Abstract
We report the crystal structure of the SARS-CoV-2 putative primase composed of the nsp7 and nsp8 proteins. We observed a dimer of dimers (2:2 nsp7-nsp8) in the crystallographic asymmetric unit. The structure revealed a fold with a helical core of the heterotetramer formed by both nsp7 and nsp8 that is flanked with two symmetry-related nsp8 β-sheet subdomains. It was also revealed that two hydrophobic interfaces one of approx. 1340 Å2 connects the nsp7 to nsp8 and a second one of approx. 950 Å2 connects the dimers and form the observed heterotetramer. Interestingly, analysis of the surface electrostatic potential revealed a putative RNA binding site that is formed only within the heterotetramer.
Keywords: SARS-CoV-2, RNA, Primase, Crystal structure
1. Introduction
The coronavirus disease 2019 (COVID-19) has recently resulted in a global pandemic affecting the lives of millions of people all along both the health and economic spectra (Huang et al., 2020, Zhu et al., 2020). The disease is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2, further referred to as CoV-2), one of only seven known human coronaviruses, which belong to either alpha or beta coronaviruses (Coronaviridae Study Group of the International Committee on Taxonomy of, V., 2020, Gralinski and Menachery, 2020). Two other genera of coronaviruses, gamma and delta, do not comprise any members that affect human health and are mostly found in other mammals (mostly pigs) or avian species (Chu et al., 2011, Torres et al., 2016, Wang et al., 2014, Woo et al., 2012). Most of the human coronavirus infections are mild and are estimated to cause up to 15% of the common cold cases (Perlman and Netland, 2009). Unfortunately, some beta-coronaviruses, namely SARS-CoV, MERS-CoV and the newly emerging CoV-2 can develop into a life threatening lower respiratory syndrome characterized by severe pneumonia with significant lethality (Huang et al., 2020, Ksiazek et al., 2003, Rota et al., 2003, Zaki et al., 2012, Zumla et al., 2015).
Coronaviruses are single-stranded RNA viruses with positive polarity (+RNA viruses), which possess very large nonsegmented genomes ranging from 27 to 32 kilobases (Ziebuhr, 2005) (the genome CoV-2 consists of almost 30 kilobases) (Wu et al., 2020). The viral RNA contains a type 1 cap on the 5′ end and polyadenyl group on the other end (Ziebuhr, 2005). Approximately two-thirds of the genome contains genetic information for just two open reading frames, ORF 1a and ORF 1b. Notably, the expression of ORF 1b requires a −1 ribosomal frameshift upstream of the ORF1a stop codon which leads to significantly lower expression of genes located in ORF 1b (Snijder et al., 2016). These ORFs encode proteins mainly of the RNA replication machinery and translate into two polyproteins (pp1a and pp1b). Both polyproteins are subsequently processed by papain-like cysteine protease (PLpro) and 3C like serine proteases (3CLpro also termed the main protease, nsp5) that are part of pp1a (Zumla et al., 2016). The ORF1a encodes for nsp1 to 11 (including both aforementioned proteases PLpro and 3CLpro), whereas the ORF1b encodes for nsps 12–16 including the most important replication enzymes, the RNA-dependent RNA polymerase (nsp12) and the helicase (nsp13) (Snijder et al., 2016). The last third of the coronavirus genome encodes for four structural proteins (S - spike glycoprotein, E - envelope protein, M - matrix protein, and N - nucleocapsid protein) (Rota et al., 2003).
In general, +RNA viruses use the RNA-dependent RNA polymerase (RdRp) as the major enzyme responsible for copying viral RNA molecules to minus-strand RNA and subsequent synthesis of multiple novel plus-strands to serve as mRNA and also for the assembly of newly formed virions (Dubankova and Boura, 2019, Hercik et al., 2017, Sebera et al., 2018). Due to the large size and complexity of its genome, RNA replication of a coronavirus involves multi-subunit replication/transcription machinery (Snijder et al., 2016). Thus, the RdRp’s catalytic domain of coronaviruses occupies approx. two-thirds of the nsp12 C-terminus, whereas the N-terminus seems to interact with several other proteins of the replicase supercomplex including nsp8 and nsp9 (Snijder et al., 2016). According to the structural and functional studies performed on the proteins form SARS-CoV, the molecules of nsp8 form hexadecameric structures with molecules of another nonstructural protein, nsp7. This nsp7-nsp8 complex forms a hollow, cylinder-like structure that can accommodate dsRNA and was suggested to possess primase activity (Snijder et al., 2016, Zhai et al., 2005). In contrast, a recent CryoEM structure of SARS-CoV nsp7-nsp8-nsp12 complex shows that the putative primase activity cannot be associated with the entire complex, since it would not be able to bring the amino acids essential for primase activity within adequate proximity of the nsp12 catalytic center. Thus, the mechanism by which coronaviruses initiates RNA synthesis remains elusive (Kirchdoerfer and Ward, 2019). However, the formation nsp7-nsp8-nsp12 complex plays an essential role in the process of activation of the coronavirus RdRp, which seems to be rather poorly active on its own in vitro (Kirchdoerfer and Ward, 2019). Association of nsp7 and nsp8 was shown to crucially enhance the activity of RdRp (te Velthuis et al., 2012). In addition, it was shown that nsp7-nsp8 complexes of SARS-CoV and feline-CoV are an RNA polymerase capable of de novo initiation and it was proposed that nsp7-nsp8 function as a primase (te Velthuis et al., 2012, Xiao et al., 2012).
Here, we report on the crystal structure of the CoV-2 nsp7-nsp8 complex. The structure revealed a dimer of dimers formed of dimers that possess an unequal conformation. The structure illustrates that the nsp7-nsp8 complex is stable in a dimeric form and suggests that an nsp7-nsp8 dimer could act as a primase for the fully assembled nsp7-nsp8-nsp12 replication complex where it probably acts as a processivity factor.
2. Results
In this study we aimed to structurally characterize the CoV-2 primase. We truncated both proteins based on secondary structure predictions and previously solved structures of the SARS virus (Fig. 1 A) and prepared them as recombinant in E. coli and purified them to homogeneity. The resulting nsp7-nsp8 protein complex was stable during the size exclusion chromatography (SEC) step of protein purification therefore we used it for crystallization trials. The crystals grew within a week, belonged to the monoclinic P2 spacegroup and diffracted to almost 2 Å resolution. We were able to solve the structure by molecular replacement and to refine it to good geometry and R factors (Rwork = 21.43% and Rfree = 23.92%, summarized in M&M section and in Table 1 ).
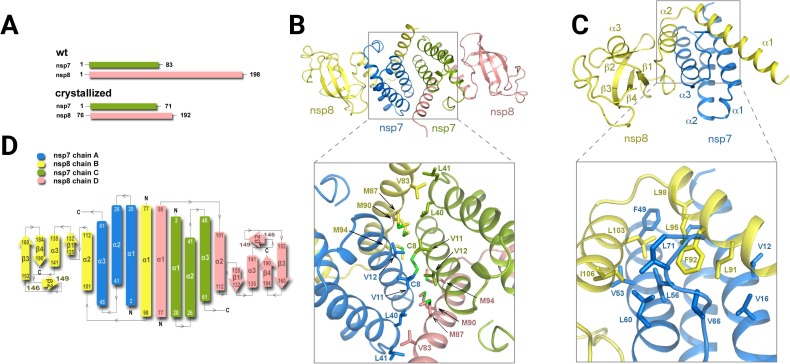
Fig. 1.
SARS-CoV-2 nsp7-nsp8 complex crystal structure A - Schematic representation of the crystallized and wild-type proteins. B - The overall view of the putative 2:2 nsp7-nsp8 heterotetrameric complex. A detailed view of the tetrameric interface between the nsp7 and nsp8 proteins is shown. C - A detailed view of the nsp7-nsp8 dimer with the dimeric interface highlighted. The protein backbones are shown in cartoon representation; the nsp7 and nsp8 proteins are depicted in blue and yellow, respectively. Selected amino acid residues involved in the nsp7-nsp8 interaction are presented in the stick representation with carbon atoms colored according to the protein assignment. D - Topology plot of the nsp7-nsp8 protein complex. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 1.
Statistics for data collection and processing, structure solution and refinement of the crystal structure of the SARS-CoV-2 nsp7-nsp8 complex. Numbers in parentheses refer to the highest resolution. R.m.s.d., root-mean-square deviation.
| Crystal | SARS-CoV-2 nsp7-nsp8 |
|---|---|
| PDB accession code | 6YHU |
| Data collection and processing | |
| Space group | P 1 21 1 |
| Cell dimensions - a, b, c (Å) | 56.4 49.7 64.8 |
| Cell dimensions - α, β, γ (°) | 90.0 106.4 90.0 |
| Resolution range (Å) | 38.79–2.0 (2.071–2.0) |
| No. of unique reflections | 23,341 (2,237) |
| Completeness (%) | 99.39 (95.15) |
| Multiplicity | 6.4 (5.0) |
| Mean I/σ(I) | 17.10 (2.88) |
| Wilson B factor (Å2) | 25.7 |
| R-merge / R-meas (%) | 7.41 (52.17) / 8.08 |
| CC1/2 | 0.999 (0.902) |
| CC* | 1.000 (0.974) |
| Structure solution and refinement | |
| R-work (%) | 21.43 (26.96) |
| R-free (%) | 23.92 (29.23) |
| R.m.s.d. - bonds (Å) / angles (°) | 0.002 / 0.60 |
| Average B factors (Å2) | 30.3 |
| Clashscore | 0.00 |
| Ramachandran favored/outliers (%) | 100 / 0 |
The crystallographic asymmetric unit contained two nsp7-nsp8 complexes forming a putative 2:2 nsp7-nsp8 heterotetramer. The structure revealed an alpha–beta mixed fold where the dimerization core is exclusively composed of the α-helices whereas the two flanking C-terminal nsp8 domains have a mixed, but predominantly β-sheet fold. The nsp7 has a three helical bundle fold (α1nsp7 Lys2-Leu20, α2nsp7 Ser26-Leu41 and α3nsp7 Thr45-Ser61) while the nsp8 subunit is comprised of two subdomains: an α-turn-α motif (α1nsp8 Glu77-Leu98 and α2nsp8 Asp101-Asp112) and a C-terminal subdomain that has a four antiparallel β strand (β1nsp8 Ala125-Ile132, β2nsp8 Thr146-Tyr149, β3nsp8 Ala152-Val160, and β4nsp8 Leu184-Arg190) with a single inserted α-helix (α3nsp8 Tyr135-Thr141) (Fig. 1). The dimerization interface is formed by the α1 and α3 helices of nsp7 and α1 and α2 helices of nsp8, with the interface area of approximately 1,340 Å2. Within the crystal, two nsp7-nsp8 dimers form a putative 2:2 nsp7-nsp8 heterotetramer. The interface area between the two nsp7-nsp8 dimers consists of approximately 950 Å2 and is formed by the α1 and α2 helices of nsp7 and the α1 helix of nsp8 (Fig. 1B, C). Interestingly, both the dimerization and tetramerization interfaces are formed almost exclusively via the hydrophobic interactions. The dimer interface resembles a leucine zipper motif, it is primarily formed by five leucine residues (nsp7 Leu56, 60, 71 and nsp8 Leu95, 103) and further stabilized by two phenylalanine residues (nsp7 Phe49 and nsp8 Phe92) and several other adjacent hydrophobic residues. Furthermore, the interaction of these two nsp7-nsp8 dimers is further stabilized by a disulfide bridge between the symmetric cysteine residues Cys8 of nsp7.
Because the nsp7-nsp8 complex was reported to synthesize RNA we analyzed its surface electrostatic potential in order to identify potential RNA binding sites. A hallmark of an RNA binding site is a highly positively charged surface patch. Surprisingly, our analysis did not identify any substantially positively charged surfaces in the dimer. However, such a place is clearly visible in the case of the tetramer (Fig. 2 ). It is located at the symmetry related dimer:dimer interface. The interior of this interface is created exclusively by hydrophobic residues, however, its surface is framed by positively charged residues including nsp7 and nsp8 lysine and arginine residues (nsp7 Lys2, 7, 43 and nsp8 Lys78, 82, 97 and Arg96). Notably, all these residues are highly conserved among different coronaviruses except for nsp7 Lys43 that is only present in SARS-CoV and SARS-CoV-2 (Fig. 2B). We refer to this place as the putative RNA binding site.
Fig. 2.
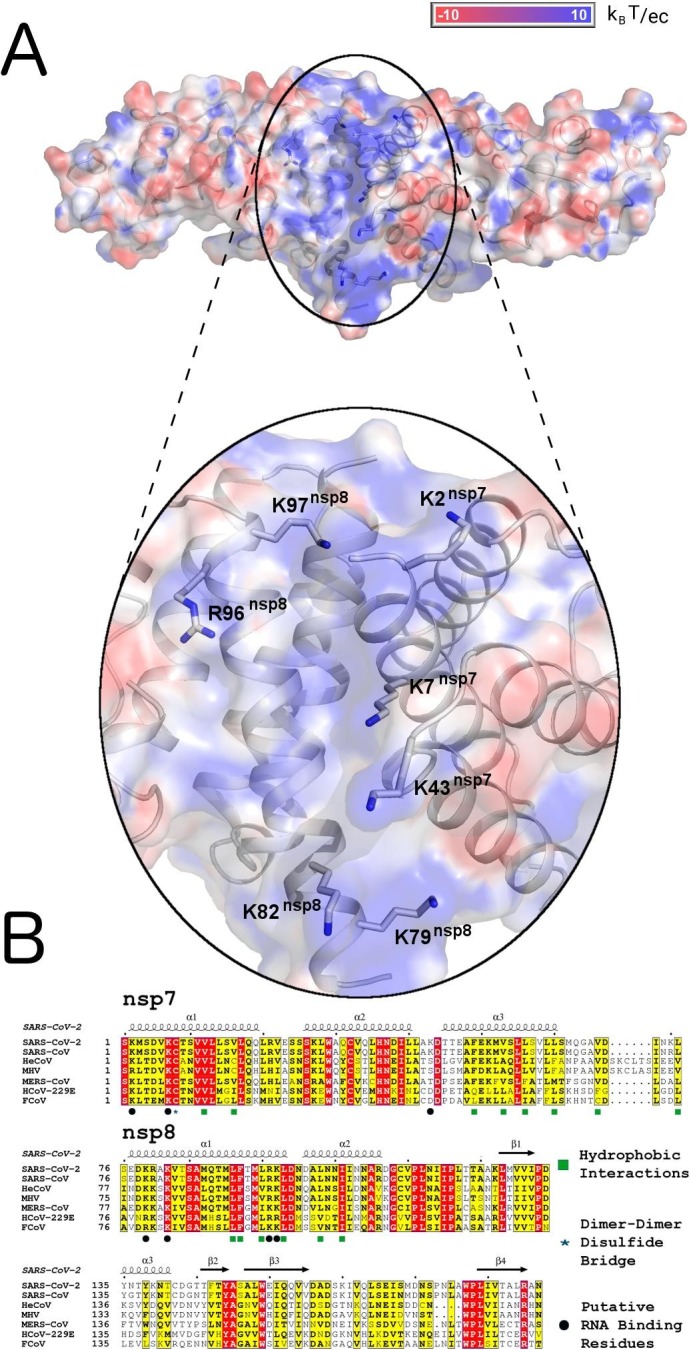
Surface and sequence analysis of the SARS-CoV-2 nsp7-nsp8 heterotetramer A - Electrostatic surface analysis. The surface is colored according to the electrostatic potential from red (negative charge) to blue (positive charge). In the lower panel, a detailed view of the positively charged groove at the nsp7-nsp8 tetrameric interface is shown with amino acid residues contributing to the positive charge of this groove highlighted in the stick representation. B - Sequence alignment of homologous coronaviral proteins. Residues in red boxes are completely conserved and those in yellow boxes are conserved more than 70%. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3. Discussion
The function of the nsp7-nsp8 complex remains unclear. Several different laboratories using nsp7-nsp8 complexes from a different coronavirus have shown that nsp7-nsp8 complex is capable of de novo short RNA synthesis (Imbert et al., 2006, Xiao et al., 2012). Such a function would imply the physiological function of the nsp7-nsp8 complex as a primase. In contrast, a hexadecameric (8x nsp7 + 8x nsp8) circular complex for the SARS nsp7-nsp8 was described (Zhai et al., 2005) and the authors proposed the ring to be wrapped around the RNA molecule and acting as a processivity factor for the RdRp. However, our study using the CoV-2 nsp7-nsp8 complex nor a structural study using a feline CoV nsp7-nsp8 (Xiao et al., 2012) observed such an intriguing arrangement of the nsp7-nsp8 complex.
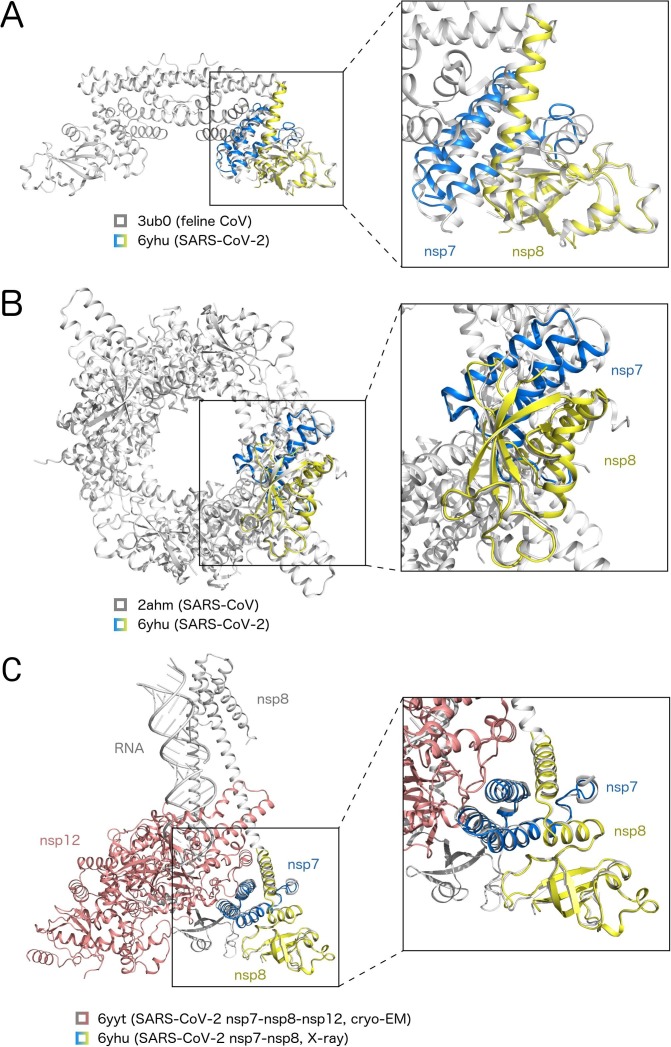
The recent cryoEM structure of the nsp7-nsp8-nsp12 fully-assembled polymerase complex suggests that the nsp7-nsp8 dimer is located too far from the nsp12 active site (Gao et al., 2020, Hillen et al., 2020) so cannot act as a primase for nsp12 at least in cis (Fig. 3 ). Notably, the conformation of the nsp7-nsp8 dimer observed here superposes rather well with subunits of higher order assemblies described for SARS and feline CoV-2 nsp7-nsp8 protein complexes (Fig. 3A, B) as well as with the cryo-EM structure of the replicating polymerase composed of nsp7-nsp8-nsp12 and RNA (Fig. 3C). Relatively small conformational changes could govern the stoichiometry of the nsp7-nsp8 protein complex. It is tempting to speculate that the CoV-2 nsp7-nsp8 complex has different functions, all biologically relevant such as processivity factor, primase or nsp12 activating factor depending on its conformation and stoichiometry. Further research has certainly been necessitated to elucidate nsp7-nsp8 assembly during CoV-2 infection and to establish its biological function during each phase of the viral replication within the host cell.
Fig. 3.
Superposition with known structures of related nsp7-nsp8 complexes Structure of the SARS-CoV-2 nsp7-nsp8 dimer presented in this study (pdb code 6yhu) was superposed with the hexameric structure of the feline CoV nsp7-nsp8 4:2 complex (A, pdb code 3ub0) or hexadecameric structure of the SARS-CoV 8:8 nsp7-nsp8 complex (B, pdb code 2ahm) or the cryo-electron microscopic structure of the SARS-CoV-2 RNA-dependent RNA polymerase complex composed of the nsp7, nsp8, and nsp12 proteins bound to two turns of RNA template-product duplex (C, pdb code 6yyt). The nsp7 and nsp8 proteins from the crystal structure are colored in blue and yellow, respectively. The nsp7 and nsp8 proteins and RNA from the cryo-EM structure are depicted in grey, while the nsp12 protein is colored in red. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Materials and methods
Protein expression and purification – The genes encoding nsp7 (GeneBank:YP_009725303, amino acid residues 1 – 71) and nsp8 (GeneBank: YP_009725304, amino acid residues 76 – 192) were commercially synthesized as codon optimized for E. coli (Invitrogen). The genes were cloned into a modified pHIS-2 vector containing N-terminal 6 X His-SUMO tag encoding sequence and expressed and purified using our protocols developed for viral polymerases (Dubankova et al., 2017). Briefly, both plasmids were co-transformed and co-expressed in E. coli (BL-21 Star (DE3)). Protein expression was induced by addition of IPTG to a final concentration of 0.6 mM, when the OD600 value of the culture reached 0.7. Then the temperature was reduced from 37 °C to 25 °C for 16 h. Bacteria were harvested by centrifugation and the cell pellet was resuspended in a lysis buffer (50 mM HEPES pH 7.5, 20 mM imidazole, 300 mM NaCl, 10% glycerol, 3 mM β-mercaptoethanol) and sonicated (Q700 Sonicator, QSonica). The lysate was cleared by centrifugation and the supernatant was incubated with an Ni-NTA agarose (Machery-Nagel), washed with lysis buffer and finally the protein was eluted with lysis buffer supplemented with 300 mM imidazole. The 6 X His-SUMO tag was digested using the Ulp1 protease at 4 °C overnight while dialyzing against 50 mM HEPES pH 7.5, 300 mM NaCl, 3 mM β-mercaptoethanol. The SUMO tag, Ulp1 protease and all uncleaved proteins were removed by Ni-NTA agarose and pure nsp7 and nsp8 proteins were then further purified by size exclusion chromatography using a Superdex 75 16/600 (GE Life Sciences) in 20 mM HEPES pH 7.5, 150 mM NaCl, 3 mM β-mercaptoethanol. Fractions containing both nsp7 and nsp8 proteins were concentrated to 5 mg/ml, flash frozen in liquid nitrogen and stored in −80 °C until needed.
Crystallization and crystallographic analysis – Crystals of the nsp7-nsp8 complex grew in seven days at 20 °C in sitting drops consisting of a 1:1 mixture (300 nl each) of the protein complex and the well solution (200 mM MgCl2, 100 mM Tris pH 8.5, 30% PEG 4000). Crystals were cryo-protected in the well solution supplemented by 20% (v/v) glycerol and flash frozen in liquid nitrogen.
The data were collected from a single frozen crystal at the home source, integrated and scaled using XDS (Kabsch, 2010). The structure of the SARS-CoV-2 nsp7 + nsp8 complex was solved by molecular replacement using the crystal structure of the SARS-CoV nsp7 + nsp8 complex (pdb code 2AHM) as a search model. The initial model was obtained with Phaser (McCoy et al., 2007) of the Phenix package (Adams et al., 2010). The model was further improved using automatic model refinement with Phenix.refine (Afonine et al., 2012) and subsequent rounds of manual model building in Coot (Emsley et al., 2010). The obtained statistics for data collection and processing, structure solution and refinement are summarized in Table 1.
Sequence analysis - Sequence alignment of coronavirus proteins homologous to SARS-CoV-2 nsp7 (YP_009725303*) and nsp8 (YP_009725304): SARS-CoV (NP_828865*, NP_828866), human enteric coronavirus strain 4408 (HeCoV; ACJ35483), murine hepatic virus strain A59 (MHV; P0C6X9), Middle East respiratory syndrome-related coronavirus (MERS-CoV; YP_009047219*, YP_009047220), human coronavirus 229E (HCoV; NP_073549), feline coronavirus (FCoV; AAY32594) was generated by ClustalW 2.1 (Thompson et al., 1994) and ESPript 3.0 (Gouet et al., 1999).
5. Accession codes
The atomic coordinates and structural factors of the crystal structure of the SARS-CoV-2 nsp7-nsp8 complex have been submitted to the Protein Data Bank (https://www.rcsb.org) and assigned the identifier 6YHU.
Author contributions
EK performed all experiments, MK interpreted the data and built the final model. RN and EB designed and supervised the project. All authors participated in preparation of the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The work was supported from European Regional Development Fund; OP RDE; Project: “Chemical biology for drugging undruggable targets (ChemBioDrug)” (No. CZ.02.1.01/0.0/0.0/16_019/0000729), by a grant from the Ministry of Health of the Czech Republic - Czech health research council (NU20-05-00472). The Academy of Sciences of the Czech Republic (RVO: 61388963) is also acknowledged. We are grateful to Michael Downey for critical reading of the manuscript.
References
- Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., McCoy A.J., Moriarty N.W., Oeffner R., Read R.J., Richardson D.C., Richardson J.S., Terwilliger T.C., Zwart P.H. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Section D, Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine P.V., Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M., Terwilliger T.C., Urzhumtsev A., Zwart P.H., Adams P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. Section D, Biol. Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D.K., Leung C.Y., Gilbert M., Joyner P.H., Ng E.M., Tse T.M., Guan Y., Peiris J.S., Poon L.L. Avian coronavirus in wild aquatic birds. J. Virol. 2011;85:12815–12820. doi: 10.1128/JVI.05838-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronaviridae Study Group of the International Committee on Taxonomy of, V., 2020. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol 5, 536-544. [DOI] [PMC free article] [PubMed]
- Dubankova A., Boura E. Structure of the yellow fever NS5 protein reveals conserved drug targets shared among flaviviruses. Antiviral Res. 2019;169 doi: 10.1016/j.antiviral.2019.104536. [DOI] [PubMed] [Google Scholar]
- Dubankova A., Humpolickova J., Klima M., Boura E. Negative charge and membrane-tethered viral 3B cooperate to recruit viral RNA dependent RNA polymerase 3D (pol) Sci. Rep. 2017;7:17309. doi: 10.1038/s41598-017-17621-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. Section D, Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Y., Yan, L., Huang, Y., Liu, F., Zhao, Y., Cao, L., Wang, T., Sun, Q., Ming, Z., Zhang, L., Ge, J., Zheng, L., Zhang, Y., Wang, H., Zhu, Y., Zhu, C., Hu, T., Hua, T., Zhang, B., Yang, X., Li, J., Yang, H., Liu, Z., Xu, W., Guddat, L.W., Wang, Q., Lou, Z., Rao, Z., 2020. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. [DOI] [PMC free article] [PubMed]
- Gouet P., Courcelle E., Stuart D.I., Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- Gralinski, L.E., Menachery, V.D., 2020. Return of the Coronavirus: 2019-nCoV. Viruses 12. [DOI] [PMC free article] [PubMed]
- Hercik K., Kozak J., Sala M., Dejmek M., Hrebabecky H., Zbornikova E., Smola M., Ruzek D., Nencka R., Boura E. Adenosine triphosphate analogs can efficiently inhibit the Zika virus RNA-dependent RNA polymerase. Antiviral Res. 2017;137:131–133. doi: 10.1016/j.antiviral.2016.11.020. [DOI] [PubMed] [Google Scholar]
- Hillen H.S., Kokic G., Farnung L., Dienemann C., Tegunov D., Cramer P. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020 doi: 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbert I., Guillemot J.C., Bourhis J.M., Bussetta C., Coutard B., Egloff M.P., Ferron F., Gorbalenya A.E., Canard B. A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO J. 2006;25:4933–4942. doi: 10.1038/sj.emboj.7601368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr. Section D, Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek, T.G., Erdman, D., Goldsmith, C.S., Zaki, S.R., Peret, T., Emery, S., Tong, S., Urbani, C., Comer, J.A., Lim, W., Rollin, P.E., Dowell, S.F., Ling, A.E., Humphrey, C.D., Shieh, W.J., Guarner, J., Paddock, C.D., Rota, P., Fields, B., DeRisi, J., Yang, J.Y., Cox, N., Hughes, J.M., LeDuc, J.W., Bellini, W.J., Anderson, L.J., Group, S.W., 2003. A novel coronavirus associated with severe acute respiratory syndrome The New England journal of medicine 348 1953 1966. [DOI] [PubMed]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Sebera J., Dubankova A., Sychrovsky V., Ruzek D., Boura E., Nencka R. The structural model of Zika virus RNA-dependent RNA polymerase in complex with RNA for rational design of novel nucleotide inhibitors. Sci. Rep. 2018;8:11132. doi: 10.1038/s41598-018-29459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Decroly E., Ziebuhr J. The nonstructural proteins directing coronavirus RNA synthesis and processing. Adv. Virus Res. 2016;96:59–126. doi: 10.1016/bs.aivir.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Velthuis A.J., van den Worm S.H., Snijder E.J. The SARS-coronavirus nsp7+nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucl. Acids Res. 2012;40:1737–1747. doi: 10.1093/nar/gkr893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl. Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres C.A., Hora A.S., Tonietti P.O., Taniwaki S.A., Cecchinato M., Villarreal L.Y., Brandao P.E. Gammacoronavirus and deltacoronavirus in quail. Avian Dis. 2016;60:656–661. doi: 10.1637/11412-032316-Reg.1. [DOI] [PubMed] [Google Scholar]
- Wang L., Byrum B., Zhang Y. Detection and genetic characterization of deltacoronavirus in pigs, Ohio, USA, 2014. Emerg. Infect. Dis. 2014;20:1227–1230. doi: 10.3201/eid2007.140296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M., Zheng B.J., Chan K.H., Yuen K.Y. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J., Sheng J., Quan L., Xia Z., Tan W., Cheng G., Jiang T. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Ma Q., Restle T., Shang W., Svergun D.I., Ponnusamy R., Sczakiel G., Hilgenfeld R. Nonstructural proteins 7 and 8 of feline coronavirus form a 2:1 heterotrimer that exhibits primer-independent RNA polymerase activity. J. Virol. 2012;86:4444–4454. doi: 10.1128/JVI.06635-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. New England J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhai Y., Sun F., Li X., Pang H., Xu X., Bartlam M., Rao Z. Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nat. Struct. Mol. Biol. 2005;12:980–986. doi: 10.1038/nsmb999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, N., Zhang, D., Wang, W., Li, X., Yang, B., Song, J., Zhao, X., Huang, B., Shi, W., Lu, R., Niu, P., Zhan, F., Ma, X., Wang, D., Xu, W., Wu, G., Gao, G.F., Tan, W., China Novel Coronavirus, I., Research, T., 2020. A Novel Coronavirus from Patients with Pneumonia in China, 2019. The New England journal of medicine 382, 727-733. [DOI] [PMC free article] [PubMed]
- Ziebuhr J. The coronavirus replicase. Curr. Top. Microbiol. Immunol. 2005;287:57–94. doi: 10.1007/3-540-26765-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla A., Hui D.S., Perlman S. Middle East respiratory syndrome. Lancet. 2015;386:995–1007. doi: 10.1016/S0140-6736(15)60454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla A., Chan J.F., Azhar E.I., Hui D.S., Yuen K.Y. Coronaviruses - drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]