Abstract
The human gut microbiome is considered an organ in its entirety and has been the subject of extensive research due to its role in physiology, metabolism, digestion, and immune regulation. Disequilibria of the normal microbiome have been associated with the development of several gastrointestinal diseases, but the exact underlying interactions are not well understood. Conventional in vivo and in vitro modelling systems fail to faithfully recapitulate the complexity of the human host–gut microbiome, emphasising the requirement for novel systems that provide a platform to study human host–gut microbiome interactions with a more holistic representation of the human in vivo microenvironment. In this review, we outline the progression and applications of new and old modelling systems with particular focus on their ability to model and to study host–microbiome cross-talk.
Keywords: biological models, biotechnology, host–microbe interactions
Introduction
The development of novel prognostic tools and effective therapeutic strategies relies upon a profound understanding of the molecular mechanisms involved in health and disease, including influences from the microbiome [1,2]. For example, the microbiome that inhabits the human gut is integral in regulating normal gut homeostasis. Such physiological functions include aiding digestion, producing metabolites from undigested fibre, regulating drug metabolism, adapting immune responses, and protecting the host from pathogens and infections [3–5]. The realisation that commensal and pathogenic bacteria play a role in dysbiosis, disease development, and chronic disorders [3–5] places an emphasis on gaining a deeper understanding of the underlying mechanisms involved in physiological and pathophysiological host–microbiome cross-talk. The complexity of a holistic understanding of these cause-and-effect relationships in humans makes human studies complicated and somewhat limited, most notably because of the challenges in accessing the human gut, making disease-modelling essential. Preclinical animal models and in vitro 2D cell culture studies have provided extensive insights into the physiological and pathophysiological processes that underpin disease. However, as they do not faithfully represent the human body, they are not suitable models to evaluate all research questions nor model drug efficacy. Moreover, the high number of therapeutic compounds that fail to translate in clinical trials [6,7] highlights the need and importance for models that are more physiologically relevant to the human body, in order to personalise treatments and better predict patient outcomes. In this review, we discuss the advantages and disadvantages of conventional modelling systems. Furthermore, we review the advantages and contributions of 3D organoid culture and systems such as organ-on-a-chip that better recapitulate the in vivo human body and provide a better understanding of cell–cell interactions and the host–gut microbiome cross-talk that occurs in health and disease. The advantages and disadvantages of the systems discussed in this review are outlined in Table 1. Figure 1 provides schematic representation of these systems.
Table 1. Advantages and disadvantages of current culturing techniques with respect to host–microbiome interaction.
| Type of assay | Advantages | Disadvantages |
|---|---|---|
| 2D ethos |
|
|
| Transwell | ||
| Organoids |
|
|
| SynVivo® (SynTumour) |
|
|
| Tissue slices |
|
|
| Organ/tissue baths |
|
|
| Organ-on-a-chip |
|
|
| Quasi-Vivo® (Kirkstall) |
|
|
| HuMiX module |
|
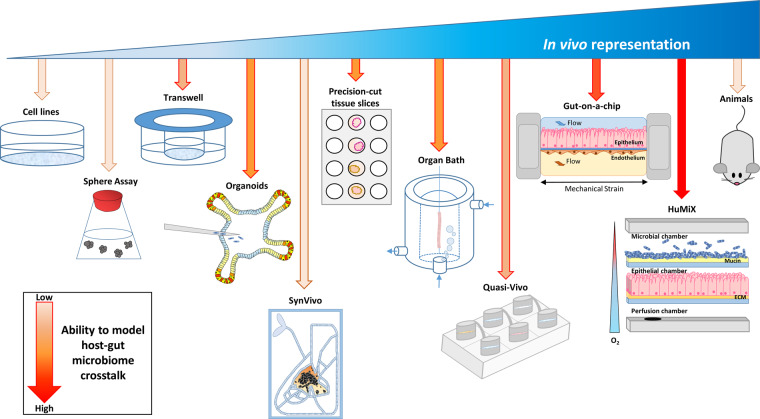
Figure 1. Current cell modelling systems and their potential to model host-microbiome cross-talk.
Schematic representation of current cell culturing techniques available and their ability to model host–microbiome interactions.
2D culture and in vivo animal models to study human disease
Human cell lines have had a fundamental role in determining the importance of pathways involved in disease and have been a good platform for screening and characterising drug therapeutics [8]. Cell lines retain many genetic, epigenetic and gene expression features of human disease [9,10]. However, they fail to represent the in vivo 3D microenvironment, induce cell differentiation [11,12], perform tissue-specific functions [11,13,14], and do not represent the natural cell heterogeneity that compose primary tumours [9]. In addition, it is often found that drug doses that are effective in 2D culture result in poor efficacy when scaled to humans [6,15–17]. Many of these limitations are addressed by genetically engineered mouse models (GEMMs). GEMMs have been an extremely useful tool for studying early and chronic stages of human disease [8], as they share similarities in anatomy, physiology, and genetics [18–22]. However, their use for investigating host–microbiome interactions is limited by the differences in lifespan, variations in diet, and microbiome populations [23]. The inability of these systems to study the cause-and-effect relationships of the host–microbiome interactions in a meaningful way is driving the development of systems to overcome this issue.
Cell culturing systems for studying the human microbiome
The gastrointestinal (GI) tract contains a flora of commensal bacteria, known as the microbiome [24]. The living gut microbiome has been of increasing interest in recent years, as its role in the regulation of the immune system, local barrier functions, homeostasis, and tissue regeneration and also in the development of certain diseases (e.g. Crohn's disease, inflammatory bowel disease, and coeliac disease) are not fully understood [3,5,25]. Additionally, it is known that the gut microbiome can influence drug absorption and metabolism [3,26]. Thus, it is paramount that host–gut microbiome interactions are elucidated.
Advancements in techniques for culturing cells have been critical for identifying and understanding cell involvement in disease initiation. In vitro sphere-forming assays were among the first techniques adopted to study these interactions. The sphere-forming assays were initially developed to study neural stem cells [27] and were later developed to culture mammary stem cells [28]. In these cultures, cells aggregate together to form 3D spheroid structures, which can then be utilised to determine stem cell behaviour of both human and animal cell populations [29,30]. Typically, the study of human GI disease has relied upon culturing intestinal epithelial cells in static well plate assays. For example, Transwell culture has been used to grow a polarised monolayer of human cell lines [31] on a nanoporous insert [3]. Microbial cells, probiotic [32–35] or pathogenic [36–39], can then be added to the apical cell surface. To gain further insights, immune cells can be simultaneously added to the basolateral side of the monolayer culture [31,40–42]. Transwell culture is limited however, as cultured epithelial cells do not produce functional specialised cell types [12,43]. Consequently, a substantial amount of research has focused on the development of a 3D culturing technique, initially using individual murine intestinal stem cells [44,45] and also patient-derived tissues and human stem cells [46–48] that grow into self-renewing organoids that encompass the structure, and all cell types of the in vivo intestinal crypt, providing a more human-relevant model.
The progression from 2D to 3D organoid culture
Long-term organoid culturing has been a major breakthrough in research and is now an extensively used tool for studying both basic and clinical biology [49–51]. Advancement in technology has enabled culturing of the stomach [52–54], pancreas [55–57], liver [58], prostate [59,60], oesophagus [46,61], gall bladder [62,63], and taste buds [64]. Additionally, CRISPR-Cas9 technology has been exploited to induce gene editing in intestinal organoids that mimics the sequential loss and gain of functions of genes involved in human colorectal cancer [65], which, after in orthotopic engraftment, could metastasise to the liver [66,67]. Thus, organoid models can be used to study advanced aspects of cancer development in a more complexed in vivo scenario. Organoid culture has been exploited to investigate host–gut microbiome interactions by microinjection of microbes to the lumen of organoids (apical surface, as this mimics aspects of microbial infection in vivo) or basally by the addition of microbes to the media [63]. The potential of organoids as a model for studying host–microbial interactions has been demonstrated using human gastric organoids and the pathogenic microbe, Helicobacter pylori [54,68,69], that is known to cause infection, ulceration, and gastric cancer in humans [70]. Apical infection of gastric organoids with H. pylori recapitulates known hallmarks of infection, including a marked increase in mucus-secreting cell expression, induction of cytokine production via up-regulation of NF-κB (known to induce gastritis), and production of the H. pylori virulence factor CagA (cytotoxin-associated gene A), which is associated with motogenesis and tumour progression [71,72]. Other studies have been able to efficiently infect human-derived organoids in a similar manner with a range of known pathogens, including Salmonella typhimurium [63], Clostridium difficile [73,74], and rotaviruses [75]. Thus, these studies present a valuable near-physiological modelling system in which to begin deciphering the complex interactions between host epithelium and infectious agents. Additionally, several recent studies have improved the human relevance of Transwell cultures. For example, Transwells have been adapted to grow human small intestinal and colonic organoids in monolayers on Transwell membranes that support microbial (Escherichia coli [42,76,77] and norovirus [78]) adherence at the cell surface, as seen in vivo. The above studies have provided an informative platform that can be exploited to use patient-derived tissues to study donor-specific disease progression, host responses to enteric pathogens, and provide a platform to develop personalised treatments.
However, like any other system, organoid culture has its limitations. In instances where microbes are added to culture medium, microbial overgrowth often occurs, which can damage the epithelium and can hinder long-term co-culture, this is also the case with co-culturing human cell lines with microbes using Transwells [3,43] and, thus, should be avoided where possible. It is not possible to exert peristalsis-like motions (which is required in vivo to sustain intestinal differentiation and confine microbial overgrowth [14]) to the cells cultured in Transwell or organoid culture. And the reductionist approach of organoid culture limits the full potential of this method as epithelial cells grow without their native microenvironment, and the true contributions from the stroma, extracellular matrix (ECM), oxygen, vasculature, immune system, and the microbiome cannot be truly assessed [49,79]. This ultimately impedes the understanding of the precise molecular mechanisms involved in transition from healthy to disease states and limits the potential efficacy of interventions when translated to clinic.
It is evident that models, which reflect the holistic environment, are crucial to better predict patient outcomes to therapy, understand the mechanisms that underpin disease and the role environmental factors, such as diet, have on physiology and pathophysiology [12]. The integration of microfluidics with living biological systems has paved the way for new techniques such as the exciting ‘organ-on-a-chip’ concepts, which aims at developing advanced in vitro models that replicate the key features of human tissues and organs [80]. The culturing methods outlined below are discussed in increasing order of their ability to represent the in vivo environment and its suitability to study host–microbe interactions.
SynVivo®
SynVivo microfluidic models have been developed to study human tissue microvasculature, cell–cell, and cell–drug interactions in a 3D setting, in the context of in vivo-relevant flow and pressures [81]. Currently, SynVivo models have four independent assays that represent the blood–brain barrier [82–84], cancer [85], inflammation, [86,87] and toxicology. SynVivo devices utilise polydimethylsiloxane (PDMS) chambers that comprise a microfluidic map of vasculature with spaces that can be lined with ECM. For example, endothelial cells can be cultured on fibronectin-coated channels and activated using cytokines. Endothelial cell interactions with white blood cells can then be assessed in real time by the injection of cells into the system [81]. Synthetic network assays can also recapitulate in vivo leaky vasculature associated with tumourigenesis and thus can be utilised to study both basic and applied research [81]. To date, SynVivo chips have not yet been developed to study microbial interactions; however, its ability to recreate a realistic vasculature network surrounding a tissue will make a profound advancement in modelling in vivo 3D environments.
Tissue slices
Precision-cut tissue slices (PCTS) are a useful tool for metabolic, pharmacology, and toxicological studies [88–90]. Predominately used to model solid organs (e.g. liver and kidney) and agarose-embedded non-solid organs such as the intestine [91,92], uniform tissue slices are cut and incubated in media in 6- or 12-well plates and exposed to atmospheric conditions [92]. PCTS are believed to have advantages over organoids and organ-on-a-chip (discussed later), as they represent all regions of the tissue [91]. It has been reported that murine intestine is maintained for 6 days in the presence of normal neuronal, muscular, and mucosal structures with contractibility functions and some immune responses, therefore, representing a more holistic intestinal organ [93]. Additionally, in the absence of antibiotic media, some aerobic commensal microbes can be maintained [93]. Although this system holds great promise to study holistically the complex mechanisms that underpin healthy gut functions and disease in a 3D physiologically relevant environment, further work is required to maintain anaerobic microbes that are predominately found within the intestinal environment [94]. In addition to maintaining tissue slices in media, epithelial integrity, neural activity, ion/nutrient/drug transport [95–97], and the effect of microbiota and pathogenic bacteria on these processes [98–101] can be examined using analytical tools, such as the Ussing chamber. As this tool compartmentalises the apical and basolateral surfaces of the tissue, each surface can be exposed to independent physiological solutions [95], representative of the in vivo microenvironment. The potential of the Ussing chamber to investigate host–pathogen interactions was highlighted when co-culture of human cell lines with the infection-causing bacteria, C. difficile, extensively impaired epithelial barrier function and elicited cytotoxic immune responses which contributes to C. difficile-mediated pathogenesis [101]. One limitation of the Ussing chamber, however, is that intestinal tissue viability is often limited to 3 h which limits long-term optimal tissue function studies [102].
Organ baths
Organ bath assays play important roles in evaluating concentration–response relationships in a variety of tissues, predominately contractile organs. The primary advantage of organ baths is that live tissue is cultured that continues to function as a whole unit with respect to contractions visualised in vivo. By retaining these physiological functions, pharmaceutical interactions can be examined to predict in vivo responses [103,104]. However, because of using relatively large samples, the core of the suspended tissue strips can often become hypoxic, which can hinder contractile readouts [104]. Another caveat with organ baths is the requirement of fresh tissue for each experiment [105,106]. This prevents analysis from a single donor or genetic background, which is achievable through 3D organoid culture. However, organ baths have been used to show that E. coli stimulates the contractility of human colonic smooth muscle cells [107]. Thus, there is scope for advancing organ bath techniques to study probiotic and pathogenic microbes ex vivo.
Organ-on-a-chip
To mimic key aspects of living organs, such as the multicellular structures, cell–cell and tissue–tissue interactions, and the native microenvironment, organ-on-a-chip systems have been developed to incorporate microfluidic and microengineering technology [7,108]. The benefit of developing such systems is that it provides a platform to study the complex physiological and pathophysiological responses of tissues at an organ level, to provide patients with quicker access to new medication. For example, microfluidics can be utilised to mimic blood and nutrient flow and maintain mucus transition in lung epithelia [108,109]. In addition, this system can be exploited to complement animal studies and also more accurately predict pharmacological effects in patients and, in the future, could be used to bypass the use of animal testing completely [110]. Organ-on-a-chip has been developed for several organs, including the lung [111,112], heart [113,114], liver [115], neuron [116], kidney [117,118], gut [14,43,119], blood vessel [120,121], tumour-on-a-chip [122], bone marrow-on-a-chip [123], liver-tumour-bone marrow-on-a-chip [124], and liver-skin-intestine-kidney-on-a-chip [125]. These systems can be extended to disease modelling, pharmaceutical analysis, drug development strategies [6,118,122], and understanding host–microbe interactions [14,111,122]. For the purpose of this review, we will discuss the use of organ-on-a-chip to study intestinal dynamics with particular focus on the gut microbiome. The potential of these systems to accurately model drug uptake and metabolism in the human body has recently been shown in the context of products produced by the microbiome. Vernetti et al. (2017) demonstrated that a liver-intestine-kidney-on-a-chip system integrated with an intact blood–brain barrier/neurovascular unit sufficiently modelled in vivo trimethylamine (a by-product of the microbiome) metabolism. Specifically, trimethylamine, microinjected into intestinal organoids, was found in the basolateral media and was subsequently metabolised to trimethylamine N-oxide by the liver module and then secreted into the lumen of the kidney module, which occurs in vivo. The study also revealed a novel finding that trimethylamine N-oxide crosses the blood–brain barrier [126], emphasising the potential of such systems to further our understanding of human physiology.
Progression to gut-on-a-chip assays
The applications of gut-on-a-chip systems are still at the early stages of development [80]; however, they hold great promise for studying the interactions between host and microbiome. Initial studies involved culturing a monolayer of human epithelial Caco-2 cells inside a microfluidic device [127]. Although this microchip provided a vascular flow over the cells, the ability of Caco-2 cells to reach confluence was hindered. This was likely caused from inherent faults within the system that did not allow large volumes of medium, and thus nutrients, to be administered at a time, and resulted in cellular damage from dead cell debris, air bubbles, and the inability to maintain a stable medium flow [127]. These problems were circumvented by identifying optimal microchannel dimensions and appropriate Caco-2 cell density, which provided sufficient nutrients and reduced shear pressures that ultimately promoted Caco-2 cellular growth and normal barrier function [127]. This study highlights the importance of chip scaling, and this should be taken into consideration when developing novel microfluidic devices in the future. More advanced microfluidic models have been subsequently developed, based on the culture of Caco-2 cells under physiologically relevant conditions, including peristalsis-like motions, intraluminal fluid flow, the use of 3D scaffolds, and support the growth of the naturally occurring microbiome without diminishing cell viability [43]. These earliest models of gut-on-a-chip were adapted from the advancements in lung-on-a-chip that enabled microfluidic systems to effectively model the cyclic breathing motions of the human lung [111]. In 2012, a biomimetic human-gut-on-a-chip microdevice was developed that consisted of two closely apposed microfluidic channels made from a self-adhesive elastomer, PDMS (that is highly permeable to oxygen), creating a luminal and capillary channel [43]. Between the channels was a thin elastic porous membrane coated with an ECM. A layer of Caco-2 cells was cultured on top of the ECM. Culture medium was allowed to flow through both channels to mimic in vivo fluid flow and shear stress. Peristaltic-like deformations were instigated by cyclic suction patterns to stretch and relax the porous membrane [43]. The group reported that this human-gut-on-a-chip system allowed intestinal epithelial cells to polarise, form 3D structures that closely resembled physical and functional features of human intestine and created a high-integrity barrier against small molecules which was not achieved when cells were cultured using Transwell models [43]. Additionally, this system allows the study of physiological gut functions in the presence of relevant cues and interactions with the microflora. Caco-2 cells have been effectively co-cultured with a naturally occurring intestinal microbe, Lactobacillus rhamnosus GG (LGG), at the luminal surface which improved intestinal barrier function [43], which has been previously reported with probiotic bacteria strains in humans [128]. Moreover, differentiated intestinal epithelium supported the growth of naturally occurring microbiota for more than 2 weeks, which is in contrast with Transwell and organoid cultures where cell viability is often lost within a few hours [12,14,43]. Taken together, these studies provide support for the use of gut-on-a-chip as a tool for studying the mechanisms involved in intestinal function and the involvement of the normal gut microbiome. This system also has the potential to be exploited to study the development of disease in a more human-relevant holistic environment, become a novel platform for drug development, and better predict responses to therapy in clinical trials.
The potential of gut-on-a-chip was proved in a proof-of-principal study in 2016 when Kim et al. studied the contribution of the microbiome to gut pathophysiology. Specifically, Caco-2 cells were co-cultured, with eight living human commensal microbes that have previously been used in clinical trials of chronic inflammatory bowel diseases [129]. Caco-2 cells displayed distinct gene expression profiles and phenotypes when cultured in the presence of commensal microbes, which closely resembled the normal human ileum. Next, the group investigated whether gut-on-a-chip was sufficient to model human intestinal inflammation, and what contribution the gut microbiome played. Culturing Caco-2 cells with immune cells and non-pathogenic bacteria, to induce pro-inflammatory cytokines, resulted in injury to the villus structure and impaired barrier function. By independently controlling the addition of each cytokine to the system, the authors were able to elucidate that only in the presence of interleukin-8 were the cytokines able to induce disease-promoting effects, which is not easy to model using GEMMs. Furthermore, the extent of villus injury and compromised barrier function was reduced and delayed by the addition of commensal probiotic microbes within the chip. Subsequently, antibiotic therapies that are commonly used in clinic to treat intestinal inflammatory disease were added to protect the epithelial cells from damage [14]. This provides direct evidence that gut-on-a-chip is efficient in mimicking the suppression of injury responses from bacteria and the immune system that are observed clinically [14]. More importantly, the gut-on-a-chip assay provides a robust platform to identify the individual and combinational effects of immune cells, commensal and pathogenic microbes, and epithelia and endothelial cells in a system that closely resembles the in vivo environment. Although still not foolproof, this system enables the study of intestinal physiology to identify mechanisms involved in gut pathophysiology and could be used to find novel therapeutics in a patient-specific manner in the future.
Quasi-Vivo® (Kirkstall)
Kirkstall have designed a system based on organ-on-a-chip models to produce a ‘system-on-a-plate’ array, named Quasi-Vivo [130]. Quasi-Vivo technology aims at producing a down-scaled system of the human body (through the use of allometrics, [131,132]), such that in vivo biochemical and physical stimuli, metabolic, volumetric and exchange rate relationships are retained in vitro [130]. Advantages of Quasi-Vivo over organ-on-a-chip devices include limited cellular damage from high wall shear stresses [130,133] and the inherent issue with bubble formation, which occurs in microfluidic systems, is overcome due to their intelligent design features and size [130]. Preliminary findings from Kirkstall show that E. coli is viable within the Quasi-Vivo system and, thus, has scope for future microbial analysis.
HuMix module
The HuMiX, human–microbial cross-talk module, composes three microfluidic chambers that are parallel to one another and separated by semipermeable membranes. The alignment of these chambers provides a microbial, epithelial, and perfusion chamber. The specific outlets and inlets of each chamber allow for the inoculation of cell lines and, through the perfusion of cell growth media, can precisely control the physiochemical conditions each chamber receives [24,134,135]. This set-up builds on previous systems such that it (1) has the ability to simultaneously establish aerobic and anaerobic conditions and (2) can monitor oxygen levels in real time [134].
Several proof-of-concept studies have highlighted the potential HuMiX to enhance our understanding of host–microbe cross-talk [134,135]. Specifically, Eain et al. report that HuMiX was sufficient to co-culture Caco-2 cells and microbial cells (LGG and Bacteroides caccae) under conditions that represented the in vivo human GI tract, i.e. the maintenance of oxygen gradients either side of the epithelium. The arterial blood supply was mimicked by the addition of an oxygen-rich media in the perfusion chamber, which provided epithelial cells with oxygen from the basal surface. In doing so, the epithelium could grow in a shear-free environment, while anaerobes were present [135]. Additionally, the group were able to co-culture immune cells via the perfusion chamber and are now focusing on developing a system, Immuno-HuMiX, to study the interactions between the immune system and the intestinal microbiota in the human gut. However, the authors note that despite the success in co-culture of human and microbial cells and the potential to extend this system to study diet, drug screening, discovery, and delivery, these cell types cannot fully represent the complex cellular makeup of the entire intestinal tract [134,135].
Bringing cell culture techniques closer to in vivo: future directions
There is an abundance of unknown information with regard to the complexity of the gut microbiome and its effect on the human host, which precludes the development of novel therapeutics for disease states. Recently, significant advancements in the scientific field have attempted to unravel these host–gut microbiome interactions. Although the reductionist approach of previous conventional modelling systems, such as static 2D and 3D cell culture, has improved our understanding of how the gut environment is relevant to both human physiology and pathophysiology, advancements in technology have paved the way to microfluidic-based devices that provide a more accurate representation of the in vivo microenvironment.
Microfluidic platforms, such as organ-on-a-chip, HuMiX, and other devices, have significant potential in advancing diagnostic, pharmaceutical, and nutritional industries with respect to earlier diagnosis, expedition of drug development, and understanding the mechanisms that underpin dietary involvement in health and disease. As these systems permit 3D co-culture of epithelial cells with underlying endothelium, immune cells, and microbes with the influence of flow and shear effects, they will be fundamental for further improving our understanding of the host–microbiome interactions holistically. Having said that, there are many challenges microfluidic technology still face today which require tweaking to better recapitulate the in vivo host–microbiome environment. Firstly, most systems heavily rely on the use of Caco-2 cells as a model of human intestinal epithelium. While this cell line has been crucial in expanding our knowledge, Caco-2 cells were originally from a human colorectal adenocarcinoma [7,24] and thus may have attributes that distinguish them from the normal gut epithelium and any cross-talk between the host–gut microbiome may not be a true representation of the native intestine. Specific issues of using Caco-2 cells to model gut epithelium are (1) Caco-2 cells, a colon cancer-derived cell line, especially in the context of gut-on-a-chip assays, are used to study small intestinal villus formation [12,14,43]. It is necessary to point out that the human colonic epithelium is not composed of villi [136] and that cancer cells can behave differently from normal epithelial cells. (2) Only loose observations can be made about gut physiology when using Caco-2 cells, as it is not clear which section of the intestine, i.e. duodenum, ileum, jejunum or colon, is being modelled. It is important to know exactly which region of the intestine is being modelled such that is mimics the architecture, cellular makeup and immune responses that differs throughout the intestine. For example, the small intestine tends to be more acidic and contains a single mucosal layer, whereas the colon consists of a protective thick double-layered mucus [137] that harbours a higher bacterial density than the small intestine [136]. Therefore, without knowing which section of intestine is being modelled, informed conclusions cannot be made. And (3) interactions between the host and microbes that primarily inhabit the human colon are analysed in the context of epithelial villus formation and barrier integrity. Again, this is not a true representation of human gut physiology and thus emphasises that Caco-2 studies should be limited when evaluating gut physiology. The progression in generating primary epithelium and organoids from stem cells holds great promise for advancing these techniques to recreate the cellular makeup and environmental conditions prevalent in the human gut, especially in light of the available organoid monolayer Transwell culturing methods that utilises donor-specific tissue and enables co-culture with microbes [42,76,77] and which has the potential to be translated to organ-on-a-chip and HuMiX modelling systems. Secondly, most microfluidic devices have been sufficient only in culturing aerobic microbes, which does not reflect the anaerobic nature of the intestinal lumen [24,94]. The development of the state-of-the-art HuMiX system has provided a platform to culture aerobic-dependent epithelium in the presence of anaerobic microbes, but further refinement of this system is required to decipher the complex interactions between the holistic gut environment (stroma, immune system, diet, microbiome, and metabolome) and the roles they play in maintaining intestinal physiology and initiating and preventing intestinal diseases. The third challenge is a technical issue. The material, PDMS, which is currently used to generate organ-on-a-chip microfluidic chambers does not faithfully represent the physicochemical properties of the ECM that occur in vivo, and can often preclude drug studies as small hydrophobic molecules are absorbed by the material [138,139]. In addition, PDMS poses challenges for large-scale reproduction of microfluidic chambers. However, studies have been performed to identify suitable alternative materials or surface coating techniques to combat this issue [140–142].
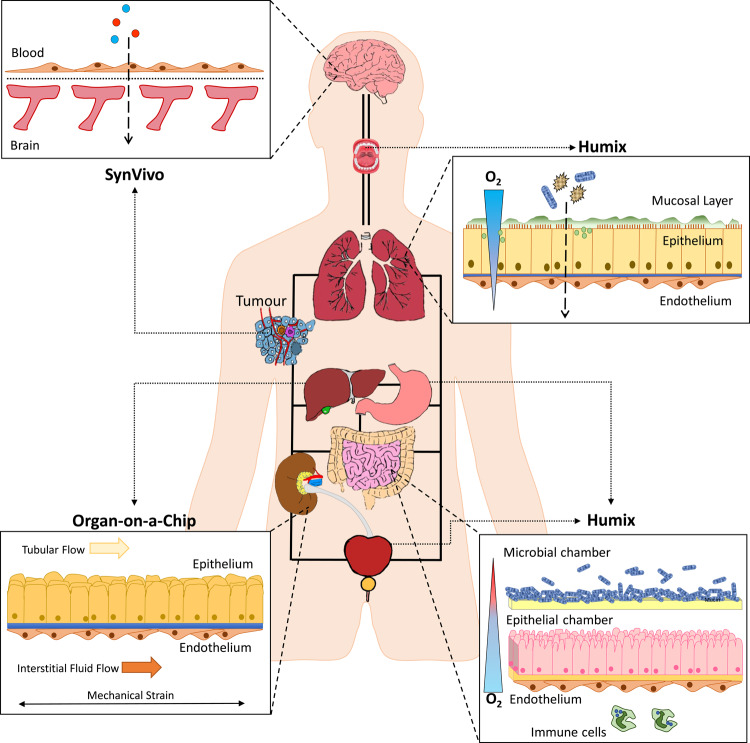
The challenge for the future is to develop a human-on-a-chip system [143,144], through the amalgamation of the technologies described herein, in which several chambers can be interconnected to represent all aspects of the human body (Figure 2), including factors such as the blood–brain barrier, aerobic–anaerobic nature of the gut epithelium, and microbiome and disease states, i.e. tumours, to functionally mimic and study the complex interactions between the circulatory, endocrine, digestive, immune, lymphatic, nervous, respiratory, and urinary systems. Moreover, such a system would provide a holistic scope to investigate pathophysiological conditions through subtle manipulations within chambers and will also provide a basis to systemically assess drug interventions in the future.
Figure 2. A proposed microfluidic chip to model host–microbiome cross-talk.
An amalgamation of SynVivo, organ-on-a-chip, and HuMiX culturing systems has the potential to model the host–microbiome interactions within the entire human body, in the presence of flow and shear stresses (representative of a circulatory system).
Summary
Understanding, holistically, the mechanisms that underpin human physiology and pathophysiology are hindered due to the difficulty in accessing organs, and a paucity of information on the interactions between the host and microbiome.
Conventional modelling systems such as 2D cell culture and Transwells fail to faithfully represent the in vivo human environment, especially in the context of studying host−microbiome cross-talk. Emphasising a need for novel modelling systems that better recapitulate the human body.
Technological advancements involving microfluidic techniques has paved the way for exciting organ-on-a-chip based culturing methods.
Novel in vitro and ex vivo culturing techniques such as patient-derived organoid culture, SynVivo®, Quasi-Vivo®, organ-on-a-chip and HuMiX systems, are beginning to provide more clinically relevant platforms in which to better model and study human host-microbiome interactions.
Abbreviations
- CagA
cytotoxin-associated gene A
- CRISPR-Cas9
clusters of regularly interspaced short palindromic repeats-CRISPR associated protein 9
- ECM
extracellular matrix
- GEMMs
genetically engineered mouse models
- GI
gastrointestinal
- LGG
Lactobacillus rhamnosus GG
- NF-κB
nuclear factor kappa-light-chain-enhancer of activated B cells
- PCTS
precision-cut tissue sections
- PDMS
polydimethylsiloxane
Author Contribution
S.M. and S.E. designed and wrote the manuscript. L.P. provided guidance, editing, and final approval of manuscript.
Funding
S.M. is supported by a philanthropic donation from the Mr Lyndon and Mrs Shirley Ann Wood from the Moorhouse Group Ltd [grant number 512990]. L.P. is supported by a Fellowship from the European Cancer Stem Cell Research Institute, Cardiff University, School of Biosciences [grant number AC1970LP01].
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript information.
References
- 1.Yissachar N., Zhou Y., Ung L., Lai N.Y., Mohan J.F., Ehrlicher A. et al. (2017) An intestinal organ culture system uncovers a role for the nervous system in microbe-immune crosstalk. Cell 168, 1135–1148.e12 10.1016/j.cell.2017.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pflughoeft K.J. and Versalovic J. (2011) Human microbiome in health and disease. Ann. Rev. Pathol. 7, 99–122 10.1146/annurev-pathol-011811-132421 [DOI] [PubMed] [Google Scholar]
- 3.Park G.-S., Park M.H., Shin W., Zhao C., Sheikh S., Oh S.J. et al. (2017) Emulating host-microbiome ecosystem of human gastrointestinal tract in vitro. Stem Cell Rev. Rep. 13, 321–334 10.1007/s12015-017-9739-z [DOI] [PubMed] [Google Scholar]
- 4.Shreiner A.B., Kao J.Y. and Young V.B. (2015) The gut microbiome in health and in disease. Curr. Opin. Gastroenterol. 31, 69–75 10.1097/MOG.0000000000000139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Round J.L. and Mazmanian S.K. (2009) The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9, 313–323 10.1038/nri2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skardal A., Shupe T. and Atala A. (2016) Organoid-on-a-chip and body-on-a-chip systems for drug screening and disease modeling. Drug Discov. Today 21, 1399–1411 10.1016/j.drudis.2016.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esch E.W., Bahinski A. and Huh D. (2015) Organs-on-chips at the frontiers of drug discovery. Nat. Rev. Drug Discov. 14, 248–260 10.1038/nrd4539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Golovko D., Kedrin D., Yilmaz Ö.H. and Roper J. (2015) Colorectal cancer models for novel drug discovery. Expert Opin. Drug Discov. 10, 1217–1229 10.1517/17460441.2015.1079618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vargo-Gogola T. and Rosen J.M. (2007) Modelling breast cancer: one size does not fit all. Nat. Rev. Cancer 7, 659–672 10.1038/nrc2193 [DOI] [PubMed] [Google Scholar]
- 10.van Staveren W.C.G., Solís D.Y.W., Hébrant A., Detours V., Dumont J.E. and Maenhaut C. (2009) Human cancer cell lines: experimental models for cancer cells in situ? For cancer stem cells? Biochim. Biophys. Acta, Rev. Cancer 1795, 92–103 10.1016/j.bbcan.2008.12.004 [DOI] [PubMed] [Google Scholar]
- 11.Takebe T., Sekine K., Enomura M., Koike H., Kimura M., Ogaeri T. et al. (2013) Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 499, 481–484 10.1038/nature12271 [DOI] [PubMed] [Google Scholar]
- 12.Kim H.J. and Ingber D.E. (2013) Gut-on-a-chip microenvironment induces human intestinal cells to undergo villus differentiation. Integr. Biol. 5, 1130–1140 10.1039/c3ib40126j [DOI] [PubMed] [Google Scholar]
- 13.Artursson P., Palm K. and Luthman K. (2001) Caco-2 monolayers in experimental and theoretical predictions of drug transport1PII of original article: S0169-409X(96)00415-2. The article was originally published in advanced drug delivery reviews 22 (1996) 67–84.1. Adv. Drug Deliv. Rev. 46, 27–43 10.1016/S0169-409X(00)00128-9 [DOI] [PubMed] [Google Scholar]
- 14.Kim H.J., Li H., Collins J.J. and Ingber D.E. (2016) Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc. Natl Acad. Sci. U.S.A. 113, E7–E15 10.1073/pnas.1522193112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kunz-Schughart L.A., Freyer J.P., Hofstaedter F. and Ebner R. (2004) The use of 3-D cultures for high-throughput screening: the multicellular spheroid model. J. Biomol. Screen. 9, 273–285 10.1177/1087057104265040 [DOI] [PubMed] [Google Scholar]
- 16.Ho W.J., Pham E.A., Kim J.W., Ng C.W., Kim J.H., Kamei D.T. et al. (2010) Incorporation of multicellular spheroids into 3-D polymeric scaffolds provides an improved tumor model for screening anticancer drugs. Cancer Sci. 101, 2637–2643 10.1111/j.1349-7006.2010.01723.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drewitz M., Helbling M., Fried N., Bieri M., Moritz W., Lichtenberg J. et al. (2011) Towards automated production and drug sensitivity testing using scaffold-free spherical tumor microtissues. Biotechnol. J. 6, 1488–1496 10.1002/biot.201100290 [DOI] [PubMed] [Google Scholar]
- 18.Chinwalla A.T., Cook L.L., Delehaunty K.D., Fewell G.A., Fulton L.A., Fulton R.S. et al. (2002) Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562 10.1038/nature01262 [DOI] [PubMed] [Google Scholar]
- 19.Mestas J. and Hughes C.C.W. (2004) Of mice and not men: differences between mouse and human immunology. J. Immunol. 172, 2731–2738 10.4049/jimmunol.172.5.2731 [DOI] [PubMed] [Google Scholar]
- 20.Corpet D.E. and Pierre F. (2005) How good are rodent models of carcinogenesis in predicting efficacy in humans? A systematic review and meta-analysis of colon chemoprevention in rats, mice and men. Eur. J. Cancer 41, 1911–1922 10.1016/j.ejca.2005.06.006 [DOI] [PubMed] [Google Scholar]
- 21.Shay T., Jojic V., Zuk O., Rothamel K., Puyraimond-Zemmour D., Feng T. et al. (2013) Conservation and divergence in the transcriptional programs of the human and mouse immune systems. Proc. Natl Acad. Sci. U.S.A. 110, 2946–2951 10.1073/pnas.1222738110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frese K.K. and Tuveson D.A. (2007) Maximizing mouse cancer models. Nat. Rev. Cancer 7, 654–658 10.1038/nrc2192 [DOI] [PubMed] [Google Scholar]
- 23.Nguyen T.L.A., Vieira-Silva S., Liston A. and Raes J. (2015) How informative is the mouse for human gut microbiota research? Dis. Models Mech. 8, 1–16 10.1242/dmm.017400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.von Martels J.Z.H., Sadaghian Sadabad M., Bourgonje A.R., Blokzijl T., Dijkstra G., Faber K.N. et al. (2017) The role of gut microbiota in health and disease: in vitro modeling of host-microbe interactions at the aerobe-anaerobe interphase of the human gut. Anaerobe 44, 3–12 10.1016/j.anaerobe.2017.01.001 [DOI] [PubMed] [Google Scholar]
- 25.Rinke C., Schwientek P., Sczyrba A., Ivanova N.N., Anderson I.J., Cheng J.-F. et al. (2013) Insights into the phylogeny and coding potential of microbial dark matter. Nature 499, 431–437 10.1038/nature12352 [DOI] [PubMed] [Google Scholar]
- 26.Wilson I.D. and Nicholson J.K. (2017) Gut microbiome interactions with drug metabolism, efficacy, and toxicity. Transl. Res. 179, 204–222 10.1016/j.trsl.2016.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reynolds B.A. and Weiss S. (1992) Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science 255, 1707–1710 10.1126/science.1553558 [DOI] [PubMed] [Google Scholar]
- 28.Dontu G., Abdallah W.M., Foley J.M., Jackson K.W., Clarke M.F., Kawamura M.J. et al. (2003) In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 17, 1253–1270 10.1101/gad.1061803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Al-Hajj M., Wicha M.S., Benito-Hernandez A., Morrison S.J. and Clarke M.F. (2003) Prospective identification of tumorigenic breast cancer cells. Proc. Natl Acad. Sci. U.S.A. 100, 3983–3988 10.1073/pnas.0530291100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ponti D., Costa A., Zaffaroni N., Pratesi G., Petrangolini G., Coradini D. et al. (2005) Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 65, 5506–5511 10.1158/0008-5472.CAN-05-0626 [DOI] [PubMed] [Google Scholar]
- 31.Haller D., Bode C., Hammes W.P., Pfeifer A.M.A., Schiffrin E.J. and Blum S. (2000) Non-pathogenic bacteria elicit a differential cytokine response by intestinal epithelial cell/leucocyte co-cultures. Gut 47, 79–87 10.1136/gut.47.1.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gratz S., Wu Q.K., El-Nezami H., Juvonen R.O., Mykkänen H. and Turner P.C. (2007) Lactobacillus rhamnosus strain GG reduces aflatoxin B1 transport, metabolism, and toxicity in Caco-2 cells. Appl. Environ. Microbiol. 73, 3958–3964 10.1128/AEM.02944-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson-Henry K.C., Donato K.A., Shen-Tu G., Gordanpour M. and Sherman P.M. (2008) Lactobacillus rhamnosus strain GG prevents enterohemorrhagic Escherichia coli O157:H7-Induced changes in epithelial barrier function. Infect. Immun. 76, 1340–1348 10.1128/IAI.00778-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roberts C.L., Keita Å.V, Duncan S.H., O'Kennedy N., Söderholm J.D., Rhodes J.M. et al. (2010) Translocation of Crohn's disease Escherichia coli across M-cells: contrasting effects of soluble plant fibres and emulsifiers. Gut 59, 1331–1339 10.1136/gut.2009.195370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madsen K., Cornish A., Soper P., McKaigney C., Jijon H., Yachimec C. et al. (2001) Probiotic bacteria enhance murine and human intestinal epithelial barrier function. Gastroenterology 121, 580–591 10.1053/gast.2001.27224 [DOI] [PubMed] [Google Scholar]
- 36.Mounier J., Vasselon T., Hellio R., Lesourd M. and Sansonetti P.J. (1992) Shigella flexneri enters human colonic Caco-2 epithelial cells through the basolateral pole. Infect. Immun. 60, 237–248 PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roselli M., Finamore A., Britti M.S., Konstantinov S.R., Smidt H., de Vos W.M. et al. (2007) The novel porcine Lactobacillus sobrius strain protects intestinal cells from enterotoxigenic Escherichia coli K88 infection and prevents membrane barrier damage. J. Nutr. 137, 2709–2716 PMID: [DOI] [PubMed] [Google Scholar]
- 38.Hu L., Tall B.D., Curtis S.K. and Kopecko D.J. (2008) Enhanced microscopic definition of Campylobacter jejuni 81-176 adherence to, invasion of, translocation across, and exocytosis from polarized human intestinal Caco-2 cells. Infect. Immun. 76, 5294–5304 10.1128/IAI.01408-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Deun K., Pasmans F., Van Immerseel F., Ducatelle R. and Haesebrouck F. (2008) Butyrate protects Caco-2 cells from Campylobacter jejuni invasion and translocation. Br. J. Nutr. 100, 480–484 10.1017/S0007114508921693 [DOI] [PubMed] [Google Scholar]
- 40.Rescigno M., Urbano M., Valzasina B., Francolini M., Rotta G., Bonasio R. et al. (2001) Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat. Immunol. 2, 361–367 10.1038/86373 [DOI] [PubMed] [Google Scholar]
- 41.Roselli M., Finamore A., Britti M.S. and Mengheri E. (2007) Probiotic bacteria Bifidobacterium animalis MB5 and Lactobacillus rhamnosus GG protect intestinal Caco-2 cells from the inflammation-associated response induced by enterotoxigenic Escherichia coli K88. Br. J. Nutr. 95, 1177–1184 10.1079/BJN20051681 [DOI] [PubMed] [Google Scholar]
- 42.Noel G., Baetz N.W., Staab J.F., Donowitz M., Kovbasnjuk O., Pasetti M.F. et al. (2017) A primary human macrophage-enteroid co-culture model to investigate mucosal gut physiology and host-pathogen interactions. Sci. Rep. 7, 45270 10.1038/srep45270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim H.J., Huh D., Hamilton G. and Ingber D.E. (2012) Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip 12, 2165–2174 10.1039/c2lc40074j [DOI] [PubMed] [Google Scholar]
- 44.Sato T., Vries R.G., Snippert H.J., van de Wetering M., Barker N., Stange D.E. et al. (2009) Single Lgr5 stem cells build crypt–villus structures in vitro without a mesenchymal niche. Nature 459, 262–266 10.1038/nature07935 [DOI] [PubMed] [Google Scholar]
- 45.Ootani A., Li X., Sangiorgi E., Ho Q.T., Ueno H., Toda S. et al. (2009) Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat. Med. 15, 701–706 10.1038/nm.1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sato T., Stange D.E., Ferrante M., Vries R.G.J., van Es J.H., van den Brink S. et al. (2011) Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology 141, 1762–1772 10.1053/j.gastro.2011.07.050 [DOI] [PubMed] [Google Scholar]
- 47.Parris A. and Williams M.R. (2015) A human colonic crypt culture system to study regulation of stem cell-driven tissue renewal and physiological function In Stem Cell Renewal and Cell-Cell Communication: Methods and Protocols (Turksen K., ed.), pp. 141–161, Springer New York, New York, NY: [DOI] [PubMed] [Google Scholar]
- 48.Mahe M.M., Sundaram N., Watson C.L., Shroyer N.F. and Helmrath M.A. (2015) Establishment of human epithelial enteroids and colonoids from whole tissue and biopsy. J. Vis. Exp. 97, 52483 10.3791/52483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hollins A.J. and Parry L. (2016) Long-term culture of intestinal cell progenitors: an overview of their development, application, and associated technologies. Curr. Pathobiol. Rep. 4, 209–219 10.1007/s40139-016-0119-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Date S. and Sato T. (2015) Mini-gut organoids: reconstitution of the stem cell niche. Ann. Rev. Cell Dev. Biol. 31, 269–289 10.1146/annurev-cellbio-100814-125218 [DOI] [PubMed] [Google Scholar]
- 51.Clevers H. (2016) Modeling development and disease with organoids. Cell 165, 1586–1597 10.1016/j.cell.2016.05.082 [DOI] [PubMed] [Google Scholar]
- 52.Barker N., Huch M., Kujala P., van de Wetering M., Snippert H.J., van Es J.H. et al. (2010) Lgr5+ve stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell 6, 25–36 10.1016/j.stem.2009.11.013 [DOI] [PubMed] [Google Scholar]
- 53.Stange D.E., Koo B.-K., Huch M., Sibbel G., Basak O., Lyubimova A. et al. (2013) Differentiated Troy+ chief cells act as reserve stem cells to generate all lineages of the stomach epithelium. Cell 155, 357–368 10.1016/j.cell.2013.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bartfeld S., Bayram T., van de Wetering M., Huch M., Begthel H., Kujala P. et al. (2015) In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology 148, 126–136.e6 10.1053/j.gastro.2014.09.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huch M., Bonfanti P., Boj S.F., Sato T., Loomans C.J.M., van de Wetering M. et al. (2013) Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J. 32, 2708–2721 10.1038/emboj.2013.204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Greggio C., De Franceschi F., Figueiredo-Larsen M., Gobaa S., Ranga A., Semb H. et al. (2013) Artificial three-dimensional niches deconstruct pancreas development in vitro. Development 140, 4452–4462 10.1242/dev.096628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boj S.F., Hwang C.-I., Baker L.A., Chio I.I.C., Engle D.D., Corbo V. et al. (2015) Organoid models of human and mouse ductal pancreatic cancer. Cell 160, 324–338 10.1016/j.cell.2014.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huch M., Gehart H., van Boxtel R., Hamer K., Blokzijl F., Verstegen Monique M. et al. (2015) Long-Term culture of genome-stable bipotent stem cells from adult human liver. Cell 160, 299–312 10.1016/j.cell.2014.11.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Karthaus W.R., Iaquinta P.J., Drost J., Gracanin A., van Boxtel R., Wongvipat J. et al. (2014) Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell 159, 163–175 10.1016/j.cell.2014.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chua C.W., Shibata M., Lei M., Toivanen R., Barlow L.J., Bergren S.K. et al. (2014) Single luminal epithelial progenitors can generate prostate organoids in culture. Nat. Cell Biol. 16, 951––961 10.1038/ncb3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.DeWard A.D., Cramer J. and Lagasse E. (2014) Cellular heterogeneity in the mouse esophagus implicates the presence of a non-quiescent epithelial stem cell population. Cell Rep. 9, 701–711 10.1016/j.celrep.2014.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lugli N., Kamileri I., Keogh A., Malinka T., Sarris M.E., Talianidis I. et al. (2016) R-spondin 1 and noggin facilitate expansion of resident stem cells from non-damaged gallbladders. EMBO Rep. 17, 769–779 10.15252/embr.201642169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Forbester J.L., Goulding D., Vallier L., Hannan N., Hale C., Pickard D. et al. (2015) Interaction of Salmonella enterica Serovar typhimurium with intestinal organoids derived from human induced pluripotent stem cells. Infect. Immun. 83, 2926–2934 10.1128/IAI.00161-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ren W., Lewandowski B.C., Watson J., Aihara E., Iwatsuki K., Bachmanov A.A. et al. (2014) Single Lgr5- or Lgr6-expressing taste stem/progenitor cells generate taste bud cells ex vivo. Proc. Natl Acad. Sci. U.S.A. 111, 16401–16406 10.1073/pnas.1409064111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Matano M., Date S., Shimokawa M., Takano A., Fujii M., Ohta Y. et al. (2015) Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat. Med. 21, 256–262 10.1038/nm.3802 [DOI] [PubMed] [Google Scholar]
- 66.Roper J., Tammela T., Cetinbas N.M., Akkad A., Roghanian A., Rickelt S. et al. (2017) In vivo genome editing and organoid transplantation models of colorectal cancer and metastasis. Nat. Biotechnol. 35, 569–576 10.1038/nbt.3836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.de Sousa e Melo F., Kurtova A.V., Harnoss J.M., Kljavin N., Hoeck J.D., Hung J. et al. (2017) A distinct role for Lgr5+ stem cells in primary and metastatic colon cancer. Nature 543, 676–680 10.1038/nature21713 [DOI] [PubMed] [Google Scholar]
- 68.McCracken K.W., Catá E.M., Crawford C.M., Sinagoga K.L., Schumacher M., Rockich B.E. et al. (2014) Modeling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 516, 400–404 10.1038/nature13863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schumacher M.A., Feng R., Aihara E., Engevik A.C., Montrose M.H., Ottemann K.M. et al. (2015) Helicobacter pylori-induced sonic hedgehog expression is regulated by NFκB pathway activation: the use of a novel in vitro model to study epithelial response to infection. Helicobacter 20, 19–28 10.1111/hel.12152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Salama N.R., Hartung M.L. and Müller A. (2013) Life in the human stomach: persistence strategies of the bacterial pathogen Helicobacter pylori. Nat. Rev. Microbiol. 11, 385–399 10.1038/nrmicro3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peek R.M. Jr, Moss S.F., Wang S., Holt P.R., Tham K.T., Pérez-Pérez G.I. et al. (1997) Helicobacter pylori cagA+ strains and dissociation of gastric epithelial cell proliferation from apoptosis. J. Natl Cancer Inst. 89, 863–868 10.1093/jnci/89.12.863 [DOI] [PubMed] [Google Scholar]
- 72.Churin Y., Al-Ghoul L., Kepp O., Meyer T.F., Birchmeier W. and Naumann M. (2003) Helicobacter pylori CagA protein targets the c-Met receptor and enhances the motogenic response. J. Cell Biol. 161, 249–255 10.1083/jcb.200208039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Leslie J.L., Huang S., Opp J.S., Nagy M.S., Kobayashi M., Young V.B. et al. (2015) Persistence and toxin production by Clostridium difficile within human intestinal organoids result in disruption of epithelial paracellular barrier function. Infect. Immun. 83, 138–145 10.1128/IAI.02561-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Engevik M.A., Engevik K.A., Yacyshyn M.B., Wang J., Hassett D.J., Darien B. et al. (2015) Human Clostridium difficile infection: inhibition of NHE3 and microbiota profile. Am. J. Physiol. Gastrointest. Liver Physiol. 308, G497–G509 10.1152/ajpgi.00090.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Finkbeiner S.R., Zeng X.-L., Utama B., Atmar R.L., Shroyer N.F. and Estes M.K. (2012) Stem cell-derived human intestinal organoids as an infection model for rotaviruses. mBio 3, e00159-12 10.1128/mBio.00159-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.VanDussen K.L., Marinshaw J.M., Shaikh N., Miyoshi H., Moon C., Tarr P.I. et al. (2015) Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut 64, 911–920 10.1136/gutjnl-2013-306651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.In J., Foulke-Abel J., Zachos N.C., Hansen A.-M., Kaper J.B., Bernstein H.D. et al. (2016) Enterohemorrhagic Escherichia coli reduces mucus and intermicrovillar bridges in human stem cell-derived colonoids. Cell. Mol. Gastroenterol. Hepatol. 2, 48–62.e3 10.1016/j.jcmgh.2015.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ettayebi K., Crawford S.E., Murakami K., Broughman J.R., Karandikar U., Tenge V.R. et al. (2016) Replication of human noroviruses in stem cell-derived human enteroids. Science 353, 1387–1393 10.1126/science.aaf5211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fatehullah A., Tan S.H. and Barker N. (2016) Organoids as an in vitro model of human development and disease. Nat. Cell Biol. 18, 246–254 10.1038/ncb3312 [DOI] [PubMed] [Google Scholar]
- 80.Pocock K., Delon L., Bala V., Rao S., Priest C., Prestidge C. et al. (2017) Intestine-on-a-chip microfluidic model for efficient in vitro screening of oral chemotherapeutic uptake. ACS Biomater. Sci. Eng. 3, 951–959 10.1021/acsbiomaterials.7b00023 [DOI] [PubMed] [Google Scholar]
- 81.Smith A.M., Prabhakarpandian B. and Pant K. (2014) Generation of shear adhesion map using SynVivo synthetic microvascular networks. J. Vis. Exp. 87, 51025 10.3791/51025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Terrell-Hall T.B., Ammer A.G., Griffith J.I.G. and Lockman P.R. (2017) Permeability across a novel microfluidic blood-tumor barrier model. Fluids Barriers CNS 14, 3 10.1186/s12987-017-0050-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Deosarkar S.P., Prabhakarpandian B., Wang B., Sheffield J.B., Krynska B. and Kiani M.F. (2015) A novel dynamic neonatal blood-brain barrier on a chip. PLoS ONE 10, e0142725 10.1371/journal.pone.0142725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Prabhakarpandian B., Shen M.-C., Nichols J.B., Mills I.R., Sidoryk-Wegrzynowicz M., Aschner M. et al. (2013) SyM-BBB: a microfluidic blood brain barrier model. Lab Chip 13, 1093–1101 10.1039/c2lc41208j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Prabhakarpandian B., Shen M.-C., Nichols J.B., Garson C.J., Mills I.R., Matar M.M. et al. (2015) Synthetic tumor networks for screening drug delivery systems. J. Control. Release 201, 49–55 10.1016/j.jconrel.2015.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lamberti G., Prabhakarpandian B., Garson C., Smith A., Pant K., Wang B. et al. (2014) Bioinspired microfluidic assay for in vitro modeling of leukocyte–endothelium interactions. Anal. Chem. 86, 8344–8351 10.1021/ac5018716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Soroush F., Zhang T., King D.J., Tang Y., Deosarkar S., Prabhakarpandian B. et al. (2016) A novel microfluidic assay reveals a key role for protein kinase C δ in regulating human neutrophil-endothelium interaction. J. Leukoc. Biol. 100, 1027–1035 10.1189/jlb.3MA0216-087R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.van de Bovenkamp M., Groothuis G.M.M., Draaisma A.L., Merema M.T., Bezuijen J.I., van Gils M.J. et al. (2005) Precision-cut liver slices as a new model to study toxicity-induced hepatic stellate cell activation in a physiologic milieu. Toxicol. Sci. 85, 632–638 10.1093/toxsci/kfi127 [DOI] [PubMed] [Google Scholar]
- 89.Martignoni M., Groothuis G. and de Kanter R. (2006) Comparison of mouse and rat cytochrome P450-mediated metabolism in liver and intestine. Drug Metab. Dispos. 34, 1047–1054 10.1124/dmd.105.009035 [DOI] [PubMed] [Google Scholar]
- 90.Groothuis G.M.M. and de Graaf I.A.M. (2013) Precision-cut intestinal slices as in vitro tool for studies on drug metabolism. Curr. Drug Metab. 14, 112–119 10.2174/138920013804545197 [DOI] [PubMed] [Google Scholar]
- 91.Li M., de Graaf I.A.M. and Groothuis G.M.M. (2016) Precision-cut intestinal slices: alternative model for drug transport, metabolism, and toxicology research. Expert Opin. Drug Metab. Toxicol. 12, 175–190 10.1517/17425255.2016.1125882 [DOI] [PubMed] [Google Scholar]
- 92.de Graaf I.A.M., Olinga P., de Jager M.H., Merema M.T., de Kanter R., van de Kerkhof E.G. et al. (2010) Preparation and incubation of precision-cut liver and intestinal slices for application in drug metabolism and toxicity studies. Nat. Protoc. 5, 1540–1551 10.1038/nprot.2010.111 [DOI] [PubMed] [Google Scholar]
- 93.Schwerdtfeger L.A., Ryan E.P. and Tobet S.A. (2016) An organotypic slice model for ex vivo study of neural, immune, and microbial interactions of mouse intestine. Am. J. Physiol. Gastrointest. Liver Physiol. 310, G240–G248 10.1152/ajpgi.00299.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Maier E., Anderson R.C. and Roy N.C. (2015) Understanding how commensal obligate anaerobic bacteria regulate immune functions in the large intestine. Nutrients 7, 45–73 10.3390/nu7010045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Clarke L.L. (2009) A guide to Ussing chamber studies of mouse intestine. Am. J. Physiol. Gastrointest. Liver Physiol. 296, G1151–G1166 10.1152/ajpgi.90649.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Forner K., Roos C., Dahlgren D., Kesisoglou F., Konerding M.A., Mazur J. et al. (2017) Optimization of the Ussing chamber setup with excised rat intestinal segments for dissolution/permeation experiments of poorly soluble drugs. Drug Dev. Ind. Pharm. 43, 338–346 10.1080/03639045.2016.1251449 [DOI] [PubMed] [Google Scholar]
- 97.Rozehnal V., Nakai D., Hoepner U., Fischer T., Kamiyama E., Takahashi M. et al. (2012) Human small intestinal and colonic tissue mounted in the Ussing chamber as a tool for characterizing the intestinal absorption of drugs. Eur. J. Pharm. Sci. 46, 367–373 10.1016/j.ejps.2012.02.025 [DOI] [PubMed] [Google Scholar]
- 98.Gustafsson J.K., Navabi N., Rodriguez-Piñeiro A.M., Alomran A.H.A., Premaratne P., Fernandez H.R. et al. (2013) Dynamic changes in mucus thickness and ion secretion during Citrobacter rodentium infection and clearance. PLoS ONE 8, e84430 10.1371/journal.pone.0084430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Llopis M., Antolin M., Carol M., Borruel N., Casellas F., Martinez C. et al. (2009) Lactobacillus casei downregulates commensals’ inflammatory signals in Crohn's disease mucosa. Inflamm. Bowel Dis. 15, 275–283 10.1002/ibd.20736 [DOI] [PubMed] [Google Scholar]
- 100.Parassol N., Freitas M., Thoreux K., Dalmasso G., Bourdet-Sicard R. and Rampal P. (2005) Lactobacillus casei DN-114 001 inhibits the increase in paracellular permeability of enteropathogenic Escherichia coli-infected T84 cells. Res. Microbiol. 156, 256–262 10.1016/j.resmic.2004.09.013 [DOI] [PubMed] [Google Scholar]
- 101.Jafari N.V., Kuehne S.A., Minton N.P., Allan E. and Bajaj-Elliott M. (2016) Clostridium difficile-mediated effects on human intestinal epithelia: modelling host-pathogen interactions in a vertical diffusion chamber. Anaerobe 37, 96–102 10.1016/j.anaerobe.2015.12.007 [DOI] [PubMed] [Google Scholar]
- 102.Anabazhagan A.N., Chatterjee I., Priyamvada S., Kumar A., Tyagi S., Saksena S. et al. (2017) Methods to study epithelial transport protein function and expression in native intestine and Caco-2 cells grown in 3D. J. Vis. Exp. 121, e55304 10.3791/55304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jespersen B., Tykocki N.R., Watts S.W. and Cobbett P.J. (2015) Measurement of smooth muscle function in the isolated tissue bath-applications to pharmacology research. J. Vis. Exp. 95, 52324 10.3791/52324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fry C.H. (2004) Experimental models to study the physiology, pathophysiology, and pharmacology of the lower urinary tract. J. Pharmacol. Toxicol. Methods 49, 201–210 10.1016/j.vascn.2004.03.002 [DOI] [PubMed] [Google Scholar]
- 105.Chan W.W. and Mashimo H. (2013) Lubiprostone increases small intestinal smooth muscle contractions through a prostaglandin E receptor 1 (EP1)-mediated pathway. J. Neurogastroenterol. Motil. 19, 312–318 10.5056/jnm.2013.19.3.312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Villa L., Buono R., Fossati N., Rigatti P., Montorsi F., Benigni F. et al. (2013) Effects by silodosin on the partially obstructed rat ureter in vivo and on human and rat isolated ureters. Br. J. Pharmacol. 169, 230–238 10.1111/bph.12123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bär F., Von Koschitzky H., Roblick U., Bruch H.P., Schulze L., Sonnenborn U. et al. (2009) Cell-free supernatants of Escherichia coli Nissle 1917 modulate human colonic motility: evidence from an in vitro organ bath study. Neurogastroenterol. Motil. 21, 559-e17 10.1111/j.1365-2982.2008.01258.x [DOI] [PubMed] [Google Scholar]
- 108.Yum K., Hong S.G., Healy K.E. and Lee L.P. (2014) Physiologically relevant organs on chips. Biotechnol. J. 9, 16–27 10.1002/biot.201300187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hu Y., Bian S., Grotberg J., Filoche M., White J., Takayama S. et al. (2015) A microfluidic model to study fluid dynamics of mucus plug rupture in small lung airways. Biomicrofluidics 9, 044119 10.1063/1.4928766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.An F., Qu Y., Liu X., Zhong R. and Luo Y. (2015) Organ-on-a-chip: new platform for biological analysis. Anal. Chem. Insights 10, 39–45 10.4137/ACI.S28905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Huh D., Matthews B.D., Mammoto A., Montoya-Zavala M., Hsin H.Y. and Ingber D.E. (2010) Reconstituting organ-level lung functions on a chip. Science 328, 1662–1668 10.1126/science.1188302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Huh D., Leslie D.C., Matthews B.D., Fraser J.P., Jurek S., Hamilton G.A. et al. (2012) A human disease model of drug toxicity-Induced pulmonary edema in a lung-on-a-chip microdevice. Sci. Transl. Med. 4, 159ra147 10.1126/scitranslmed.3004249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nguyen M.-D., Tinney J.P., Ye F., Elnakib A.A., Yuan F., El-Baz A. et al. (2015) Effects of physiologic mechanical stimulation on embryonic chick cardiomyocytes using a microfluidic cardiac cell culture model. Anal. Chem. 87, 2107–2113 10.1021/ac503716z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chen H., Cornwell J., Zhang H., Lim T., Resurreccion R., Port T. et al. (2013) Cardiac-like flow generator for long-term imaging of endothelial cell responses to circulatory pulsatile flow at microscale. Lab Chip 13, 2999–3007 10.1039/c3lc50123j [DOI] [PubMed] [Google Scholar]
- 115.Bhise N.S., Manoharan V., Massa S., Tamayol A., Ghaderi M., Miscuglio M. et al. (2016) A liver-on-a-chip platform with bioprinted hepatic spheroids. Biofabrication 8, 014101 10.1088/1758-5090/8/1/014101 [DOI] [PubMed] [Google Scholar]
- 116.Shamloo A., Heibatollahi M. and Mofrad M.R.K. (2015) Directional migration and differentiation of neural stem cells within three-dimensional microenvironments. Integr. Biol. 7, 335–344 10.1039/C4IB00144C [DOI] [PubMed] [Google Scholar]
- 117.Jang K. and Suh K. (2010) A multi-layer microfluidic device for efficient culture and analysis of renal tubular cells. Lab Chip 10, 36–42 10.1039/b907515a [DOI] [PubMed] [Google Scholar]
- 118.Jang K.-J., Mehr A.P., Hamilton G.A., McPartlin L.A., Chung S., Suh K.-Y. et al. (2013) Human kidney proximal tubule-on-a-chip for drug transport and nephrotoxicity assessment. Integr. Biol. 5, 1119–1129 10.1039/c3ib40049b [DOI] [PubMed] [Google Scholar]
- 119.Kimura H., Yamamoto T., Sakai H., Sakai Y. and Fujii T. (2008) An integrated microfluidic system for long-term perfusion culture and on-line monitoring of intestinal tissue models. Lab Chip 8, 741–746 10.1039/b717091b [DOI] [PubMed] [Google Scholar]
- 120.Tsai M., Kita A., Leach J., Rounsevell R., Huang J.N., Moake J. et al. (2012) In vitro modeling of the microvascular occlusion and thrombosis that occur in hematologic diseases using microfluidic technology. J. Clin. Invest. 122, 408–418 10.1172/JCI58753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Theberge A.B., Yu J., Young E.W.K., Ricke W.A., Bushman W. and Beebe D.J. (2015) Microfluidic multiculture assay to analyze biomolecular signaling in angiogenesis. Anal. Chem. 87, 3239–3246 10.1021/ac503700f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Walsh C.L., Babin B.M., Kasinskas R.W., Foster J.A., McGarry M.J. and Forbes N.S. (2009) A multipurpose microfluidic device designed to mimic microenvironment gradients and develop targeted cancer therapeutics. Lab Chip 9, 545–554 10.1039/b810571e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Torisawa Y.-s., Spina C.S., Mammoto T., Mammoto A., Weaver J.C., Tat T. et al. (2014) Bone marrow–on–a–chip replicates hematopoietic niche physiology in vitro. Nat. Methods 11, 663–669 10.1038/nmeth.2938 [DOI] [PubMed] [Google Scholar]
- 124.Sung J.H., Kam C. and Shuler M. (2010) A microfluidic device for a pharmacokinetic–pharmacodynamic (PK–PD) model on a chip. Lab Chip 10, 446 10.1039/b917763a [DOI] [PubMed] [Google Scholar]
- 125.Maschmeyer I., Lorenz A., Ramme A., Hasenberg T., Schimek K., Hübner J. et al. (2015) A microfluidic four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents. Toxicol. Lett. 238, 176 10.1016/j.toxlet.2015.08.512 [DOI] [PubMed] [Google Scholar]
- 126.Vernetti L., Gough A., Baetz N., Blutt S., Broughman J.R., Brown J.A. et al. (2017) Functional coupling of human microphysiology systems: intestine, liver, kidney proximal tubule, blood-brain barrier and skeletal muscle. Sci. Rep. 7, 42296 10.1038/srep42296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Imura Y., Asano Y., Sato K. and Yoshimura E. (2009) A microfluidic system to evaluate intestinal absorption. Anal. Sci. 25, 1403–1407 10.2116/analsci.25.1403 [DOI] [PubMed] [Google Scholar]
- 128.Dai C., Zhao D.-H. and Jiang M. (2012) VSL#3 probiotics regulate the intestinal epithelial barrier in vivo and in vitro via the p38 and ERK signaling pathways. Int. J. Mol. Med. 29, 202–208 10.3892/ijmm.2011.839 [DOI] [PubMed] [Google Scholar]
- 129.Chapman T.M., Plosker G.L. and Figgitt D.P. (2007) Spotlight on VSL#3 probiotic mixture in chronic inflammatory bowel diseases. BioDrugs 21, 61–63 10.2165/00063030-200721010-00007 [DOI] [PubMed] [Google Scholar]
- 130.Sbrana T. and Ahluwalia A. (2012) Engineering quasi-vivo in vitro organ models. Adv. Exp. Med. Biol. 745, 138–153 10.1007/978-1-4614-3055-1_9 [DOI] [PubMed] [Google Scholar]
- 131.Lindstedt S.L. and Schaeffer P.J. (2002) Use of allometry in predicting anatomical and physiological parameters of mammals. Lab. Anim. 36, 1–19 10.1258/0023677021911731 [DOI] [PubMed] [Google Scholar]
- 132.Ahluwalia A. (2017) Allometric scaling in-vitro. Sci. Rep. 7, 42113 10.1038/srep42113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Nithiananthan S., Crawford A., Knock J.C., Lambert D.W. and Whawell S.A. (2017) Physiological fluid flow moderates fibroblast responses to TGF-β1. J. Cell. Biochem. 118, 878–890 10.1002/jcb.25767 [DOI] [PubMed] [Google Scholar]
- 134.Shah P., Fritz J.V., Glaab E., Desai M.S., Greenhalgh K., Frachet A. et al. (2016) A microfluidics-based in vitro model of the gastrointestinal human-microbe interface. Nat. Commun. 7, 11535 10.1038/ncomms11535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Eain M.M.G., Baginska J., Greenhalgh K., Fritz J.V., Zenhausern F. and Wilmes P. (2017) Engineering solutions for representative models of the gastrointestinal human-microbe interface. Engineering 3, 60–65 10.1016/J.ENG.2017.01.011 [DOI] [Google Scholar]
- 136.Donaldson G.P., Lee S.M. and Mazmanian S.K. (2016) Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 14, 20–32 10.1038/nrmicro3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Atuma C., Strugala V., Allen A. and Holm L. (2001) The adherent gastrointestinal mucus gel layer: thickness and physical state in vivo. Am. J. Physiol. Gastrointest. Liver Physiol. 280, G922–G929 PMID: [DOI] [PubMed] [Google Scholar]
- 138.van der Meer A.D. and van den Berg A. (2012) Organs-on-chips: breaking the in vitro impasse. Integr. Biol. 4, 461–470 10.1039/c2ib00176d [DOI] [PubMed] [Google Scholar]
- 139.Berthier E., Young E.W.K. and Beebe D. (2012) Engineers are from PDMS-land, biologists are from Polystyrenia. Lab Chip 12, 1224–1237 10.1039/c2lc20982a [DOI] [PubMed] [Google Scholar]
- 140.Wong I. and Ho C.-M. (2009) Surface molecular property modifications for poly(dimethylsiloxane) (PDMS) based microfluidic devices. Microfluidics Nanofluidics 7, 291–306 10.1007/s10404-009-0443-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Domansky K., Leslie D.C., McKinney J., Fraser J.P., Sliz J.D., Hamkins-Indik T. et al. (2013) Clear castable polyurethane elastomer for fabrication of microfluidic devices. Lab Chip 13, 3956–3964 10.1039/c3lc50558h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.van Midwoud P.M., Janse A., Merema M.T., Groothuis G.M.M. and Verpoorte E. (2012) Comparison of biocompatibility and adsorption properties of different plastics for advanced microfluidic cell and tissue culture models. Anal. Chem. 84, 3938–3944 10.1021/ac300771z [DOI] [PubMed] [Google Scholar]
- 143.Esch M.B., Mahler G.J., Stokol T. and Shuler M.L. (2014) Body-on-a-chip simulation with gastrointestinal tract and liver tissues suggests that ingested nanoparticles have the potential to cause liver injury. Lab Chip 14, 3081–3092 10.1039/C4LC00371C [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Perestrelo A.R., Águas A.C.P., Rainer A. and Forte G. (2015) Microfluidic organ/body-on-a-chip devices at the convergence of biology and microengineering. Sensors 15, 31142–31170 10.3390/s151229848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Justus C.R., Leffler N., Ruiz-Echevarria M. and Yang L.V. (2014) In vitro cell migration and invasion assays. J. Vis. Exp. 88, 51046 10.3791/51046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Castellone R.D., Leffler N.R., Dong L. and Yang L.V. (2011) Inhibition of tumor cell migration and metastasis by the proton-sensing GPR4 receptor. Cancer Lett. 312, 197–208 10.1016/j.canlet.2011.08.013 [DOI] [PubMed] [Google Scholar]
- 147.Man S., Ubogu E.E., Williams K.A., Tucky B., Callahan M.K. and Ransohoff R.M. (2008) Human brain microvascular endothelial cells and umbilical vein endothelial cells differentially facilitate leukocyte recruitment and utilize chemokines for T cell migration. Clin. Dev. Immunol. 2008, 384982 10.1155/2008/384982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Antoni D., Burckel H., Josset E. and Noel G. (2015) Three-dimensional cell culture: a breakthrough in vivo. Int. J. Mol. Sci. 16, 5517–5527 10.3390/ijms16035517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Edmondson R., Broglie J.J., Adcock A.F. and Yang L. (2014) Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev. Technol. 12, 207–218 10.1089/adt.2014.573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Cox M.C., Reese L.M., Bickford L.R. and Verbridge S.S. (2015) Toward the broad adoption of 3D tumor models in the cancer drug pipeline. ACS Biomater. Sci. Eng. 1, 877–894 10.1021/acsbiomaterials.5b00172 [DOI] [PubMed] [Google Scholar]
- 151.Hill D.R. and Spence J.R. (2017) Gastrointestinal organoids: understanding the molecular basis of the host–microbe interface. Cell. Mol. Gastroenterol. Hepatol. 3, 138–149 10.1016/j.jcmgh.2016.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Grivel J.-C. and Margolis L. (2009) Use of human tissue explants to study human infectious agents. Nat. Protoc. 4, 256–269 10.1038/nprot.2008.245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kim S. and Takayama S. (2015) Organ-on-a-chip and the kidney. Kidney Res. Clin. Pract. 34, 165–169 10.1016/j.krcp.2015.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Aziz U.A., Geng C., Fu M., Yu X., Qin K. and Liu B. (2017) The role of microfluidics for organ on chip simulations. Bioengineering 4, E39 10.3390/bioengineering4020039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Valencia P.M., Farokhzad O.C., Karnik R. and Langer R. (2012) Microfluidic technologies for accelerating the clinical translation of nanoparticles. Nat. Nanotechnol. 7, 623–629 10.1038/nnano.2012.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Bhatia S.N. and Ingber D.E. (2014) Microfluidic organs-on-chips. Nat. Biotechnol. 32, 760–772 10.1038/nbt.2989 [DOI] [PubMed] [Google Scholar]
- 157.Pagliari S., Tirella A., Ahluwalia A., Duim S., Goumans M.-J., Aoyagi T. et al. (2014) A multistep procedure to prepare pre-vascularized cardiac tissue constructs using adult stem cells, dynamic cell cultures, and porous scaffolds. Front. Physiol. 5, 210 10.3389/fphys.2014.00210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mazzei D., Guzzardi M.A., Giusti S. and Ahluwalia A. (2010) A low shear stress modular bioreactor for connected cell culture under high flow rates. Biotechnol. Bioeng. 106, 127–137 10.1002/bit.22671 [DOI] [PubMed] [Google Scholar]