Abstract
Single-cell genome sequencing of individual archaeal and bacterial cells is a vital approach to decipher the genetic makeup of uncultured microorganisms. With this review, we describe single-cell genome analysis with a focus on the unique properties of single-cell sequence data and with emphasis on quality assessment and assurance.
Keywords: assembly, genome sequence, quality control, single cell
Introduction
The planet's biological diversity is overwhelmingly microbial. However, much of this diversity has evaded detection through traditional microbiological approaches, largely as a result of our inability to cultivate most microorganisms in a laboratory setting. Since the development of molecular-based, cultivation-independent tools, we have witnessed a burst in the detection of previously elusive microbial taxa. This was initially driven by the widespread adoption of high-throughput 16S rRNA gene sequencing where studies now span ecological gradients [1] and, in some cases, cross-biome comparisons [2,3]. However, 16S rRNA gene PCR-based surveys are limited due to constraints inherent to single-gene surveys. In many instances, single-gene surveys would have missed entire clades [4,5]. More recently, the genomes of novel phylogenetic groups have been uncovered with single-cell [6–8] and metagenomic sequencing techniques [4,9].
Single-cell genomics (Figure 1) and metagenomics are two techniques that provide access to microbial genomes without the requirement of cultivation. Sequencing all DNA from a bulk sample, also known as metagenomics, has become a powerful technique where hundreds and sometimes thousands of genomes can be extracted from an individual environmental sample [9]. Alternatively, single-cell genomics has more recently emerged as an approach that provides genomic information for an individual cell [10–12]. This simplifies some of the challenges associated with metagenome assembly and provides a direct link between the genome and any additional cellular DNA, such as phages or plasmids (Figure 2). For example, single-cell genomics has uniquely linked viruses with their host cells in uncultivated clades of bacteria [13,14] and revealed organismal interactions in protists by associating single-cell protist DNA with intracellular bacterial and ssDNA viral sequences [15].
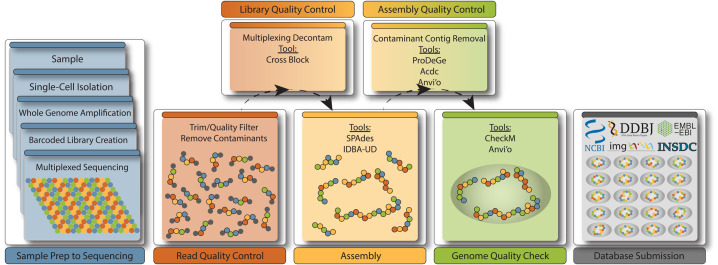
Figure 1. A schematic representation of the single-cell workflow with a focus on the analysis following sequencing.
Left panels with blue background represent the production of single-cell genomes, while the rest of the workflow relates specifically to the analysis of single-cell sequence data going from raw reads to public database submission. The bottom row of analysis boxes refers to the steps that are considered mandatory to any single-cell analysis pipeline, while the top row can be considered context-dependent. For example, if multiplexing was not performed, poolmate decontamination is not necessary (Library Quality Control). However, in nearly all cases, an SAG will benefit from contamination screening (Assembly Quality Control), as even the cleanest SAGs may contain a few contaminating contigs, and if not, this step can serve as validation of a clean SAG that is nearly ready for submission to the public databases.
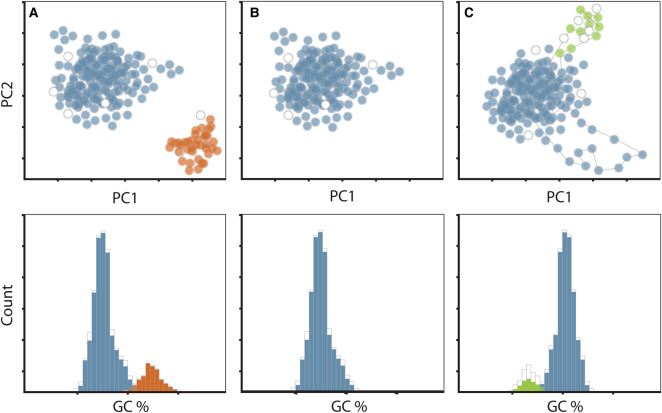
Figure 2. Tetranucleotide principle component analysis (top) and GC content analysis (bottom) of target SAGs (blue) alongside additional contaminating sequence (red) and integrated phage sequences (green).
Target SAG containing contamination (A); target SAG where contamination was removed (B); target SAG with an outlying rRNA gene and an integrated phage (C). (C) Chromosomal elements such as the highly conserved rRNA genes (blue outlying points) often have tetranucleotide frequencies that differ from the main genome. Integrated phage genes can also appear as outlying points with distinct nucleotide composition and unique taxonomy (green outlying points). Each point in the plot represents fragments of contigs that split into 5000 bp fragments. Colored points (top panel) and bars (bottom panel) represent contigs that can be taxonomically classified, whereas white points or bars represent contigs with no taxonomic assignment.
While the preparation of single-cell genomes, or ‘single amplified genomes’ (SAGs), is technically challenging, advances in isolation techniques, sequencing technologies and bioinformatics capabilities have greatly increased throughput and data quality. The analysis of SAG sequence data typically includes the following discrete steps: quality assurance of raw reads, genome assembly using a single-cell-specific assembler, automated and/or manual contaminant identification and removal, annotation, genome quality inspection and categorization according to the minimum information about a single amplified genome (MISAG) standards [16], and database submission (Figure 1). These individual steps can be assembled into a semi-automated workflow.
In this review, we focus on the unique properties of single-cell sequence data, make recommendations for data handling, including raw data quality control, suggest SAG specific assembly tools, discuss important contamination identification and removal procedures, and finally, review standards for reporting and submission of SAGs to the public repositories.
Properties of single-cell sequence data
The generation of single-cell genome sequences includes the following major steps: sample preservation and preparation, single-cell isolation, cell lysis, whole genome amplification (WGA), library preparation, sequencing and data analysis [10–12,17] (Figure 1). Given the extremely low yield of DNA from a single microbial cell (∼1–6 fg) [18], laboratory cleanliness needs to be one of the main considerations when preparing single cells for sequencing. The target DNA should be free of contaminating DNA molecules, as even the most minuscule amount of contaminant DNA will co-amplify during the WGA step and will be difficult to remove since single-cell assemblers now include low coverage regions. Although WGA can be a source of contaminant DNA, this step is essential because libraries cannot yet be prepared with DNA from a single cell [19]. Alternative methods for WGA have been under rapid development over the last several years [20–22], yet multiple displacement amplification (MDA) [23] remains the most commonly used and dependable method for WGA for bacteria and archaea. However, biases typical of MDA include high coverage variation [24], the production of chimeric sequences [25] and a shift in overall GC content [26]. It is largely these biases that contribute to the downstream challenges associated with the analysis of single-cell genomic data.
Quality assurance of single-cell sequence data
Genomes produced from single cells comprise distinct challenges due to the chimeric, biased and potentially contaminated nature of the underlying data, as discussed above. SAG sequence data thus require thorough quality control and specialized data handling.
Read-level quality assessment
Assessing the quality of a single-cell genomic dataset typically begins with a cleanup step prior to assembly. Such read-level quality assessment includes read trimming, quality filtering and read-based contamination identification and removal (Figure 1). For example, adapters are removed and reads are filtered to include only those reads above a specific base call quality score. Reads should also be checked against a microbial contaminant database specific to the laboratory where the SAGs were generated, in addition to genomes for microbial organisms that have been identified as common contaminants in the literature, such as Pseudomonas, Delftia [31] and skin-associated bacteria such as Propionibacterium, Streptococcus and Staphylococcus [32]. It is good practice to map reads against human, dog and cat databases. Tools for read-based decontamination include DeconSeq [33] and modules from the BBTools bioinformatics package (https://sourceforge.net/projects/bbmap/). These tools can map reads against a sequence database of common contaminants and then remove the resulting hits from the dataset.
As discussed above, MDA leads to highly uneven coverage. Variability in coverage can be normalized, which is beneficial to assemblers, as normalization decreases runtime and memory requirements. However, the normalization step is becoming increasingly unnecessary as single-cell-specific assemblers are now publicly available, such as SPAdes [28] and IDBA-UD [30]. These assembly algorithms make use of multiple coverage cutoffs as opposed to a single coverage threshold, resulting in the inclusion of a larger fraction of the data when compared with traditional assemblers. These approaches avoid reconstructing a string of k-mers with static read coverage thresholds; SPADes uses k-bimmers to build a topology of coverage and lengths before assigning a sequence, and IDBA-UD iteratively adjusts coverage thresholds. In addition, the use of reads from either end of a chimera is enabled without direct linkage.
Assessment of poolmate cross-contamination
The quality control described above does not take into account multiplex sequencing of single-cell genomes on high-throughput platforms. Depending on the capacity of the sequencing facility, SAG library preparation can take place in multi-welled plates, generating barcoded libraries for multiplexed sequencing of library pools. Multiplexing samples, specifically biased MDA'd samples, for sequencing on the Illumina platform, however, can cause significant ‘bleed over’ between poolmates. For example, Sinha et al. [34] showed that 5–10% of reads can be assigned to the wrong sample based on low levels of index-free primers present in the multiplexed pool, when using the HiSeq platform. More stringent library cleanup procedures, the use of dual indexes [35] and quality filtering [36] are all methods that can reduce this effect. Poolmate cross-contamination should also be detected during sequence analysis, since even a low fraction of cross-talk between multiplexed libraries (i.e. 0.01%) can have large effects on the assembly, an effect of the unevenness of amplification coverage of WGA methods like MDA. To assess poolmate cross-bleeding, it is good practice to map the reads of a given library to all assemblies across a plate, in an all-vs-all fashion. CrossBlock is a module available in the BBTools software package (https://sourceforge.net/projects/bbmap/) that performs this type of analysis. The program compares the coverage of contigs from one library to the coverage of all other libraries in a pool. However, this approach is only applicable when library pools contain different organisms, as highly similar organisms would produce a high fraction of false positives, which would be flagged as contaminants. To our knowledge, this is the only currently available tool specifically designed for this analysis, though other similar approaches have been performed: for example, searching the contigs of a genome against all other genomes in the pool using blastn and removing those contigs that match above user-defined identity and length thresholds [37].
Contig-level quality assurance
Following assembly, small contigs are removed, as these are more likely to contain assembly errors. At the U.S. Department of Energy's Joint Genome Institute, contigs <2 kb in length are removed from all SAG datasets. After removal of small contigs, screening for additional contaminating contigs originating from organismal DNA not representative of the target cell is typically performed (Figure 2). Identification and removal of contaminants following assembly can be performed with many currently available semi-automated and automated tools. Generally, assembly-based contaminant screening tools scan for outlying features of an SAG, including unusual 16S rRNA genes and protein-coding genes, abnormal k-mer frequencies and/or variation in GC content (Figure 2). These features can be identified interactively within the IMG interface [38] (tutorial available here: https://img.jgi.doe.gov/er/doc/SingleCellDataDecontamination.pdf) and within the recently developed analysis and visualization platform, Anvi'o [39]. Both tools provide interactive platforms, facilitating the removal of contaminating contigs from an assembly based on outlying genomic signatures. Anvi'o and another recently developed software package, CheckM, estimate genome completeness and contamination based on the presence of single-copy marker genes. ProDeGe [40] and acdc [41] are additional tools that perform automated contamination screens returning separate fasta files for clean and contaminant contigs. These tools can be used in combination with tools like CheckM and Anvi'o, especially on large sets of SAGs where manual curation is challenging. As such, automated screening methods can be performed on SAGs with high contamination estimates, then checked for completeness and contamination using CheckM and/or Anvi'o, followed by additional rounds of cleanup if necessary. Figure 2 displays a schematic of a target SAG (blue) that is contaminated with contigs derived from another cell (red) (panel A), the same SAG following contaminant removal (panel B) and a different target SAG (blue) exhibiting features that could be flagged as contamination (panel C) (i.e. false positives). The situation depicted in Figure 2C can happen when an SAG contains rRNA genes with variable nucleotide composition and/or an integrated phage that is embedded within a contig that contains regions of deviating tetramer composition (Figure 2C). Because all automated methods produce false positives and negatives, we highly recommend manual evaluation of all SAGs prior to making biological inferences and submitting to the public databases.
Genome quality reporting
To avoid making biological inferences with contaminated SAGs, it is critical to confirm and report SAG quality before performing comparative genome analysis. Reporting SAG quality also informs other researchers that retrieve SAG data from public databases for their own analyses. For quality reporting, we suggest following the MISAG standards [16]. These are simple standards that require a minimal set of mandatory genome quality criteria such as the reporting of basic metadata, assembly statistics, and genome completeness and contamination estimates. Additional mandatory reporting standards include fields specific to laboratory production (e.g. cell isolation, cell lysis and WGA), taxonomic identification of SAGs, identification of ribosomal RNA genes and software used for assembly and contamination detection and removal. We strongly suggest following these guidelines, as the criteria outlined in MISAG will be valuable for future comparative genomic studies as users of public databases can filter genomes based on the genome quality required of a particular downstream analysis.
Downstream single-cell genomic analysis
Once a single-cell genome is curated, it can be analyzed together with additional genomic sequences to place it into a larger evolutionary, ecological and functional context. For example, phylogenetically informative genes such as the 16S rRNA gene and sets of conserved protein-coding marker genes have been used to assess intra- and inter-phylum-level relationships of microbial dark matter lineages [7,42]. When closely related isolate genomes are unavailable, SAGs can be used as reference genomes to recruit metagenomic reads for quantifying abundance patterns across temporal and spatial gradients [7,43–45]. SAGs have further shown utility in the analysis of recombination frequencies in bacterial populations, such as freshwater bacteria of the SAR11 clade [46] and for the determination of the overall genetic heterogeneity within discrete populations in honey bee gut symbionts [47] and wild Prochlorococcus [48]. Unlike MAGs, single-cell datasets are particularly powerful in linking phage sequences to their host [13,14] (represented schematically in Figure 2C) or deciphering eukaryote multipartite associations [15]. As such, single-cell sequence data offer a broad array of downstream analyses, depending on the research questions to be addressed.
Conclusion
Single-cell sequencing of individual bacterial and archaeal cells is becoming an important tool available to the microbiologist as single-cell sequencing is highly complementary to other approaches including traditional culture-based approaches and metagenomic sequencing. Single-cell sequencing has demonstrated its utility across disciplines including microbial ecology, evolutionary biology, agriculture and medicine. With this review, we provide suggestions for single-cell analysis workflows going from raw sequence data to the submission of single-cell genomes to public databases. As technical advancements continue and bioinformatic tools are refined, our ability to resolve whole microbial communities down to the genetic differences defining individual strains will improve and, undoubtedly, benefit from the production and analysis of DNA sequences originating from an individual cell.
Summary
Single-cell genome sequencing has become an important complement to metagenomics, facilitating the direct extraction of genomes from environmental samples in the absence of cultivation, yet requiring amplification of the DNA.
Due to the unique nature of the resulting single cell sequence data, it is of value to outline recommendations for the analysis of single-cell genomes, specifically describing a start to finish pipeline, from the assessment of read and contig quality to database submission.
Thoughtful consideration and execution of each step in a single-cell genome analysis pipeline is critically important for the reporting and deposition of single-cell genomes to the public databases.
Abbreviations
- MDA
multiple displacement amplification
- MISAG
minimum information about a single amplified genome
- SAGs
single amplified genomes
- WGA
whole genome amplification.
Author Contribution
R.M.B., D.F.R.D. and T.W. wrote the paper.
Funding
The work conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported under contract no. DE-AC02-05CH11231. DFRD was supported under the LBNL Microbes to Biomes LDRD entitled ‘Tackling microbial-mediated plant carbon decomposition using “function-driven” genomics’.
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
References
- 1.Monard C., Gantner S., Bertilsson S., Hallin S. and Stenlid J. (2016) Habitat generalists and specialists in microbial communities across a terrestrial-freshwater gradient. Sci. Rep. 6, 37719 10.1038/srep37719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowers R.M., Sullivan A.P., Costello E.K., Collett J.L., Knight R. and Fierer N. (2011) Sources of bacteria in outdoor air across cities in the midwestern United States. Appl. Environ. Microbiol. 77, 6350–6356 10.1128/AEM.05498-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fierer N., Leff J.W., Adams B.J., Nielsen U.N., Bates S.T., Lauber C.L. et al. (2012) Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc. Natl Acad. Sci. U.S.A. 109, 21390–21395 10.1073/pnas.1215210110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown C.T., Olm M.R., Thomas B.C. and Banfield J.F. (2016) Measurement of bacterial replication rates in microbial communities. Nat. Biotechnol. 34, 1256–1263 10.1038/nbt.3704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eloe-Fadrosh E.A., Paez-Espino D., Jarett J., Dunfield P.F., Hedlund B.P., Dekas A.E. et al. (2016) Global metagenomic survey reveals a new bacterial candidate phylum in geothermal springs. Nat. Commun. 7, 10476 10.1038/ncomms10476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marcy Y., Ouverney C., Bik E.M., Lösekann T., Ivanova N., Martin H.G. et al. (2007) Dissecting biological ‘dark matter’ with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc. Natl Acad. Sci. U.S.A. 104, 11889–11894 10.1073/pnas.0704662104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rinke C., Schwientek P., Sczyrba A., Ivanova N.N., Anderson I.J., Cheng, J.-F. et al. (2013) Insights into the phylogeny and coding potential of microbial dark matter. Nature 499, 431–437 10.1038/nature12352 [DOI] [PubMed] [Google Scholar]
- 8.Youssef N.H., Blainey P.C., Quake S.R. and Elshahed M.S. (2011) Partial genome assembly for a candidate division OP11 single cell from an anoxic spring (Zodletone Spring, Oklahoma). Appl. Environ. Microbiol. 77, 7804–7814 10.1128/AEM.06059-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anantharaman K., Brown C.T., Hug L.A., Sharon I., Castelle C.J., Probst A.J. et al. (2016) Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat. Commun. 7, 13219 10.1038/ncomms13219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blainey P.C. (2013) The future is now: single-cell genomics of bacteria and archaea. FEMS Microbiol. Rev. 37, 407–427 10.1111/1574-6976.12015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lasken R.S. (2012) Genomic sequencing of uncultured microorganisms from single cells. Nat. Rev. Microbiol. 10, 631–640 10.1038/nrmicro2857 [DOI] [PubMed] [Google Scholar]
- 12.Stepanauskas R. (2012) Single cell genomics: an individual look at microbes. Curr. Opin. Microbiol. 15, 613–620 10.1016/j.mib.2012.09.001 [DOI] [PubMed] [Google Scholar]
- 13.Labonté J.M., Swan B.K., Poulos B., Luo H., Koren S., Hallam S.J. et al. (2015) Single-cell genomics-based analysis of virus-host interactions in marine surface bacterioplankton. ISME J. 9, 2386–2399 10.1038/ismej.2015.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roux S., Hawley A.K., Torres Beltran M., Scofield M., Schwientek P., Stepanauskas R. et al. (2014) Ecology and evolution of viruses infecting uncultivated SUP05 bacteria as revealed by single-cell- and meta-genomics. eLife 3, e03125 10.7554/eLife.03125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoon H.S., Price D.C., Stepanauskas R., Rajah V.D., Sieracki M.E., Wilson W.H. et al. (2011) Single-cell genomics reveals organismal interactions in uncultivated marine protists. Science 332, 714–717 10.1126/science.1203163 [DOI] [PubMed] [Google Scholar]
- 16.Bowers R.M., Kyrpides N.C., Stepanauskas R., Harmon-Smith M., Doud D., Reddy T.B.K. et al. (2017) Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat. Biotechnol. 35, 725–731 10.1038/nbt.3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rinke C., Lee J., Nath N., Goudeau D., Thompson B., Poulton N. et al. (2014) Obtaining genomes from uncultivated environmental microorganisms using FACS-based single-cell genomics. Nat. Protoc. 9, 1038–1048 10.1038/nprot.2014.067 [DOI] [PubMed] [Google Scholar]
- 18.Hutchison C.A. and Venter J.C. (2006) Single-cell genomics. Nat. Biotechnol. 24, 657–658 10.1038/nbt0606-657 [DOI] [PubMed] [Google Scholar]
- 19.Bowers R.M., Clum A., Tice H., Lim J., Singh K., Ciobanu D. et al. (2015) Impact of library preparation protocols and template quantity on the metagenomic reconstruction of a mock microbial community. BMC Genomics 16, 856 10.1186/s12864-015-2063-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoeijmakers W.A.M., Bártfai R., Françoijs K.-J. and Stunnenberg H.G. (2011) Linear amplification for deep sequencing. Nat. Protoc. 6, 1026–1036 10.1038/nprot.2011.345 [DOI] [PubMed] [Google Scholar]
- 21.Duhaime M.B., Deng L., Poulos B.T. and Sullivan M.B. (2012) Towards quantitative metagenomics of wild viruses and other ultra-low concentration DNA samples: a rigorous assessment and optimization of the linker amplification method. Environ. Microbiol. 14, 2526–2537 10.1111/j.1462-2920.2012.02791.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zong C., Lu S., Chapman A.R. and Xie X.S. (2012) Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science 338, 1622–1626 10.1126/science.1229164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dean F.B., Hosono S., Fang L., Wu X., Faruqi A.F., Bray-Ward P. et al. (2002) Comprehensive human genome amplification using multiple displacement amplification. Proc. Natl Acad. Sci. U.S.A. 99, 5261–5266 10.1073/pnas.082089499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martínez Martínez J., Swan B.K. and Wilson W.H. (2014) Marine viruses, a genetic reservoir revealed by targeted viromics. ISME J. 8, 1079–1088 10.1038/ismej.2013.214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lasken R.S. (2007) Single-cell genomic sequencing using multiple displacement amplification. Curr. Opin. Microbiol. 10, 510–516 10.1016/j.mib.2007.08.005 [DOI] [PubMed] [Google Scholar]
- 26.Yilmaz S., Allgaier M. and Hugenholtz P. (2010) Multiple displacement amplification compromises quantitative analysis of metagenomes. Nat. Methods 7, 943–944 10.1038/nmeth1210-943 [DOI] [PubMed] [Google Scholar]
- 27.Lasken R.S. and Stockwell T.B. (2007) Mechanism of chimera formation during the multiple displacement amplification reaction. BMC Biotechnol. 7, 19 10.1186/1472-6750-7-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S. et al. (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nurk S., Bankevich A., Antipov D., Gurevich A.A., Korobeynikov A., Lapidus A. et al. (2013) Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J. Comput. Biol. 20, 714–737 10.1089/cmb.2013.0084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng Y., Leung H.C.M., Yiu S.M. and Chin F.Y.L. (2012) IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28, 1420–1428 10.1093/bioinformatics/bts174 [DOI] [PubMed] [Google Scholar]
- 31.Woyke T., Sczyrba A., Lee J., Rinke C., Tighe D., Clingenpeel S. et al. (2011) Decontamination of MDA reagents for single cell whole genome amplification. PLoS ONE 6, e26161 PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salter S.J., Cox M.J., Turek E.M., Calus S.T., Cookson W.O., Moffatt M.F. et al. (2014) Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 12, 87 10.1186/s12915-014-0087-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmieder R. and Edwards R. (2011) Fast identification and removal of sequence contamination from genomic and metagenomic datasets. PLoS ONE 6, e17288 10.1371/journal.pone.0017288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sinha R., Stanley G., Gulati G.S., Ezran C., Travaglini K.J., Wei E. et al. (2017) Index switching causes ‘Spreading-of-Signal’ among multiplexed samples in Illumina HiSeq 4000 DNA sequencing. bioRxiv 10.1101/125724 [DOI] [Google Scholar]
- 35.Kircher M., Sawyer S. and Meyer M. (2012) Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 40, e3 10.1093/nar/gkr771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wright E.S. and Vetsigian K.H. (2016) Quality filtering of Illumina index reads mitigates sample cross-talk. BMC Genomics 17, 876 10.1186/s12864-016-3217-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Utturkar S.M., Cude W.N., Robeson M.S., Yang Z.K., Klingeman D.M., Land M.L. et al. (2016) Enrichment of root endophytic bacteria from populus deltoides and single-cell-genomics analysis. Appl. Environ. Microbiol. 82, 5698–5708 10.1128/AEM.01285-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Markowitz V.M., Chen, I.-M.A., Chu K., Szeto E., Palaniappan K., Pillay M. et al. (2014) IMG/m 4 version of the integrated metagenome comparative analysis system. Nucleic Acids Res. 42(Database issue), D568–D573 10.1093/nar/gkt919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eren A.M., Esen Ö.C., Quince C., Vineis J.H., Morrison H.G., Sogin M.L. et al. (2015) Anvi'o: an advanced analysis and visualization platform for ‘omics data. PeerJ 3, e1319 10.7717/peerj.1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tennessen K., Andersen E., Clingenpeel S., Rinke C., Lundberg D.S., Han J. et al. (2015) Prodege: a computational protocol for fully automated decontamination of genomes. ISME J. 10, 269–272 10.1038/ismej.2015.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lux M., Krüger J., Rinke C., Maus I., Schlüter A., Woyke T. et al. (2016) acdc — automated contamination detection and confidence estimation for single-cell genome data. BMC Bioinf. 17, 543 10.1186/s12859-016-1397-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hedlund B.P., Dodsworth J.A., Murugapiran S.K., Rinke C. and Woyke T. (2014) Impact of single-cell genomics and metagenomics on the emerging view of extremophile ‘microbial dark matter’. Extremophiles 18, 865–875 10.1007/s00792-014-0664-7 [DOI] [PubMed] [Google Scholar]
- 43.Hedlund B.P., Murugapiran S.K., Alba T.W., Levy A., Dodsworth J.A., Goertz G.B. et al. (2015) Uncultivated thermophiles: current status and spotlight on ‘Aigarchaeota’. Curr. Opin. Microbiol. 25, 136–145 PMID: [DOI] [PubMed] [Google Scholar]
- 44.Woyke T., Xie G., Copeland A., González J.M., Han C., Kiss H. et al. (2009) Assembling the marine metagenome, one cell at a time. PLoS ONE 4, e5299 10.1371/journal.pone.0005299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swan B.K., Tupper B., Sczyrba A., Lauro F.M., Martinez-Garcia M., González J.M. et al. (2013) Prevalent genome streamlining and latitudinal divergence of planktonic bacteria in the surface ocean. Proc. Natl Acad. Sci. U.S.A. 110, 11463–11468 10.1073/pnas.1304246110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zaremba-Niedzwiedzka K., Viklund J., Zhao W., Ast J., Sczyrba A., Woyke T. et al. (2013) Single-cell genomics reveal low recombination frequencies in freshwater bacteria of the SAR11 clade. Genome Biol. 14, R130 10.1186/gb-2013-14-11-r130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Engel P., Stepanauskas R. and Moran N.A. (2014) Hidden diversity in honey bee gut symbionts detected by single-cell genomics. PLoS Genet. 10, e1004596 10.1371/journal.pgen.1004596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kashtan N., Roggensack S.E., Rodrigue S., Thompson J.W., Biller S.J., Coe A. et al. (2014) Single-cell genomics reveals hundreds of coexisting subpopulations in wild Prochlorococcus. Science 344, 416–420 10.1126/science.1248575 [DOI] [PubMed] [Google Scholar]