Abstract
NADH:ubiquinone oxidoreductase, more commonly referred to as mitochondrial complex I (CI), is the largest discrete enzyme of the oxidative phosphorylation system (OXPHOS). It is localized to the mitochondrial inner membrane. CI oxidizes NADH generated from the tricarboxylic acid cycle to NAD+, in a series of redox reactions that culminates in the reduction of ubiquinone, and the transport of protons from the matrix across the inner membrane to the intermembrane space. The resulting proton-motive force is consumed by ATP synthase to generate ATP, or harnessed to transport ions, metabolites and proteins into the mitochondrion. CI is also a major source of reactive oxygen species. Accordingly, impaired CI function has been associated with a host of chronic metabolic and degenerative disorders such as diabetes, cardiomyopathy, Parkinson’s disease (PD) and Leigh syndrome. Studies on Drosophila have contributed to our understanding of the multiple roles of CI in bioenergetics and organismal physiology. Here, we explore and discuss some of the studies on Drosophila that have informed our understanding of this complex and conclude with some of the open questions about CI that can be resolved by studies on Drosophila.
Keywords: Mitochondria, NADH:ubiquinone oxidoreductase, Complex I assembly, OXPHOS, Supercomplex, Drosophila
Configuration of CI
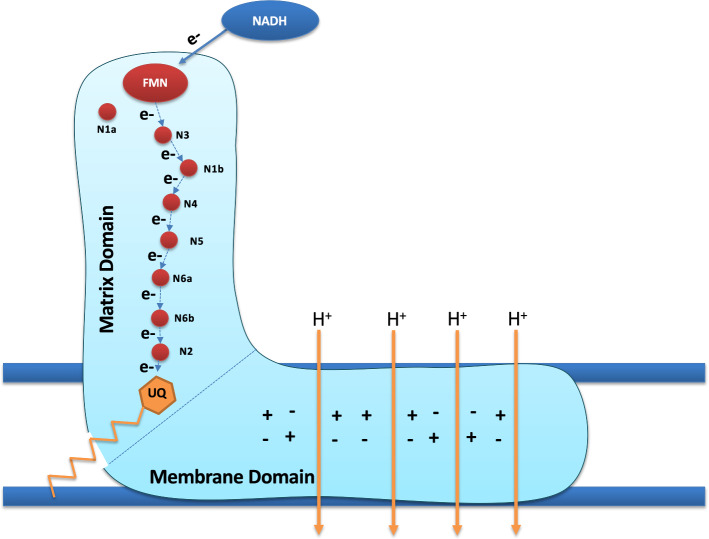
With a molecular mass approaching 1MDa (reviewed in [1]), mitochondrial CI is the largest holoenzyme of the OXPHOS. The primary bioenergetic role of CI is to transfer electrons from NADH to ubiquinone, and to move protons from the mitochondrial matrix into the mitochondrial intermembrane space. Although these two functions of CI are coupled, they are largely segregated in the complex, as they take place in two distinct domains of the complex—a hydrophilic matrix arm and a hydrophobic membrane arm. These two domains of CI are oriented almost orthogonally to each other, resulting in a boot-shaped complex (Fig. 1). The redox reactions of CI occur in the matrix domain; which is named as such because it extends into the mitochondrial matrix. The redox reactions involve oxidation of NADH by a flavin mononucleotide (FMN) prosthetic group, transfer of electrons along a chain of Fe–S clusters, and ultimately, ubiquinone reduction. Proton translocation occurs in the hydrophobic membrane domain, which is integrated into the mitochondrial inner membrane. It has been hypothesized to require a central axis of polar residues in the membrane domain that is conserved from bacteria to humans (Fig. 1) [2–5]. However, the precise molecular mechanism by which redox-coupled proton translocation occurs remains inexplicable.
Fig. 1.
A representation of CI showing the hydrophilic matrix and hydrophobic membrane arms/domains oriented almost perpendicularly to each other. NADH donates its electrons to the flavin mononucleotide (FMN) prosthetic group located at the tip of the matrix arm. The matrix arm has eight Fe–S clusters, seven of which are involved in transferring electrons from FMN to Ubiquinone. The Fe–S cluster adjacent to the FMN molecule (cluster N1a) has been proposed to be essential for maintaining the structure around the FMN site, but is not involved in electron transport. A central axis of polar residues that have been hypothesized to be involved in driving proton translocation across the membrane arm is also shown. The + and − signs in the membrane domain denote the presence of polar amino acids and are not meant to show the number or distribution pattern of charged amino acids in the central axis
Mammalian CI has 44 distinct subunits, but a total of 45 subunits, because one of the subunits (NDUFAB1) appears twice in the complex (Table 1). Human CI subunits encoded by the nucleus are labeled with the NDUF prefix, followed by an abbreviated description of their predicted function or location within the complex. This was mostly based on studies where mild chaotropic agents were used to dissociate CI into four subcomplexes (denoted as Iα, Iβ, Iγ and Iλ). Subcomplex Iα is made up predominantly of hydrophilic subunits that project into the mitochondrial matrix, while most of the subunits in subcomplex Iβ are hydrophobic and localized to the membrane arm of CI. Subunits with the prefix NDUFA (NDUFA1-3 and NDUFA5-13) are part of the Iα subcomplex, whereas the NDUFB subunits (NDUFB1-NDUFB11) are part of the Iβ subcomplex. The NDUFA4 subunit, although initially isolated as part of the 1α subcomplex, is not considered a CI subunit, as several reports have established that it is a CIV subunit [6–9]. The NDUFAB1 subunit has been identified in both the Iα and Iβ subcomplexes. This observation has been supported by recent high-resolution structures showing that NDUFAB1 appears twice in the complex. In addition, subunits that are found in the vicinity of the eight Fe–S clusters (NDUFS) or FMN molecule (NDUFV) are also localized in the matrix [4, 5, 10]. The seven mtDNA-encoded subunits have the p.MT-ND prefix followed by a number. The nomenclature of CI subunits varies between organisms; but to facilitate comparisons between the human subunits and their Drosophila orthologs, in this review we will use the human nomenclature. Drosophila orthologs of the human CI subunits will have the dNDUF or dND prefix, depending on whether they are nuclear or mitochondria-encoded subunits, respectively. Their actual annotation identifiers (CG numbers) are shown in Table 1.
Table 1.
Complex I subunits in different model organisms
| Homo sapiens | Bos taurus | Yarrowia lipolytica | Drosophila melanogaster | Escherichia coli |
|---|---|---|---|---|
| Core subunits | ||||
| MT-ND1 | MT-ND1 | NU1M | CG34092 | NuoH |
| MT-ND2 | MT-ND2 | NU2M | CG34063 | NuoN |
| MT-ND3 | MT-ND3 | NU3M | CG34076 | NuoA |
| MT-ND4 | MT-ND4 | NU4M | CG34085 | NuoM |
| MT-ND4L | MT-ND4L | NULM | CG34086 | NuoK |
| MT-ND5 | MT-ND5 | NU5M | CG34083 | NuoL |
| MT-ND6 | MT-ND6 | NU6M | CG34089 | NuoJ |
| NDUFS1 | 75 kDa | NUAM | CG2286 | NuoG |
| NDUFS2 | 49 kDa | NUCM | CG1970, CG11913a | NuoD |
| NDUFS3 | 30 kDa | NUGM | CG12079 | NuoC |
| NDUFS7 | PSST | NUKM | CG9172, CG2014a | NuoB |
| NDUFS8 | TYKY | NUIM | CG3944 | NuoI |
| NDUFV1 | 51 kDa | NUBM | CG9140, CG11423a, CG8102a | NuoF |
| NDUFV2 | 24 kDa | NUHM | CG5703, CG6485a | NuoE |
| Accessory subunits | ||||
| NDUFS4 | AQDQ/18 kDa | NUYM | CG12203 | |
| NDUFS5 | PFFD/15 kDa | NIPM | CG11455 | |
| NDUFS6 | 13 kDa | NUMM | CG8680 | |
| NDUFV3 | 10 kDa | CG11752 | ||
| NDUFC1 | KFYI | |||
| NDUFC2 | B14.5b | CG12400 | ||
| NDUFA1 | MWFE | NIMM | CG34439 | |
| NDUFA2 | B8 | NI8M | CG15434 | |
| NDUFA3 | B9 | NI9M | ||
| NDUFA5 | B13 | NUFM | CG6463 | |
| NDUFA6 | B14 | NB4M | CG7712 | |
| NDUFA7 | B14.5a | NUZM | CG3621, CG6914a | |
| NDUFA8 | PGIV | NUPM | CG3683 | |
| NDUFA9 | 39 kDa | NUEM | CG6020 | |
| NDUFA10 | 42 kDa | CG6343 | ||
| NDUFA11 | B14.7 | NUJM | CG9350 | |
| NDUFA12 | B17.2 | N7BM | CG3214 | |
| NDUFA13 | B16.6 | NB6M | CG3446 | |
| NDUFAB1 | SDAPα, SDAPβ | ACPM1, ACPM2 | CG9160 | |
| NDUFB1 | MNLL | CG18624 | ||
| NDUFB2 | AGGG | CG40002, CG40472a | ||
| NDUFB3 | B12 | NB2M | CG10320 | |
| NDUFB4 | B15 | NB5M | CG12859 | |
| NDUFB5 | SGDH | CG9762 | ||
| NDUFB6 | B17 | CG13240 | ||
| NDUFB7 | B18 | NB8M | CG5548 | |
| NDUFB8 | ASHI | NIAM | CG3192 | |
| NDUFB9 | B22 | NI2M | CG9306 | |
| NDUFB10 | PDSW | NIDM | CG8844 | |
| NDUFB11 | ESSS | NESM | CG6008 | |
| NUXM | ||||
| NEBM | ||||
| NUNM | ||||
| NUUM | ||||
| ST1 | ||||
aThe various paralogs for a specific CI subunit. Where multiple paralogs were obtained, the paralog with the most extensive sequence similarity with the human ortholog was listed first in the table
Fourteen of the subunits form the catalytic centers of the enzyme as they are directly involved in transferring electrons from NADH to ubiquinone, or for generation of the membrane potential. These 14 subunits are referred to as the core or central subunits and are conserved from the ancestral enzyme in bacteria to mammals [2–5]. Seven core subunits are encoded by the nuclear genome (i.e., NDUFS1, NDUFS2, NDUFS3, NDUFS7, NDUFS8, NDUFV1 and NDUFV2), while the other seven are encoded by mtDNA (i.e., p.MT-ND1, p.MT-ND2, p.MT-ND3, p.MT-ND4, p.MT-ND4L, p.MT-ND5 and p.MT-ND6). The 31 (30 distinct) remaining subunits are also encoded by nuclear DNA and are referred to as accessory or supernumerary subunits (Fig. 2). The accessory subunits are not directly involved in catalysis and are expressed to varying extents among eukaryotes (Table 1) [2–5]. All the NDUFA, NDUFB and NDUFC subunits are accessory subunits. Although orthologs of human accessory subunits are not found in the primitive CI found in E. coli (Table 1), orthologs of NDUFS4, NDUFS6 and NDUFA12 are found in the nitrate-reducing bacterium, Paracoccus denitrificans. In addition, the thermophilic bacterium, Thermus thermophilus, has two unique accessory subunits that are not present in eukaryotes [2].
Fig. 2.
Architecture of mammalian CI showing the approximate relative positions of the 45 subunits based on recent high-resolution CI structures from the Hirst and Sazanov groups. The first four letters of the nuclear-encoded subunit names have been eliminated for clarity. For instance, NDUFS1 has been abbreviated to S1, NDUFA12 to A12, etc. NDUFS6 and NDUFA9 contain Zn2+ and NADPH as co-factors, respectively, and have been proposed to serve as redox sensors for the complex. The nuclear-encoded core subunits, mitochondria-encoded subunits and accessory subunits are color coded in brown, blue and pink, respectively. Readily identifiable orthologs of the subunits denoted with an asterisk (NDUFA3 and NDUFC1) are not found in Drosophila
Role of accessory subunits
It has long been proposed that the accessory subunits may be required for stabilizing the complex. This has generally been corroborated by current high-resolution structures of CI showing that the majority of accessory subunits are wrapped around the core subunits. Moreover, many of the accessory subunits form extended structures containing α-helices, coils and disulfide bonds which allow them to intertwine extensively with other subunits, and perhaps stabilize the whole complex [3–5]. In addition, several phospholipid molecules have been found to interact with various portions of CI [5, 11]. These include cardiolipin, a highly abundant mitochondrial phospholipid, but also phosphatidylethanolamine and phosphatidylcholine. As the phospholipids occupy various cavities between the hydrophobic subunits, they have been proposed to stabilize the complex. However, the integrity of cardiolipin in the inner membrane is also dependent on the assembly of OXPHOS complexes [12].
More specialized roles have been proposed for some of the accessory subunits based on their association with specific co-factors or the presence of domains with specific biochemical functions (Fig. 2). For instance, NDUFA9 contains a tightly bound non-catalytic NADPH molecule that interacts with arginine-178 in NDUFS7 [5]. NDUFS7 interacts with one of the Fe–S clusters. As a consequence, redox-dependent fluctuations in the NADPH/NADP ratio may trigger conformational changes in both NDUFA9 and NDUFS7, causing disruptions in the Fe–S clusters redox reactions, which can possibly serve as a mechanism for sensing oxidative stress. Similarly, NDUFS6 contains a Zn2+ ion that is localized in close proximity to Fe–S clusters [5]. As proteins that harbor Zn2+ ions tend to be sensitive to oxidative stress, and mutating the Zn-binding residues of NDUFS6 impairs CI assembly, NDUFS6 could also conceivably function as an oxidative stress sensor [13, 14]. NDUFA2 is a small globular subunit that adopts a thioredoxin fold. This thioredoxin fold is likely functional as its disulfide bond has a redox potential that is comparable to that of disulfides in other thioredoxin-like proteins [15]. NDUFA2 interacts with NDUFS1 on the matrix side of the complex and is unique, in that it is the only matrix subunit that can possibly form a disulfide bond (between cysteine-23 and cysteine-57 in the ovine enzyme) [5]. The cysteine residues are in the reduced state in the mitochondrial matrix, making them prime targets for oxidation [11]. Interestingly, NDUFA2 is lost from the complex in Parkinson’s disease patients and this loss correlates with the extent of oxidative damage to the complex [16]. Thus, NDUFA2 may also serve as a sensor of oxidative stress. Therefore, it appears CI may be equipped with several mechanisms for sensing oxidative stress to provide a highly efficient mechanism to modulate CI function in response to oxidative stress.
In addition, the two NDUFAB1 subunits contain phosphopantetheine molecules that engage in stable interactions with NDUFA6 and NDUFB9 [5]. Considering that the NDUFAB1/NDUFA6 interaction depends on sequestration of the acyl side chain by an acyl carrier protein, it has been posited that this may provide a regulatory link between fatty acid biosynthesis and OXPHOS activity in mitochondria. Furthermore, there are long (50 kDa, NDUFV3L) and short (10 kDa, NDUFV3S) isoforms of NDUFV3 in the mammalian enzyme that have been proposed to compete for a common binding site on CI in the vicinity of FMN, and as a result may modulate CI function [17–19]. Indeed, the long NDUFV3 isoform is transcriptionally induced during cortical ischemia, lending credence to the hypothesis that it may have a regulatory role [20]. Moreover, phosphorylation of NDUFA10 at serine-250 is necessary for coenzyme Q reduction by CI [21], and NDUFA10 has a nucleoside kinase domain with unknown function. Finally, NDUFB11, NDUFA1, NDUFA7, NDUFC2 and NDUFA13 have phosphorylated residues that may regulate various aspects of mitochondrial function [22]. Evidently, future studies will likely uncover additional regulatory roles for the accessory subunits.
At least 28 of the 30 distinct mammalian accessory subunits have readily identifiable orthologs in Drosophila (Table 1 and Fig. 2), making it a suitable model organism for exploring the role of CI in health and organismal physiology. Here, we provide a synopsis of some of the chemical and genetic approaches that have been used in Drosophila to dissect the function of CI and its mechanism of assembly. We conclude by highlighting some of the key open questions about CI biology that can be resolved by studies in Drosophila.
Modeling CI dysfunction in Drosophila with chemical approaches
CI produces superoxide primarily at the FMN site in the matrix arm [23]. Once formed, the superoxide radical can be converted by the mitochondria-localized superoxide dismutase 2 to hydrogen peroxide. Both hydrogen peroxide and the superoxide radical are highly reactive and can further be converted into the hydroxyl radical. Collectively, these products are referred to as reactive oxygen species (ROS). Rotenone and paraquat (PQ) (1,1′-dimethyl-4-4′-bipyridinium dichloride) are two compounds known to stimulate CI-mediated ROS production in vitro [24–26]. Fittingly, various markers of oxidative stress and mitochondrial dysfunction are elevated in a dose-dependent manner in flies exposed to varying concentrations of PQ [27].
Consequently, Drosophila geneticists have performed a number of studies with rotenone and PQ to either evaluate their effects on various physiological parameters, or assess the efficacy of various compounds in rescuing their deleterious effects in Drosophila. For instance, it has been shown that the omega-3 fatty acids, eicosapentaenoic (EPA) and docosahexaenoic (DHA) acids, are protective against PQ-induced neuronal and mitochondrial dysfunction in flies [28]. Similarly, the anti-inflammatory sesquiterpene alcohol (-)-α-bisabolol (BISA) protects flies from rotenone-induced lethality and locomotory defects [29]. While the exact mechanism of BISA protection is unclear, the authors indicated that this could possibly be due to the scavenging of superoxide generated from rotenone-induced mitochondrial dysfunction. In addition, the ability of curcumin and melatonin to protect flies from rotenone and PQ-induced toxicity, respectively, have also been reported [30, 31]. Other reports have shown that overexpressing various proteins, such as the actin-binding protein afadin-6, or the mitochondrial uncoupling protein (UCP2), also protect flies from the damaging effects of rotenone [32–34]. Altogether, these findings demonstrate how studies on flies can be used to uncover various compounds or treatments that can mitigate the deleterious effects of CI-mediated oxidative stress.
Modeling CI disorders in Drosophila with genetic approaches
Because inhibition of CI with rotenone and PQ usually triggers acute, instead of the chronic responses that are typically found in patients with mitochondria disorders, many Drosophila researchers have preferred modeling CI dysfunction using genetic approaches. Additionally, genetic modeling of CI dysfunction affords the ability to examine the effect of disease-causing mutations on specific CI subunits or CI assembly factors (CIAFs). With 44 distinct human CI subunits and a dozen or so CIAFs, CI is a hot spot for mutations. Mutations in at least 33 CI subunits and assembly factors have been reported [35].
NDUFS4 is one of the accessory subunits of CI and is required for proper CI assembly [36, 37]. Mutations in NDUFS4 have been linked to Leigh syndrome and cardiomyopathy [38]. A Drosophila model where dNDUFS4 expression was knocked down using transgenic RNAi displayed progressive neurodegeneration, locomotory defects and a severely shortened life span; thus recapitulating cardinal phenotypes observed in NDUFS4-mutated patients [39]. Importantly, the dNDUFS4-knockdown flies had severe feeding difficulties—a feature of mitochondrial disorders that is usually observed in patients, but is often overlooked in animal models of mitochondrial diseases. Additional studies comparing the phenotypes of flies with ubiquitous knockdown of dNDUFS4, dNDUFS7 and dNDUFV1 established a compendium of phenotypes, such as eclosion defects and alterations in life span, with varying degrees of severity [40]. Intriguingly, some of the phenotypes are amenable to moderate- to high-throughput drug screening. Consequently, we anticipate that these disease-relevant models of CI deficiency may be suitable for screening for new compounds that suppress CI disorders.
A Drosophila model of CI deficiency has also been created for the mitochondria-encoded subunit, dND2 [41]. The dND2 mutant flies are derived from a 9-nucleotide homoplasmic mutation in mtDNA. These dND2 mutants display many of the hallmarks of mitochondrial disease, including reduced life span, multiple signs of neurodegeneration, and lower levels of ATP [41]. Although structural information available suggests that ND2 likely acts as a proton-conducting channel in the mitochondrial inner membrane, further analyses of this dND2 mutant definitively showed that coupling between electron transfer and proton pumping was impaired. This study revealed that the dND2 subunit is required for proton translocation across the mitochondrial inner membrane in vivo.
A frequently observed feature in some mouse models of CI deficiency is the hyperactivation of the target of the rapamycin (TOR) pathway. Markedly, phosphorylation of the TOR target p70S6K is elevated in dND2 mutant flies, relative to wild-type controls [42]. Because dND2 mutant flies display several additional readily discernible phenotypes—such as flight deficits and sensitivity to multiple stressors—they can be fed various drugs and easily assessed for suppression of the dND2-dependent adverse phenotypes. As a consequence, the effect of manipulating TOR signaling on disease progression in dND2 mutants has been explored by feeding flies with rapamycin [42]. Treatment with rapamycin extends the shortened life span of the dND2 mutant flies. Paradoxically, although TOR inhibition causes the induction of autophagy, and many longevity-promoting factors in Drosophila have been linked to facilitation of autophagy; the life span-extending effect of rapamycin on the dND2 mutant does not involve enhanced autophagy. Rapamycin treatment did not rescue the short-term paralysis induced by mechanically induced stress, indicating that the life span and mobility defects in dND2 mutants are not linked to each other. As in mice, the fat storage defect in the dND2 mutant flies is suppressed by rapamycin [43, 44]. These results further highlight the potential of TOR inhibition as a therapeutic strategy for mitochondrial disorders, and for elucidating the mechanistic underpinnings of treating mitochondrial CI diseases with rapamycin.
A number of reports have addressed the contribution of both neuronal and glial cells to neurodegeneration triggered by inhibiting CI. NDUFS1 is the largest subunit of CI. Using RNAi-mediated disruption of dNDUFS1 in either neurons or glia to assess their relative contribution to neurodegeneration, it was revealed that while both cell populations were important, glia play a critical non-cell autonomous role [45]. A Drosophila model where dNDUFS8 expression was disrupted in neurons or glial cells via transgenic RNAi expression has been created [46]. As with many other instances of CI disruption in Drosophila, the major phenotypes were shortened life span and impaired locomotory ability, but signs of neurodegeneration were by and large restricted to photoreceptors. Intriguingly, the neuronal dNDUFS8 RNAi-mediated phenotypes were suppressed by overexpressing the human glucose transporter, hGluT3. These results indicate that enhancing glucose metabolism in neurons may be sufficient to counteract the effect of reduced mitochondrial function. In contrast to what was observed in neurons, knockdown of dNDUFS8 in glia did not significantly affect longevity or locomotory ability. However, significant neurodegeneration in the brain was observed. This was coupled with a massive accumulation of lipid droplets at the cortex–neuropile boundaries, hinting at a perturbation of lipid metabolism in glia. Forced expression of hGluT3 did not rescue any of the phenotypes observed in glia. These results demonstrate that the neuropathology of CI disorders is due to disruption of metabolism in both neuronal and glial cells. A similar glia–neuron dichotomy has been observed for dNDUFS7B (CG2014) as RNAi-mediated knockdown of this CI protein in neurons causes increased aggression, but knockdown in glia has no effect [47].
Reports of serious complications occurring during and after anesthetic exposure in patients with mitochondrial disorders exist [48, 49]. However, the number of patients for whom such a phenomenon has been observed is relatively modest; and some patients with mitochondrial disease are not susceptible to anesthesia-induced complications, making it difficult to draw firm conclusions about causality. Accordingly, the effects of the general anesthetics isoflurane and sevoflurane have been studied in a Drosophila model of CI deficiency [50]. Wild-type flies exposed to isoflurane take a longer time to recover from the anesthesia-induced torpor, than wild-type flies exposed to sevoflurane; but flies carrying a mutation in dNDUFS8 are more sensitive to both anesthetics than wild-type flies [50]. Hence, flies were used to essentially confirm that genetic variations in CI proteins affect the pharmacokinetic and pharmacodynamic profiles of some anesthetics.
Regulation of CI assembly in Drosophila
The eight Fe–S clusters and single FMN molecule of CI must be incorporated together with the multiple subunits to form a functioning enzyme. For this reason, CI assembly is a highly regulated process, with many of the mechanistic molecular details still unclear. CI biogenesis proceeds through multiple steps and involves a number of assembly intermediates that ultimately merge with each other or other subunits to form the mature complex. The assembly intermediates generally correspond to partial or complete domains of the three functional modules of CI. The NADH dehydrogenase module (N module) contains the FMN molecule and is located at the tip of the matrix arm. The N module is the site of NADH oxidation. Situated between the N module and the membrane domain is the Q module. The Q module houses the majority of the Fe–S clusters, and is responsible for ubiquinone reduction. The proton-conducting P module is situated in the membrane arm and can be subdivided into a proximal PP module and distal PD module. The PP module connects with the Q module and contains p.MT-ND1, p.MT-ND2, p.MT-ND3, p.MT-ND4L and p.MT-ND6, while the PD module contains p.MT-ND4 and p.MT-ND5 (Fig. 3).
Fig. 3.
A schematic of mitochondrial CI assembly. An initiating assembly intermediate consisting of NDUFS2 and NDUFS3 combines with NDUFS7 and NDUFS8 to ultimately form the Q module. The Q module is anchored to the membrane by combining with subunits that are part of the proximal part of the P module (PP). The Q + PP module combines with the distal part of the P module (PD) to form an assembly intermediate consisting of the complete Q + P modules. Finally, the assembly intermediate consisting of the Q + P modules associates with the independently formed N module to form the fully assembled CI
Moreover, about a dozen or so CIAFs have been identified in mammalian systems. CIAFs are proteins that assist with the assembly process, but are not found in the fully assembled complex. Some CIAFs function as chaperones to stabilize specific CI assembly intermediates, or assist with the combination of two assembly intermediates to form a larger assembly intermediate. Others have more specific roles. The Drosophila ortholog of the human CI assembly factor NDUFAF1 (CG7598, henceforth referred to as dNDUFAF1) is required for proper CI assembly in Drosophila [51]. When grown at 25 °C, fruit flies take just about 10 days to proceed through development. Fruit flies develop through a number of well-defined stages: the embryonic, larval and pupal stages, before finally eclosing as adults. Flies with a mutation in dNDUFAF1 proceed through the initial stages of development and become largely arrested at the pupal stage. However, the few that eclose as adults have severely degenerated mitochondria. RNAi-mediated global knockdown of dNDUFAF1 produce larvae with impaired CI assembly; and the adults that eclose are sensitive to a number of stressors [51]. The role of another CIAF, dNDUFAF6 (CG15738) has also been studied in Drosophila [52]. dNDUFAF6 interacts with Hsp90 in the cytosol to chaperone the CI subunit dNDUFA10, in the cytoplasm. Disruption of dNDUFAF6 leads to impaired CI activity and ROS production is elevated. Thus, dNDUFAF6 regulates CI assembly by escorting dNDUFA10 in the cytoplasm prior to its import into the mitochondrion. As Drosophila orthologs exist for most of the other CIAFs described in humans, it will be interesting to further explore their function in Drosophila and examine how their function is linked with aging and other aspects of Drosophila physiology.
Until recently, studies of eukaryotic CI assembly had been performed primarily in plants, the fungus Neurospora crassa, aerobic yeast Yarrowia lipolytica and in various rodent and human cell lines [53–57]. However, because there are drawbacks to using any model system for studying CI assembly, additional model systems for studying CI assembly are required to complement the current systems available. For instance, about 25% of the accessory subunits in humans are not conserved in Y. lipolytica and several accessory subunits in Y. lipolytica CI are not found in the human enzyme (Table 1). Similarly, CI in Arabidopsis thaliana has a carbonic anhydrase domain and several additional subunits that are not present in the human enzyme [58]. In addition, there are notable differences between the N. crassa model of CI assembly and the CI assembly pathway in mammalian systems [59, 60]. In particular, the kinds of CI assembly intermediates observed in N. crassa are different from those seen in higher eukaryotes [61]. Moreover, CIA84, one of the CIAFs identified in N. crassa more than two decades ago, has not been shown to play a similar role in higher eukaryotes [62].
Investigating CI assembly in human or other mammalian cell lines is a major step forward; and a number of elegant studies in mammalian systems have defined the mechanism of CI assembly in cell lines [36, 37]. But the transformed or embryonic cells used to study this phenomenon tend to be very glycolytic and may rely on the OXPHOS only minimally. Cells grown in growth media containing galactose have an increased reliance on the OXPHOS system for ATP generation [63]. Consequently, in many instances where CI assembly has been studied in human cell lines, it has been necessary to treat the human cells with galactose, to “force” them to use the OXPHOS system [37, 64].
Many of these challenges can be circumvented by studying CI assembly in Drosophila flight muscles, which are highly enriched with mitochondria. Studying CI assembly in Drosophila has the added advantage of being in an in vivo context, where the effects of both developmental cues and environmental stressors can be explored. Accordingly, we recently described the role of several nuclear-encoded CI subunits in CI assembly in Drosophila muscles [6]. Using the Gal4/UAS system to selectively knock down CI transcripts in flight muscles, we found that many of the subunits (both core and accessory subunits) regulate specific steps in the assembly process in vivo [65]. Consequently, when their level of expression is reduced, CI activity is diminished due to impaired CI assembly. In addition, CI biogenesis in Drosophila proceeds through the same assembly intermediates described in mammalian systems, and the overall mechanism is essentially the same as what has been reported in mammalian systems.
Since many of the accessory subunits that associate with the membrane domain are single pass transmembrane domain (SPTD) subunits, they have been proposed to assist with organizing the transmembrane helices of the very hydrophobic mtDNA-encoded subunits during CI biogenesis. We found that when accessory subunits in the matrix domain are disrupted, an assembly intermediate consisting of the PD module stalls and accumulates in blue native gels. Mass spectrometry analyses revealed that the PD module is essentially the part of the membrane domain consisting of dND4, dND5 and their associated accessory subunits. Thus its accumulation, in spite of the disruption of the matrix domain accessory subunits provides further proof that the various modules of CI are assembled independently of each other. However, RNAi-mediated disruption of many of the SPTD subunits prevented the accumulation of the PD module in the gel, which supports the hypothesis that they may regulate membrane insertion of dND4 and dND5 [6]. Altogether, these analyses revealed that at least one function of many of the accessory subunits is to regulate the formation or stability of specific assembly intermediates at specific stages of the assembly process. Furthermore, they show that Drosophila is an important genetically pliable model organism for addressing questions relevant to mammalian CI biogenesis in vivo.
The current model for CI assembly in mammalian systems, which is essentially the same in Drosophila, begins with the formation of an initiating assembly intermediate containing NDUFS2 and NDUFS3, which combines with NDUFS7 and NDUFS8 (Fig. 3). This assembly intermediate is the primary component of the Q module and ultimately combines with the PP module to form an assembly intermediate that is anchored to the mitochondrial inner membrane. The Q + PP assembly intermediate combines with an independently formed assembly intermediate consisting of the PD module to form another assembly intermediate consisting of the complete Q + P modules. Finally, an independently formed assembly intermediate consisting of NDUFS1, NDUFV1, NDUFV2, NDUFV3, NDUFS4, NDUFS6 and NDUFA12, which together form the N module, is added to the Q + P assembly intermediate to produce the ~ 950 kDa CI holoenzyme (reviewed in [66].
Future directions and concluding remarks of CI studies in Drosophila
We have presented some insights from fruit flies on mitochondrial CI that has underscored the potential of studies of CI biology in Drosophila. However, what has currently been gleaned from CI studies in Drosophila can be reckoned as just the tip of the iceberg. For instance, while high-resolution CryoEM structures of CI, either as a discrete complex or in association with other OXPHOS complexes have now been described for bacteria, Y. lipolytica and various mammalian systems, the structure of Drosophila CI remains to be determined [3–5, 11, 67–74]. Although it may be argued that the structure of CI is now well known, critical details about the mechanism and regulation of catalysis remain obscure. In addition, a comparison of the bovine and Yarrowia structures of CI has revealed differences between the location of some of the transmembrane helices of the mtDNA-encoded subunits; and in some instances transmembrane helices of core subunits have been substituted by various segments of accessory subunits. Nevertheless, studies of how CI has evolved from Y. lipolytica to mammalian systems have been hampered by the evolutionary chasm between Y. lipolytica and mammals. Because of the genetic pliability of Drosophila, these and many other questions about CI biology can be resolved by first obtaining a high-resolution structure of Drosophila CI.
While more than a dozen CIAFs have been identified, the mechanism by which most of them regulate CI assembly has not been determined. For instance, a detailed mechanism has been described for the CIAF, NDUFAF7; which regulates CI assembly by acting as a methyltransferase that dimethylates arginine-85 in NDUFS2. This methylation step occurs during the initial stages of CI assembly and is required to stabilize the Q module [75, 76]. Similarly, NDUFAF5 is required for hydroxylating arginine-73 in the NDUFS7 subunit of human CI [77]. However, for many of the remaining CIAFs, all that is known about their mechanism of regulating CI assembly are the kinds of assembly intermediates that accumulate when they are disrupted. Interestingly, it was recently shown in Drosophila that when various CI subunits are disrupted, stalled assembly intermediates accumulate in blue native gels; which can be excised for subsequent mass spectrometry analyses [6]. Indeed, proteomic analyses revealed that many CIAFs were found to be associated with these stalled assembly intermediates. As a result, we anticipate that once a structure of Drosophila CI is described, subsequent studies aimed at identifying the structures of stalled assembly intermediates in association with various CIAFs may provide further details about the exact molecular mechanism by which many CIAFs regulate CI assembly.
An imperative issue in CI biology is the need to identify novel regulators of CI assembly. This need can be met, at least in part, by performing genetic screens in model organisms for new regulators of CI assembly and analyzing whether any candidate identified CI regulators regulate CI assembly in mammalian systems. The ideal model organism for discovering novel regulators of CI assembly will have to satisfy at least four criteria:
It should be highly enriched with mitochondria to enable the examination of the effects of thousands of candidate genes on CI assembly rather easily. The flight muscles of Drosophila are highly enriched with mitochondria.
The genetic tool kit in such an organism should be significantly advanced to the point where the effects of disrupting thousands of candidate genes on CI assembly can rapidly be tested. This condition is also satisfied by Drosophila, as it has a vast arsenal of genetic tools that coupled with its relatively short life span and high fertility allow both loss- and gain-of-function experiments to be performed rather easily.
It should be possible to analyze CI assembly in vivo where it is subject to both developmental and environmental signals. CI assembly can be analyzed in vivo, in the highly oxidative flight muscles of Drosophila.
Finally, the mechanism of CI assembly should closely mimic that of the human enzyme. It has recently been shown that the current mechanism of CI assembly described in vertebrate systems is conserved in Drosophila (see [6]).
As Drosophila is a genetically tractable organism that fulfills all four criteria described, we envisage that going forward, genetic screens performed in flies will lead to the discovery of novel regulators of CI assembly.
CI can exist as a discrete complex (also referred to as the holoenzyme) or in stable interactions with other complexes to form supercomplexes (SCs). Soon after the initial protocols for isolating discrete complexes were published more than half a century ago, evidence began to mount showing that some of the complexes engage in stable interactions with each other to form what are now referred to as SCs [78, 79]. Subsequently, blue native polyacrylamide gel electrophoresis was used to show that mild detergents can be used to resolve OXPHOS complexes into both individual units and SCs [80]. Additional studies using fractionation by sucrose density gradients and electron cryotomography of intact mitochondria confirmed that SCs were not artifacts of mild detergent solubilization [81, 82]. SCs with various stoichiometries have now been observed in multiple organisms (reviewed in [83]). The most widely described CI-containing SCs are an SC of CI and the complex III (CIII) dimer (CI:CIII2) and an SC consisting of CI, a dimer of CIII and between one and four copies of CIV (CI:CIII2:CIV1–4) (reviewed in [83]). CI:CIII2:CIV SC has been referred to as the respirasome because it was shown to contain all the constituents required for transferring electrons from NADH to O2 and is capable of oxygen consumption [81]. Additionally, a higher-order assembly consisting of CI2:CIII2:CIV2, referred to as the megacomplex, has also been reported [70]. While the existence of SCs is now well established, their exact function(s) is still a matter of intense debate. Because CI-containing SCs have also been observed in Drosophila, the stage is set for future studies in Drosophila to define their precise roles in physiology and mechanism of assembly, and identify novel SC assembly factors [6, 84, 85].
Many studies on CI function and ROS generation in Drosophila have been performed using PQ. However, using exogenous compounds such as PQ to study mitochondrial superoxide generation in vivo is confounded by the fact that PQ can generate ROS in many cell compartments. To circumvent this challenge, a derivative of PQ, dubbed MitoParaquat (MitoPQ), which is targeted specifically to the mitochondrial matrix has recently been shown to selectively increase superoxide production therein [86, 87]. MitoPQ-dependent production of superoxide occurs by redox cycling at the FMN site of CI. MitoPQ can enhance mitochondrial superoxide production in isolated mitochondria and cultured cells several orders of magnitude more effectively than untargeted PQ. MitoPQ is also more toxic than PQ in Drosophila, indicating that it can be administered and studied in vivo. Going forward, this novel ROS-generating tool will be useful for exploring the specific effects of mitochondrial superoxide generation in organismal physiology, redox signaling and aging.
In conclusion, we have described a Drosophila perspective on mitochondrial CI and highlighted some of the cardinal studies that show the potential of CI studies in this organism. CI in many of the genetically tractable organisms currently used to study CI biology has significantly diverged from human CI, to the point where sometimes critical accessory subunits such as NDUFA10 are missing. However, of the 44 distinct CI subunits, at least 42 have readily distinguishable orthologs in Drosophila. Accordingly, we anticipate that the ease of isolating copious amounts of mitochondria from various Drosophila tissues, extensive arsenal of tools for genetic analyses, relatively short generation time and limited gene redundancy in Drosophila are assets that should facilitate discoveries in Drosophila that should improve our understanding of CI biology in physiology and disease.
Acknowledgements
We thank members of the Owusu-Ansah lab for various discussions related to complex I. Work in the authors’ lab is funded by the following National Institute of Health (NIH) grants (R21 DK112074 and R35 GM124717).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hirst J. Mitochondrial complex I. Annu Rev Biochem. 2013;82:551–575. doi: 10.1146/annurev-biochem-070511-103700. [DOI] [PubMed] [Google Scholar]
- 2.Baradaran R, Berrisford JM, Minhas GS, Sazanov LA. Crystal structure of the entire respiratory complex I. Nature. 2013;494:443–448. doi: 10.1038/nature11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vinothkumar KR, Zhu J, Hirst J. Architecture of mammalian respiratory complex I. Nature. 2014;515:80–84. doi: 10.1038/nature13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu J, Vinothkumar KR, Hirst J. Structure of mammalian respiratory complex I. Nature. 2016;536:354–358. doi: 10.1038/nature19095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fiedorczuk K, et al. Atomic structure of the entire mammalian mitochondrial complex I. Nature. 2016;538:406–410. doi: 10.1038/nature19794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia CJ, Khajeh J, Coulanges E, Chen EI, Owusu-Ansah E. Regulation of mitochondrial complex I biogenesis in Drosophila flight muscles. Cell Rep. 2017;20:264–278. doi: 10.1016/j.celrep.2017.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zong S, et al. Structure of the intact 14-subunit human cytochrome c oxidase. Cell Res. 2018;28:1026–1034. doi: 10.1038/s41422-018-0071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pitceathly RD, et al. NDUFA4 mutations underlie dysfunction of a cytochrome c oxidase subunit linked to human neurological disease. Cell Rep. 2013;3:1795–1805. doi: 10.1016/j.celrep.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balsa E, et al. NDUFA4 is a subunit of complex IV of the mammalian electron transport chain. Cell Metab. 2012;16:378–386. doi: 10.1016/j.cmet.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 10.Marelja Z, Leimkühler S, Missirlis F. Iron sulfur and molybdenum cofactor enzymes regulate the Drosophila life cycle by controlling cell metabolism. Front Physiol. 2018;9:50. doi: 10.3389/fphys.2018.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Agip AA, et al. Cryo-EM structures of complex I from mouse heart mitochondria in two biochemically defined states. Nat Struct Mol Biol. 2018;25:548–556. doi: 10.1038/s41594-018-0073-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Y, et al. Assembly of the complexes of oxidative phosphorylation triggers the remodeling of cardiolipin. Proc Natl Acad Sci USA. 2019;116:11235–11240. doi: 10.1073/pnas.1900890116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oteiza PI. Zinc and the modulation of redox homeostasis. Free Radic Biol Med. 2012;53:1748–1759. doi: 10.1016/j.freeradbiomed.2012.08.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kmita K, et al. Accessory NUMM (NDUFS6) subunit harbors a Zn-binding site and is essential for biogenesis of mitochondrial complex I. Proc Natl Acad Sci USA. 2015;112:5685–5690. doi: 10.1073/pnas.1424353112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brockmann C, et al. The oxidized subunit B8 from human complex I adopts a thioredoxin fold. Structure. 2004;12:1645–1654. doi: 10.1016/j.str.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 16.Keeney PM, Xie J, Capaldi RA, Bennett JP. Parkinson’s disease brain mitochondrial complex I has oxidatively damaged subunits and is functionally impaired and misassembled. J Neurosci. 2006;26:5256–5264. doi: 10.1523/JNEUROSCI.0984-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bridges HR, Mohammed K, Harbour ME, Hirst J. Subunit NDUFV3 is present in two distinct isoforms in mammalian complex I. Biochim Biophys Acta Bioenerg. 2017;1858:197–207. doi: 10.1016/j.bbabio.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dibley MG, Ryan MT, Stroud DA. A novel isoform of the human mitochondrial complex I subunit NDUFV3. FEBS Lett. 2017;591:109–117. doi: 10.1002/1873-3468.12527. [DOI] [PubMed] [Google Scholar]
- 19.Guerrero-Castillo S, Cabrera-Orefice A, Huynen MA, Arnold S. Identification and evolutionary analysis of tissue-specific isoforms of mitochondrial complex I subunit NDUFV3. Biochim Biophys Acta Bioenerg. 2017;1858:208–217. doi: 10.1016/j.bbabio.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 20.Schneider A, Fischer A, Krüger C, Aronowski J. Identification of regulated genes during transient cortical ischemia in mice by restriction-mediated differential display (RMDD) Brain Res Mol Brain Res. 2004;124:20–28. doi: 10.1016/j.molbrainres.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 21.Morais VA, et al. PINK1 loss-of-function mutations affect mitochondrial complex I activity via NdufA10 ubiquinone uncoupling. Science. 2014;344:203–207. doi: 10.1126/science.1249161. [DOI] [PubMed] [Google Scholar]
- 22.Palmisano G, Sardanelli AM, Signorile A, Papa S, Larsen MR. The phosphorylation pattern of bovine heart complex I subunits. Proteomics. 2007;7:1575–1583. doi: 10.1002/pmic.200600801. [DOI] [PubMed] [Google Scholar]
- 23.Hirst J, Roessler MM. Energy conversion, redox catalysis and generation of reactive oxygen species by respiratory complex I. Biochim Biophys Acta. 2016;1857:872–883. doi: 10.1016/j.bbabio.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Degli Esposti M. Inhibitors of NADH-ubiquinone reductase: an overview. Biochim Biophys Acta. 1998;1364:222–235. doi: 10.1016/s0005-2728(98)00029-2. [DOI] [PubMed] [Google Scholar]
- 25.Fato R, et al. Differential effects of mitochondrial Complex I inhibitors on production of reactive oxygen species. Biochim Biophys Acta. 2009;1787:384–392. doi: 10.1016/j.bbabio.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cochemé HM, Murphy MP. Complex I is the major site of mitochondrial superoxide production by paraquat. J Biol Chem. 2008;283:1786–1798. doi: 10.1074/jbc.M708597200. [DOI] [PubMed] [Google Scholar]
- 27.Hosamani R. Muralidhara, Acute exposure of Drosophila melanogaster to paraquat causes oxidative stress and mitochondrial dysfunction. Arch Insect Biochem Physiol. 2013;83:25–40. doi: 10.1002/arch.21094. [DOI] [PubMed] [Google Scholar]
- 28.de Oliveira Souza A, et al. Neuroprotective action of Eicosapentaenoic (EPA) and Docosahexaenoic (DHA) acids on Paraquat intoxication in Drosophila melanogaster. Neurotoxicology. 2019;70:154–160. doi: 10.1016/j.neuro.2018.11.013. [DOI] [PubMed] [Google Scholar]
- 29.Leite GO, et al. Protective effect of (-)-α-bisabolol on rotenone-induced toxicity in Drosophila melanogaster. Can J Physiol Pharmacol. 2018;96:359–365. doi: 10.1139/cjpp-2017-0207. [DOI] [PubMed] [Google Scholar]
- 30.Pandareesh MD, Shrivash MK, Naveen Kumar HN, Misra K, Srinivas Bharath MM. Curcumin monoglucoside shows improved bioavailability and mitigates rotenone induced neurotoxicity in cell and Drosophila models of Parkinson’s disease. Neurochem Res. 2016;41:3113–3128. doi: 10.1007/s11064-016-2034-6. [DOI] [PubMed] [Google Scholar]
- 31.Medina-Leendertz S, et al. Longterm melatonin administration alleviates paraquat mediated oxidative stress in Drosophila melanogaster. Invest Clin. 2014;55:352–364. [PubMed] [Google Scholar]
- 32.Hwang RD, et al. The neuroprotective effect of human uncoupling protein 2 (hUCP2) requires cAMP-dependent protein kinase in a toxin model of Parkinson’s disease. Neurobiol Dis. 2014;69:180–191. doi: 10.1016/j.nbd.2014.05.032. [DOI] [PubMed] [Google Scholar]
- 33.Islam R, et al. A neuroprotective role of the human uncoupling protein 2 (hUCP2) in a Drosophila Parkinson’s disease model. Neurobiol Dis. 2012;46:137–146. doi: 10.1016/j.nbd.2011.12.055. [DOI] [PubMed] [Google Scholar]
- 34.Basil AH, et al. AF-6 protects against dopaminergic dysfunction and mitochondrial abnormalities in Drosophila models of Parkinson’s disease. Front Cell Neurosci. 2017;11:241. doi: 10.3389/fncel.2017.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mayr JA, et al. Spectrum of combined respiratory chain defects. J Inherit Metab Dis. 2015;38:629–640. doi: 10.1007/s10545-015-9831-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guerrero-Castillo S, et al. The assembly pathway of mitochondrial respiratory chain complex I. Cell Metab. 2017;25:128–139. doi: 10.1016/j.cmet.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 37.Stroud DA, et al. Accessory subunits are integral for assembly and function of human mitochondrial complex I. Nature. 2016;538:123–126. doi: 10.1038/nature19754. [DOI] [PubMed] [Google Scholar]
- 38.Fassone E, Rahman S. Complex I deficiency: clinical features, biochemistry and molecular genetics. J Med Genet. 2012;49:578–590. doi: 10.1136/jmedgenet-2012-101159. [DOI] [PubMed] [Google Scholar]
- 39.Foriel S, et al. Feeding difficulties, a key feature of the Drosophila NDUFS4 mitochondrial disease model. Dis Model Mech. 2018 doi: 10.1242/dmm.032482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Foriel S, et al. A Drosophila mitochondrial complex I deficiency phenotype array. Front Genet. 2019;10:245. doi: 10.3389/fgene.2019.00245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burman JL, et al. A Drosophila model of mitochondrial disease caused by a complex I mutation that uncouples proton pumping from electron transfer. Dis Model Mech. 2014;7:1165–1174. doi: 10.1242/dmm.015321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang A, Mouser J, Pitt J, Promislow D, Kaeberlein M. Rapamycin enhances survival in a Drosophila model of mitochondrial disease. Oncotarget. 2016;7:80131–80139. doi: 10.18632/oncotarget.12560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson SC, et al. mTOR inhibition alleviates mitochondrial disease in a mouse model of Leigh syndrome. Science. 2013;342:1524–1528. doi: 10.1126/science.1244360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson SC, et al. Dose-dependent effects of mTOR inhibition on weight and mitochondrial disease in mice. Front Genet. 2015;6:247. doi: 10.3389/fgene.2015.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hegde VR, Vogel R, Feany MB. Glia are critical for the neuropathology of complex I deficiency in Drosophila. Hum Mol Genet. 2014;23:4686–4692. doi: 10.1093/hmg/ddu188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cabirol-Pol MJ, Khalil B, Rival T, Faivre-Sarrailh C, Besson MT. Glial lipid droplets and neurodegeneration in a Drosophila model of complex I deficiency. Glia. 2018;66:874–888. doi: 10.1002/glia.23290. [DOI] [PubMed] [Google Scholar]
- 47.Li-Byarlay H, Rittschof CC, Massey JH, Pittendrigh BR, Robinson GE. Socially responsive effects of brain oxidative metabolism on aggression. Proc Natl Acad Sci USA. 2014;111:12533–12537. doi: 10.1073/pnas.1412306111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Casta A, Quackenbush EJ, Houck CS, Korson MS. Perioperative white matter degeneration and death in a patient with a defect in mitochondrial oxidative phosphorylation. Anesthesiology. 1997;87:420–425. doi: 10.1097/00000542-199708000-00030. [DOI] [PubMed] [Google Scholar]
- 49.Grattan-Smith PJ, Shield LK, Hopkins IJ, Collins KJ. Acute respiratory failure precipitated by general anesthesia in Leigh’s syndrome. J Child Neurol. 1990;5:137–141. doi: 10.1177/088307389000500214. [DOI] [PubMed] [Google Scholar]
- 50.Olufs ZPG, Loewen CA, Ganetzky B, Wassarman DA, Perouansky M. Genetic variability affects absolute and relative potencies and kinetics of the anesthetics isoflurane and sevoflurane in Drosophila melanogaster. Sci Rep. 2018;8:2348. doi: 10.1038/s41598-018-20720-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cho J, Hur JH, Graniel J, Benzer S, Walker DW. Expression of yeast NDI1 rescues a Drosophila complex I assembly defect. PLoS One. 2012;7:e50644. doi: 10.1371/journal.pone.0050644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang K, et al. The C8ORF38 homologue Sicily is a cytosolic chaperone for a mitochondrial complex I subunit. J Cell Biol. 2013;200:807–820. doi: 10.1083/jcb.201208033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cardol P, Matagne RF, Remacle C. Impact of mutations affecting ND mitochondria-encoded subunits on the activity and assembly of complex I in Chlamydomonas. Implication for the structural organization of the enzyme. J Mol Biol. 2002;319:1211–1221. doi: 10.1016/S0022-2836(02)00407-2. [DOI] [PubMed] [Google Scholar]
- 54.Remacle C, Duby F, Cardol P, Matagne RF. Mutations inactivating mitochondrial genes in Chlamydomonas reinhardtii. Biochem Soc Trans. 2001;29:442–446. doi: 10.1042/bst0290442. [DOI] [PubMed] [Google Scholar]
- 55.Duarte M, Sousa R, Videira A. Inactivation of genes encoding subunits of the peripheral and membrane arms of neurospora mitochondrial complex I and effects on enzyme assembly. Genetics. 1995;139:1211–1221. doi: 10.1093/genetics/139.3.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Antonicka H, et al. Identification and characterization of a common set of complex I assembly intermediates in mitochondria from patients with complex I deficiency. J Biol Chem. 2003;278:43081–43088. doi: 10.1074/jbc.M304998200. [DOI] [PubMed] [Google Scholar]
- 57.Ugalde C, et al. Human mitochondrial complex I assembles through the combination of evolutionary conserved modules: a framework to interpret complex I deficiencies. Hum Mol Genet. 2004;13:2461–2472. doi: 10.1093/hmg/ddh262. [DOI] [PubMed] [Google Scholar]
- 58.Fromm S, Senkler J, Zabaleta E, Peterhansel C, Braun HP. The carbonic anhydrase domain of plant mitochondrial complex I. Physiol Plant. 2016;157:289–296. doi: 10.1111/ppl.12424. [DOI] [PubMed] [Google Scholar]
- 59.Nehls U, Friedrich T, Schmiede A, Ohnishi T, Weiss H. Characterization of assembly intermediates of NADH:ubiquinone oxidoreductase (complex I) accumulated in Neurospora mitochondria by gene disruption. J Mol Biol. 1992;227:1032–1042. doi: 10.1016/0022-2836(92)90519-p. [DOI] [PubMed] [Google Scholar]
- 60.Tuschen G, et al. Assembly of NADH: ubiquinone reductase (complex I) in Neurospora mitochondria. Independent pathways of nuclear-encoded and mitochondrially encoded subunits. J Mol Biol. 1990;213:845–857. doi: 10.1016/S0022-2836(05)80268-2. [DOI] [PubMed] [Google Scholar]
- 61.Vartak RS, Semwal MK, Bai Y. An update on complex I assembly: the assembly of players. J Bioenerg Biomembr. 2014;46:323–328. doi: 10.1007/s10863-014-9564-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kuffner R, Rohr A, Schmiede A, Krull C, Schulte U. Involvement of two novel chaperones in the assembly of mitochondrial NADH: Ubiquinone oxidoreductase (complex I) J Mol Biol. 1998;283:409–417. doi: 10.1006/jmbi.1998.2114. [DOI] [PubMed] [Google Scholar]
- 63.Marroquin LD, Hynes J, Dykens JA, Jamieson JD, Will Y. Circumventing the Crabtree effect: replacing media glucose with galactose increases susceptibility of HepG2 cells to mitochondrial toxicants. Toxicol Sci. 2007;97:539–547. doi: 10.1093/toxsci/kfm052. [DOI] [PubMed] [Google Scholar]
- 64.Lim SC, et al. Loss of the mitochondrial fatty acid beta-oxidation protein medium-chain acyl-coenzyme A dehydrogenase disrupts oxidative phosphorylation protein complex stability and function. Sci Rep. 2018;8:153. doi: 10.1038/s41598-017-18530-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 66.Formosa LE, Dibley MG, Stroud DA, Ryan MT. Building a complex complex: assembly of mitochondrial respiratory chain complex I. Semin Cell Dev Biol. 2018;76:154–162. doi: 10.1016/j.semcdb.2017.08.011. [DOI] [PubMed] [Google Scholar]
- 67.Zickermann V, et al. Structural biology. Mechanistic insight from the crystal structure of mitochondrial complex I. Science. 2015;347:44–49. doi: 10.1126/science.1259859. [DOI] [PubMed] [Google Scholar]
- 68.Gu J, et al. The architecture of the mammalian respirasome. Nature. 2016;537:639–643. doi: 10.1038/nature19359. [DOI] [PubMed] [Google Scholar]
- 69.Letts JA, Fiedorczuk K, Sazanov LA. The architecture of respiratory supercomplexes. Nature. 2016;537:644–648. doi: 10.1038/nature19774. [DOI] [PubMed] [Google Scholar]
- 70.Guo R, Zong S, Wu M, Gu J, Yang M. Architecture of human mitochondrial respiratory megacomplex I. Cell. 2017;170:1247–1257.e1212. doi: 10.1016/j.cell.2017.07.050. [DOI] [PubMed] [Google Scholar]
- 71.Wu M, Gu J, Guo R, Huang Y, Yang M. Structure of mammalian respiratory supercomplex I. Cell. 2016;167:1598–1609.e1510. doi: 10.1016/j.cell.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 72.Blaza JN, Vinothkumar KR, Hirst J. Structure of the deactive state of mammalian respiratory complex I. Structure. 2018;26:312–319.e313. doi: 10.1016/j.str.2017.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Parey K, et al. Cryo-EM structure of respiratory complex I at work. Elife. 2018;7:e39213. doi: 10.7554/eLife.39213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Agip AA, Blaza JN, Fedor JG, Hirst J. Mammalian respiratory complex I through the lens of cryo-EM. Annu Rev Biophys. 2019;49:165–184. doi: 10.1146/annurev-biophys-052118-115704. [DOI] [PubMed] [Google Scholar]
- 75.Rhein VF, Carroll J, Ding S, Fearnley IM, Walker JE. NDUFAF7 methylates arginine 85 in the NDUFS2 subunit of human complex I. J Biol Chem. 2013;288:33016–33026. doi: 10.1074/jbc.M113.518803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zurita Rendón O, Silva Neiva L, Sasarman F, Shoubridge EA. The arginine methyltransferase NDUFAF7 is essential for complex I assembly and early vertebrate embryogenesis. Hum Mol Genet. 2014;23:5159–5170. doi: 10.1093/hmg/ddu239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rhein VF, Carroll J, Ding S, Fearnley IM, Walker JE. NDUFAF5 hydroxylates NDUFS7 at an early stage in the assembly of human complex I. J Biol Chem. 2016;291:14851–14860. doi: 10.1074/jbc.M116.734970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hatefi Y, Haavik AG, Griffiths DE. Reconstitution of the electron transport system. I. Preparation and properties of the interacting enzyme complexes. Biochem Biophys Res Commun. 1961;4:441–446. doi: 10.1016/0006-291x(61)90305-9. [DOI] [PubMed] [Google Scholar]
- 79.Hatefi Y, Haavik AG, Griffiths DE. Studies on the electron transfer system. XL. Preparation and properties of mitochondrial DPNH-coenzyme Q reductase. J Biol Chem. 1962;237:1676–1680. [PubMed] [Google Scholar]
- 80.Schägger H, Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000;19:1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Acín-Pérez R, Fernández-Silva P, Peleato ML, Pérez-Martos A, Enriquez JA. Respiratory active mitochondrial supercomplexes. Mol Cell. 2008;32:529–539. doi: 10.1016/j.molcel.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 82.Davies KM, et al. Macromolecular organization of ATP synthase and complex I in whole mitochondria. Proc Natl Acad Sci USA. 2011;108:14121–14126. doi: 10.1073/pnas.1103621108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Letts JA, Sazanov LA. Clarifying the supercomplex: the higher-order organization of the mitochondrial electron transport chain. Nat Struct Mol Biol. 2017;24:800–808. doi: 10.1038/nsmb.3460. [DOI] [PubMed] [Google Scholar]
- 84.Copeland JM, et al. Extension of Drosophila life span by RNAi of the mitochondrial respiratory chain. Curr Biol. 2009;19:1591–1598. doi: 10.1016/j.cub.2009.08.016. [DOI] [PubMed] [Google Scholar]
- 85.Pareek G, Thomas RE, Vincow ES, Morris DR, Pallanck LJ. Lon protease inactivation in Drosophila causes unfolded protein stress and inhibition of mitochondrial translation. Cell Death Discov. 2018;4:51. doi: 10.1038/s41420-018-0110-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Robb EL, et al. Selective superoxide generation within mitochondria by the targeted redox cycler MitoParaquat. Free Radic Biol Med. 2015;89:883–894. doi: 10.1016/j.freeradbiomed.2015.08.021. [DOI] [PubMed] [Google Scholar]
- 87.Antonucci S, et al. Selective mitochondrial superoxide generation in vivo is cardioprotective through hormesis. Free Radic Biol Med. 2019;134:678–687. doi: 10.1016/j.freeradbiomed.2019.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]