Abstract
Purpose:
Hans and coworkers previously developed an immunohistochemical algorithm with ~80% concordance with the gene expression profiling (GEP) classification of diffuse large B-cell lymphoma (DLBCL) into the germinal center B-cell–like (GCB) and activated B-cell–like (ABC) subtypes. Since then, new antibodies specific to germinal center B-cells have been developed, which might improve the performance of an immunostain algorithm.
Experimental Design:
We studied 84 cases of cyclophosphamide-doxorubicin-vincristine-prednisone (CHOP)–treated DLBCL (47 GCB, 37 ABC) with GCET1, CD10, BCL6, MUM1, FOXP1, BCL2, MTA3, and cyclin D2 immunostains, and compared different combinations of the immunostaining results with the GEP classification. A perturbation analysis was also applied to eliminate the possible effects of interobserver or intraobserver variations. A separate set of 63 DLBCL cases treated with rituximab plus CHOP (37 GCB, 26 ABC) was used to validate the new algorithm.
Results:
A new algorithm using GCET1, CD10, BCL6, MUM1, and FOXP1 was derived that closely approximated the GEP classification with 93% concordance. Perturbation analysis indicated that the algorithm was robust within the range of observer variance. The new algorithm predicted 3-year overall survival of the validation set [GCB (87%) versus ABC (44%); P < 0.001], simulating the predictive power of the GEP classification. For a group of seven primary mediastinal large B-cell lymphoma, the new algorithm is a better prognostic classifier (all “GCB”) than the Hans' algorithm (two GCB, five non-GCB).
Conclusion:
Our new algorithm is significantly more accurate than the Hans' algorithm and will facilitate risk stratification of DLBCL patients and future DLBCL research using archival materials. (Clin Cancer Res 2009;15(17):5494–502)
Gene expression profiling (GEP) studies have shown that diffuse large B-cell lymphoma (DLBCL) can be reproducibly divided into the prognostically important subtypes of germinal center B-cell–like (GCB), activated B-cell–like (ABC), and unclassified DLBCL (1-3). When treated with a regimen containing cyclophosphamide, doxorubicin, vincristine, and prednisone (CHOP) or other CHOP-like regimens (CHOP-treated), patients with GCB-DLBCL have a better survival independent of the International Prognostic Index (1, 2, 4). The prognostic value of GEP classification remains significant for DLBCL patients treated with rituximab plus CHOP or CHOP-like therapy (R-CHOP–treated; ref. 5). The GCB and ABC subtypes have different pathogenetic mechanisms that will impact the development of targeted therapies (6). Other GEP studies have also characterized primary mediastinal large B-cell lymphoma (PMBL) as distinct from the other subtypes of DLBCL (7, 8), and with a good prognosis similar to GCB-DLBCL (7).
Despite the robustness of GEP in subclassifying DLBCL, substantial time, technological expertise, and resources are required. To facilitate translational application of the GEP classification for prognostication using formalin-fixed, paraffin-embedded tissues, we have previously proposed an immunohistochemical (IHC) stain algorithm (Hans' algorithm) using three antibodies [CD10, multiple myeloma oncogene 1 (MUM1), and polyclonal B-cell lymphoma 6 (BCL6)] to classify DLBCL into GCB and non-GCB (ABC and unclassified) subtypes (9). It resulted in a concordance of ~80% when compared with the GEP classification (9), and subsequent studies have yielded conflicting results regarding its prognostic prediction (10-23). Most of these studies included patients from the pre-rituximab era (10-15, 17-23), but the controversy has extended to R-CHOP–treated patients as well (15, 19, 22, 23). However, only one of these studies had corresponding GEP data, and showed that the Hans' algorithm and GEP were concordant in ~70% of the cases (11).
We postulated that the mixed results of these subsequent studies might be due to the suboptimal identification of GCB-DLBCL cases secondary to an insufficient number of specific GC markers in that algorithm. Given the prognostic importance of GEP classification in DLBCL, and the availability of new antibodies to GC markers, such as germinal center B-cell expressed transcript 1 (GCET1; ref. 24) and metastasis-associated gene 3 (MTA3; ref. 25), we attempted to derive a better IHC stain algorithm to subclassify DLBCL using archival materials.
Materials and Methods
Case selection.
One hundred and ten cases of de novo, CHOP-treated DLBCL were obtained from the Nebraska Lymphoma Study Group, British Columbia Cancer Agency, University of Wurzburg, Norwegian Radium Hospital, and the University of Arizona. They represented a subset of the cohort described by Rosenwald et al. (2), and were classified into the GCB (47 of 110; 43%) or ABC (37 of 110; 34%) subtypes, PMBL (7 of 110; 6%) or unclassified (19 of 110; 17%), by GEP using the Lymphochip cDNA microarray (1), and the Bayesian algorithm described by Wright et al. (3). The 84 cases of GCB- and ABC-DLBCL were used as the training set for developing a new algorithm.
In addition, 68 cases of de novo, R-CHOP-treated DLBCL (37 GCB, 26 ABC, 1 PMBL, and 4 unclassified by GEP) were also obtained from the Nebraska Lymphoma Study Group, Norwegian Radium Hospital, and the Oregon Health and Science University for validation of the new algorithm. The GeneChip Human Genome U133 Plus 2.0 Array (Affymetrix, Inc.) was used for GEP study of these 68 cases, along with the Bayesian algorithm (3). The 63 cases of GCB- and ABC-DLBCL were used as the validation set.
The GEP data of both the training and validation sets were normalized before processing by the Bayesian classifier. The training set was rerun on the GeneChip Human Genome U133 Plus 2.0 Array, and the classifier distinguished ABC and GCB in the same manner. Additionally, the set of genes used to distinguish the two groups was the same on both platforms (5). This study was approved by the institutional review boards of the respective institutions, and all patients gave written informed consent.
Immunohistochemistry.
Tissue microarrays were constructed and sections were cut and immunostained as previously described (9). Monoclonal antibodies to CD20, CD3, GCET1 (24), CD10, BCL6, MUM1 (26), and Forkhead box-P1 (FOXP1; ref. 27) were applied to the training and validation sets. The training set was also stained with a monoclonal antibody to B-cell lymphoma 2 (BCL2), and polyclonal antibodies to MTA3 (25), BCL6, and cyclin D2 (Supplementary Table S1). A rabbit antimouse amplification kit (Ventana Medical Systems, Inc.) was used to enhance staining for monoclonal BCL6, GCET1, CD10, and FOXP1. An endogenous biotin-blocking kit (Ventana) was used for polyclonal BCL6 to decrease background staining. Following antigen retrieval and antibody incubation, staining was done on a Ventana Benchmark XT instrument and developed using the iView 3,3′-diaminobenzidine detection kit (Ventana) for all but one antibody (MTA3), according to manufacturer's instructions. Staining for MTA3 was done manually as previously described (25). All of the assays were validated with proper positive and negative controls.
The percentages of positive cells were scored in 10% increments for each antibody, and the highest percentage was recorded for each case. The scoring was done independently by three hematopathologists (W.W.L.C., C.P.H., and D.D.W.), who were blinded to the GEP data at the time of the scoring, and discrepancies were resolved over a multiheaded microscope.
Construction of the new algorithm.
The percentages of positive cells for the training set were tabulated on a spreadsheet alongside the corresponding GEP classification using Microsoft Office Excel 2003 (Microsoft Corporation). We first sorted the cases by the percentages of positive cells for each immunostain using the data-sort function of the program. The proportions of GCB and ABC cases positive for a particular stain at different cutoff percentages were thus obtained, and the cutoffs with the highest sensitivity or specificity for either the GCB or ABC subtype were identified. We then focused on those stains that could achieve high specificity (≥90%) for either the GCB or ABC subtype because the main aim of the study was to achieve a new IHC algorithm that replicated the GEP classification with high concordance. Because sensitivity usually fell as specificity increased, we aimed at a sensitivity of ≥50% for a particular stain while obtaining optimal specificity. Three immunostains fulfilled this criterion for either the GCB (GCET1 and CD10) or ABC subtype (MUM1). GCET1 at >60% achieved 92% specificity with 64% sensitivity for the GCB subtype. The optimal cutoffs for MUM1 (≥70%, with sensitivity and specificity for the ABC subtype of 58% and 94%, respectively) and CD10 (≥20%, with sensitivity and specificity for the GCB subtype of 60% and 95%, respectively) were similarly determined. These three immunostains were chosen for the initial decision points in the algorithm. By comparing different combinations of these three using the data-sort function, we found that use of GCET1 followed by MUM1 or CD10 could classify 42 of the 84 DLBCL (50%) with high concordance (97%) with the GEP classification. For the remaining 42 cases (9 GCB and 33 ABC), we again used the data-sort function and found that sequential use of either monoclonal or polyclonal BCL6 (≥30%) followed by FOXP1 (≥80%) classified 37 (88%) of the cases correctly. Monoclonal BCL6 was chosen because it does not require additional procedures to decrease background staining, and it was the BCL6 antibody most commonly used in recent studies (10-21, 23). Whereas changing the cutoffs of monoclonal BCL6 or FOXP1 in this algorithm led to worse overall accuracy, we found that changing the cutoffs for GCET1 and MUM1 to ≥80%, and CD10 to ≥30%, did not affect the concordance with the GEP classification. The cutoffs for GCET1, MUM1, and CD10 were thus changed to simplify the whole algorithm by reducing the number of cutoffs to two. Therefore, we finalized the new algorithm with the use of five immunostains, five decision points, and two cutoffs for the immunostains (Supplementary Fig. 1A). Some other possible algorithms had slightly better overall concordance with the GEP classification, but were more complex requiring more immunostains and decision points with different cutoffs, and were not adopted.
For the five immunostains used in the new algorithm, the cases that were scored differently for >10% positive cells among the hematopathologists ranged from 5% (CD10) to 18% (monoclonal BCL6). To further address the impact of intra-observer and inter-observer variations, a computer perturbation and simulation program was used to introduce random noise to each IHC marker on a given case. Because the vast majority of the differences among the hematopathologists were at 10% level (data not shown), we introduced random noise of + 10%, −10%, and 0% with equal probability to each marker on a given case. The computer program then randomly selected one of the three possible values for each stain on each case, and re-applied the classification algorithm on the “new” data set. The classification of each of these perturbed data set was then compared with the GEP classification as the gold standard. We repeated the perturbation procedure 10,000 times, and obtained the means, standard deviations (SD), and confidence intervals (CI) of the concordance rate of the perturbed new algorithm classification and the GEP classification in both the training and validation sets.
Statistical analysis.
The χ2 test was used to compare patient characteristics according to GEP subtypes. The Kaplan-Meier method was used to estimate the overall and event-free survival (EFS) distributions (28). Overall survival (OS) was calculated as the time from diagnosis to the date of death or last contact. EFS was calculated as the time from diagnosis to either the date of progression, relapse, death, or last contact. Patients who were alive at last contact were censored for OS analysis. Patients who were alive and had not progressed were censored for EFS analysis. Patients with unavailable disease progression data were excluded for EFS analysis. For the training set, the survival rates were 5-y estimates, whereas those for the validation set were 3-y estimates because of a shorter median follow-up (2.6 y) compared with the training set (6.4 y). The log-rank test was used to compare survival distributions of GCB and ABC cases classified by GEP or the new IHC stain algorithm. Multivariate analysis was done in the validation set using Cox regression with OS and EFS as the outcomes, and the new IHC stain algorithm and the International Prognostic Index as potential predictors in a backward selection model (P = 0.05). The statistical analysis was conducted using the SAS software (SAS Institute, Inc.).
Results
Clinical features of the training and validation sets.
The training set included 46 males (56%) and 38 females (44%) with a median age of 63 years (range, 24-84 years). With a median follow-up of 6.4 years (range, 0.8-21.8 years), 45 patients (54%) had died and 39 patients (46%) were alive at last contact. The validation set comprised 29 males (46%) and 34 females (54%) with a median age of 62 years (range, 22-92 years). With a median follow-up of 2.6 years (range, 0.2-10.3 years), 17 patients (27%) had died and 46 (73%) were alive at last contact.
For the training set, the clinical features of the patients with GEP-defined GCB and ABC subtypes were not significantly different, except that the GCB patients were more likely to have a higher Karnofsky score (better performance status), and more extranodal involvement (Supplementary Table S2). For the validation set, patients with GEP-defined GCB and ABC subtypes were not significantly different for any of the clinical characteristics (Supplementary Table S3).
Classification results in the training and validation sets.
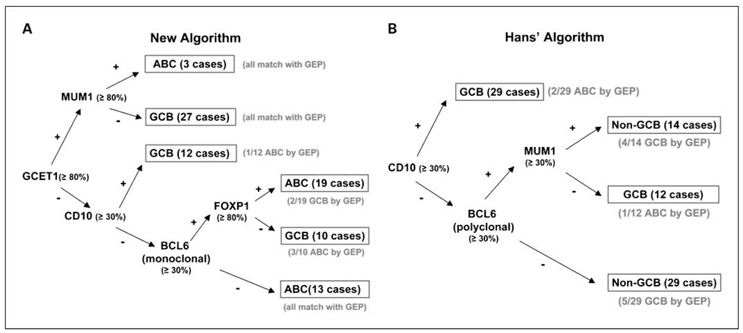
The percentages of cases that were positive for each immunostain are shown in Supplementary Table S4, and the staining patterns of a typical GCB-DLBCL and a typical ABC-DLBCL are shown in the Supplementary Figure. Sequential use of GCET1, followed by MUM1 or CD10, classified 42 of the 84 cases of DLBCL (50%) with high concordance (97%) with the GEP classification (Fig. 1A). Sequential use of monoclonal BCL6 and FOXP1 further separated the remaining 42 cases into GCB and ABC subtypes with high concordance (88%) with the GEP classification (Table 1; Fig. 1A). Two GEP-defined GCB cases and four GEP-defined ABC cases were misclassified by the new algorithm, whereas nine GEP-defined GCB cases and three GEP-defined ABC cases were misclassified by the Hans' algorithm. The overall concordance between the new IHC stain algorithm and GEP was thus 93% (78 of 84), which was better than that of the Hans' algorithm (86%; Table 1; Fig. 1B).
Fig. 1.
The new algorithm and the Hans' algorithm. The new IHC algorithm (A) uses five markers with 78 of 84 cases concordant (93%) when compared with the GEP classification, whereas the Hans' algorithm (B) had 72 of 84 cases concordant (86%) in the same training set.
Table 1.
Performance statistics of the new algorithm and the Hans' algorithm in classifying the 84 cases in the training set
| DLBCL subtypes by GEP | New algorithm (n = 84) |
Hans' algorithm (n = 84) |
||
|---|---|---|---|---|
| GCB (n = 47) | ABC (n = 37) | GCB (n = 47) | ABC (n = 37) | |
| Sensitivity (%) | 96 | 89 | 81 | 92 |
| Specificity (%) | 89 | 96 | 92 | 81 |
| Positive predictive value (%) | 92 | 94 | 93 | 79 |
| Negative predictive value (%) | 94 | 92 | 79 | 93 |
| Concordant cases (n) | 45 | 33 | 38 | 34 |
| Concordance rate (%) | 93 | 86 | ||
Because GEP-unclassified cases will not be excluded in the practice setting, the performance statistics of the new algorithm was also evaluated with the GEP-defined GCB and ABC cases combining with the GEP-unclassified cases. The new algorithm again showed comparable performance in the R-CHOP–treated cases (88% concordance), and maintained its high sensitivity, specificity, and negative predictive values in the CHOP-treated cases. The lower positive predictive values and overall concordance rate in the CHOP-treated cases reflected the relatively large number of 19 unclassified cases in this earlier cohort (Table 2). The computer perturbation and simulation program yielded a mean classification concordance rate of 90% (SD, 1.66%; 95% CI, 87-93%) for the training set, and 86% (SD, 1.53%; 95% CI, 84-88%) for the validation set with the four GEP-unclassified cases included in the analysis.
Table 2.
Performance statistics of the new algorithm of all the CHOP-treated cases and all the R-CHOP–treated cases with GEP-unclassified cases included
| DLBCL subtypes by GEP | Training set plus unclassified cases (all CHOP-treated cases) (n = 103) |
Validation set plus unclassified cases (all R-CHOP–treated cases) (n = 67) |
||||
|---|---|---|---|---|---|---|
| GCB (n = 47) |
ABC (n = 37) |
Unclassified (n = 19) |
GCB (n = 37) |
ABC (n = 26) |
Unclassified (n = 4) |
|
| Sensitivity (%) | 96 | 89 | Not applicable | 100 | 85 | Not applicable |
| Specificity (%) | 89 | 96 | Not applicable | 85 | 100 | Not applicable |
| Positive predictive value (%) | 80 | 70 | Not applicable | 88 | 88 | Not applicable |
| Negative predictive value (%) | 96 | 93 | Not applicable | 100 | 90 | Not applicable |
| Concordant cases (n) | 45 | 33 | Not applicable | 37 | 22 | Not applicable |
| Concordance rate for GCB and ABC subtypes among total (%) | 76 | 88 | ||||
NOTE: The sensitivity, specificity, and positive and negative predictive values were not applicable to the unclassified cases because the new IHC algorithm was designed to classify DLBCL cases into GCB and ABC subtypes only.
Survival analysis of the training and validation sets.
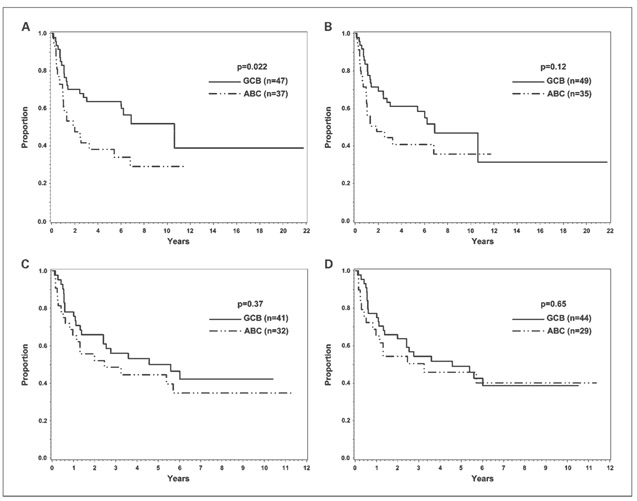
In the training set, the 5-year OS of the GEP-defined GCB and ABC cases (CHOP-treated) was 64% and 38%, respectively (P = 0.022; Fig. 2A). Eleven (6 GCB and 5 ABC) of the 84 cases did not have disease progression data and were excluded for EFS analysis. The 5-year EFS for the GEP-defined GCB and ABC cases was 50% and 45%, respectively (P = 0.37; Fig. 2C). The 5-year EFS of the ABC cases seemed slightly better than the 5-year OS because the five patients with ABC-DLBCL excluded from the EFS analysis died early. Classifying the training set with the new algorithm gave similar OS (5-year OS for GCB, 61%; 5-year OS for ABC, 41%; Fig. 2B) and EFS curves (5-year EFS for GCB, 49%; 5-year EFS for ABC, 46%; Fig. 2D), and a trend for superior OS in the GCB cases (P = 0.12; Fig. 2B).
Fig. 2.
Survival analysis of the training set. OS and EFS of the 84 cases in the training set classified by GEP (A and C) and the new algorithm (B and D). Eleven of the 84 cases did not have disease progression data and were excluded for EFS analysis.
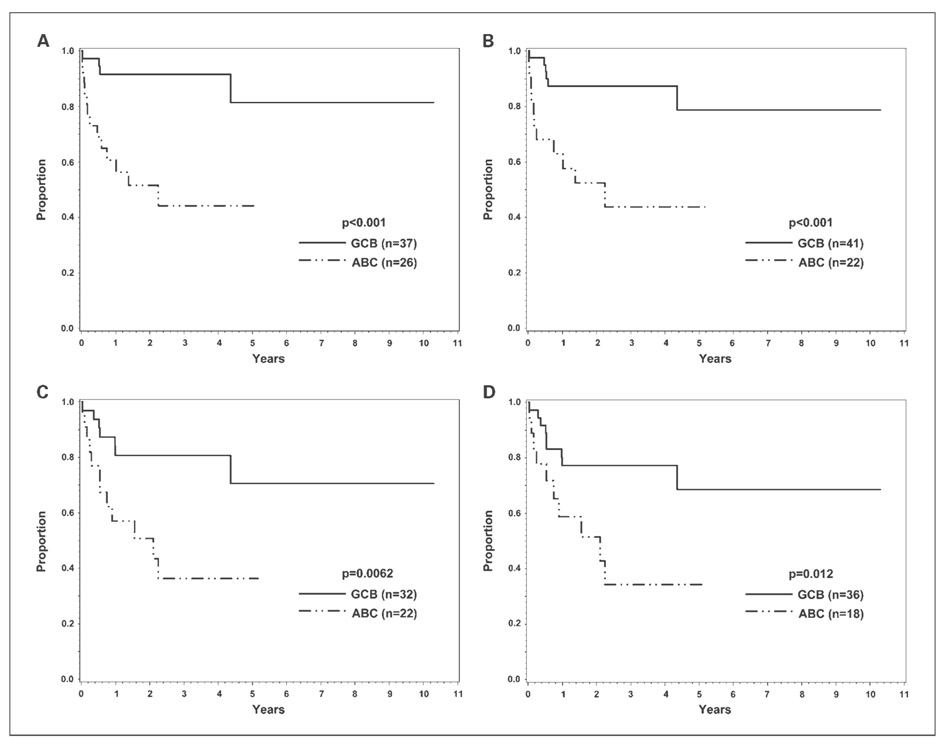
In the validation set, the 3-year OS of the GEP-defined GCB and ABC cases (R-CHOP treated) was 92% and 44%, respectively (P < 0.001; Fig. 3A). Nine of the 63 cases did not have disease progression data and were excluded for EFS analysis. The 3-year EFS for the GCB and ABC cases was 81% and 36%, respectively (P = 0.0062; Fig. 3C). Classifying the validation set with the new algorithm also resulted in a significantly different 3-year OS [87% (GCB) versus 44% (ABC), P < 0.001; Fig. 3B] and EFS [77% (GCB) versus 34% (ABC); P = 0.012; Fig. 3D] between the GCB and ABC cases. Multivariate analysis showed that a high International Prognostic Index score (hazard ratio, 4.5; 95% CI, 1.3-15.0; P = 0.015) and ABC classification by the new algorithm (hazard ratio, 5.9; 95% CI, 1.7-20.3; P = 0.005) were both significant adverse predictors of OS, but the CIs were wide due to the small sample size. Multivariate analysis also identified ABC classification by the new algorithm as the only significant predictor of EFS (hazard ratio, 3.0; 95% CI, 1.2-7.5; P = 0.017).
Fig. 3.
Survival analysis of the validation set. OS and EFS of the 63 cases in the validation set classified by GEP (A and C) and the new algorithm (B and D). Nine of the 63 cases did not have disease progression data and were excluded for EFS analysis.
Application of the new IHC stain algorithm to unclassified DLBCL and PMBL.
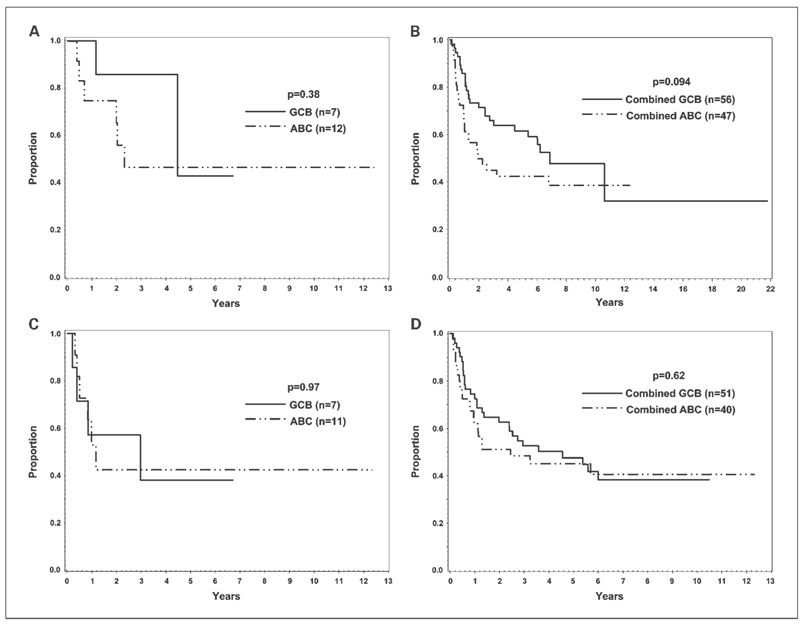
The 19 cases of CHOP-treated, GEP-unclassified DLBCL were assigned to either the GCB (7 cases) or ABC (12 cases) subtypes by the new algorithm. In comparison, the Hans' algorithm classified four cases into the “GCB” and 15 cases into the “non-GCB” subtypes. Using the Fisher's exact test, the two algorithms did not divide the GEP-unclassified cases differently (P = 0.48). The seven GCB cases had positive rates for CD10 and monoclonal BCL6 comparable with the GEP-defined GCB cases, whereas the 12 ABC cases showed low rates of positivity for all of the stains (Supplementary Table S4). Patients with unclassified DLBCL with a GCB phenotype had a better, but not statistically significant, OS compared with patients with unclassified DLBCL with an ABC phenotype, but EFS for both groups was similar (Fig. 4A and C). Applying the new algorithm to classify the combined set of 19 GEP-unclassified and the 84 GEP-defined GCB and ABC training set cases yielded survival curves (Fig. 4B and D) that were similar to those of the latter group alone (Fig. 2B and D).
Fig. 4.
Survival analysis of the GEP-unclassified cases. OS (A) and EFS (C) of the 19 GEP-unclassified cases when classified by the new algorithm. One of the 19 cases did not have disease progression data and was excluded for EFS analysis. Also shown are the OS (B) and EFS (D) for the combined group of GEP-unclassified and GEP-defined GCB and ABC cases in the training set as classified by the new algorithm. Twelve of the 103 cases in the combined group did not have disease progression data and were excluded for EFS analysis.
Similarly, four cases of R-CHOP–treated, GEP-unclassified DLBCL were classified as ABC (three cases) or GCB (one case) subtypes by the new algorithm. One of the ABC cases was negative for all the five stains, one was positive for FOXP1 only, and the other one was positive for both MUM1 and FOXP1. The GCB case was positive for monoclonal BCL6 only. Combining these cases with the rest of the validation set did not alter the significant differences in survival between the GCB and ABC subtypes (3-year OS for GCB, 88% versus 3-year OS for ABC, 51%; P = 0.004; 3-year EFS for GCB, 65% versus 3-year EFS for ABC, 44%; P = 0.072).
All seven cases of PMBL collected along with the CHOP-treated training set were classified by the new algorithm as the GCB subtype, in contrast with the Hans' algorithm, which classified five of these seven cases as the non-GCB subtype. The one case of PMBL that was collected along with the R-CHOP–treated validation set was also assigned to the GCB category by the new algorithm.
Discussion
Apart from the Hans' algorithm (9), several other IHC stain algorithms using up to eight markers have been proposed to classify (12, 29, 30) and/or predict prognosis in DLBCL (12, 29-32), but none of these studies had corresponding GEP data. Assessing protein expression by IHC staining provides information similar to that obtained by GEP and can be done on archival formalin-fixed, paraffin-embedded tissues. The IHC staining methods are already available in most pathology laboratories, making a simple IHC stain model more practical for widespread use. Additionally, direct evaluation of the tumor cells by microscopy avoids the inclusion of nonneoplastic elements, which is an inevitable drawback of GEP and reverse transcription-PCR.
The new algorithm places less weight on the older GC-specific marker BCL6 that shows poor interlaboratory agreement (33). Thus, it is an improvement over the Hans' algorithm. In this study, we tested two GC-specific markers, GCET1 and MTA3. We acknowledged that there has been an emergence of other GC-specific markers in recent years, which included GCET2 (ref. 34; also known as human germinal center-associated lymphoma; ref. 35) and LIM domain only 2 (LMO2; ref. 36). On examination of GEP data, GCET1, GCET2, and MTA3 showed high GC specificity but LIM domain only 2 was less specific (7), although its protein expression by IHC was reported to be more GC-specific and also associated with significantly better survival (19, 36). Antibodies for GCET1 and MTA3 were much more robust than the GCET2 antibody from our previous experience. Therefore, GCET1 and MTA3 were finally chosen as the two new GC-specific markers tested. Further studies should be conducted in future to assess whether addition of new GC-specific antibodies will result in a more refined and improved algorithm.
GCET1 (also called centerin or serpin A9; refs. 34, 37) and FOXP1 antibodies were used in the new but not the Hans' algorithm. The GCET1 transcript is highly associated with the GCB subtype of DLBCL (2, 34), and the GCET1 protein was later found to be restricted to B-cell neoplasms with a presumed GC B-cell origin (24). Our study clearly shows that GCET1 antibody can be used to improve the discrimination of the GCB and ABC subtypes of DLBCL. The FOXP1 gene on 3p14.1 encodes a member of the FOX family of transcription factors (27, 38). Two alternatively spliced FOXP1 mRNA isoforms are highly expressed in ABC-DLBCL (39), and some studies showed that uniform, high expression of FOXP1 protein is associated with an inferior survival in DLBCL patients (40, 41). In this study, we found that a high cutoff (≥80%) for FOXP1 was needed to achieve high specificity for the ABC subtype.
We only used GEP-defined GCB and ABC cases to derive the new algorithm because GEP-unclassified DLBCL is not a well-defined biological entity. Previous studies have shown that GEP-unclassified DLBCL is associated with poor survival similar to the ABC subtype (2, 5). Our observations, together with the relatively small number of unclassified cases in GEP studies (2, 5), suggest that we can apply the new IHC stain algorithm to all DLBCL cases in everyday practice without significantly jeopardizing the prognostic value of the new algorithm. It is also interesting to note that the majority (10 of 16) of GEP-unclassified cases with a high probability (70-90%) of being GCB (3 cases) or ABC (7 cases) by the Bayesian algorithm had a matched classification by the new IHC algorithm. Whether IHC can separate the unclassified cases into prognostic groups similar to the GCB and ABC subtypes requires further studies.
The seven PMBL cases collected along with the training set were classified differently by the Hans' algorithm (two “GCB” and five “non-GCB”) and the new algorithm (all “GCB”). The low CD10 positivity (1 of 7; refs. 42-44) and the lower BCL6 positivity with the polyclonal antibody (3 of 7, compared with 5 of 7 with the monoclonal BCL6 antibody) of PMBL, as well as the use of CD10 as a major decision point in the Hans' algorithm contributed to this discrepant allocation of PMBL by the two algorithms. The labeling of PMBL cases as “GCB” by the new algorithm is an improvement over the Hans' algorithm and is acceptable for prognostication because both PMBL and the GCB subtype of DLBCL have good prognosis (7). Additional IHC markers will be needed if different treatment options are considered for these two entities (45).
Because the number of cases studied was small, the outcome data were used mainly for comparing the IHC and GEP classifications. The more robust demonstrations of survival difference between the GEP-defined GCB-DLBCL and ABC-DLBCL were reported separately (2, 5). However, we have shown clearly that the IHC-based classification provided very similar outcome prediction compared with the GEP-based classification. Specifically, the computer perturbation and simulation program showed that the new algorithm was still highly predictive when reasonable intra-observer and inter-observer variations were taken into consideration.
In summary, we have derived and validated a new IHC stain algorithm that closely replicates the GEP classification of DLBCL into GCB and ABC subtypes, and which represents an improvement from the Hans' algorithm. This new algorithm will facilitate future research in DLBCL using archival materials and could be used to evaluate patients for novel or experimental therapies. Future incorporation of other prognostic markers into this new algorithm may provide additional prognostic value to this new classifier.
Supplementary Material
Translational Relevance.
Diffuse large B-cell lymphoma (DLBCL) can be classified into the prognostically favorable subtype of germinal center B-cell–like, and the prognostically unfavorable subtypes of activated B-cell–like, and unclassified DLBCL by gene expression profiling. To facilitate the application of this gene expression profiling (GEP) classification in therapeutic trials of DLBCL, which may ultimately lead to targeted therapies of individual molecular subtypes of DLBCL, we derived a robust immunohistochemical algorithm with a high concordance rate with the GEP classification. This immunohistochemical algorithm will therefore provide a simpler surrogate or alternative to GEP in providing useful prognostic information and in stratifying DLBCL patients in clinical trials, even when frozen lymphoma samples are not available for GEP analysis.
Acknowledgments
We thank Dr. Paul Wade for providing the MTA3 antibody; Dr. George Wright for analyzing the GEP data; Greg Cochran and Cissy Geigerman for performing the IHC staining; and Martin Bast, Kathryn Dufek, and Kim Klinetobe for clerical support.
Grant support: National Cancer Institute Grants U01CA114778 (W.C. Chan) and CA36727 (W.C. Chan, K. Fu, T.C. Greiner, J.M. Vose, and D.D. Weisenburger), and the Leukaemia Research Fund of the United Kingdom (A.H. Banham).
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000;403: 503–11. [DOI] [PubMed] [Google Scholar]
- 2.Rosenwald A, Wright G, Chan WC, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med 2002;346:1937–47. [DOI] [PubMed] [Google Scholar]
- 3.Wright G, Tan B, Rosenwald A, Hurt EH, Wiestner A, Staudt LM. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc Natl Acad Sci USA 2003;100:9991–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.A predictive model for aggressive non-Hodgkin's lymphoma. The International Non-Hodgkin's Lymphoma Prognostic Factors Project. N Engl J Med 1993;329:987–94. [DOI] [PubMed] [Google Scholar]
- 5.Lenz G, Wright G, Dave SS, et al. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med 2008;359:2313–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lossos IS. Molecular pathogenesis of diffuse large B-cell lymphoma. J Clin Oncol 2005;23: 6351–7. [DOI] [PubMed] [Google Scholar]
- 7.Rosenwald A, Wright G, Leroy K, et al. Molecular diagnosis of primary mediastinal B cell lymphoma identifies a clinically favorable subgroup of diffuse large B cell lymphoma related to Hodgkin lymphoma. J Exp Med 2003;198: 851–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Savage KJ, Monti S, Kutok JL, et al. The molecular signature of mediastinal large B-cell lymphoma differs from that of other diffuse large B-cell lymphomas and shares features with classical Hodgkin lymphoma. Blood 2003;102: 3871–9. [DOI] [PubMed] [Google Scholar]
- 9.Hans CP, Weisenburger DD, Greiner TC, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohisto-chemistry using a tissue microarray. Blood 2004; 103:275–82. [DOI] [PubMed] [Google Scholar]
- 10.Berglund M, Thunberg U, Amini RM, et al. Evaluation of immunophenotype in diffuse large B-cell lymphoma and its impact on prognosis. Mod Pathol 2005;18:1113–20. [DOI] [PubMed] [Google Scholar]
- 11.Haarer CF, Roberts RA, Frutiger YM, Grogan TM, Rimsza LM. Immunohistochemical classification of de novo, transformed, and relapsed diffuse large B-cell lymphoma into germinal center B-cell and nongerminal center B-cell subtypes correlates with gene expression profile and patient survival. Arch Pathol Lab Med 2006;130: 1819–24. [DOI] [PubMed] [Google Scholar]
- 12.Muris JJ, Meijer CJ, Vos W, et al. Immunohistochemical profiling based on Bcl-2, CD10 and MUM1 expression improves risk stratification in patients with primary nodal diffuse large B cell lymphoma. J Pathol 2006;208:714–23. [DOI] [PubMed] [Google Scholar]
- 13.Sjo LD, Poulsen CB, Hansen M, Moller MB, Ralfkiaer E. Profiling of diffuse large B-cell lymphoma by immunohistochemistry: identification of prognostic subgroups. Eur J Haematol 2007; 79:501–7. [DOI] [PubMed] [Google Scholar]
- 14.van Imhoff GW, Boerma EJ, van der Holt B, et al. Prognostic impact of germinal center-associated proteins and chromosomal breakpoints in poor-risk diffuse large B-cell lymphoma. J Clin Oncol 2006;24:4135–42. [DOI] [PubMed] [Google Scholar]
- 15.Nyman H, Adde M, Karjalainen-Lindsberg ML, et al. Prognostic impact of immunohistochemically defined germinal center phenotype in diffuse large B-cell lymphoma patients treated with immunochemotherapy. Blood 2007;109: 4930–5. [DOI] [PubMed] [Google Scholar]
- 16.Amara K, Trimeche M, Ziadi S, et al. Presence of simian virus 40 in diffuse large B-cell lymphomas in Tunisia correlates with germinal center B-cell immunophenotype, t(14;18) translocation, and P53 accumulation. Mod Pathol 2008;21:282–96. [DOI] [PubMed] [Google Scholar]
- 17.De Paepe P, Achten R, Verhoef G, et al. Large cleaved and immunoblastic lymphoma may represent two distinct clinicopathologic entities within the group of diffuse large B-cell lymphomas. J Clin Oncol 2005;23:7060–8. [DOI] [PubMed] [Google Scholar]
- 18.Dupuis J, Gaulard P, Hemery F, et al. Respective prognostic values of germinal center phenotype and early 18fluorodeoxyglucose-positron emission tomography scanning in previously untreated patients with diffuse large B-cell lymphoma. Haematologica 2007;92:778–83. [DOI] [PubMed] [Google Scholar]
- 19.Natkunam Y, Farinha P, Hsi ED, et al. LMO2 protein expression predicts survival in patients with diffuse large B-cell lymphoma treated with anthracycline-based chemotherapy with and without rituximab. J Clin Oncol 2008;26: 447–54. [DOI] [PubMed] [Google Scholar]
- 20.Veelken H, Vik Dannheim S, Schulte Moenting J, Martens UM, Finke J, Schmitt-Graeff A. Immunophenotype as prognostic factor for diffuse large B-cell lymphoma in patients undergoing clinical risk-adapted therapy. Ann Oncol 2007; 18:931–9. [DOI] [PubMed] [Google Scholar]
- 21.Amen F, Horncastle D, Elderfield K, et al. Absence of cyclin-D2 and Bcl-2 expression within the germinal centre type of diffuse large B-cell lymphoma identifies a very good prognostic subgroup of patients. Histopathology 2007;51: 70–9. [DOI] [PubMed] [Google Scholar]
- 22.Molina TJ, Gaulard P, Jais J-P, et al. Germinal center phenotype determined by immunohisto-chemistry on tissue microarray does not correlate with outcome in diffuse large B-cell lymphoma patients treated with immunochemotherapy in the randomized trial LNH98–5. A GELA study [abstract 51]. Blood 2007;110:24a. [Google Scholar]
- 23.Fu K, Weisenburger DD, Choi WW, et al. Addition of rituximab to standard chemotherapy improves the survival of both the germinal center B-cell-like and non-germinal center B-cell-like subtypes of diffuse large B-cell lymphoma. J Clin Oncol 2008;26:4587–94. [DOI] [PubMed] [Google Scholar]
- 24.Montes-Moreno S, Roncador G, Maestre L, et al. Gcet1 (centerin), a highly restricted marker for a subset of germinal center-derived lymphomas. Blood 2008;111:351–8. [DOI] [PubMed] [Google Scholar]
- 25.Fujita N, Jaye DL, Kajita M, Geigerman C, Moreno CS, Wade PA. MTA3, a Mi-2/NuRD complex subunit, regulates an invasive growth pathway in breast cancer. Cell 2003;113:207–19. [DOI] [PubMed] [Google Scholar]
- 26.Falini B, Fizzotti M, Pucciarini A, et al. A monoclonal antibody (MUM1p) detects expression of the MUM1/IRF4 protein in a subset of germinal center B cells, plasma cells, and activated T cells. Blood 2000;95:2084–92. [PubMed] [Google Scholar]
- 27.Banham AH, Beasley N, Campo E, et al. The FOXP1 winged helix transcription factor is a novel candidate tumor suppressor gene on chromosome 3p. Cancer Res 2001;61:8820–9. [PubMed] [Google Scholar]
- 28.Kaplan E, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc 1958;53:457–81. [Google Scholar]
- 29.Chang CC, McClintock S, Cleveland RP, et al. Immunohistochemical expression patterns of germinal center and activation B-cell markers correlate with prognosis in diffuse large B-cell lymphoma. Am J Surg Pathol 2004;28:464–70. [DOI] [PubMed] [Google Scholar]
- 30.Barrans SL, Carter I, Owen RG, et al. Germinal center phenotype and bcl-2 expression combined with the International Prognostic Index improves patient risk stratification in diffuse large B-cell lymphoma. Blood 2002;99:1136–43. [DOI] [PubMed] [Google Scholar]
- 31.Saez AI, Saez AJ, Artiga MJ, et al. Building an outcome predictor model for diffuse large B-cell lymphoma. Am J Pathol 2004;164:613–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Colomo L, Lopez-Guillermo A, Perales M, et al. Clinical impact of the differentiation profile assessed by immunophenotyping in patients with diffuse large B-cell lymphoma. Blood 2003;101: 78–84. [DOI] [PubMed] [Google Scholar]
- 33.de Jong D, Rosenwald A, Chhanabhai M, et al. Immunohistochemical prognostic markers in diffuse large B-cell lymphoma: validation of tissue microarray as a prerequisite for broad clinical applications-a study from the Lunenburg Lymphoma Biomarker Consortium. J Clin Oncol 2007;25:805–12. [DOI] [PubMed] [Google Scholar]
- 34.Pan Z, Shen Y, Du C, et al. Two newly characterized germinal center B-cell-associated genes, GCET1 and GCET2, have differential expression in normal and neoplastic B cells. Am J Pathol 2003;163:135–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lossos IS, Alizadeh AA, Rajapaksa R, Tibshirani R, Levy R. HGAL is a novel interleukin-4-inducible gene that strongly predicts survival in diffuse large B-cell lymphoma. Blood 2003; 101:433–40. [DOI] [PubMed] [Google Scholar]
- 36.Natkunam Y, Zhao S, Mason DY, et al. The oncoprotein LMO2 is expressed in normal germinalcenter B cells and in human B-cell lymphomas. Blood 2007;109:1636–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frazer JK, Jackson DG, Gaillard JP, et al. Identification of centerin: a novel human germinal center B cell-restricted serpin. Eur J Immu-nol 2000;30:3039–48. [DOI] [PubMed] [Google Scholar]
- 38.Shu W, Yang H, Zhang L, Lu MM, Morrisey EE. Characterization of a new subfamily of winged-helix/forkhead (Fox) genes that are expressed in the lung and act as transcriptional repressors. J Biol Chem 2001;276:27488–97. [DOI] [PubMed] [Google Scholar]
- 39.Brown PJ, Ashe SL, Leich E, et al. Potentially oncogenic B-cell activation-induced smaller isoforms of FOXP1 are highly expressed in the activated B cell-like subtype of DLBCL. Blood 2008;111:2816–24. [DOI] [PubMed] [Google Scholar]
- 40.Banham AH, Connors JM, Brown PJ, et al. Expression of the FOXP1 transcription factor is strongly associated with inferior survival in patients with diffuse large B-cell lymphoma. Clin Cancer Res 2005;11:1065–72. [PubMed] [Google Scholar]
- 41.Barrans SL, Fenton JA, Banham A, Owen RG, Jack AS. Strong expression of FOXP1 identifies a distinct subset of diffuse large B-cell lymphoma (DLBCL) patients with poor outcome. Blood 2004;104:2933–5. [DOI] [PubMed] [Google Scholar]
- 42.Calaminici M, Piper K, Lee AM, Norton AJ. CD23 expression in mediastinal large B-cell lymphomas. Histopathology 2004;45:619–24. [DOI] [PubMed] [Google Scholar]
- 43.de Leval L, Ferry JA, Falini B, Shipp M, Harris NL. Expression of bcl-6 and CD10 in primary mediastinal large B-cell lymphoma: evidence for derivation from germinal center B cells? Am J Surg Pathol 2001;25:1277–82. [DOI] [PubMed] [Google Scholar]
- 44.Pileri SA, Gaidano G, Zinzani PL, et al. Primary mediastinal B-cell lymphoma: high frequency of BCL-6 mutations and consistent expression of the transcription factors OCT-2, BOB.1, and PU.1 in the absence of immunoglobulins. Am J Pathol 2003;162:243–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodig SJ, Savage KJ, LaCasce AS, et al. Expression of TRAF1 and nuclear c-Rel distinguishes primary mediastinal large cell lymphoma from other types of diffuse large B-cell lymphoma. Am J Surg Pathol 2007;31:106–12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.