Abstract
Initially discovered in bacteria and archaea, CRISPR–Cas9 is an adaptive immune system found in prokaryotes. In 2012, scientists found a way to use it as a genome editing tool. In 2013, its application in plants was successfully achieved. This breakthrough has opened up many new opportunities for researchers, including the opportunity to gain a better understanding of plant biological systems more quickly. The present study reviews agricultural applications related to the use of CRISPR systems in plants from 52 peer-reviewed articles published since 2014. Based on this literature review, the main use of CRISPR systems is to achieve improved yield performance, biofortification, biotic and abiotic stress tolerance, with rice (Oryza sativa) being the most studied crop.
Keywords: agriculture, CRISPR, gRNA, plant genome editing
Introduction
CRISPR/Cas systems have proved to be important tools for genome editing in model plants and crops. On the one hand, CRISPR/Cas or tools derived from this technology have permitted rapid and straightforward determination of the function of different coding and non-coding DNA sequences in model plants [1–5]. On the other hand, numerous studies describe applications of CRISPR systems for the development of new traits in crops and could be considered as proof-of-concept studies.
The CRISPR/Cas gene-editing system is able to generate heritable, targeted mutations and also to address concerns over the presence of foreign DNA sequences as it can generate transgene-free plants. Therefore, it offers advantages in giving precision that was previously not possible and in allowing the induction of mutations without the presence of transgenes in the final plants. Articles focusing on these areas are considered, together with the analysis of the agricultural opportunities offered by the technology within specific geographic areas. We built up the following bibliographic research quest to gather scientific peer-reviewed articles specifically dealing with trait improvement in crops: ‘CRISPR’ OR ‘clustered regularly interspaced short palindromic’ OR ‘cas9’ OR ‘cas 9’ AND (Plant* OR vegetal OR Spermatophyt* OR algae OR Dicot* OR Monocot* OR Legume* OR Cereal* OR crop*). Then, we submitted this research request in databases such as Infodoc, Sciencedirect, BiblioVie, EBSCO, BergeRicrochGMlibrary and Web of Science to perform this systematic literature review. These search terms may not have captured every relevant article; however, those captured clearly identify key trends.
Induction of heritable targeted mutations and generation of ‘transgene-free’ plants
Regarding economic perspectives and social acceptance of CRISPR/Cas systems, a major concern is the heritability of the gene-induced mutations and the generation of transgene-free plants. Various articles have reported the induction and stable inheritance of single- [6–8] and multiple-targeted mutations [2], studying the T0 plants and T1 and T2 progenies. Generally speaking, the mutants of interest are selected by segregation [9].
Regarding the heritability concern, Pan et al. [8] used a visually interesting tool to demonstrate the inheritance of mutations induced in the PDS gene of Solanum lycopersicum. SlPDS encodes the phytoene desaturase which is a key enzyme in carotenoid biosynthesis. The silencing of this gene causes photobleaching or albino phenotypes. The authors were, thus, able to monitor mutation and inheritance patterns with a visual indicator associated with genotyping and sequencing. They demonstrated that the CRISPR/Cas system can induce heritable mutations in tomato plants (from T0 to T2 generation plants) and that homozygous and biallelic mutants were generated even in the first generation. In addition, a classical study on Arabidopsis thaliana provided a general scheme regarding the heritability of mutations induced by CRISPR/Cas using Agrobacterium-mediated transformation [10].
To generate transgene-free plants, it is necessary to obtain a stable production of CRISPR/Cas-mutated lines without the presence of the CRISPR DNA expression cassettes in the final mutant plants. This can be achieved in many ways. Most studies using Agrobacterium-mediated transformation intend to generate final mutant plants without transferred DNA (T-DNA). As specific primers are used to detect the presence of transgenes encoding CRISPR/Cas components, scientists showed that transgene-free, T2 mutant lines could be obtained by genetic segregation: the targeted mutations were stably passed on in transgene-free plants [Table 1, column ‘Transgene-free plants studied (Yes/No)’]. Other studies show how to generate transgene-free plants using alternative delivery methods [11,12]. These methods include ways of introducing the CRISPR components in a transient fashion such that integration is unlikely, for example using protoplast systems [12]. Alternatively, it is possible to avoid the presence of foreign DNA at any stage of the process, therefore avoiding the possibility of foreign DNA insertion. This can be achieved by the introduction of RNA or a ribonucleoprotein complex [13].
Table 1. Agricultural applications of the use of CRISPR systems in the 52 articles studied (2014–2017).
| Plant species | Application perspectives | Targeted sequence(s) | Molecular functions | Delivery method//main strategy | Transgene-free plants studied (yes/no) | Reference |
|---|---|---|---|---|---|---|
| BIOTIC STRESS TOLERANCE | ||||||
| Virus stress tolerance | ||||||
| Model plants | ||||||
| Arabidopsis thaliana | Potyvirus resistance (TuMV) | eIF(iso)4E, member of the eukaryotic translation initiation factor | Recessive resistance alleles against various potyviruses | Agrobacterium-mediated transformation with a Cas9/gRNA recombinant plasmid binary vector (floral dipping) // gene knockout with Cas9/gRNA | Yes | [9] |
| Arabidopsis thaliana and Nicotiana benthamiana | Beet severe curly top virus (BSCTV) tolerance | 43 candidate sites in coding or non-coding sequences of the BSCTV genome for transient expression assays and selection of two sites for transgenic lines induction | Virus replication mechanism | Agrobacterium-mediated transformation of leaves with Cas9/gRNA expression plasmid vectors // gene knockout with Cas9/gRNA | No | [7] |
| Nicotiana benthamiana | Tomato yellow leaf curl virus (TYLCV) resistance | Coding and non-coding sequences of TYLCV | Virus replication mechanism | Agrobacterium-mediated transformation of leaves with a TRV RNA replicon for the delivery of gRNAs into Cas9 overexpressing plants // gene knockout with Cas9/gRNA | No | [14] |
| Virus tolerance | AGO2 gene | Contribution to antiviral immunity (virus-specific antiviral role of AGO2 gene) | Agrobacterium-mediated transformation of leaves with Cas9/gRNA expression plasmid vectors // gene knockout with Cas9/gRNA | No | [20] | |
| Crops | ||||||
| Cucumis sativus | Ipomovirus immunity, tolerance to the Zucchini yellow mosaic virus and Papaya ring spot mosaic virus-W | eIF4E (eukaryotic translation initiation factors 4E) | Host factors for RNA viruses, recessive resistance alleles against viruses | Agrobacterium-mediated transformation of cut cotyledons (without embryo) with binary vector containing Cas9/gRNA // gene knockout with Cas9/gRNA | Yes | [21] |
| Fungus stress tolerance | ||||||
| Crops | ||||||
| Oriza sativa | Blast (caused by Magnaporthe oryzae) tolerance | Ethylene responsive factor ERF transcription factor gene OsERF922 | Involved in the modulation of multiple stress tolerance | Agrobacterium-mediated transformation of embryogenic calli with Cas9/gRNA-expressing binary vectors // single and multiplex gene knockout with Cas9/gRNA | Yes | [22] |
| Solanum lycopersicum | Powdery mildew resistance | SlMlo gene | Major contributor to powdery mildew susceptibility | Agrobacterium-mediated transformation of cotyledons with Cas9/gRNA expression plasmid vectors // gene knockout with Cas9/gRNA | Yes | [23] |
| Triticum aestivum | Powdery mildew (Blumeria graminis f. sp. Tritici) resistance | One of the three mildew-resistance locus (MLO) homeologs in bread wheat: TaMLO-A1 allele | Encode a protein that was shown to repress defenses against powdery mildew diseases | Particle bombardment with Cas9/gRNA expressing plasmid into immature wheat embryos // gene knockout with Cas9/gRNA | Yes | [6] |
| Bacteria stress tolerance | ||||||
| Crops | ||||||
| Citrus paradisi | Citrus canker (caused by Xanthomonas citri subspecies citri (Xcc)) tolerance | PthA4 effector binding elements (EBEs) in the Type I CsLOB1 promoter (EBEPthA4-CsLOBP) of the CsLOB1 (Citrus sinensis lateral organ boundaries) gene | CsLOB1: susceptibility gene for citrus canker CsLOB1 gene expression induced by the binding of the pathogenicity factor PthA4 to the EBEPthA4-CsLOBP | Agrobacterium-mediated transformation of epicotyl with Cas9/gRNA expression plasmid vectors // gene knockout with Cas9/gRNA | No | [24] |
| Citrus sinensis Osbeck | Canker resistance | CsLOB1 promoter | Susceptibility gene CsLOB1 promoter in citrus | Agrobacterium-mediated epicotyl transformation // gene knockout with Cas9/gRNA | No | [25] |
| Oryza sativa | Bacterial blight (caused by Xanthomonas oryzae pv. oyzae) tolerance | Sucrose transporter gene OsSWEET13 | Disease-susceptibility gene for PthXo2 (TAL effector gene of X. oryzae pv. oryzae) | Agrobacterium-mediated transformation of embryogenic callus with Cas9/gRNA expression plasmid vectors // gene knockout with Cas9/gRNA | No | [26] |
| ABIOTIC STRESS TOLERANCE | ||||||
| Herbicide tolerance | ||||||
| Model plants | ||||||
| Arabidopsis thaliana | Cold, salt and drought stress tolerance | UDP-glycosyltransferases UGT79B2 and UGT79B3 | UGT family responsible for transferring sugar moieties onto a variety of small molecules and control many metabolic processes; UGT79B2 and UGT79B3 could be induced by various abiotic stresses | Agrobacterium-mediated transformation with a Cas9/gRNA recombinant plasmid binary vector via floral dipping // gene knockout with Cas9/gRNA | No | [27] |
| Glufosinate resistance and reduced trichomes formation | BAR gene GL1 gene |
BAR gene confers glufosinate resistance. GL1 gene is required for trichomes formation. |
Agrobacterium-mediated transformation with Cas9/gRNA plasmid vectors (floral dipping) // gene knockout with Cas9/gRNA | Yes | [28] | |
| Lotus japonicus | Bioavailability of soil organic nitrogen and capability to accommodate nitrogen-fixing bacteria intracellularly to fix its own nitrogen | Single and multiple symbiotic nitrogen fixation (SNF) genes: simbiosis receptor-like kinase (SYMRK), leghemglobin loci (LjLb1, LjLb2, LjLb3) | Involved in symbiotic nitrogen fixation | A. tumefaciens and A. rhizogenes-mediated transformation containing the appropriate plasmids // gene knockout with Cas9/gRNA | No | [3] |
| Crops | ||||||
| Linum usitatissimum | Glyphosate tolerance | 5′-Enolpyruvylshikimate-3-phosphate synthase (EPSPS) | EPSPS genes encode a protein in the Shikimate pathway that participates in the biosynthesis of aromatic amino acids; EPSPS is a target for the glyphosate where it acts as a competitive inhibitor of the binding site for phosphoenolpyruvate | Protoplast transfection with ssODN and CRISPR-Cas9 plasmid // gene replacement | No | [15] |
| Oryza sativa | Herbicide resistance | C287 gene | The C287Tgene mutation endows rice plants with resistance to the herbicide imazamox (IMZ) | Agrobacterium-mediated transformation // CRISPR-Cas9-mediated multiplex genome editing | No | [29] |
| Herbicide tolerance | Acetolactate synthase (ALS) gene | Involved in the ALS biosynthesis (amino acid biosynthesis) | Co-transformation of rice calli through particle bombardment with Cas9/gRNA expression plasmid vector and oligonucleotide donor // gene replacement with a donor template | No | [16] | |
| Glyphosate tolerance | 5′-Enolpyruvylshikimate-3-phosphate synthase (EPSPS) | Involved in the biosynthesis of aromatic amino acids | Co-transformation of rice calli through particle bombardment with Cas9/gRNA expression plasmid and donor plasmid // gene insertion and replacement with a donor template | Yes | [30] | |
| Solanum tuberosum | Reduced susceptibility to ALS-inhibiting herbicides | Acetolactate synthase 1 (ALS1) | Involved in the acetolactate synthase biosynthesis (amino acid biosynthesis) | Agrobacterium-mediated transformation for GVR-mediated delivery of CRISPR–Cas9 system and donor template // gene knockout and replacement | No | [18] |
| Salt stress tolerance | ||||||
| Crops | ||||||
| Oryza sativa | Salt stress tolerance | GT-1 element in the salt induction of OsRAV2 (key regulatory regions in its promoter) | RAV subfamily involved in developmental processes such as the brassinosteroid response, leaf senescence and flowering time and also in plant responses to abiotic stress including high salinity | Agrobacterium-mediated transformation of leaves with Cas9gRNA plasmid expression vector // gene knockout with Cas9/gRNA | No | [1] |
| Drought stress tolerance | ||||||
| Crops | ||||||
| Zea mays | Improved grain yield under field drought stress conditions | ARGOS8 | Negative regulator of ethylene responses | Co-transformation of immature embryos by particle bombardment with DNA repair template Cas9-sgRNA expression plasmids // gene insertion or replacement with a donor template | No | [17] |
| YIELD, BIOFORTIFICATION AND CONSERVATION PARAMETERS | ||||||
| Yield | ||||||
| Crops | ||||||
| Brassica oleracea and Hordeum vulgare | Pod shatter and control of dormancy | HvPM19 BolC.GA4.a |
Positive regulator of grain dormancy Involved in pod valve margin development |
Agrobacterium-mediated transformation // gene knockout with Cas9/gRNA | Yes | [31] |
| Dendrobium officinale | Lignocellulose biosynthesis | C3H, C4H, 4CL, CCR and IRX genes | Target genes are involved in the lignocellulose biosynthesis pathway | Agrobacterium-mediated transformation // gene knockout with Cas9/gRNA | No | [32] |
| Nicotiana tabacum, sylvestris and tomentosiformis | Regulation of axillary bud growth | NtPIN4 gene | Involved in auxin biosynthesis | Agrobacterium-mediated transformation of leaves // gene knockout with Cas9/gRNA | Yes | [33] |
| Oryza sativa | Starch synthesis pathway in rice pollen | Plastific large subunit of ADP-glucose pyrophosphorylase (OsAGPL4) | Involved in the starch synthesis pathway | Agrobacterium-mediated transformation with Cas9/gRNA plasmid expression vector // gene knockout with Cas9/gRNA | No | [34] |
| Regulation of pollen tube growth and integrity | Rice member of plant-specific receptor-like kinase CrRLK1LS subfamily, ruptured pollen tube (RUPO) | Receptor-like kinase (RUPO) as a regulator of high-affinity potassium transporters via phosphorylation-dependent interaction | Agrobacterium-mediated transformation of embryo-derived rice callus with Cas9/gRNA expression plasmids // gene knockout with Cas9/gRNA | No | [35] | |
| Grain yield performance | Grain size3 (GS3) and Grain number 1a (Gn1a) | Grain yield QTLs identified to regulate grain size and grain number | Agrobacterium-mediated transformation with Cas9/gRNA plasmid expression vector // gene knockout with Cas9/gRNA | Yes | [36] | |
| Grain weight | Grain width 2 (GW2), grain width 5 (GW5) and thousand-grain weight (TGW6) | Three major genes that negatively regulate rice grain weight | Agrobacterium-mediated transformation with Cas9/gRNAs plasmid expression vector // CRISPR–Cas9-mediated multiplex genome editing | Yes | [37] | |
| Development of japonica photo-sensitive genic male sterile rice lines | Carbon starved anther (CSA) | One important locus for regulating photoperiod-controlled male sterility in japonica rice | Agrobacterium-mediated transformation with two plasmids into calli // gene knockout with Cas9/gRNA | Yes | [38] | |
| Enhanced grain number, dense erect panicles, larger grain size | Cytokinin dehydrogenase2 (Gn1a), γ-subunit of G protein (DEP1), γ-subunit of G protein (GS3) and squamosa promoter binding protein (IPA1) | Regulators of grain number, panicle architecture, grain size and plant architecture | Agrobacterium-mediated transformation with Cas9/gRNA plasmid expression vectors // gene knockout with Cas9/gRNA | Yes | [39] | |
| Maintenance and determinacy of the flower meristem | FLORAL ORGAN NUMBER2 (FON2) gene OsMADS3 gene |
Involved in meristem maintenance and in stamen specification | Agrobacterium-mediated transformation of calli // gene knockout with Cas9/gRNA | No | [40] | |
| Rice caryopsis development | OsSWEET11 gene | Sugar transporter | Agrobacterium-mediated transformation of leaves // gene knockout with Cas9/gRNA | No | [41] | |
| Stomatal developmental | EPFL9 gene | Positive regulator of stomatal developmental pathway | Agrobacterium-mediated transformation of immature embryos // gene knockout with CRISPR–Cas9/Cpf1 system | Yes | [42] | |
| Developing marker-free transgenic plants | GUS gene | Marker gene | Agrobacterium or gene gun with a construct expressing Cas9 and two gRNAs // gene knockout with Cas9/gRNA | No | [43] | |
| Rice development | MPK1 and MPK6 gnes | Essential genes for rice development | Agrobacterium-mediated transformation of rice calli // gene knockout with Cas9/gRNA | Yes | [5] | |
| Regulation of seed development | MEGs and PEGs genes | Involved in the regulation of nutrient metabolism and endosperm development | Agrobacterium-mediated transformation // gene knockout with Cas9/gRNA | No | [44] | |
| Breeding of early-maturing rice cultivars | Hd2, Hd4 and Hd5 genes | Flowering suppressors in Ehd1-dependent photoperiodic flowering pathway and major genes that negatively control the heading date of rice varieties grown in the north of China | Agrobacterium-mediated transformation // gene knockout with Cas9/gRNA | No | [45] | |
| Solanum lycopersicum | Generation of parthenocarpic tomato plants | SlIAA9 gene | A key gene controlling parthenocarpy | Agrobacterium-mediated transformation of leaves // gene knockout with Cas9/gRNA | No | [46] |
| Taraxacum kok-saghyz | Rubber biosynthesis in hairy roots | TK 1-FFT (fructan:fructan 1-fructosyltransferase) | Implicated in inulin biosynthesis (antagonist of rubber production) | TK plantlets inoculated with Agrobacterium rhizogenes harbouring a plasmid encoding Cas9/gRNA (wounded surface of the plantlets dipping) // gene knockout with Cas9/gRNA | No | [47] |
| Zea mays | High-frequency targeted mutagenesis | Argonaute 18 (ZmAgo18a and ZmAgo18b), dihydroflavonol 4-reductase or anthocyaninless genes (a1 and a4) | Involved in sporogenesis and anthocyanin biosynthesis | Agrobacterium-mediated transformation | No | [48] |
| Reduction of the linkage drag during breeding procedure | LG1 gene | Genetic basis for the upright architecture of maize leaves | Agrobacterium-mediated transformation of immature embryos // gene knockout with Cas9/gRNA | No | [49] | |
| Biofortification | ||||||
| Crops | ||||||
| Camelina sativa | Enhancement of seed oil (fatty acid) composition in seeds | Fatty acid desaturase 2 (FAD2) genes | Key gene involved in the synthesis of polyunsaturated fatty acids [insertion of a double bond at the delta-12 (omega-6) position of oleic acid to obtain linoleic acid] | Agrobacterium-mediated transformation with Cas9/gRNA plasmid vectors (floral dipping) // gene knockout with Cas9/gRNA | No | [50] |
| Reduced levels of polyunsaturated fatty acids and increased accumulation of oleic acid in the oil | Fatty acid desaturase 2 (FAD2) | Key gene involved in the synthesis of polyunsaturated fatty acids [insertion of a double bond at the delta-12 (omega-6) position of oleic acid to obtain linoleic acid] | Agrobacterium-mediated transformation with Cas9/gRNA plasmid vectors (floral dipping) // gene knockout with Cas9/gRNA | No | [51] | |
| Seed oil biosynthesis | CsDGAT1 or CsPDAT1 homeologous genes | Involved in triacylglycerol (TAG) synthesis in developing seeds | Agrobacterium-mediated floral vacuum infiltration method // CRISPR–Cas9-mediated multiplex genome editing | No | [52] | |
| Hordeum vulgare cv. “Golden Promise” | N-glycans modification in cereal grains | The putative endogenous barley ENGase gene | Involved in N-glycans biosynthesis | Co-bombarding selected combinations of sgRNA with wild-type cas9 using separate plasmids, or by co-infection with separate Agrobacterium tumefaciens cultures // CRISPR–Cas9-mediated multiplex genome editing | No | [53] |
| Nicotiana tabacum | Production of biotherapeutic proteins | XylT gene FucT gene |
Involved in glycans biosynthesis | Agrobacterium-mediated transformation // gene knockout with Cas9/gRNA | No | [54] |
| Production of biotherapeutic proteins | Beta(1,2)-xylosyltransferase (XylT) and alpha(1,3)fucosyltransferase (FucT). | Involved in glycans biosynthesis | Agrobacterium-mediated transformation // CRISPR-Cas9-mediated multiplex genome editing | No | [55] | |
| Oryza sativa | Generation of high-amylose rice | SBEI and SBEIIb genes | Starch branching enzyme (SBE) genes involved in starch biosynthesis | Agrobacterium-mediated transformation // gene knockout with Cas9/gRNA | Yes | [56] |
| Papaver somniferum | Biosynthesis of Benzylisoquinoline alkaloids (BIAs): medical biomolecules | 3′-hydroxyl-N-methylcoclaurine 4′-O-methyltransferase isoform 2 (4′ OMT2) gene | Implicated in the regulation of the biosythesis of benzylisoquinoline alkaloids (BIAs, e.g. morphine, thebaine) | Agrobacterium-mediated transformation of leaves with TRV-based synthetic plasmids expressing gRNA and a Cas9-encoding synthetic vector // gene knockout with Cas9/gRNA | No | [19] |
| Solanum tuberosum | Starch quality (amylopectin potato starch) | Three different regions of the gene encoding granule-bound starch synthase (GBSS) | Enzyme responsible for the synthesis of amylose (encoded by a single locus) | PEG-mediated protoplast transfection with CRISPR-Cas9 expression plasmid constructs // gene knockout with Cas9/gRNA | Yes | [12] |
| Salvia miltiorrhiza | Knock out the committed diterpene synthase gene | Diterpene synthase gene SmCPS1 | Involved in tanshinone biosynthesis | Agrobacterium rhizogenes-mediated transformation // gene knockout with Cas9/gRNA | No | [57] |
| Conservation parameters | ||||||
| Crops | ||||||
| Solanum lycopersicum | Inhibition of tomato fruit ripening | Three regions within the RIN gene (ripening inhibitor) | Master regulator gene for tomato fruit ripening; encodes a MADS-box transcription factor regulating fruit ripening | Agrobacterium-mediated transformation with Cas9/sgRNA-expressing plasmid vectors // CRISPR–Cas9-mediated multiplex genome editing | Yes | [58] |
The heritability and the transgene-free character of the generated plants were demonstrated in several studies, confirming that these areas should no longer be a concern for agricultural applications. This opens up many opportunities for different agricultural and industrial applications of CRISPR systems and below we focus on those that were developed in proof-of-concept studies.
Agricultural and industrial proof-of-concept studies
To study the agricultural and industrial applications of CRISPR/Cas systems in plants, 52 articles dealing with trait improvement of crops were selected to assess how scientists are directing their use. The use of CRISPR/Cas systems covers various applications, from biotic stress tolerance to abiotic stress tolerance, and also includes the achievements of improved yield performance, biofortification and enhancement of plant quality (Table 1 and Figure 1). Table 1 summarizes the main information found in these applied research articles, with a view to
- understanding the main applications of CRISPR/Cas systems in plant genome editing;
- looking at whether the production of transgene-free plants was addressed in the studies and
- detailing the main strategy used and method of delivery of the CRISPR components.
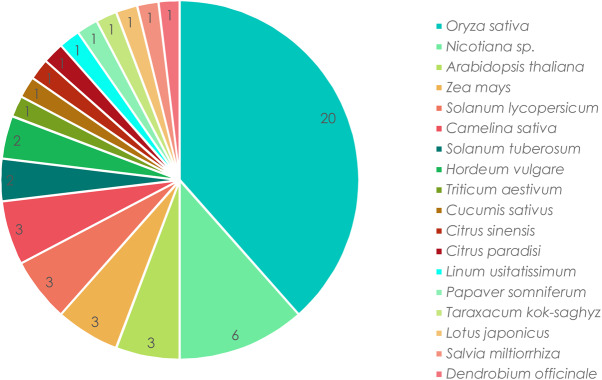
Figure 1. Plant species studied.
Plant species studied in articles with agricultural applications (2014–2017).
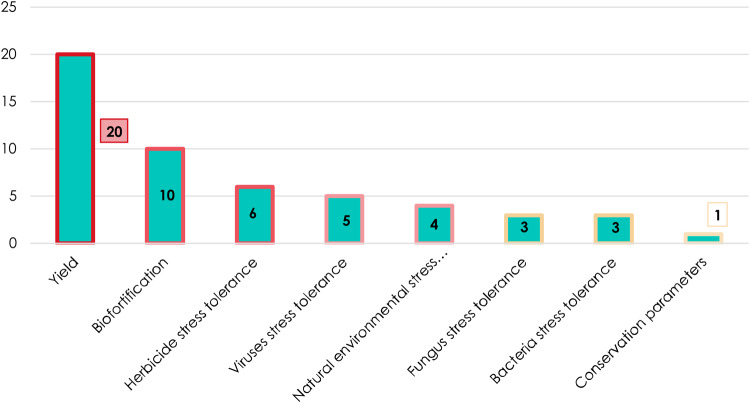
First of all, the application of CRISPR/Cas systems is mainly achieved directly in crops: 42 out of 52 articles studying 15 crops (Figure 1). Few studies use model plants for transient assays before studying the stable and heritable patterns of CRISPR-induced mutations in the target crop(s). Several trends can be observed with regard to the scope of applications of CRISPR/Cas systems in plant genome editing. The most important group of target applications relates to yield traits followed by the achievement of biotic or abiotic stress tolerance (Figure 2). Biotic stress tolerance includes induced tolerance to viral, fungal and bacterial diseases with a higher number of articles exploring plant tolerances to viral disease (Figure 2). As for abiotic stress tolerance, the two main objectives are to achieve herbicide and natural environmental stress tolerances (Figure 2). Environmental stress includes cold, salt, drought and nitrogen stress. All of these trait improvements are related to economic and agronomic challenges faced by farmers as pathogens, and environmental conditions are important threats that need to be dealt with in agriculture. Furthermore, plant breeders are continually trying to increase yield performances. The most studied crop is rice (Oryza sativa) (Figure 1) followed by other major crops: maize (Zea mays), tomato (S. lycopersicum), potato (Solanum tuberosum), barley (Hordeum vulgare) and wheat (Triticum aestivum) (Figure 1). Finally, the emergence of biofortification in the list of applications can be related to that of metabolic engineering in the 1990s.
Figure 2. CRISPR applications.
Relative importance of the different applications of CRISPR systems in terms of the number of articles (2014–2017).
For the achievement of viral disease resistance, two main strategies are observed:
- the integration of CRISPR-coding sequence in the host plant genome that targets and interferes with the virus genome once it is incorporated in the plant: the aim is to establish a CRISPR-like immune system in the host genome [7,14] and
- the induction of a CRISPR-mediated targeted mutation in the host plant genome that will confer improved virus resistance traits [9].
As an example, Ji et al. [7] demonstrated resistance to the Geminivirus, beet severe curly top virus using a CRISPR/Cas-based approach in the model plants Arabidopsis and Nicotiana benthamiana. The resulting plants were highly resistant to the virus. An extensive knowledge of plant biology and gene functionalities is required before using CRISPR/Cas systems in a specific species for a particular application. The application of CRISPR/Cas gene editing requires the precise definition of the target DNA sequence and the availability of good genome sequence data of the studied species in order to allow design of single-guide RNAs (sgRNA). The presence of a PAM sequence (protospacer-adjacent motif) upstream of the sequence complementary to the sgRNA is also required and it is necessary to search for putative off-target sites.
Once the decision is taken to employ CRISPR/Cas systems for a given application, scientists need to choose adapted delivery methods and strategies to fulfill their objectives. Table 1 lists a selection of articles with agricultural applications that could be considered as proof-of-concept studies for future commercial application of CRISPR/Cas systems in plants. It shows that conventional Agrobacterium-mediated transformation using plasmid vectors containing, for example, Cas9/sgRNA expression cassettes is mainly used to deliver the system to plants. However, additional delivery methods have also been implemented such as
- protoplast transfection in Linum usitatissimum and S. tuberosum [12,15];
- biolistic delivery in T. aestivum, O. sativa and Z. mays [6,16,17], and
- use of reconstituted viral replicons in N. benthamiana, S. tuberosum and Papaver somniferum [14,18,19,59,60].
Table 1 also describes how CRISPR/Cas systems can be used not only for site-directed mutagenesis (gene knockout) but also for gene insertion or replacement and multiplex genome editing (column ‘Delivery method//Main strategy’).
Geographic distribution of studied articles
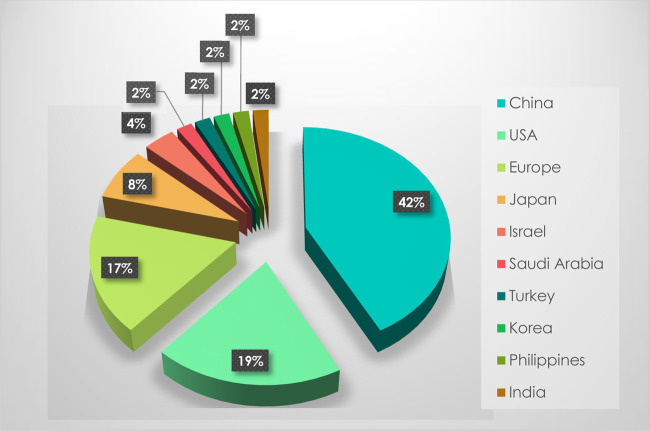
As there are distinct research, economic and regulatory contexts in the world, it was interesting to focus on the importance of the use of CRISPR/Cas systems in plant genome editing depending on the country where studies were carried out. Regarding articles with agricultural applications, Figure 3 shows that China and the U.S.A. are ranked first with 22 (42%) and second with 10 articles (19%), respectively. Europe, which includes the U.K., Sweden, France, Hungary, Germany, Austria and Belgium, had 9 articles (17%). Four studies were carried out in Japan and two in Israel. Five studies were carried out in each of the following countries: Saudi Arabia, Turkey, Korea, Philippines and India. This figure is consistent with the globalized economic, regulatory and research contexts and can be partly explained by the uncertain regulatory framework in Europe that may be holding back work towards commercial application (Figure 3).
Figure 3. CRISPR studies by country.
Number of articles studying the use of CRISPR systems in plant genome editing with agricultural applications according to the country of the research team (2014–2017).
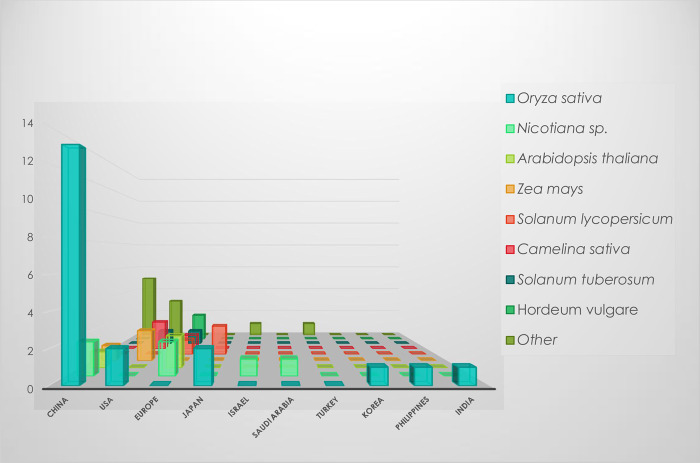
Regarding the plant species studied according to the country of the research team (Figure 4), the dominance of rice (O. sativa) is again observed, and mainly in China, which is in accordance with the Chinese research and economic contexts. Additionally, the application of CRISPR/Cas systems in maize (Z. mays) seems to be mainly studied in the U.S.A. Efficient systems for genome editing in soybean have also been reported for example [61]. Other crops that were studied include vegetables and industrial plants:
- Cucumis sativus, Citrus paradisi and Citrus sinensis,
- L. usitatissimum, P. somniferum, Taraxacum kok-saghyz, Salvia miltiorrhiza and Dendrobium officinale and
- the model plant Lotus japonicus.
Figure 4. Plant species studied by country.
Plant species studied in articles using CRISPR systems in plant genome editing with agricultural applications according to the country of the research team (2014–2017).
In terms of methods, the generation of transgene-free plants (that is to say plants in which the Cas9/sgRNA-expressing sequence was not integrated) is important to examine in relation to the geographic location of the research team describing the specific agricultural application. Although the number of reviewed articles is low, one trend worth noting is that only one of the studies carried out in the U.S.A. addressed the generation of transgene-free plants (Figure 5). In contrast, Chinese and European studies paid particular attention to the generation of transgene-free plants. This is likely to be linked to GMO regulatory requirements and intellectual property considerations that differ from country to country.
Figure 5. Generation of transgene-free plants by country.
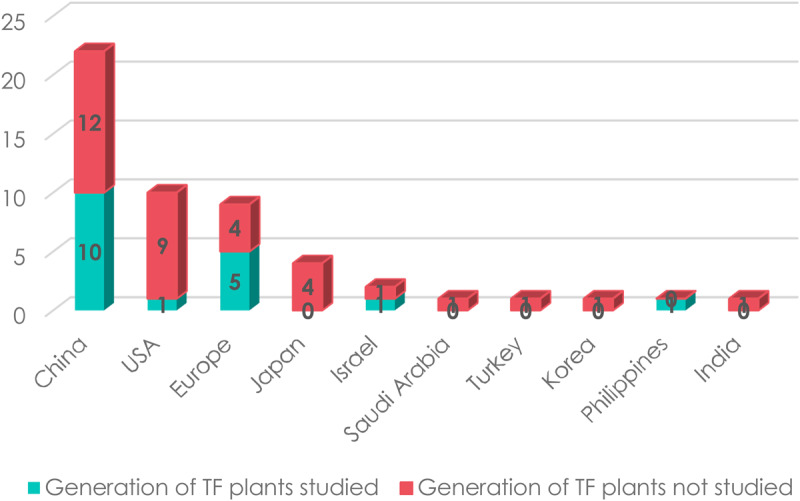
Sorting of the 52 articles according to the country of the research teams showing whether the generation of transgene-free plants was studied (green) or not (red).
Conclusion
Since 2013, considerable progress has been made in plant genome editing thanks to CRISPR/Cas systems. This technology has allowed straightforward, cost-effective and efficient gene editing compared with previous technologies, including zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs), making it accessible to many researchers. However, this emerging method is still developing and scientific efforts must continue to be made in order to obtain a mature technology and to realize the full potential of the technology. CRISPR/Cas-based technologies are, however, advancing at a rapid pace with the description of many new technological advances. Such advances are often first described in animal systems and then transferred to plants. A recent example is the application of ‘base editing’ in a crop where a specific base change was achieved in wheat rather than the usual mutation at a specific site involving a small insertion or deletion [62].
Concerns have been raised over the relationships that may exist between the use of this method and GMOs, and the many studies related to the generation of transgene-free plants [63] show that scientists aim to demonstrate that this technology is distinct from GM technology. In the U.S.A., the legal status of CRISPR/Cas-induced mutations is that they are exempt from GMO laws. In Europe, in October 2016, the French Council of State asked the European Court of Justice whether CRISPR/Cas and other site-directed mutagenesis tools should fall under the EU GMO legislation. The European Court of Justice has 18 months to reply. Moreover, ethical concerns could also emerge regarding the impact on public health and the environment of using CRISPR/Cas in plants. However, the use of this system already represents an emerging market, with CRISPR/Cas applications spanning a wide range of industries including research, agricultural and biomedical [64]. The agricultural applications described in this literature review represent only the very first, initial uses of this exciting technology, and we can expect many more valuable opportunities for agriculture in the near future.
Summary
A systematic review of 52 scientific articles from 2014 to mid-2017 regarding the use of CRISPR systems for agricultural applications.
The principal species studied is rice. The main applications are yield performance, biofortification and tolerance to abiotic and biotic stress (virus, fungi and bacteria). China published most articles in this area followed by the U.S.A. and Europe.
The heritability of the induced mutations and the development of transgene-free plants are the most studied areas.
Acknowledgements
The authors thank Jacqueline Martin-Laffon (CNRS, France) for providing scientific literature and Lamya Sajjaa (University of Paris-Sud, France) for her help in article sorting.
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
References
- 1.Duan Y.-B., Li J., Qin R.-Y., Xu R.-F., Li H., Yang Y.-C. et al. (2016) Identification of a regulatory element responsible for salt induction of rice OsRAV2 through ex situ and in situ promoter analysis. Plant Mol. Biol. 90, 49–62 10.1007/s11103-015-0393-z [DOI] [PubMed] [Google Scholar]
- 2.Yan W., Chen D. and Kaufmann K. (2016) Efficient multiplex mutagenesis by RNA-guided Cas9 and its use in the characterization of regulatory elements in the AGAMOUS gene. Plant Methods 12, 23 10.1186/s13007-016-0125-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang L., Wang L., Tan Q., Fan Q., Zhu H., Hong Z. et al. (2016) Efficient inactivation of symbiotic nitrogen fixation related genes in Lotus japonicus using CRISPR-Cas9. Front Plant. Sci. 7, 1333 10.3389/fpls.2016.01333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meng Y., Hou Y., Wang H., Ji R., Liu B., Wen J. et al. (2017) Targeted mutagenesis by CRISPR/Cas9 system in the model legume Medicago truncatula. Plant Cell Rep. 36, 371–374 10.1007/s00299-016-2069-9 [DOI] [PubMed] [Google Scholar]
- 5.Minkenberg B., Xie K. and Yang Y. (2017) Discovery of rice essential genes by characterizing a CRISPR-edited mutation of closely related rice MAP kinase genes. Plant J. 89, 636–648 10.1111/tpj.13399 [DOI] [PubMed] [Google Scholar]
- 6.Wang Y., Cheng X., Shan Q., Zhang Y., Liu J., Gao C. et al. (2014) Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951 10.1038/nbt.2969 [DOI] [PubMed] [Google Scholar]
- 7.Ji X., Zhang H., Zhang Y., Wang Y. and Gao C. (2015) Establishing a CRISPR–Cas-like immune system conferring DNA virus resistance in plants. Nat. Plants 1, 15144 10.1038/nplants.2015.144 [DOI] [PubMed] [Google Scholar]
- 8.Pan C., Ye L., Qin L., Liu X., He Y., Wang J. et al. (2016) CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations. Sci. Rep. 6, 24765 10.1038/srep24765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pyott D.E., Sheehan E. and Molnar A. (2016) Engineering of CRISPR/Cas9-mediated potyvirus resistance in transgene-free Arabidopsis plants. Mol. Plant Pathol. 17, 1276–1288 10.1111/mpp.12417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fauser F., Schiml S. and Puchta H. (2014) Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 79, 348–359 10.1111/tpj.12554 [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y., Liang Z., Zong Y., Wang Y., Liu J., Chen K. et al. (2016) Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 7, 12617 10.1038/ncomms12617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andersson M., Turesson H., Nicolia A., Fält A.-S., Samuelsson M. and Hofvander P. (2016) Efficient targeted multiallelic mutagenesis in tetraploid potato (Solanum tuberosum) by transient CRISPR-Cas9 expression in protoplasts. Plant Cell Rep. 36, 117–128 10.1007/s00299-016-2062-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang Z., Chen K., Li T., Zhang Y., Wang Y., Zhao Q. et al. (2017) Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat. Commun. 8, 14261 10.1038/ncomms14261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ali Z., Abulfaraj A., Idris A., Ali S., Tashkandi M. and Mahfouz M.M. (2015) CRISPR/Cas9-mediated viral interference in plants. Genome Biol. 16, 238 10.1186/s13059-015-0799-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sauer N.J., Narváez-Vásquez J., Mozoruk J., Miller R.B., Warburg Z.J., Woodward M.J. et al. (2016) Oligonucleotide-mediated genome editing provides precision and function to engineered nucleases and antibiotics in plants. Plant Physiol. 170, 1917–1928 10.1104/pp.15.01696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun Y., Zhang X., Wu C., He Y., Ma Y., Hou H. et al. (2016) Engineering herbicide-resistant rice plants through CRISPR/Cas9-mediated homologous recombination of acetolactate synthase. Mol. Plant 9, 628–631 10.1016/j.molp.2016.01.001 [DOI] [PubMed] [Google Scholar]
- 17.Shi J., Gao H., Wang H., Lafitte H.R., Archibald R.L., Yang M. et al. (2017) ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 15, 207–216 10.1111/pbi.12603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butler N.M., Baltes N.J., Voytas D.F. and Douches D.S. (2016) Geminivirus-mediated genome editing in potato (Solanum tuberosum L.) using sequence-specific nucleases. Front. Plant Sci. 7, 1045 10.3389/fpls.2016.01045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alagoz Y., Gurkok T., Zhang B. and Unver T. (2016) Manipulating the biosynthesis of bioactive compound alkaloids for next-generation metabolic engineering in opium poppy using CRISPR-Cas 9 genome editing technology. Sci. Rep. 6, 30910 10.1038/srep30910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ludman M., Burgyán J. and Fátyol K. (2017) CRISPR/Cas9 mediated inactivation of argonaute 2 reveals its differential involvement in antiviral responses. Sci. Rep. 7, 1010 10.1038/s41598-017-01050-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chandrasekaran J., Brumin M., Wolf D., Leibman D., Klap C., Pearlsman M. et al. (2016) Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol. Plant Pathol. 17, 1140–1153 10.1111/mpp.12375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang F., Wang C., Liu P., Lei C., Hao W., Gao Y. et al. (2016) Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922. PLoS ONE 11, e0154027 10.1371/journal.pone.0154027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nekrasov V., Wang C., Win J., Lanz C., Weigel D. and Kamoun S. (2017) Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci. Rep. 7, 482 10.1038/s41598-017-00578-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jia H., Orbovic V., Jones J.B. and Wang N. (2016) Modification of the PthA4 effector binding elements in type I CsLOB1 promoter using Cas9/sgRNA to produce transgenic Duncan grapefruit alleviating XccΔpthA4:dCsLOB1.3 infection. Plant Biotechnol. J. 14, 1291–1301 10.1111/pbi.12495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng A., Chen S., Lei T., Xu L., He Y., Wu L. et al. (2017) Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol. J. 10.1111/pbi.12733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou J., Peng Z., Long J., Sosso D., Liu B., Eom J.-S. et al. (2015) Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice. Plant J. 82, 632–643 10.1111/tpj.12838 [DOI] [PubMed] [Google Scholar]
- 27.Li P., Li Y.-J., Zhang F.-J., Zhang G.-Z., Jiang X.-Y., Yu H.-M. et al. (2017) The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. Plant J. 89, 85–103 10.1111/tpj.13324 [DOI] [PubMed] [Google Scholar]
- 28.Hahn F., Mantegazza O., Greiner A., Hegemann P., Eisenhut M. and Weber A.P.M. (2017) An efficient visual screen for CRISPR/Cas9 activity in Arabidopsis thaliana. Front. Plant Sci. 8, 39 10.3389/fpls.2017.00039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimatani Z., Kashojiya S., Takayama M., Terada R., Arazoe T., Ishii H. et al. (2017) Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat. Biotechnol. 35, 441–443 10.1038/nbt.3833 [DOI] [PubMed] [Google Scholar]
- 30.Li J., Meng X., Zong Y., Chen K., Zhang H., Liu J. et al. (2016) Gene replacements and insertions in rice by intron targeting using CRISPR–Cas9. Nat. Plants 2, 16139 10.1038/nplants.2016.139 [DOI] [PubMed] [Google Scholar]
- 31.Lawrenson T., Shorinola O., Stacey N., Li C., Østergaard L., Patron N. et al. (2015) Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA-guided Cas9 nuclease. Genome Biol. 16, 258 10.1186/s13059-015-0826-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kui L., Chen H., Zhang W., He S., Xiong Z., Zhang Y. et al. (2017) Building a genetic manipulation tool box for orchid biology: identification of constitutive promoters and application of CRISPR/Cas9 in the orchid, Dendrobium officinale. Front. Plant Sci. 7, 2036 10.3389/fpls.2016.02036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie X., Qin G., Si P., Luo Z., Gao J., Chen X. et al. (2017) Analysis of Nicotiana tabacum PIN genes identifies NtPIN4 as a key regulator of axillary bud growth. Physiol. Plant. 160, 222–239 10.1111/ppl.12547 [DOI] [PubMed] [Google Scholar]
- 34.Lee S.-K., Eom J.-S., Hwang S.-K., Shin D., An G., Okita T.W. et al. (2016) Plastidic phosphoglucomutase and ADP-glucose pyrophosphorylase mutants impair starch synthesis in rice pollen grains and cause male sterility. J. Exp. Bot. 67, 5557–5569 10.1093/jxb/erw324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu L., Zheng C., Kuang B., Wei L., Yan L. and Wang T. (2016) Receptor-like kinase RUPO interacts with potassium transporters to regulate pollen tube growth and integrity in rice. PLOS Genet. 12, e1006085 10.1371/journal.pgen.1006085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen L., Wang C., Fu Y., Wang J., Liu Q., Zhang X. et al. (2016) QTL editing confers opposing yield performance in different rice varieties. J. Integr. Plant Biol. 10.1111/jipb.12501 [DOI] [PubMed] [Google Scholar]
- 37.Xu R., Yang Y., Qin R., Li H., Qiu C., Li L. et al. (2016) Rapid improvement of grain weight via highly efficient CRISPR/Cas9-mediated multiplex genome editing in rice. J. Genet. Genomics 43, 529–532 10.1016/j.jgg.2016.07.003 [DOI] [PubMed] [Google Scholar]
- 38.Li Q., Zhang D., Chen M., Liang W., Wei J., Qi Y. et al. (2016) Development of japonica photo-sensitive genic male sterile rice lines by editing carbon starved anther using CRISPR/Cas9. J. Genet. Genomics 43, 415–419 10.1016/j.jgg.2016.04.011 [DOI] [PubMed] [Google Scholar]
- 39.Li M., Li X., Zhou Z., Wu P., Fang M., Pan X. et al. (2016) Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system. Front. Plant Sci. 7, 377 10.3389/fpls.2016.00377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yasui Y., Tanaka W., Sakamoto T., Kurata T. and Hirano H.-Y. (2017) Genetic enhancer analysis reveals that FLORAL ORGAN NUMBER2 and OsMADS3 co-operatively regulate maintenance and determinacy of the flower meristem in rice. Plant Cell Physiol. 58, 893–903 10.1093/pcp/pcx038 [DOI] [PubMed] [Google Scholar]
- 41.Ma L., Zhang D., Miao Q., Yang J., Xuan Y. and Hu Y. (2017) Essential role of sugar transporter OsSWEET11 during the early stage of rice grain filling. Plant Cell Physiol. 58, 863–873 10.1093/pcp/pcx040 [DOI] [PubMed] [Google Scholar]
- 42.Yin X., Biswal A.K., Dionora J., Perdigon K.M., Balahadia C.P., Mazumdar S. et al. (2017) CRISPR-Cas9 and CRISPR-Cpf1 mediated targeting of a stomatal developmental gene EPFL9 in rice. Plant Cell Rep. 36, 745–757 10.1007/s00299-017-2118-z [DOI] [PubMed] [Google Scholar]
- 43.Srivastava V., Underwood J.L. and Zhao S. (2017) Dual-targeting by CRISPR/Cas9 for precise excision of transgenes from rice genome. Plant Cell Tissue Organ Culture 129, 153–160 10.1007/s11240-016-1166-3 [DOI] [Google Scholar]
- 44.Yuan J., Chen S., Jiao W., Wang L., Wang L., Ye W. et al. (2017) Both maternally and paternally imprinted genes regulate seed development in rice. New Phytol. 216, 373–387 10.1111/nph.14510 [DOI] [PubMed] [Google Scholar]
- 45.Li X., Zhou W., Ren Y., Tian X., Lv T., Wang Z. et al. (2017) High-efficiency breeding of early-maturing rice cultivars via CRISPR/Cas9-mediated genome editing. J. Genet. Genomics 44, 175–178 10.1016/j.jgg.2017.02.001 [DOI] [PubMed] [Google Scholar]
- 46.Ueta R., Abe C., Watanabe T., Sugano S.S., Ishihara R., Ezura H. et al. (2017) Rapid breeding of parthenocarpic tomato plants using CRISPR/Cas9. Sci. Rep. 7, 507 10.1038/s41598-017-00501-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iaffaldano B., Zhang Y. and Cornish K. (2016) CRISPR/cas9 genome editing of rubber producing dandelion Taraxacum kok-saghyz using Agrobacterium rhizogenes without selection. Ind. Crops Prod. 89, 356–362 10.1016/j.indcrop.2016.05.029 [DOI] [Google Scholar]
- 48.Char S.N., Neelakandan A.K., Nahampun H., Frame B., Main M., Spalding M.H. et al. (2017) An Agrobacterium-delivered CRISPR/Cas9 system for high-frequency targeted mutagenesis in maize. Plant Biotechnol. J. 15, 257–268 10.1111/pbi.12611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li C., Liu C., Qi X., Wu Y., Fei X., Mao L. et al. (2017) RNA-guided Cas9 as an in vivo desired-target mutator in maize. Plant Biotechnol. J. 10.1111/pbi.12739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiang W.Z., Henry I.M., Lynagh P.G., Comai L., Cahoon E.B. and Weeks D.P. (2017) Significant enhancement of fatty acid composition in seeds of the allohexaploid, Camelina sativa, using CRISPR/Cas9 gene editing. Plant Biotechnol. J. 15, 648–657 10.1111/pbi.12663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morineau C., Bellec Y., Tellier F., Gissot L., Kelemen Z., Nogué F. et al. (2016) Selective gene dosage by CRISPR-Cas9 genome editing in hexaploid Camelina sativa. Plant Biotechnol. 15, 729–739 10.1111/pbi.12671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aznar-Moreno J.A. and Durrett T.P. (2017) Simultaneous targeting of multiple gene homeologs to alter seed oil production in Camelina sativa. Plant Cell Physiol. 58, 1260–1267 10.1093/pcp/pcx058 [DOI] [PubMed] [Google Scholar]
- 53.Kapusi E., Corcuera-Gómez M., Melnik S. and Stoger E. (2017) Heritable genomic fragment deletions and small indels in the putative ENGase gene induced by CRISPR/Cas9 in barley. Front. Plant Sci. 8, 540 10.3389/fpls.2017.00540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hanania U., Ariel T., Tekoah Y., Fux L., Sheva M., Gubbay Y. et al. (2017) Establishment of a tobacco BY2 cell line devoid of plant-specific xylose and fucose as a platform for the production of biotherapeutic proteins. Plant Biotechnol. J. 15, 1120–1129 10.1111/pbi.12702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mercx S., Smargiasso N., Chaumont F., De Pauw E., Boutry M. and Navarre C. (2017) Inactivation of the β(1,2)-xylosyltransferase and the α(1,3)-fucosyltransferase genes in nicotiana tabacum BY-2 cells by a multiplex CRISPR/Cas9 strategy results in glycoproteins without plant-specific glycans. Front. Plant Sci. 8, 403 10.3389/fpls.2017.00403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun Y., Jiao G., Liu Z., Zhang X., Li J., Guo X. et al. (2017) Generation of high-amylose rice through CRISPR/Cas9-mediated targeted mutagenesis of starch branching enzymes. Front. Plant Sci. 8, 298 10.3389/fpls.2017.00298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li B., Cui G., Shen G., Zhan Z., Huang L., Chen J. et al. (2017) Targeted mutagenesis in the medicinal plant Salvia miltiorrhiza. Sci. Rep. 7, 43320 10.1038/srep43320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ito Y., Nishizawa-Yokoi A., Endo M., Mikami M. and Toki S. (2015) CRISPR/Cas9-mediated mutagenesis of the RIN locus that regulates tomato fruit ripening. Biochem. Biophys. Res. Commun. 467, 76–82 10.1016/j.bbrc.2015.09.117 [DOI] [PubMed] [Google Scholar]
- 59.Čermák T., Baltes B., Čegan R., Zhang Y. and Voytas D. F. (2015) High-frequency, precise modification of the tomato genome. Genome Biol. 16, 232 10.1186/s13059-015-0796-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baltes N.J., Gil-Humanes J., Cermak T., Atkins P.A. and Voytas D. F. (2014) DNA replicons for plant genome engineering. Plant Cell 26, 151–163 10.1105/tpc.113.119792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Du H., Zeng X., Zhao M., Cui X., Wang Q., Yang H. et al. (2016) Efficient targeted mutagenesis in soybean by TALENS and CRISPR/Cas9. J. Biotechnol. 217, 90–97 10.1016/j.jbiotec.2015.11.005 [DOI] [PubMed] [Google Scholar]
- 62.Zong Y., Wang Y., Li C., Zhang R., Chen K., Ran Y. et al. (2017) Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat. Biotechnol. 35, 438–440 10.1038/nbt.3811 [DOI] [PubMed] [Google Scholar]
- 63.Feng Z., Mao Y., Xu N., Zhang B., Wei P., Yang D.-L. et al. (2014) Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc. Natl Acad. Sci. U.S.A. 111, 4632–4637 10.1073/pnas.1400822111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van Erp P.B., Bloomer G., Wilkinson R. and Wiedenheft B. (2015) The history and market impact of CRISPR RNA-guided nucleases. Curr. Opin. Virol. 12, 85–90 10.1016/j.coviro.2015.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]