobox 1 expression by interacting with TAp63 and promotes cell metastasis and invasion in Lung squamous cell carcinoma. Biomed Pharmacother. 2017;90:650–58. doi: 10.1016/j.biopha.2017.03.104. [DOI] [PubMed] [Google Scholar]