Abstract
Ribosome profiling has revealed pervasive but largely uncharacterized translation outside of canonical coding sequences (CDSs). Here, we exploit a systematic CRISPR-based screening strategy to identify hundreds of non-canonical CDSs that are essential for cellular growth and whose disruption elicit specific, robust transcriptomic and phenotypic changes in human cells. Functional characterization of the encoded microproteins reveals distinct cellular localizations, specific protein binding partners, and hundreds that are presented by the HLA system. Interestingly, we find multiple microproteins encoded in upstream open reading frames, which form stable complexes with the main, canonical protein encoded on the same mRNA, thus revealing the diverse use of functional bicistronic operons in mammals. Together, our results point to a family of functional human microproteins that play critical and diverse cellular roles.
Sentence Summary
Systematic interrogation of unannotated open reading frames reveals extensive translation of novel microproteins with important functional roles, expanding our understanding of the proteome.
Main Text
Efforts to bioinformatically discover and annotate protein-coding open reading frames (ORFs) in genomes, termed coding sequences (CDSs), have traditionally relied on rules such as amino acid conservation and homology, translation initiation from an AUG start codon, and minimum length (e.g. 100 amino acids)(1). The widespread adoption of these rules has been based on the assumption that short peptides are unlikely to fold into stable structures to perform functions. However, the generality of these rules has been challenged. For example, the ribosomal protein RPL41 is a 25 amino acid (aa) peptide; Sarcolipin (SLN, 31 aa) and Phospholamban (PLN, 52 aa) bind to and regulate the sarcoplasmic Ca2+ transport ATPase SERCA(2, 3). In addition, MYC can be translated from a non-canonical start codon CUG(4), demonstrating that non-AUG initiation can produce functional proteins. Recent studies have added a handful of remarkable examples of short proteins, or “microproteins” (also called “micropeptides” or just peptides), performing diverse functions(5–18), some encoded on transcripts annotated as long non-coding RNAs (lncRNAs). Finally, upstream ORFs (uORFs), located in the 5’ untranslated regions (UTR) of mRNAs, have long been implicated in cis-acting translational control of the main, canonical CDS(19–21), though it has remained unclear whether they can generate stable, functional peptides.
Systematic identification of functional short CDSs remains challenging. Recent ribosome profiling (deep sequencing of ribosome protected fragments) and mass spectrometry (MS) studies have identified thousands of previously unannotated CDSs(22–25) across bacteria, yeast, viruses, and mammalian cells. However, for most cases, the cellular function of these identified CDSs or their peptide products remains unexplored. We reasoned that the advent of CRISPR and its ability to precisely disrupt protein-coding regions(26), when combined with ribosome profiling, provides a unique opportunity to define and characterize empirically the functional protein-coding capacity of a given genome. Here, we applied a diversity of approaches, including ribosome profiling, mass spectrometry, and multiple CRISPR-based techniques, to systematically discover non-canonical CDSs encoded in the human genome and validate their critical roles in diverse cellular pathways.
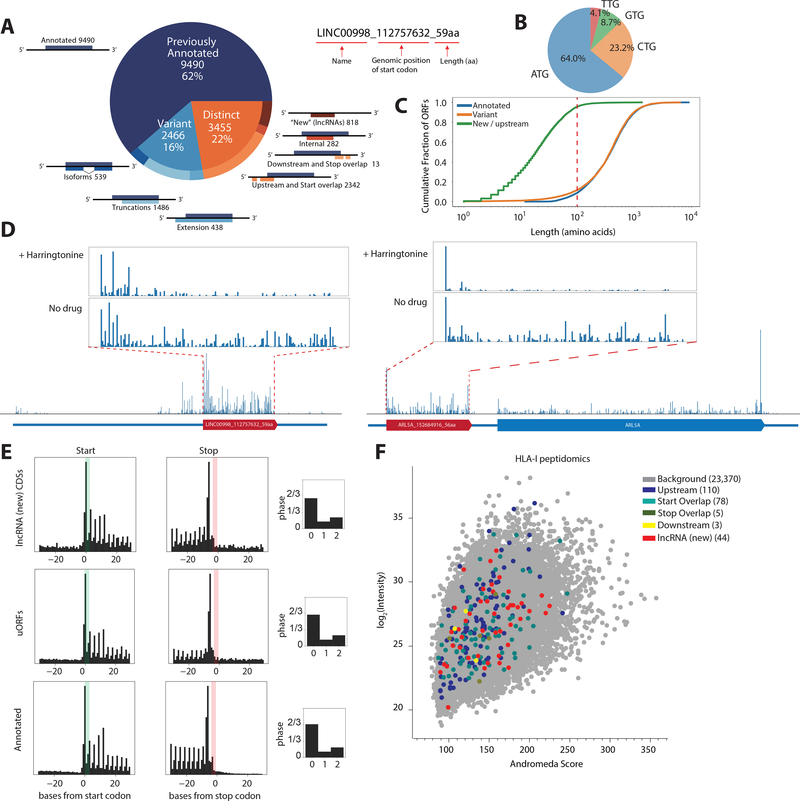
To annotate potential CDSs comprehensively and accurately, we first investigated genome-wide translation by ribosome profiling across multiple cell types and conditions, including human induced pluripotent stem cells (iPSCs), iPSC-derived cardiomyocytes, human foreskin fibroblasts (HFFs), and HFFs infected with cytomegalovirus(27, 28) (fig. S1A). We leveraged the ORF-RATER algorithm to annotate ORFs(27), incorporating multiple lines of evidence to identify ORFs undergoing active translation. This included consideration of accumulation of ribosome densities at the start and stop codons, three-nucleotide periodicity, and additional experimental results such as data from harringtonine-treated cells, in which ribosomes are stalled at initiation sites(27). In iPSCs and cardiomyocytes, in addition to 9490 annotated CDSs (62% of the identified CDSs), we identified 3455 distinct, non-canonical CDSs (22%, i.e. with no in-frame overlap with previously annotated CDSs) and 2466 variant CDSs of annotated proteins (16%) in our high statistical confidence set (Fig. 1A, see Methods and (27)). Among the distinct CDSs, 818 were CDSs on transcripts lacking prior protein-coding annotations (“new”, i.e. lncRNAs), 2342 were upstream CDSs (i.e. uORFs or “start overlaps”, CDSs that overlap annotated start codons in a different reading frame), and only 13 were downstream CDSs. Similar numbers of CDSs were present in HFFs (fig. S1B), with 75% of the CDSs shared between the two cell types. 96% of the distinct CDSs are less than 100 aa in length, and 36% of the CDSs use non-AUG start codons (Fig. 1B, C, and see fig. S2 for further characterizations).
Fig. 1. Ribosome profiling reveals translation of unannotated CDSs.
(A) ORF-RATER analysis of ribosome profiling data: 62% are previously annotated coding sequences, while 16% are variants of canonical coding sequences that share portions of the coding sequence, and 22% are distinct from annotated coding sequences. The naming convention of the identified ORFs is shown on the right. (B) Start-codon usage of the identified CDSs. (C) Cumulative distribution of CDS length. For distinct CDSs, 96% are smaller than 100 amino acids. (D) Example ribosome profiling traces of a lncRNA peptide from LINC00998 and a uORF peptide from ARL5A displaying the hallmarks of translation, including peaks of density around the start codon following harringtonine treatment and three nucleotide periodicities along the coding region. (E) Metagene analysis shows that the signatures of translation, including three-nucleotide periodicity in the expected reading frame, for uORFs and lncRNA CDSs are similar to annotated coding regions. (F) Identification of more than 200 non-canonical CDS peptides from HLA-I peptidomics, cross-validating their existence across the whole abundance range, with a mean Andromeda score of 141 compared to a total mean Andromeda score of 144. See Methods.
Multiple lines of evidence suggested that the non-canonical CDSs are actively translated. The average ribosome density (metagene) of the lncRNA CDSs and of the translated uORFs closely mirrors footprints from that of annotated coding regions with strong 3-nucleotide periodicities, a hallmark of active translation, as exemplified by traces from the lncRNA LINC00998 transcript and a uORF of ARL5A (Fig. 1D, E and see fig. S3). Our analysis also successfully recapitulated well-characterized short ORFs, such as the uORF on ATF4(29), and the recently discovered lncRNA-encoded microproteins MOXI/mitoregulin(11, 12) and NoBody(10). Bona fide lncRNAs such as XIST, HOTAIR, and NEAT1 were not identified to be protein coding (fig. S3E). Moreover, many of the CDSs were differentially translated during iPSC differentiation or viral infection (fig. S3F), providing evidence for translational control in different cell states.
MS-based proteomics in iPSCs and major Human Leukocyte Antigen Class I (HLA-I) peptidomics confirmed the stable expression of hundreds of non-canonical CDS peptides (Fig. 1F, fig. S4–5). HLA-I peptidomics identified 240 non-canonical peptides, suggesting that these peptides enter the HLA-I presentation pathway and contribute to the antigen repertoire and possible immunogenicity (Fig. 1F)(30). HLA class I prediction analysis cross-validated strong binding (Kd ≤ 50 mM) of non-canonical CDS HLA-I peptides to their respective allotype (fig. S6)(30). MS-based proteomics based on tryptic digestion identified far fewer non-canonical peptides, which may be due to challenges in detecting the trypsin-digested products from short, non-canonical CDSs, or possibly due to more rapid turnover of these non-canonical peptides (fig. S7).
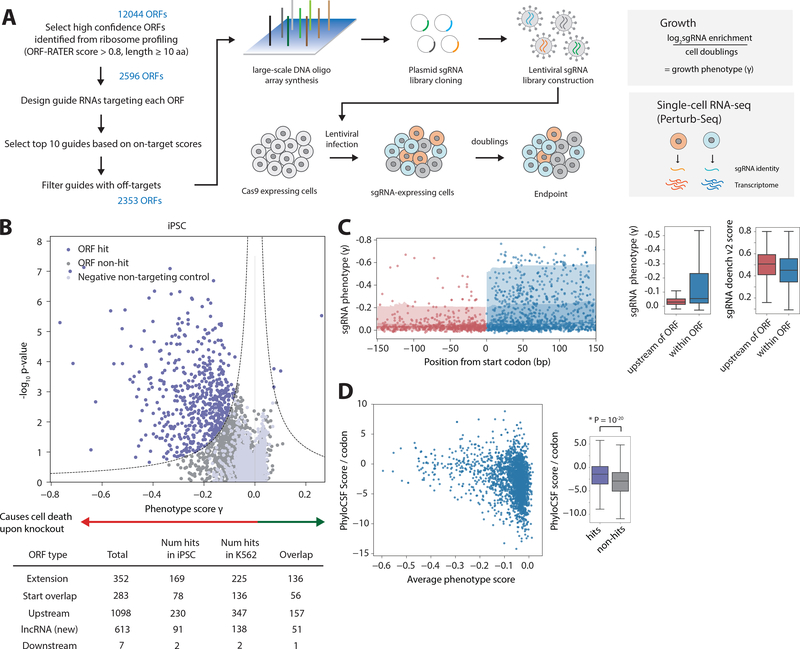
To test whether translation of the non-canonical CDSs are important for cell growth and potentially yield functional peptides, we measured the growth phenotypes resulting from CRISPR-mediated ORF knockout in pooled screens(26). We designed a Cas9 ORF single-guide RNA (sgRNA) library to specifically knock out thousands of the non-canonical CDSs identified by ribosome profiling (Fig. 2A, see Methods)(31, 32), targeting 1098 uORFs, 613 lncRNA CDSs, 352 extensions of annotated coding regions, 283 “start overlaps”, and 7 downstream CDSs. We performed pooled Cas9 knockout screens in iPSC and K562 chronic myeloid leukemia cells expressing Cas9 and the sgRNA library, akin to conventional pooled screens for essential proteins(26, 31). We measured sgRNA abundance in the cell populations shortly after library transduction and after 10 additional population doublings by deep sequencing to quantify the fitness defect conferred by each sgRNA. We then calculated a “phenotype” score (γ) and confidence (P-value) for each ORF from the relative enrichment or depletion of sgRNAs targeting a particular ORF (Fig. 2B and Methods). In iPSCs, our screen identified over 500 ORF knockout “hits” that result in statistically significant phenotypes. The hits include 169 genes that are variants of annotated proteins, 78 “start overlap” hits, 230 uORF hits, 91 lncRNA CDS hits, and 2 downstream CDS hits. iPSC and K562 cells had 401 shared hits, suggesting housekeeping or general cellular roles, as well as CDSs that may play cell-specific functions (fig. S8). A fraction of the uORF hits do not have main, canonical CDSs with fitness defects upon knockout, suggesting an independent function of the uORFs or that disruption of the uORFs result in increases in main CDS expression that results in the growth phenotype (fig. S8E). Thus, unannotated CDSs with important functions across multiple cell types are an abundant feature of the genome.
Fig. 2. Genome-scale CRISPR screens to identify functional, non-canonical CDSs.
(A) Schematic of CRISPR library design and screening strategies, either by growth screens or Perturb-Seq. For growth screens, frequencies of cells expressing a given sgRNA are determined by next-generation sequencing, and phenotype scores are quantified with the formula shown. For Perturb-Seq, single-cell transcriptomes and sgRNA identities were obtained by single-cell RNA-Seq. (B) Volcano plot summarizing knockout phenotypes and statistical significance (Mann-Whitney U test) for ORFs targeted in the pooled screen in iPSCs. Each dot represents a targeted ORF, and ORF hits are labeled in purple, with a more negative phenotype score indicating a stronger growth defect. See Methods. (C) Plot of the sgRNA phenotypes and distance from the start codon, across all ORF hits. sgRNAs targeting the genome immediately upstream of the ORF (shown in red) have significantly lower phenotype than sgRNAs targeting within the ORF (shown in blue). Note the axis is increasingly negative (stronger) phenotype. The difference is not due to differences in sgRNA on-target efficiencies, as quantified by the Doench v2 score. (D) The PhyloCSF Score per codon (higher is more conserved across the Euarchontoglires) is generally higher for ORF hits (P = 10−20, Kolmogorov–Smirnov test) and ORFs with a stronger phenotype. Note that lack of a growth phenotype does not necessarily imply a low PhyloCSF score.
Several lines of evidence further suggested that our screen reports specifically on the phenotypes of the selected ORFs. First, the phenotypes of control sgRNAs targeted directly upstream of each ORF in the genome (Fig. 2C) were significantly weaker than those of sgRNAs targeting within the ORF (P = 10−26, Mann-Whitney test). Second, sgRNA phenotypes are independent of distance to other annotated proteins, splice sites, or transcriptional start sites (fig. S9A). Functionally, ORF hits are on average more phylogenetically conserved, with a higher conservation score than non-hits (PhyloCSF score per codon, P = 10−20, Kolmogorov–Smirnov test, Fig. 2D)(33), and have other distinguishing sequence features (e.g. enrichment for Kozak consensus sequence) (fig. S10). Though, interestingly, the non-canonical CDSs on average have lower PhyloCSF scores compared to canonical proteins (fig. S2B). Finally, sgRNAs targeting ORF hits versus non-hits have indistinguishable off-target and on-target scores (fig. S9B)(32). We then performed validation follow-ups with individual sgRNAs, which recapitulated the growth phenotypes from our genome-scale screen (fig. S8D). Sequencing of the targeted genomic regions revealed indels of <50 bp (fig. S9C, D). Together, these analyses independently supported the conclusion that our screen phenotypes result specifically from the disruption of the target ORFs.
To survey function of the non-canonical CDSs at scale, we combined CRISPR screening with single-cell RNA sequencing (Perturb-seq) (34, 35). Disruptions of the various non-canonical CDSs resulted in broad and diverse changes in RNA-seq profiles across a variety of critical pathways, suggesting that the candidate CDSs play diverse cellular roles (fig. S11). As an example, disruption of the CDS on LINC00998 resulted in differentially expressed genes related to glycosylation (P-value < 10−10), suggesting a function at the Golgi or ER (see below). Importantly, the transcriptional phenotype also allowed us to functionally profile CDSs that are not essential for robust growth (fig. S11C). Furthermore, we found that CRISPR-targeted transcripts did not show detectable changes in abundance that might result from processes such as nonsense-mediated decay, indicating that the phenotypes we observed were not due to decreasing the abundance of the entire transcript (fig. S11D). Thus, similar to screens for essential protein-coding genes(26, 31), our screen for non-canonical CDSs required for robust cell growth underestimated the true number of functional CDSs in the genome, further underscoring the pervasiveness of functional, unannotated CDSs in the genome affecting a wide range of cellular activities.
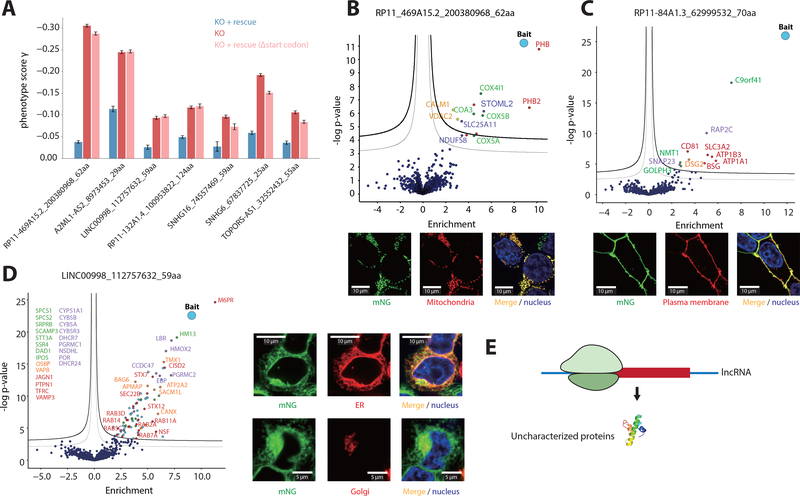
We next explored the functional role of the peptides encoded by the non-canonical CDSs identified from our screen, first focusing on lncRNA CDSs. For seven lncRNAs, we ectopically expressed the transcript encoding for the peptide and found in all cases knockout-induced growth defect was partially or completely rescued. This rescue was abrogated by the removal of the initiating start codon (Δstart codon) (Fig. 3A), suggesting an essential role of the peptide itself in cell growth. To further interrogate the specific functions of the non-canonical microproteins, we adopted a split-fluorescent protein (FP) approach using mNeonGreen (mNG), in which we fused each peptide with a minimally disruptive 16 aa tag (mNG11). Co-expression of the tagged peptide with the remainder of the mNG protein (mNG1–10) results in a fluorescence signal upon complementation (36, 37), creating both a fluorescent reporter to detect stable expression and cellular localization, and a handle for co-immunoprecipitation (co-IP) and MS to define interaction partners(36) (fig. S12A). We probed the functions of six essential lncRNA CDSs and found five to form specific complexes that were consistent with their subcellular localization. For example, the 62 aa peptide encoded by lncRNA RP11_469A15.2 specifically localized to the mitochondria. The peptide has a predicted transmembrane domain and co-immunoprecipitates with the cytochrome c oxidase (COX) complex and the mitochondrial Prohibitin complex (Fig. 3B). Moreover, the 70 aa peptide encoded by RP11–84A1.3 localizes to the plasma membrane and interacts with various cell surface proteins (Fig. 3C). Thirdly, the 59 aa peptide encoded by lncRNA LINC00998, which contains two predicted transmembrane domains, localizes specifically to both the ER and Golgi, and co-immunoprecipitates with lysosomal and vesicular transport proteins (Fig. 3D). Finally, the 55 aa peptide encoded on TOPORS-AS1 and the 124 aa peptide on RP11–132A1.4 also form functional complexes consistent with their cellular localization (fig. S12C, D and fig. S13). Consistent with prior studies(5–18), these examples demonstrated that lncRNAs can encode uncharacterized proteins, and highlight the need to fully extend the annotation of lncRNAs and the proteome.
Fig. 3. Short lncRNA CDSs encode functional microproteins.
(A) Rescue of lncRNA CDS knockout growth phenotypes by the ectopic expression of the transcript encoding the peptide, as well as controls where the initiating start codon is removed (Δstart codon). Error bars represent standard deviation of triplicates. P < 0.05 for all comparisons between knockout (KO) and KO + rescue. (B-D) Microscopy images and volcano plots of the co-IP MS of three example lncRNA-encoded microproteins tagged with mNG11, expressed ectopically (in the native transcript context) in a HEK293T cell line expressing mNG1–10. Green is mNG, red is the indicated organelle localization, and blue is Hoechst 33342, which stains for the nucleus. Scale bar dimensions are labeled. Significant interactors are shown in the top, right corner of the volcano plots. Thick threshold line is 1% FDR (false discovery rate), and the thin threshold line is 5% FDR. The bait (the tagged peptide) is labeled in blue. The interactors are colored according to their functional groups. (E) lncRNA-encoded microproteins are uncharacterized proteins that may play important regulatory roles in cells.
We next explored the functional effects of uORF translation, which is complicated by the fact that phenotypes can in principle be mediated by the peptide product (24, 38–41) or the impact of uORF translation on expression of the main, canonical CDS(20), or both. To distinguish between these possibilities, we first separately tagged the uORF and the main CDS, and used Western blot to confirm the independent expression of uORF peptides from the canonical protein (fig. S14). Furthermore, we established that ectopic expression of a transcript encoding only the uORF peptide was able to at least partially rescue the growth phenotype caused by disruption of the endogenous uORF. Critically, in all cases this rescue is dependent on the initiating start codon in the ectopically expressed message, demonstrating that the rescue is due to production of the expressed peptide (Fig. 4A). Consistent with this, in all cases we tested, deleting the start codon for the uORFs only minimally increased (around 20% - 60%) the expression of the main CDS, suggesting that the growth defect seen is mediated by the peptide and not increased expression of the canonical protein (fig. S14E, F). Taken together, these findings established that uORFs could function through the peptide they produce independently of any cis regulatory effects.
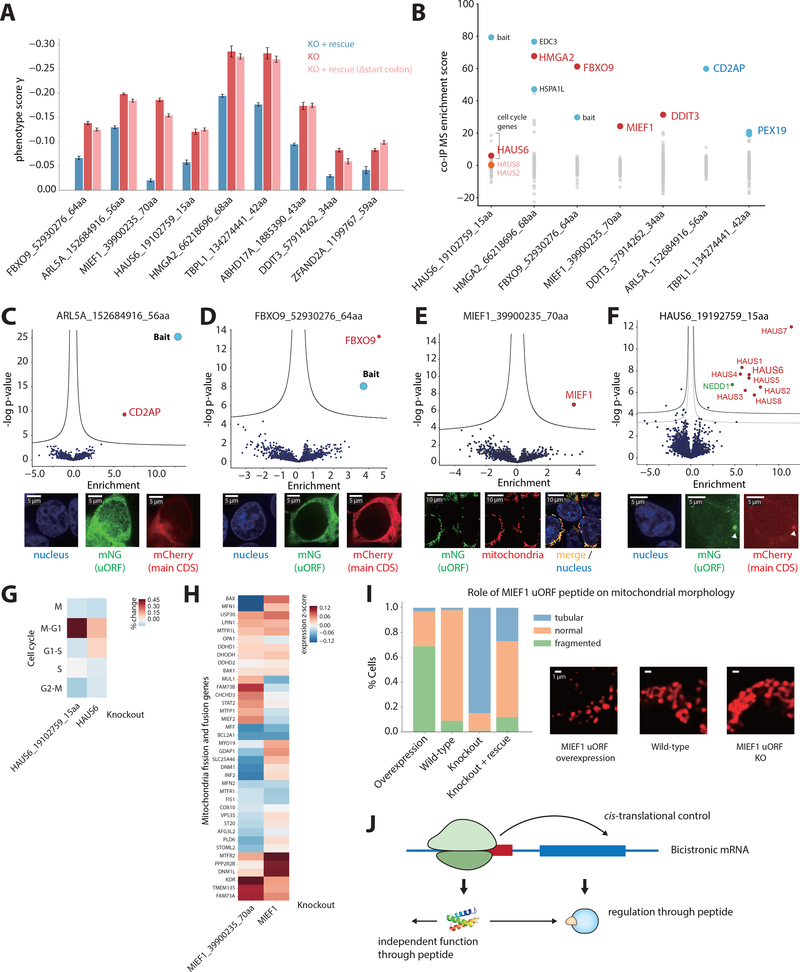
Fig. 4. Bicistronic mRNAs can encode uORF peptides that function in trans.
(A) Rescue of uORF knockout growth phenotypes by the ectopic expression of a transcript encoding the uORF peptide alone, as well as a controls where the initiating start codon is removed (Δstart codon). Error bars represent standard deviation of triplicates. P < 0.05 for all comparisons between KO and KO + rescue. (B) Summary of co-IP MS interactions, showing five uORF peptides that interact with their downstream-encoded protein (shown in red). Other significant interactors are shown in blue. (C-E) Examples of uORF peptides tagged with mNG11, expressed alone ectopically (in the native transcript context) in a HEK293T cell line expressing mNG1–10. Volcano plot of co-IP MS reveals significant interactors with uORF peptides. Threshold line is 1% FDR. The bait (the tagged peptide) is labeled in blue. For microscopy in C and D, the main, canonical protein tagged with mCherry (red) is co-expressed. For E, the mNG11-tagged MIEF1 uORF peptide (green) localizes to the mitochondria (red). (F) Volcano plot of co-IP MS from endogenously mNG11-tagged HAUS6 uORF. For microscopy, the mNG11-tagged uORF is expressed alone ectopically (green), and the canonical HAUS6 tagged with mCherry (red) is co-expressed. (G) Percent change for each cell cycle state for HAUS6 knockout (KO) and HAUS6 uORF KO, compared to control cells. (H) Transcriptome response of the MIEF1 uORF KO compared with the main CDS KO from Perturb-Seq. (I) Quantification of mitochondria morphology upon MIEF1 uORF peptide overexpression and knockout, as well as rescue of knockout phenotype. Representative microscopy images of the different mitochondria morphologies are shown to the right. (J) Possible model of uORF peptide functions and regulatory roles in cells.
To explore the functions of uORF-encoded microproteins, we examined their localization and protein binding partners by tagging the uORF peptides with mNG11. Out of the 10 uORF peptides further tested by co-IP MS, we failed to detect statistically significant interaction partners for three of the tagged peptides. Two peptides, encoded by the uORFs of TBPL1 and ARL5A, localize generally to the cytoplasm, while the main CDS proteins exhibit different cellular localization patterns. Consistent with our observed cellular localizations, these two uORF peptides immunoprecipitate specifically proteins with function independent of the main CDS protein (Fig. 4B, C and fig. S12). Thus, these uORF peptides and their main CDS protein have independent functions.
Interestingly, we found that five of the ten uORF peptides co-localized and formed a stable physical complex with the downstream-encoded, canonical protein on their shared mRNA. These include MIEF1, DDIT3, FBXO9, HMGA2, and HAUS6 (Fig. 4B, D, E and fig. S12). In all the cases, we expressed the tagged peptides in their native transcript context but without the downstream CDS, eliminating the possibility of stop codon read-through. We further confirmed this interaction by co-IP of the canonical protein and immunoblotting for the uORF peptide (fig. S12F), as well as with endogenously tagged clonal lines (fig. S15, Fig. 4F). This physical interaction between the proteins encoded by the uORF and the canonical CDS on the same transcript is particularly interesting(39, 42, 43), because it implies an additional layer of regulation beyond the propensity of uORFs to modulate translation of downstream CDSs.
We further explored the function of uORF-expressed microproteins by HAUS6 and MIEF1. In both cases, disrupting the uORF led to minimal increase in the expression of the main CDS protein, and the ectopic expression of a peptide-encoding transcript rescued the knockout-induced growth phenotype (Fig. 4A and fig. S14). mNG11-tagged HAUS6 uORF expressed from its endogenous locus efficiently pulled down key components of the HAUS6 complex, localized to the centrosome, and knockout of the uORF caused cells to arrest in the G1 stage, consistent with the role of HAUS6 microtubule attachment to the kinetochore and central spindle formation (Fig. 4F, G, fig. S12, fig. S15). Similarly, the MIEF1 uORF peptide localized to the mitochondria, consistent with the localization of the MIEF1 protein (Fig. 4E), which regulates mitochondrial fission and fusion (44). The MIEF1 uORF peptide knockout induced differential expression of mitochondrial fusion and fission genes, with a distinct transcriptional signature from that seen in the knockout of the MIEF1 protein (Fig. 4H). We observed that overexpression of the MIEF1 uORF peptide alone induced a fragmented mitochondrial phenotype (increased fission), whereas a clonal knockout of the MIEF1 uORF (with the sequence disrupted but nonetheless preserving an upstream ORF, see fig. S15) resulted in a tubular and more elongated mitochondrial phenotype (increased fusion). Importantly, this knockout morphology could be rescued by the exogenous expression of the MIEF1 uORF peptide (Fig. 4I). Together, our results indicated a possible role of the uORF-encoded peptide in regulating the downstream-encoded protein thereby challenging the monocistronic assumption in mammalian genomes. We speculated that this type of genomic architecture may be general, opening the doors to the investigation of the cooperative and regulatory nature of bicistronic human mRNAs. Indeed, a number of stress-regulated alternate translation initiation factors can modulate translation initiation site choice and uORF usage suggesting that regulation of bicistronic expression could play important roles in both normal biology and diseases states(21, 45).
In summary, we described a strategy that combines ribosome profiling, MS-based proteomics, microscopy and CRISPR-based genetic screens to discover and characterize widespread translation of functional microproteins and define the protein-coding potential of complex genomes. We identified a subset of lncRNAs that can encode stable, functional proteins, suggesting that they may be mis-annotated RNAs or potentially have dual roles at the RNA and protein levels. Furthermore, we provided examples of uORFs encoding functional peptides, highlighting the diverse cellular roles that uORFs may play beyond translational control. Intriguing, we also identified uORF-encoded peptides binding to the downstream-encoded protein on the same mRNA. Thus, our data highlighted an unappreciated complexity to the functional mammalian proteome, as well as the full spectrum of antigens presented by the HLA system.
Supplementary Material
Acknowledgments
We thank Weissman lab members, particularly T. M. Norman, M. Jost, M. J. Shurtleff, M. A. Horlbeck, L. A. Gilbert, M. Y. Hein, S. E. Torres, C. R. Liem, D. A. Santos, J. M. Replogle, & A. Xu; B. R. Conklin and M. P. Olvera (Gladstone Institute) for iPSC culturing; N. Cho (Chan-Zuckerberg Biohub) for endogenous tagging; S. E. O’Leary and I W. Lin for discussions.
Funding: This work was funded by NIH (RM1 HG009490) & DARPA (HR0011–17-2–0043), as well as the Chan-Zuckerberg Initiative. J.S.W. is a HHMI Investigator. A.B. and M.M. are supported by the Max Planck Society. J.C. is funded by the Jane Coffin Childs Memorial Fund for Medical Research and the NIH K99/R00 Pathway to Independence Award (K99 GM134154). J.K.N. is a fellow of the Hanna H. Gray Fellows Program. Oligonucleotide pools were courtesy of the Innovative Genomics Institute.
Footnotes
Competing interests: J.S.W. consults for and holds equity in KSQ Therapeutics and Maze Therapeutics, and consults for 5AM Ventures. B.A. is an advisory board member and has restricted stock in Celsius Therapeutics, Inc.
Data and materials availability: Raw sequencing data are deposited on NCBI Gene Expression Omnibus database with accession number GSE131650. MS-based proteomics data are deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD014031. Processed data are included as supplementary tables.
References and Notes
- 1.Basrai MA, Hieter P, Boeke JD, Small open reading frames: beautiful needles in the haystack. Genome research 7, 768–771 (1997). [DOI] [PubMed] [Google Scholar]
- 2.Odermatt A, Taschner PE, Scherer SW, Beatty B, Khanna VK, Cornblath DR, Chaudhry V, Yee WC, Schrank B, Karpati G, Breuning MH, Knoers N, MacLennan DH, Characterization of the gene encoding human sarcolipin (SLN), a proteolipid associated with SERCA1: absence of structural mutations in five patients with Brody disease. Genomics 45, 541–553 (1997). [DOI] [PubMed] [Google Scholar]
- 3.MacLennan DH, Kranias EG, Phospholamban: a crucial regulator of cardiac contractility. Nat Rev Mol Cell Biol 4, 566–577 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Hann SR, King MW, Bentley DL, Anderson CW, Eisenman RN, A non-AUG translational initiation in c-myc exon 1 generates an N-terminally distinct protein whose synthesis is disrupted in Burkitt’s lymphomas. Cell 52, 185–195 (1988). [DOI] [PubMed] [Google Scholar]
- 5.Jackson R, Kroehling L, Khitun A, Bailis W, Jarret A, York AG, Khan OM, Brewer JR, Skadow MH, Duizer C, Harman CCD, Chang L, Bielecki P, Solis AG, Steach HR, Slavoff S, Flavell RA, The translation of non-canonical open reading frames controls mucosal immunity. Nature 564, 434–438 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kondo T, Plaza S, Zanet J, Benrabah E, Valenti P, Hashimoto Y, Kobayashi S, Payre F, Kageyama Y, Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science 329, 336–339 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Nelson BR, Makarewich CA, Anderson DM, Winders BR, Troupes CD, Wu F, Reese AL, McAnally JR, Chen X, Kavalali ET, Cannon SC, Houser SR, Bassel-Duby R, Olson EN, A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 351, 271–275 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson DM, Anderson KM, Chang CL, Makarewich CA, Nelson BR, McAnally JR, Kasaragod P, Shelton JM, Liou J, Bassel-Duby R, Olson EN, A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 160, 595–606 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Magny EG, Pueyo JI, Pearl FM, Cespedes MA, Niven JE, Bishop SA, Couso JP, Conserved regulation of cardiac calcium uptake by peptides encoded in small open reading frames. Science 341, 1116–1120 (2013). [DOI] [PubMed] [Google Scholar]
- 10.D’Lima NG, Ma J, Winkler L, Chu Q, Loh KH, Corpuz EO, Budnik BA, Lykke-Andersen J, Saghatelian A, Slavoff SA, A human microprotein that interacts with the mRNA decapping complex. Nature chemical biology 13, 174–180 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stein CS, Jadiya P, Zhang X, McLendon JM, Abouassaly GM, Witmer NH, Anderson EJ, Elrod JW, Boudreau RL, Mitoregulin: A lncRNA-Encoded Microprotein that Supports Mitochondrial Supercomplexes and Respiratory Efficiency. Cell reports 23, 3710–3720 e3718 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Makarewich CA, Baskin KK, Munir AZ, Bezprozvannaya S, Sharma G, Khemtong C, Shah AM, McAnally JR, Malloy CR, Szweda LI, Bassel-Duby R, Olson EN, MOXI Is a Mitochondrial Micropeptide That Enhances Fatty Acid beta-Oxidation. Cell reports 23, 3701–3709 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsumoto A, Pasut A, Matsumoto M, Yamashita R, Fung J, Monteleone E, Saghatelian A, Nakayama KI, Clohessy JG, Pandolfi PP, mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature 541, 228–232 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Slavoff SA, Heo J, Budnik BA, Hanakahi LA, Saghatelian A, A human short open reading frame (sORF)-encoded polypeptide that stimulates DNA end joining. The Journal of biological chemistry 289, 10950–10957 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bi P, Ramirez-Martinez A, Li H, Cannavino J, McAnally JR, Shelton JM, Sanchez-Ortiz E, Bassel-Duby R, Olson EN, Control of muscle formation by the fusogenic micropeptide myomixer. Science 356, 323–327 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang JZ, Chen, Chen M, Gao XC, Zhu S, Huang H, Hu M, Zhu H, Yan GR, A Peptide Encoded by a Putative lncRNA HOXB-AS3 Suppresses Colon Cancer Growth. Molecular cell 68, 171–184 e176 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Zhang Q, Vashisht AA, O’Rourke J, Corbel SY, Moran R, Romero A, Miraglia L, Zhang J, Durrant E, Schmedt C, Sampath SC, The microprotein Minion controls cell fusion and muscle formation. Nature communications 8, 15664 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pauli A, Norris ML, Valen E, Chew GL, Gagnon JA, Zimmerman S, Mitchell A, Ma J, Dubrulle J, Reyon D, Tsai SQ, Joung JK, Saghatelian A, Schier AF, Toddler: an embryonic signal that promotes cell movement via Apelin receptors. Science 343, 1248636 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnstone TG, Bazzini AA, Giraldez AJ, Upstream ORFs are prevalent translational repressors in vertebrates. The EMBO journal 35, 706–723 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chew GL, Pauli A, Schier AF, Conservation of uORF repressiveness and sequence features in mouse, human and zebrafish. Nature communications 7, 11663 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Starck SR, Tsai JC, Chen K, Shodiya M, Wang L, Yahiro K, Martins-Green M, Shastri N, Walter P, Translation from the 5’ untranslated region shapes the integrated stress response. Science 351, aad3867 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ingolia NT, Brar GA, Stern-Ginossar N, Harris MS, Talhouarne GJ, Jackson SE, Wills MR, Weissman JS, Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell reports 8, 1365–1379 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ingolia NT, Lareau LF, Weissman JS, Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slavoff SA, Mitchell AJ, Schwaid AG, Cabili MN, Ma J, Levin JZ, Karger AD, Budnik BA, Rinn JL, Saghatelian A, Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nature chemical biology 9, 59–64 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bazzini AA, Johnstone TG, Christiano R, Mackowiak SD, Obermayer B, Fleming ES, Vejnar CE, Lee MT, Rajewsky N, Walther TC, Giraldez AJ, Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J 33, 981–993 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang T, Wei JJ, Sabatini DM, Lander ES, Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80–84 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fields AP, Rodriguez EH, Jovanovic M, Stern-Ginossar N, Haas BJ, Mertins P, Raychowdhury R Hacohen N, Carr SA, Ingolia NT, Regev A, Weissman JS, A Regression-Based Analysis of Ribosome-Profiling Data Reveals a Conserved Complexity to Mammalian Translation. Molecular cell 60, 816–827 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stern-Ginossar N, Weisburd B, Michalski A, Le VT, Hein MY, Huang SX, Ma M, Shen B, Qian SB, Hengel H, Mann M, Ingolia NT, Weissman JS, Decoding human cytomegalovirus. Science 338, 1088–1093 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vattem KM, Wek RC, Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America 101, 11269–11274 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bassani-Sternberg M, Pletscher-Frankild S, Jensen LJ, Mann M, Mass spectrometry of human leukocyte antigen class I peptidomes reveals strong effects of protein abundance and turnover on antigen presentation. Mol Cell Proteomics 14, 658–673 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, Qi LS, Kampmann M, Weissman JS, Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell 159, 647–661 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perez AR, Pritykin Y, Vidigal JA, Chhangawala S, Zamparo L, Leslie CS, Ventura A, GuideScan software for improved single and paired CRISPR guide RNA design. Nature biotechnology 35, 347–349 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin MF, Jungreis I, Kellis M, PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics 27, i275–282 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adamson B, Norman TM, Jost M, Cho MY, Nunez JK, Chen Y, Villalta JE, Gilbert LA, Horlbeck MA, Hein MY, Pak RA, Gray AN, Gross CA, Dixit A, Parnas O, Regev A, Weissman JS, A Multiplexed Single-Cell CRISPR Screening Platform Enables Systematic Dissection of the Unfolded Protein Response. Cell 167, 1867–1882 e1821 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Datlinger P, Rendeiro AF, Schmidl C, Krausgruber T, Traxler P, Klughammer J, Schuster LC, Kuchler A, Alpar D, Bock C, Pooled CRISPR screening with single-cell transcriptome readout. Nature methods 14, 297–301 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leonetti MD, Sekine S, Kamiyama D, Weissman JS, Huang B, A scalable strategy for high-throughput GFP tagging of endogenous human proteins. Proceedings of the National Academy of Sciences of the United States of America 113, E3501–3508 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng S, Sekine S, Pessino V, Li H, Leonetti MD, Huang B, Improved split fluorescent proteins for endogenous protein labeling. Nature communications 8, 370 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ji Z, Song R, Regev A, Struhl K, Many lncRNAs, 5’UTRs, and pseudogenes are translated and some are likely to express functional proteins. eLife 4, e08890 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Samandi S, Roy AV, Delcourt V, Lucier JF, Gagnon J, Beaudoin MC, Vanderperre B, Breton MA, Motard J, Jacques JF, Brunelle M, Gagnon-Arsenault I, Fournier I, Ouangraoua A, Hunting DJ, Cohen AA, Landry CR, Scott MS, Roucou X, Deep transcriptome annotation enables the discovery and functional characterization of cryptic small proteins. eLife 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rathore A, Chu Q, Tan D, Martinez TF, Donaldson CJ, Diedrich JK, Yates JR 3rd, Saghatelian A, MIEF1 Microprotein Regulates Mitochondrial Translation. Biochemistry 57, 5564–5575 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Delcourt V, Brunelle M, Roy AV, Jacques JF, Salzet M, Fournier I, Roucou X, The Protein Coded by a Short Open Reading Frame, Not by the Annotated Coding Sequence, Is the Main Gene Product of the Dual-Coding Gene MIEF1. Mol Cell Proteomics 17, 2402–2411 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bergeron D, Lapointe C, Bissonnette C, Tremblay G, Motard J, Roucou X, An out-of-frame overlapping reading frame in the ataxin-1 coding sequence encodes a novel ataxin-1 interacting protein. The Journal of biological chemistry 288, 21824–21835 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee CF, Lai HL, Lee YC, Chien CL, Chern Y, The A2A adenosine receptor is a dual coding gene: a novel mechanism of gene usage and signal transduction. The Journal of biological chemistry 289, 1257–1270 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu R, Liu T, Jin SB, Ning C, Lendahl U, Nister M, Zhao J, MIEF½ function as adaptors to recruit Drp1 to mitochondria and regulate the association of Drp1 with Mff. Scientific reports 7, 880 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sendoel A, Dunn JG, Rodriguez EH, Naik S, Gomez NC, Hurwitz B, Levorse J, Dill BD, Schramek D, Molina H, Weissman JS, Fuchs E, Translation from unconventional 5’ start sites drives tumour initiation. Nature 541, 494–499 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mandegar MA, Huebsch N, Frolov EB, Shin E, Truong A, Olvera MP, Chan AH, Miyaoka Y, Holmes K, Spencer CI, Judge LM, Gordon DE, Eskildsen TV, Villalta JE, Horlbeck MA, Gilbert LA, Krogan NJ, Sheikh SP, Weissman JS, Qi LS, So PL, Conklin BR, CRISPR Interference Efficiently Induces Specific and Reversible Gene Silencing in Human iPSCs. Cell stem cell 18, 541–553 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, Azarin SM, Raval KK, Zhang J, Kamp TJ, Palecek SP, Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proceedings of the National Academy of Sciences of the United States of America 109, E1848–1857 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McGlincy NJ, Ingolia NT, Transcriptome-wide measurement of translation by ribosome profiling. Methods 126, 112–129 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lareau LF, Hite DH, Hogan GJ, Brown PO, Distinct stages of the translation elongation cycle revealed by sequencing ribosome-protected mRNA fragments. eLife 3, e01257 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL, Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes & development 25, 1915–1927 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, Poliakov A, Cao X, Dhanasekaran SM, Wu YM, Robinson DR, Beer DG, Feng FY, Iyer HK, Chinnaiyan AM, The landscape of long noncoding RNAs in the human transcriptome. Nature genetics 47, 199–208 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dunn JG, Weissman JS, Plastid: nucleotide-resolution analysis of next-generation sequencing and genomics data. BMC genomics 17, 958 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M, Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nature methods 11, 319–324 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Kulak NA, Geyer PE, Mann M, Loss-less Nano-fractionator for High Sensitivity, High Coverage Proteomics. Mol Cell Proteomics 16, 694–705 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Andreatta M, Alvarez B, Nielsen M, GibbsCluster: unsupervised clustering and alignment of peptide sequences. Nucleic acids research 45, W458–W463 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Andreatta M, Nielsen M, Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics 32, 511–517 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu H, Xiao T, Chen CH, Li W, Meyer CA, Wu Q, Wu D, Cong L, Zhang F, Liu JS, Brown M, Liu XS, Sequence determinants of improved CRISPR sgRNA design. Genome research 25, 1147–1157 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, Virgin HW, Listgarten J, Root DE, Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nature biotechnology 34, 184–191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Horlbeck MA, Gilbert LA, Villalta JE, Adamson B, Pak RA, Chen Y, Fields AP, Park CY, Corn JE, Kampmann M, Weissman JS, Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. eLife 5, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu SJ, Horlbeck MA, Cho SW, Birk HS, Malatesta M, He D, Attenello FJ, Villalta JE, Cho MY, Chen Y, Mandegar MA, Olvera MP, Gilbert LA, Conklin BR, Chang HY, Weissman JS, Lim DA, CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 355, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR, Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nature methods 12, 767–772 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, Maitland A, Mostafavi S, Montojo J, Shao Q, Wright G, Bader GD, Morris Q, The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic acids research 38, W214–220 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chan MM, Smith ZD, Grosswendt S, Kretzmer H, Norman TM, Adamson B, Jost M, Quinn JJ, Yang D, Jones MG, Khodaverdian A, Yosef N, Meissner A, Weissman JS, Molecular recording of mammalian embryogenesis. Nature, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jost M, Chen Y, Gilbert LA, Horlbeck MA, Krenning L, Menchon G, Rai A, Cho MY, Stern JJ, Prota AE, Kampmann M, Akhmanova A, Steinmetz MO, Tanenbaum ME, Weissman JS, Combined CRISPRi/a-Based Chemical Genetic Screens Reveal that Rigosertib Is a Microtubule-Destabilizing Agent. Molecular cell 68, 210–223 e216 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Palmer CS, Osellame LD, Laine D, Koutsopoulos OS, Frazier AE, Ryan MT, MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO reports 12, 565–573 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bassik MC, Kampmann M, Lebbink RJ, Wang S, Hein MY, Poser I, Weibezahn J, Horlbeck MA, Chen S, Mann M, Hyman AA, Leproust EM, McManus MT, Weissman JS, A systematic mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell 152, 909–922 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mas-Ponte D, Carlevaro-Fita J, Palumbo E, Hermoso Pulido T, Guigo R, Johnson R, LncATLAS database for subcellular localization of long noncoding RNAs. RNA 23, 1080–1087 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tyanova S, Temu T, Cox J, The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc 11, 2301–2319 (2016). [DOI] [PubMed] [Google Scholar]
- 69.Xu D, Zhang Y, Ab initio protein structure assembly using continuous structure fragments and optimized knowledge-based force field. Proteins 80, 1715–1735 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kozakov D, Hall DR, Xia B, Porter KA, Padhorny D, Yueh C, Beglov D, Vajda S, The ClusPro web server for protein-protein docking. Nat Protoc 12, 255–278 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.