Abstract
Mechanotransduction via the extracellular matrix (ECM)–myosin pathway is involved in determining cell morphology during development and in coupling external transient mechanical stimuli to the reorganization of the cytoskeleton. Here, we present a review on the molecular mechanisms involved in this pathway and how they influence cellular development and organization. We investigate key proteins involved in the ECM–myosin pathway and discuss how specific binding events and conformational changes under force are related to mechanical signaling. We connect these molecular mechanisms with observed morphological changes at the cellular and organism level. Finally, we propose a model encompassing the biomechanical signals along the ECM–myosin pathway and how it could be involved in cell adhesion, cell migration, and tissue architecture.
Keywords: extracellular matrix, mechanotransduction, myosins, protein unfolding, talin
Introduction
Mechanotransduction and the role of physical force in mediating the unfolding of proteins to generate the signals required for development has been studied for more than a century. Proteins that mediate mechanotransduction serve as part of the ‘molecular clutch’ between the extracellular matrix (ECM) and the actomyosin cytoskeleton, first proposed in 1988 by Mitchison and Kirschner [1]. While there are over 50 proteins involved in mechanotransduction [2], here we will focus our attention on the main molecular components of the ECM–myosin pathway, and how protein unfolding and mechanical forces play a role in controlling focal adhesions and the contractile activity within cells. The ECM–myosin pathway is one of many mechanisms that drive cellular mechanotransduction. This pathway is still relatively poorly understood, but extremely important for comprehending key steps in organism development, and false activation during cancer. Here, we will take an in-depth look at some of the molecular components regulating this mechanotransducing path and their effects on cell shape and organism development. Finally, we propose a model on how this interaction might work in vivo.
Molecular mechanisms
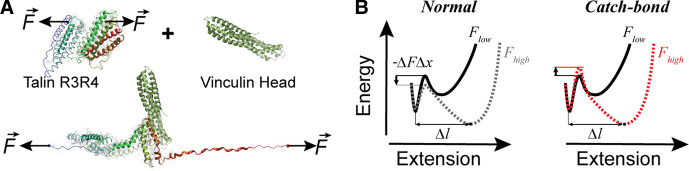
Proteins operating under force are generally segregated into several domains with varying stabilities [3]. Mechanical perturbations can lead to unfolding and extension of some of these protein domains. A profound new concept is currently gaining acceptance, that protein unfolding is a mechanism used to signal and interpret local mechanical cues, by exposing cryptic binding sites that are otherwise hidden from the solvent environment. As discussed below, this process was first demonstrated for the recruitment of vinculin by talin1 (referred to as talin from here on) in a force-dependent manner [4] (Figure 1A).
Figure 1. Mechanical unfolding and extension of proteins in vivo acts as a signaling pathway during integrin activation.
(A) Schematics showing how force triggers the partial unfolding of the talin R3R4 domains, activating the binding of vinculin head. Adapted with permissions from ref. [4]. (B) Diagram of the energy landscape projection on the pulling coordinate, depicting how force drives the unfolding and extension of a protein domain. Left: Typically, mechanical force decreases the energy barrier with a value equal to the performed work [5] (which is the product between the force F and the distance to transition state Δx). Right: Catch-bonds show an increase with force of the barrier which separates the folded and unfolded states [6]. Following unfolding, proteins extend due to the entropic elasticity of the polypeptide chain [3]. We argue that this extension process is more important than the unfolding itself, as it allows for exposure of cryptic binding sites, such as those required by vinculin to bind to unfolded talin rod domains.
The best way to understand the effect of force on the structure and response of a protein is through its energy landscape [7], which represents frequently occupied states as minima (Figure 1B). Mechanical forces have a two-fold effect on the energy landscape of a protein. First, force performs mechanical work on the energy barrier, which decreases proportionally [5] (Figure 1B, left). An important exception to this rule is given by catch-bonds [6], where force-activation locks secondary structure elements in a clamp-like conformation [8] (Figure 1B, right). When catch-bonds form, there is an initial increase with force of the barrier height separating the folded and unfolded states. Further increase in force will eventually drive the system to slip-bond kinetics, resulting in a non-monotonic catch-to-slip switching behavior [9,10]. Second, force shifts the position of the folded and unfolded minima along the energy and extension coordinates. This last change in position was only recently recognized as important for protein unfolding, and is given by the entropic response of the denatured polypeptide chain [11]. Similar to a spring which lengthens proportionally with force, a peptide chain also unfolds and extends more under a higher mechanical perturbation. Indeed, as measured experimentally, the extension of the unfolded polypeptide chain depends on the experienced force and is independent of the amino acid sequence [3,12].
Because proteins are exposed to a dynamic environment, where the thermal motion of the surrounding solvent molecules can have transient directions, protein unfolding and refolding is a probabilistic event. When repeatedly exposing a single protein to a given force, the same molecule takes a slightly different time to transition from the folded to the unfolded state. This variation in the dwell time can be explained by the multidimensionality of the folding energy landscape of a protein [10,13]. This non-deterministic behavior means that, while less frequent, proteins will still unfold at low or zero forces, while this process, in the absence of catch-bonds, will be more frequent as force increases [14]. The main difference between low and high force is not the unfolding transition itself, but the extension reached following this process (Figure 1). Many studies try to identify the experienced force-per-protein during mechanotransduction, to determine if domain unfolding and refolding takes place in vivo. If one accepts the fact that protein unfolding at the molecular level is a game of odds, the question should not be if a protein domain unfolds in vivo, but more importantly, how does the extension following unfolding of a protein domain under force drive the mechanotransduction process in one direction or another.
Cellular and tissue response
Cellular context is key to understanding mechanotransduction in vivo, as different cell types express different proteins that respond to force. In our opinion, two structures stand out as great models to study and understand vertebrate mechanotransduction in vivo: one is the formation of the highly conserved midbrain–hindbrain boundary (MHB) [15–17] and the other is the invagination of the optic cup [18,19] (Figure 2). Each structure originates from the neuroepithelium and depends on specific and unique aspects of the ECM–myosin mechanotransduction pathway.
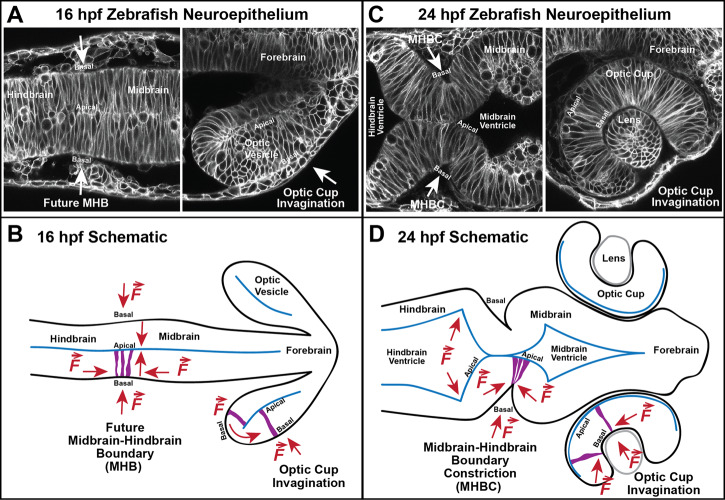
Figure 2. Live confocal images of MHB and optic cup development in the zebrafish neuroepithelium with schematics of localized mechanotransduction forces.
(A) Live scanning confocal images of the zebrafish neuroepithelium at 16 hpf of the future MHB (left) and optic vesicle after it has evaginated from the neural tube (right). (B) Diagram of the 16 hpf zebrafish embryo. A specialized form of the ECM, the basement membrane, lines the entire basal surface of the tissues, separating the neuroepithelium from its surroundings. Black outlines indicate the basement membrane surrounding the tissue. Arrows indicate hypothesized orientation of forces that signal from the outside-in and inside-out to generate cell and tissue shape changes. (C) Live scanning confocal images of the zebrafish neuroepithelium at 24 hpf. The MHBC after the tissue has folded and the brain ventricles have inflated (left) and the optic cup and lens following invagination (right). (D) Diagram of a 24 hpf zebrafish embryo after the MHB and optic cup have folded basally. The MHB develops into a sharp constriction on the basal surface forming the MHBC. This is generated by the shortening of cells in the apical–basal direction, constriction of cells on their basal side, and expansion of cells on their apical side in the anterior–posterior direction. Force from ventricle lumen inflation is also hypothesized to be required for the acute tissue angle [15,20]. The optic cup develops into a U-shaped structure that surrounds the developing lens. Cell migration around the rim of the optic cup into the neural retina and constriction of cells on their basal side bend the tissue. The live images were obtained using wild-type embryos injected at the single-cell stage with CAAX-GFP mRNA, to localize GFP to the cell membranes. At these stages of development, the tissues are composed of single layers of pseudostratified epithelial cells exhibiting apical–basal polarity. Anterior is to the right in all images.
Zebrafish are an excellent model for studying these morphogenetic events due to their fast development time, large sample sizes, and available genetic tools. Importantly, transparent zebrafish embryos allow for live imaging at both the cell and tissue level to study the mechanotransduction pathway in vivo. The MHB is a basal tissue fold in the embryonic brain that later becomes the cerebellum and part of the tectum [21]. In the zebrafish, between 16 somite stage (ss) (∼17 hours post fertilization, hpf) and primordium 6 (∼24 hpf), specific cell shape changes form the MHB constriction (MHBC). First, cells at the MHBC shorten in the apical–basal direction and narrow in the anterior–posterior direction [17], then MHBC cells constrict basally and expand apically [15]. Together, these cell shape changes result in a basally folded tissue (Figure 2). Zebrafish optic cup morphogenesis occurs between 12 and 24 hpf, overlapping with MHB morphogenesis. At 16 hpf, the retinal neuroepithelium begins to invaginate and folds basally. This invagination process requires both epithelial cell migration around the rim of the optic cup (rim involution) and basal constriction of the retinal neuroepithelial cells [18,19,22–24]. Although the neuroepithelium folds basally to form the MHB and the optic cup, each structure has a unique shape. The MHB fold develops into a sharp, acute angle of constricted tissue, while the optic cup folds into a curved U-shape (Figure 2). Mechanotransduction pathway components have been implicated in mediating both of these processes and will be discussed below, including comparisons of overlapping and distinct mechanisms utilized to form these unique structures.
Pathway proteins
Extracellular matrix
The ECM (extracellular matrix) is formed from secreted modular proteins, proteoglycans, and polysaccharides and is known to play major roles in cellular support and differentiation [25]. The modular proteins inside the ECM can respond to forces by unfolding and extending their domains and actively regulate the elasticity of tissues [26]. Force spectroscopy measurements have shown that the fibronectin domains (which contain the Arg-Gly-Asp or RGD site) unfold under forces ranging from 80 to 200 pN [27]. The measured extension of fibronectin indicates a force-per-molecule of >120 pN, in the absence of any change in contour length [28,29]. While unfolding was not yet directly measured in vivo for fibronectin, it is likely to happen if such high forces are reached. Owing to its randomly cross-linked multicomponent organization, the study of the ECM elasticity requires novel instrumentation and theoretical approaches [30].
The basement membrane, a specialized ECM, surrounds the developing neuroepithelium. One main component of this ECM is the heterotrimeric protein laminin, where the predominant isoform during early development is laminin-111 [31,32]. During MHB morphogenesis, laminin is required for basal constriction and loss of laminin results in abnormal distribution of actin at the constriction point [15]. Loss of laminin in the optic cup results in multiple structural defects during morphogenesis [23,24,33,34] and, as in the MHB, laminin is required for basal constriction of the tissue at the retina–lens interface [24,34]. In contrast with the formation of the MHB, optic cup formation requires migration of epithelial cells (Figure 2), and this process is laminin-dependent [18,23,35]. These migratory cells use lamellipodia to attach to the ECM and form dynamic basal adhesions to generate force for movement [23]. The ECM also has specific spatiotemporal roles during optic cup morphogenesis. During optic stalk constriction, lam1a promotes focal adhesions, but during invagination, it inhibits focal adhesions [18,23], demonstrating the localized specificity of the mechanotransduction pathway during complex morphogenetic events. Interestingly, in zebrafish, proliferation is not required for proper optic cup morphogenesis, but is needed to form the MHB [36] and in the chick, tissue growth constrained by the ECM drives invagination of the optic cup [37].
Integrins
Integrins are the transmembrane proteins responsible for recognizing and binding specific extracellular ligands (Figure 3A). These glycoprotein receptors are formed of two non-covalently linked subunits: α and β. There currently are 24 known complexes, made from a pool of 18 α and 8 β subunits, which bind to ECM components [38]. These transmembrane complexes can be activated by binding to an extracellular ligand such as laminin (known as outside-in activation), or by the β-integrin subunit attachment to the cytoplasmic protein talin (known as inside-out activation).
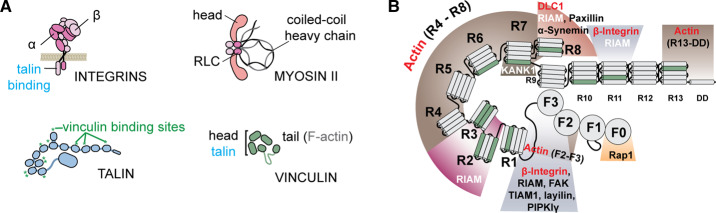
Figure 3. Organization of some multidomain proteins involved in mechanotransduction.
(A) Schematics of several key proteins involved in the ECM–myosin pathway, all shown in their inhibited form. Intergrins form heterodimeric transmembrane complexes and link ECM to talin. Talin attaches between the cytoplasmic side of β-integrin and actin filaments and recruits vinculin by unfolding its rod domains. Vinculin links exposed talin binding sites with actin filaments, and can reinforce the talin–actin connection. NMII forms a dimer via its coiled-coil heavy chain, and its activation is controlled through phosphorylation of the RLC. (B) Detailed schematics of talin protein, composed of four head (FERM) domains and 13 rod (R) domains, and terminated with a dimerization domain (DD). Vinculin binding sites are marked in green and hidden inside the protein structure. The proteins involved in the ECM–myosin pathway are marked in red.
The activation of integrins is fascinating, as it has one of the largest known molecular ranges of motion, which gets transmitted over a length of 15–20 nm through the plasma membrane and leads to a 7 nm increase in separation between the cytoplasmic ends [39]. The force generated by a single integrin complex on the ECM is still subject to debate, and was reported to be as high as 54 pN [40]. A catch-bond mechanism was recently reported between α5β1 integrins and their fibronectin ligand, with a maximum value at ∼35 pN [41]. An interesting mechanism was also shown for the interaction of the αvβ6 integrins with the pro-domain of the Transforming Growth Factor (isoform β1, pro-TGF-β1) [42]. Similar to a feed-forward loop, this integrin complex binds to the ECM and starts pulling on the bowtie-like structure of pro-TGF-β1, it partially unfolds it and makes it release the growth factor (GF). The uncaging of the GF from the ECM is followed by its capture via its specific membrane receptors, which can lead to activation of more integrins through inside-out activation.
The role for integrins during eye formation has been established in a variety of vertebrates including teleosts, chick, and in human embryonic stem cell cultures [19,43,44]. One study in medaka defined a role for integrin-mediated adhesion to the ECM for proper optic cup morphogenesis where trafficking and regulation of integrin endocytosis at the retinal epithelial basal surface is essential for optic cup invagination [45]. In addition, dynamic foci of integrin-β1 was shown on the basal side of cells during rim cell migration [23]. The role of integrins in MHB formation has yet to be examined, but is hypothesized to be critical for the sharp tissue fold.
Talin
Talin is the molecular computer of cellular mechanotransduction, which accepts as inputs mechanical and biochemical signals, and, as output, drives cytoskeleton assembly and formation of focal adhesions. In its inactive form, talin is self-inhibited, with domain F3 from the FERM section being attached to rod domain R9 [46] (Figure 3). Talin activates upon tethering to at least one β-integrin and one actin filament [47,48]. Talin can bind to the β subunit of the integrin complex through its N-terminal FERM domain F3, as well as through its R11 domain [49]. There are three actin binding sites, one in the F2–F3 region of the FERM domains, and two in the rod region: R4–R8 and R12-DD (dimerization domain). There are many binding partners which act as biochemical inputs for talin and dock in various regions along the molecule. An interesting architecture is seen for the talin R7–R8 domains. The structure of the R8 domain is sequestered in-between the first two and last two helixes of the R7 domain. Hence, R8 is mechanically protected by R7 [47,48], and will only experience force when the R7 domain unfolds, acting as an ‘antenna’ domain. This feature makes R8 one of the most ‘popular’ domains [50], and the only binding site on talin for DLC1. Interestingly, zebrafish talin mutants demonstrate neural crest cell migration defects [51] and later structural defects in the eye where the choroid fissure fails to fuse [52]. Additional careful investigation into the role for talin in MHB formation and optic cup invagination in these mutants is warranted.
Vinculin
Vinculin, similar to talin, is found in solution in an auto-inhibited form (Figure 3A) and becomes activated when it is tethered between an actin filament and a talin, α-actinin, or α/β-catenin molecule [53–56]. The force-activated binding between talin and vinculin was the first reported example where unfolding and extension due to local mechanical force of talin rod domains leads to exposure of vinculin binding sites, which otherwise stay latent inside the folded structure [4,57] (Figure 1A). There are a total of 11 vinculin binding sites buried inside the structure of nine talin rod domains (marked with green in Figure 3B). It was shown experimentally that the average force required to unfold most talin rod domains lies between 5 and 25 pN, at loading rates between 5 and 40 pN/s. [47,48], while helix bundles have a stability between 25 and 35 pN [58]. Hence, a low force, below <5 pN, would not suffice to unfold and extend talin domains, while a high force, >35 pN, would result in the breaking of the alpha helical structures needed for vinculin to bind. Vinculin was also recently reported to form a directional catch bond with its actin substrate [59]. The maximum bond strength for the vinculin–actin interaction was measured under constant force conditions at ∼8 pN [59]. This interaction favors a precise directional binding toward the negative end of the polar actin filament [59]. It was recently reported that in zebrafish, zygotic vinculin is not essential for early development, although phenotypes, where both vinculin A and vinculin B isoforms were knocked out, failed to survive after embryonic stages [60]. The authors reported defects in craniofacial development, which would implicate a role for vinculin in neural crest cell migration and the mechanotransduction pathway. Therefore, it is likely that cellular level defects are present in vinculin double mutants, but were not detected using the techniques presented. Furthermore, in laminin mutants, vinculin was increased at the lens-retina interface during optic cup invagination [34], and it was hypothesized that laminin may be critical to limit the stiffness of the ECM network in this region.
Deleted in liver cancer-1
Deleted in liver cancer-1 (DLC1) protein was first discovered as deleted or down-regulated in liver cancer patients [61] and it was first shown to bind and activate Rho-GTPase protein RhoA [62]. DLC1 is a RhoGAP protein that acts as a down-regulator for focal adhesions, as it inhibits the activation of Rho-GTPase by accelerating their intrinsic GTPase activity [63,64]. DLC1 was also shown to bind talin at the R8 antenna site [65,66]. An interesting single molecule study recently showed that recruitment of DLC1 by talin enhances the assembly and contraction of myosin motors [67]. In this case, the authors have engineered a disulfide bond inside the R7 protective structure, to prevent its unfolding and enhance the R8-DLC1 interaction, which decreased the myosin contractile activity. To date, DLC1 has not been examined during MHB or optic cup morphogenesis; however, it is likely that DLC1 also plays an important role in the ECM–myosin mechanotransduction pathway that mediates these tissue folds and should be investigated.
Rhoa/ROCK
The small GTPase, RhoA, is a member of the Ras homology family of GTPases and has many critical cellular functions [68]. RhoA is a molecular switch that oscillates between an active (GTP bound) and an inactive (GDP bound) states, where activation occurs with the exchange of a GDP for GTP via the guanine nucleotide exchange factors (GEFs). The inactivation is mediated by GTPase activating proteins (GAPs) by increasing GTP hydrolysis. There are more than 70 GEFs and 70 GAPs that are known to mediate Rho activity. Each is tightly regulated with cell-type specificity [69,70]. While these molecules themselves do not appear to unfold under force, they are critical signaling mediators in mechanotransduction. RhoA activation, by intrinsic or extrinsic cues, can initiate a wide variety of signaling cascades to mediate morphogenetic events in response to force [71]. In its active state, RhoA transduces its signal to a variety of proteins including Rho-associated kinase (ROCK). ROCK phosphorylates and activates myosin regulatory light chain (RLC), resulting in actomyosin contraction via activation of non-muscle myosin II (NMII) [72]. In addition, ROCK phosphorylates the myosin phosphatase target subunit 1 (Mypt1), and this phosphorylation allows for continued NMII contraction [72]. In zebrafish mypt1 mutants, where NMII is overactive, hyper-contracted neuroepithelial cells result in abnormal brain morphogenesis [17,73].
Non-muscle myosin II
NMIIs are known to be important for cell adhesion, cell motility, and cellular architecture, all of which are critical for cell shape and cell migration during development [74–76]. NMIIs are hexameric proteins composed of three pairs of peptides that includes two heavy chains, two RLCs, and two essential light chains (ELCs) (see also Figure 2). They interact with themselves and actin filaments to generate contractile forces [77]. To tether actin and drive contraction, NMII heavy chains assemble into bipolar filaments. NMII activity is regulated through reversible phosphorylation by kinases such as ROCK on specific amino acids present in its RLC, or via phosphorylation of the heavy chain itself, which prevents the filament assembly [74]. Conformational changes during myosin light chain phosphorylation increase the total ATPase activity and enable the assembly of NMII into bipolar filaments, without affecting the myosin head–actin interaction [74]. During the power stroke, myosin takes discrete steps of ∼11 nm and generates forces of 3–4 pN per head [78]. In zebrafish, NMII proteins are required for MHB morphogenesis and NMII isoforms have differential roles in mediating the cell shape changes required to fold the neuroepithelium. NMIIA (encoded by myh9b) is required for regulation of apical–basal cell length and NMIIB (encoded by myh10) is required for regulation of anterior–posterior cell width [17]. Actin levels at the MHBC are also NMII-dependent [17]. However, the molecular mechanisms that determine differential function and activation of NMIIA versus NMIIB within the same cells in an in vivo system remain unknown. In the optic cup, retinal neuroepithelial cells accumulate myosin RLC and actin basally [23,24] and cells exhibit actomyosin pulsatile behavior, which correlate with the episodic contractions to basally constrict the cells [24]. Laminin knockdown appeared to stabilize myosin RLC foci, which correlated with decreased basal contractility [24]. Chemical inhibition of ROCK slowed the myosin kinetics; however, it did not completely prevent optic cup invagination [23], suggesting that invagination of the optic cup can also occur via compensatory mechanisms. These studies of the optic cup invagination did not examine NMII isoform-specific roles.
Model for the ECM–myosin pathway
The elasticity and biochemical environment of the ECM influences and is influenced by the neighboring cells. Similar to a tug-of-war, cells need to balance the mechanical forces developed with the ECM through changes in their internal structure (Figure 4). During outside-in activation, binding of integrin to an extracellular ligand, such as the RGD sequence of fibronectin, triggers the cytoplasmic recruitment of talin and formation of connections to the actin-based cytoskeleton [79,80]. As the force between matrix and cytoskeleton increases, mechanical unfolding of the rod domains of talin exposes up to eleven vinculin binding sites, which cross-links talin to more actin filaments [4,56,81]. Unfolding has a two-fold effect. First, as the force on talin increases, the release of contour length due to unfolding acts as a ‘safety valve’, resetting the force to a low value [48]. Second, the new talin–actin connections via vinculin reduce the overall force, which is now distributed over more connections. Hence, the talin–vinculin–actin pathway maintains the overall force at a relatively constant value.
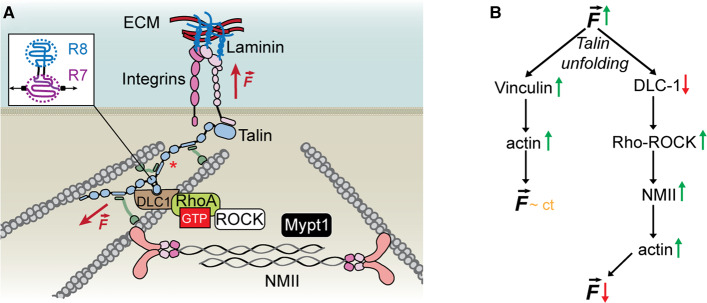
Figure 4. Schematics of the proposed mechanism for the ECM–myosin mechanotransduction pathway.
(A) Binding of integrins to an extracellular ligand on the ECM triggers opening of the integrin complex and talin recruitment at the plasma membrane. Activated talin attaches to actin and responds to mechanical force by unfolding and extending its rod domains. These unfolding events trigger vinculin recruitment, which reinforces the connection between talin and actin. Folded R8 domain can bind the DLC1–RhoA–ROCK complex, which down-regulates the activity of any non-muscle myosin motor molecules present in its proximity. (B) Schematics of the ECM–myosin pathway. Talin unfolding and vinculin reinforcement maintain the experienced force in a narrow (almost constant) range. Mechanically triggered unbinding of DLC1 can allow for active RhoA-ROCK to activate NMII contractility. This results in the movement of more actin filaments close to the plasma membrane, leading to a decrease in the experienced force.
A unique location on the talin protein is the rod domain R8, which is protected from force by its only neighbor, R7, and can act as a molecular antenna. As long as R8 is folded, it can bind to DLC1 [66,67,82]. As DLC1 down-regulates local Rho-GTPase by accelerating their intrinsic GTPase activity [63,64], its absence due to unfolding of R8 under force will increase the catalytic activity of Rho-GTPase/ROCK complex and lead to activation of NMII through phosphorylation. Through its contractile action, NMII can bring actin filaments closer to the plasma membrane and closer together. Through this mechanism, the force on the talin–vinculin–actin junctions can decrease, if enough actin filaments are present close to the plasma membrane and limit the growth of the actin network. This decrease in force can ultimately lead to refolding of R8, recruitment of DLC1 and inactivation of myosin. A second pathway may also be possible: a bound DLC1 close to the plasma membrane can act as a local off switch. In this case, as myosin molecules approach the plasma membrane and encounter an active talin–DLC1 complex, ROCK will be deactivated, preventing further myosin contraction.
Conclusion and future directions
Discerning the ECM–myosin mechanotransduction pathway is key for elucidating the mechanisms of how genetic mutations and environmental factors can ultimately lead to detrimental multifactorial diseases, birth defects, or cancer; however, our understanding of this pathway is currently limited. Many basic developmental processes require this pathway, including the cell shape changes and cell migratory mechanisms that form the brain, eye, and facial structures. Talin and vinculin mutants demonstrate neural crest cell migration defects that can lead to abnormal craniofacial development [51], and talin mutants also exhibit ocular colobomas [52]. In addition, talin overexpression was also related to prostate cancer [83]. As evidence of the involvement of the ECM–myosin pathway, numerous cancers are characterized by the deletion of the gene coding for DLC1 [82]. DLC1 was shown to act as a tumor suppressor both in vivo and in vitro, but its mechanism of action in this process is not currently understood [63,84]. Further investigation into this pathway also has the potential for increasing our understanding of how human mutations in the gene that encodes for a non-muscle myosin IIA (MYH9), an NMII isoform, leads to MYH9-Related diseases [85]. Hence, understanding the control on actin recruitment and cytoskeleton remodeling will become a valuable tool to elucidate the etiology of genetic mutations, birth defects, and cancer metastasis.
Summary
The ECM–myosin pathway for mechanotransduction is responsible for determining cell morphology during development.
Unfolding and extension under force of several multidomain proteins, such as talin and vinculin, allow cells to arrange their cytoskeleton in response to the elasticity of the ECM.
Recruitment of DLC1 close to the plasma membrane by activated talin molecules acts as a molecular turn-off switch for myosin contraction.
Understanding the ECM–myosin pathway will help better understand the cause of detrimental multifactorial diseases, birth defects, or cancer.
Acknowledgements
I.P. and J.H.G. acknowledge support from UWM Collaborative Research Team Development Award.
Abbreviations
- DLC1
deleted in liver cancer-1
- ECM
extracellular matrix
- ELC
essential light chains
- GAPs
GTPase activating proteins
- GEFs
guanine nucleotide exchange factors
- GF
growth factor
- hpf
hours post fertilization
- MHB
midbrain–hindbrain boundary
- MHBC
midbrain–hindbrain boundary constriction
- Mypt1
myosin phosphatase target subunit 1
- NMII
non-muscle myosin II
- RLC
regulatory light chains
- ROCK
Rho-associated kinase
- ss
somite stage
Funding
This work was supported by grants to I.P. from National Science Foundation, Major Research Instrumentation Program [Grant No. PHY-1626450], Greater Milwaukee Foundation (Shaw Award), and University of Wisconsin System (Applied Research Grant), and to J.H.G. from the University of Wisconsin-Milwaukee Research Growth Initiative.
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
References
- 1.Mitchison T. and Kirschner M. (1988) Cytoskeletal dynamics and nerve growth. Neuron 1, 761–772 10.1016/0896-6273(88)90124-9 [DOI] [PubMed] [Google Scholar]
- 2.Zamir E. and Geiger B. (2001) Molecular complexity and dynamics of cell-matrix adhesions. J. Cell Sci. 114, 3583–3590 [DOI] [PubMed] [Google Scholar]
- 3.Valle-Orero J., Rivas-Pardo J.A. and Popa I. (2017) Multidomain proteins under force. Nanotechnology 28, 174003 10.1088/1361-6528/aa655e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.del Rio A., Perez-Jimenez R., Liu R., Roca-Cusachs P., Fernandez J.M. and Sheetz M.P. (2009) Stretching single talin rod molecules activates vinculin binding. Science 323, 638–641 10.1126/science.1162912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell G.I. (1978) Models for specific adhesion of cells to cells. Science 200, 618–627 10.1126/science.347575 [DOI] [PubMed] [Google Scholar]
- 6.Marshall B.T., Long M., Piper J.W., Yago T., McEver R.P. and Zhu C. (2003) Direct observation of catch bonds involving cell-adhesion molecules. Nature 423, 190–193 10.1038/nature01605 [DOI] [PubMed] [Google Scholar]
- 7.Valle-Orero J., Eckels E.C., Stirnemann G., Popa I., Berkovich R. and Fernandez J.M. (2015) The elastic free energy of a tandem modular protein under force. Biochem. Biophys. Res. Commun. 460, 434–438 10.1016/j.bbrc.2015.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schoeler C., Malinowska K.H., Bernardi R.C., Milles L.F., Jobst M.A., Durner E. et al. (2014) Ultrastable cellulosome-adhesion complex tightens under load. Nat. Commun. 5, 5635 10.1038/ncomms6635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rakshit S., Zhang Y., Manibog K., Shafraz O. and Sivasankar S. (2012) Ideal, catch, and slip bonds in cadherin adhesion. Proc. Natl Acad. Sci. U.S.A. 109, 18815–18820 10.1073/pnas.1208349109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan G., Le S., Yao M., Qian H., Zhou X., Yan J. et al. (2017) Elasticity of the transition state leading to an unexpected mechanical stabilization of titin immunoglobulin domains. Angew. Chem. Int. Ed. Engl. 56, 5490–5493 10.1002/anie.201700411 [DOI] [PubMed] [Google Scholar]
- 11.Berkovich R., Garcia-Manyes S., Klafter J., Urbakh M. and Fernández J.M. (2010) Hopping around an entropic barrier created by force. Biochem. Biophys. Res. Commun. 403, 133–137 10.1016/j.bbrc.2010.10.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Popa I., Rivas-Pardo J.A., Eckels E.C., Echelman D.J., Badilla C.L., Valle-Orero J. et al. (2016) A HaloTag anchored ruler for week-long studies of protein dynamics. J. Am. Chem. Soc. 138, 10546–10553 10.1021/jacs.6b05429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berkovich R., Mondal J., Paster I. and Berne B.J. (2017) Simulated force quench dynamics shows GB1 protein is not a two state folder. J. Phys. Chem. B 121, 5162–5173 10.1021/acs.jpcb.7b00610 [DOI] [PubMed] [Google Scholar]
- 14.Popa I., Kosuri P., Alegre-Cebollada J., Garcia-Manyes S. and Fernandez J.M. (2013) Force dependency of biochemical reactions measured by single-molecule force-clamp spectroscopy. Nat. Protoc. 8, 1261–1276 10.1038/nprot.2013.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gutzman J.H., Graeden E.G. Lowery L.A., Holley H.S. and Sive H. (2008) Formation of the zebrafish midbrain-hindbrain boundary constriction requires laminin-dependent basal constriction. Mech. Dev. 125, 974–983 10.1016/j.mod.2008.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gibbs H.C., Chang-Gonzalez A., Hwang W., Yeh A.T. and Lekven A.C. (2017) Midbrain–hindbrain boundary morphogenesis: at the intersection of Wnt and Fgf signaling. Front. Neuroanat. 11, 64 10.3389/fnana.2017.00064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gutzman J.H., Sahu S.U. and Kwas C. (2015) Non-muscle myosin IIA and IIB differentially regulate cell shape changes during zebrafish brain morphogenesis. Dev. Biol. 397, 103–115 10.1016/j.ydbio.2014.10.017 [DOI] [PubMed] [Google Scholar]
- 18.Kwan K.M., Otsuna H., Kidokoro H., Carney K.R., Saijoh Y. and Chien C.-B. (2012) A complex choreography of cell movements shapes the vertebrate eye. Development 139, 359–372 10.1242/dev.071407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez-Morales J.R. and Wittbrodt J. (2009) Shaping the vertebrate eye. Curr. Opin. Genet. Dev. 19, 511–517 10.1016/j.gde.2009.08.003 [DOI] [PubMed] [Google Scholar]
- 20.Navis A. and Bagnat M. (2015) Developing pressures: fluid forces driving morphogenesis. Curr. Opin. Genet. Dev. 32, 24–30 10.1016/j.gde.2015.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Louvi A., Alexandre P., Metin C., Wurst W. and Wassef M. (2003) The isthmic neuroepithelium is essential for cerebellar midline fusion. Development 130, 5319–5330 10.1242/dev.00736 [DOI] [PubMed] [Google Scholar]
- 22.Heermann S., Schütz L., Lemke S., Krieglstein K. and Wittbrodt J. (2015) Eye morphogenesis driven by epithelial flow into the optic cup facilitated by modulation of bone morphogenetic protein. eLife 4 10.7554/eLife.05216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sidhaye J. and Norden C. (2017) Concerted action of neuroepithelial basal shrinkage and active epithelial migration ensures efficient optic cup morphogenesis. eLife 6 10.7554/eLife.22689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicolás-Pérez M., Kuchling F., Letelier J., Polvillo R., Wittbrodt J. and Martínez-Morales J.R. (2016) Analysis of cellular behavior and cytoskeletal dynamics reveal a constriction mechanism driving optic cup morphogenesis. eLife 5 10.7554/eLife.15797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frantz C., Stewart K.M. and Weaver V.M. (2010) The extracellular matrix at a glance. J. Cell Sci. 123, 4195–4200 10.1242/jcs.023820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Erickson H.P. (1994) Reversible unfolding of fibronectin type III and immunoglobulin domains provides the structural basis for stretch and elasticity of titin and fibronectin. Proc. Natl Acad. Sci. U.S.A. 91, 10114–10118 10.1073/pnas.91.21.10114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oberhauser A.F., Badilla-Fernandez C., Carrion-Vazquez M. and Fernandez J.M. (2002) The mechanical hierarchies of fibronectin observed with single-molecule AFM. J. Mol. Biol. 319, 433–447 10.1016/S0022-2836(02)00306-6 [DOI] [PubMed] [Google Scholar]
- 28.Früh S.M., Schoen I., Ries J. and Vogel V. (2015) Molecular architecture of native fibronectin fibrils. Nat. Commun. 6, 7275 10.1038/ncomms8275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Erickson H.P., Carrell N. and McDonagh J. (1981) Fibronectin molecule visualized in electron microscopy: a long, thin, flexible strand. J. Cell Biol. 91, 673–678 10.1083/jcb.91.3.673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khoury L.R., Nowitzke J., Shmilovich K. and Popa I. (2018) Study of biomechanical properties of protein-based hydrogels using force-clamp rheometry. Macromolecules 51, 1441–1452 10.1021/acs.macromol.7b02160 [DOI] [Google Scholar]
- 31.Colognato H. and Yurchenco P.D. (2000) Form and function: the laminin family of heterotrimers. Dev. Dyn. 218, 213–234 [DOI] [PubMed] [Google Scholar]
- 32.Miner J.H. and Yurchenco P.D. (2004) Laminin functions in tissue morphogenesis. Annu. Rev. Cell Dev. Biol. 20, 255–284 10.1146/annurev.cellbio.20.010403.094555 [DOI] [PubMed] [Google Scholar]
- 33.Lee J. and Gross J.M. (2007) Laminin beta1 and gamma1 containing laminins are essential for basement membrane integrity in the zebrafish eye. Invest. Ophthalmol. Vis. Sci. 48, 2483–2490 10.1167/iovs.06-1211 [DOI] [PubMed] [Google Scholar]
- 34.Bryan C.D., Chien C.-B. and Kwan K.M. (2016) Loss of laminin alpha 1 results in multiple structural defects and divergent effects on adhesion during vertebrate optic cup morphogenesis. Dev. Biol. 416, 324–337 10.1016/j.ydbio.2016.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Picker A., Cavodeassi F., Machate A., Bernauer S., Hans S., Abe G. et al. (2009) Dynamic coupling of pattern formation and morphogenesis in the developing vertebrate retina. PLoS Biol. 7, e1000214 10.1371/journal.pbio.1000214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lowery L.A. and Sive H. (2005) Initial formation of zebrafish brain ventricles occurs independently of circulation and requires the nagie oko and snakehead/atp1a1a.1 gene products. Development 132, 2057–2067 10.1242/dev.01791 [DOI] [PubMed] [Google Scholar]
- 37.Oltean A., Huang J., Beebe D.C. and Taber L.A. (2016) Tissue growth constrained by extracellular matrix drives invagination during optic cup morphogenesis. Biomech. Model. Mechanobiol. 15, 1405–1421 10.1007/s10237-016-0771-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Srichai M.B. and Zent R. (2010) In Cell-Extracellular Matrix Interactions in Cancer (Zent R. and Pozzi A., eds), Springer [Google Scholar]
- 39.Springer T.A. and Dustin M.L. (2012) Integrin inside-out signaling and the immunological synapse. Curr. Opin. Cell. Biol. 24, 107–115 10.1016/j.ceb.2011.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X.F., Sun J., Xu Q., Chowdhury F., Roein-Peikar M., Wang Y.X. et al. (2015) Integrin molecular tension within motile focal adhesions. Biophys. J. 109, 2259–2267 10.1016/j.bpj.2015.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kong F., García A.J., Mould A.P., Humphries M.J. and Zhu C. (2009) Demonstration of catch bonds between an integrin and its ligand. J. Cell Biol. 185, 1275–1284 10.1083/jcb.200810002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dong X., Zhao B., Iacob R.E., Zhu J., Koksal A.C., Lu C. et al. (2017) Force interacts with macromolecular structure in activation of TGF-β. Nature 542, 55–59 10.1038/nature21035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Svennevik E. and Linser P.J. (1993) The inhibitory effects of integrin antibodies and the RGD tripeptide on early eye development. Invest. Ophthalmol. Vis. Sci. 34, 1774–1784 [PubMed] [Google Scholar]
- 44.Nakano T., Ando S., Takata N., Kawada M., Muguruma K., Sekiguchi K. et al. (2012) Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell 10, 771–785 10.1016/j.stem.2012.05.009 [DOI] [PubMed] [Google Scholar]
- 45.Bogdanović O., Delfino-Machin M., Nicolás-Pérez M., Gavilán M.P., Gago-Rodrigues I., Fernández-Miñán A. et al. (2012) Numb/Numbl-Opo antagonism controls retinal epithelium morphogenesis by regulating integrin endocytosis. Dev. Cell 23, 782–795 10.1016/j.devcel.2012.09.004 [DOI] [PubMed] [Google Scholar]
- 46.Zhang H., Chang Y.-C., Huang Q., Brennan M.L. and Wu J. (2016) Structural and functional analysis of a talin triple-domain module suggests an alternative talin autoinhibitory configuration. Structure 24, 721–729 10.1016/j.str.2016.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haining A.W.M., von Essen M., Attwood S.J., Hytönen V.P. and Hernandez A.D. (2016) All subdomains of the talin rod are mechanically vulnerable and may contribute to cellular mechanosensing. ACS Nano 10, 6648–6658 10.1021/acsnano.6b01658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yao M., Goult B.T., Klapholz B., Hu X., Toseland C.P., Guo Y. et al. (2016) The mechanical response of talin. Nat. Commun. 7, 11966 10.1038/ncomms11966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gingras A.R., Ziegler W.H., Bobkov A.A., Joyce M.G., Fasci D., Himmel M. et al. (2009) Structural determinants of integrin binding to the talin rod. J. Biol. Chem. 284, 8866–8876 10.1074/jbc.M805937200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haining A.W., Lieberthal T.J. and Del Río Hernández A. (2016) Talin: a mechanosensitive molecule in health and disease. FASEB J. 30, 2073–2085 10.1096/fj.201500080R [DOI] [PubMed] [Google Scholar]
- 51.Ishii K., Mukherjee K., Okada T. and Liao E.C. (2018) Genetic requirement of talin1 for proliferation of cranial neural crest cells during palate development. Plast. Reconstr. Surg. Glob. Open 6, e1633 10.1097/GOX.0000000000001633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.James A., Lee C., Williams A.M., Angileri K., Lathrop K.L. and Gross J.M. (2016) The hyaloid vasculature facilitates basement membrane breakdown during choroid fissure closure in the zebrafish eye. Dev. Biol. 419, 262–272 10.1016/j.ydbio.2016.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Izard T. and Vonrhein C. (2004) Structural basis for amplifying vinculin activation by talin. J. Biol. Chem. 279, 27667–27678 10.1074/jbc.M403076200 [DOI] [PubMed] [Google Scholar]
- 54.Dumbauld D.W., Lee T.T., Singh A., Scrimgeour J., Gersbach C.A., Zamir E.A. et al. (2013) How vinculin regulates force transmission. Proc. Natl Acad. Sci. U.S.A. 110, 9788–9793 10.1073/pnas.1216209110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yao M., Qiu W., Liu R., Efremov A.K., Cong P., Seddiki R. et al. (2014) Force-dependent conformational switch of alpha-catenin controls vinculin binding. Nat. Commun. 5, 4525 10.1038/ncomms5525 [DOI] [PubMed] [Google Scholar]
- 56.Yao M., Goult B.T., Chen H., Cong P., Sheetz M.P. and Yan J. (2014) Mechanical activation of vinculin binding to talin locks talin in an unfolded conformation. Sci. Rep. 4, 4610 10.1038/srep04610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hytönen V.P. and Vogel V. (2008) How force might activate talin's vinculin binding sites: SMD reveals a structural mechanism. Plos Comput. Biol. 4, e24 10.1371/journal.pcbi.0040024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rief M., Pascual J., Saraste M. and Gaub H.E. (1999) Single molecule force spectroscopy of spectrin repeats: low unfolding forces in helix bundles. J. Mol. Biol. 286, 553–561 10.1006/jmbi.1998.2466 [DOI] [PubMed] [Google Scholar]
- 59.Huang D.L., Bax N.A., Buckley C.D., Weis W.I. and Dunn A.R. (2017) Vinculin forms a directionally asymmetric catch bond with F-actin. Science 357, 703–706 10.1126/science.aan2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Han M.K.L., van der Krogt G.N.M. and de Rooij J. (2017) Zygotic vinculin is not essential for embryonic development in zebrafish. PLoS ONE 12, e0182278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yuan B.Z., Miller M.J., Keck C.L., Zimonjic D.B., Thorgeirsson S.S. and Popescu N.C. (1998) Cloning, characterization, and chromosomal localization of a gene frequently deleted in human liver cancer (DLC-1) homologous to rat RhoGAP. Cancer Res. 58, 2196–2199 [PubMed] [Google Scholar]
- 62.Kim T.Y., Lee J.W., Kim H.-P., Jong H.-S., Kim T.-Y., Jung M. et al. (2007) DLC-1, a GTPase-activating protein for Rho, is associated with cell proliferation, morphology, and migration in human hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 355, 72–77 10.1016/j.bbrc.2007.01.121 [DOI] [PubMed] [Google Scholar]
- 63.Durkin M.E., Avner M.R., Huh C.-G., Yuan B.-Z., Thorgeirsson S.S. and Popescu N.C. (2005) DLC-1, a Rho GTPase-activating protein with tumor suppressor function, is essential for embryonic development. Febs Lett. 579, 1191–1196 10.1016/j.febslet.2004.12.090 [DOI] [PubMed] [Google Scholar]
- 64.Kim T.Y., Healy K.D., Der C.J., Sciaky N., Bang Y.-J. and Juliano R.L. (2008) Effects of structure of Rho GTPase-activating protein DLC-1 on cell morphology and migration. J. Biol. Chem. 283, 32762–32770 10.1074/jbc.M800617200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zheng H., Gao L., Feng Y., Yuan L., Zhao H. and Cornelius L.A. (2009) Down-regulation of Rap1GAP via promoter hypermethylation promotes melanoma cell proliferation, survival, and migration. Cancer Res. 69, 449–457 10.1158/0008-5472.CAN-08-2399 [DOI] [PubMed] [Google Scholar]
- 66.Li G.R., Du X.L., Vass W.C., Papageorge A.G., Lowy D.R. and Qian X.L. (2011) Full activity of the deleted in liver cancer 1 (DLC1) tumor suppressor depends on an LD-like motif that binds talin and focal adhesion kinase (FAK). Proc. Natl Acad. Sci. U.S.A. 108, 17129–17134 10.1073/pnas.1112122108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Haining A.W.M., Rahikainen R., Cortes E., Lachowski D., Rice A., von Essen M. et al. (2018) Mechanotransduction in talin through the interaction of the R8 domain with DLC1. PLoS Biol. 16, e2005599 10.1371/journal.pbio.2005599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Narumiya S. and Thumkeo D. (2018) Rho signaling research: history, current status and future directions. Febs Lett. 592, 1763–1776 10.1002/1873-3468.13087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cherfils J. and Zeghouf M. (2013) Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol. Rev. 93, 269–309 10.1152/physrev.00003.2012 [DOI] [PubMed] [Google Scholar]
- 70.Braun A.C. and Olayioye M.A. (2015) Rho regulation: DLC proteins in space and time. Cell Signal. 27, 1643–1651 10.1016/j.cellsig.2015.04.003 [DOI] [PubMed] [Google Scholar]
- 71.Marjoram R.J., Lessey E.C. and Burridge K. (2014) Regulation of RhoA activity by adhesion molecules and mechanotransduction. Curr. Mol. Med. 14, 199–208 10.2174/1566524014666140128104541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Julian L. and Olson M.F. (2014) Rho-associated coiled-coil containing kinases (ROCK): structure, regulation, and functions. Small GTPases 5, e29846 10.4161/sgtp.29846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gutzman J.H. and Sive H. (2010) Epithelial relaxation mediated by the myosin phosphatase regulator Mypt1 is required for brain ventricle lumen expansion and hindbrain morphogenesis. Development 137, 795–804 10.1242/dev.042705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vicente-Manzanares M., Ma X., Adelstein R.S. and Horwitz A.R. (2009) Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 10, 778–790 10.1038/nrm2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Munjal A. and Lecuit T. (2014) Actomyosin networks and tissue morphogenesis. Development 141, 1789–1793 10.1242/dev.091645 [DOI] [PubMed] [Google Scholar]
- 76.Newell-Litwa K.A., Horwitz R. and Lamers M.L. (2015) Non-muscle myosin II in disease: mechanisms and therapeutic opportunities. Dis. Model. Mech. 8, 1495–1515 10.1242/dmm.022103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Odronitz F. and Kollmar M. (2007) Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome Biol. 8, R196 10.1186/gb-2007-8-9-r196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Finer J.T., Simmons R.M. and Spudich J.A. (1994) Single myosin molecule mechanics: piconewton forces and nanometre steps. Nature 368, 113–119 10.1038/368113a0 [DOI] [PubMed] [Google Scholar]
- 79.Bouvard D., Pouwels J., De Franceschi N. and Ivaska J. (2013) Integrin inactivators: balancing cellular functions in vitro and in vivo. Nat. Rev. Mol. Cell Biol. 14, 430–442 10.1038/nrm3599 [DOI] [PubMed] [Google Scholar]
- 80.Hoffman B.D., Grashoff C. and Schwartz M.A. (2011) Dynamic molecular processes mediate cellular mechanotransduction. Nature 475, 316–323 10.1038/nature10316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Roberts G.C.K. and Critchley D.R. (2009) Structural and biophysical properties of the integrin-associated cytoskeletal protein talin. Biophys. Rev. 1, 61–69 10.1007/s12551-009-0009-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zacharchenko T., Qian X., Goult B.T., Jethwa D., Almeida T.B., Ballestrem C. et al. (2016) LD motif recognition by talin: structure of the talin-DLC1 complex. Structure 24, 1130–1141 10.1016/j.str.2016.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sakamoto S., McCann R.O., Dhir R. and Kyprianou N. (2010) Talin1 promotes tumor invasion and metastasis via focal adhesion signaling and anoikis resistance. Cancer Res. 70, 1885–1895 10.1158/0008-5472.CAN-09-2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu X., Pan Y.J., Zheng J.N. and Pei D.S. (2017) The role of tumor suppressor DLC-1: far from clear. Anticancer Agents Med. Chem. 17, 896–901 PMID: [DOI] [PubMed] [Google Scholar]
- 85.Pecci A., Ma X., Savoia A. and Adelstein R.S. (2018) MYH9: structure, functions and role of non-muscle myosin IIA in human disease. Gene 664, 152–167 10.1016/j.gene.2018.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]