Abstract
Introduction
Recent studies showed that SARS-CoV-2 RNA may be found in fecal specimens of COVID-19 patients, but the sample size is limited. This systematic review and meta-analysis examined the detection rate of SARS-CoV-2 RNA in fecal specimens of these patients according to their clinical characteristics.
Methods
MEDLINE, Embase, Scopus, and three Chinese biomedical databases were searched up to 25 March 2020 with no language restriction. We included original observational studies that reported the detection rate of SARS-CoV-2 RNA in fecal specimens of COVID-19 patients. Two separate reviewers conducted the review. Metaprop was adopted to conduct a meta-analysis of prevalence with variances stabilized by Freeman-Tukey Double Arcsine Transformation. A random-effects model was used. Heterogeneity across different studies was computed using Cochran's Q test and chi square statistics.
Results
From 17 studies, the pooled detection rate of fecal SARS-CoV-2 RNA was 43.7% (95% CI 32.6%-55.0%) and 33.7% (95% C.I. 33.7%, 95% C.I. 20.1%-48.8%) by patient and number of specimens as a unit count, respectively. Female individuals (59.6% vs. 53.5%), those who presented with gastrointestinal symptoms (77.1% vs. 57.7%), and patients with more severe disease (68.3% vs. 34.6%) tended to have a higher detection rate.
Discussion
A significant proportion of COVID-19 patients carry SARS-CoV-2 in their intestinal tract. Feces being a self-collected specimen bears a potential to improve case identification in community, especially for young children where proper respiratory sampling at home is difficult. Specific infection control strategies focusing on spread via fecal contamination and faulty toilet drainage are urgently needed.
Keywords: COVID-19, SARS-CoV-2, detection rate, faecal specimen, meta-analysis
Study Highlights
WHAT IS KNOWN
-
•
Recent studies showed that SARS-CoV-2 RNA may be found in fecal specimens of patients diagnosed with COVID-19, but the sample size is limited.
-
•
There is no systematic review and meta-analysis that have evaluated the detection rate of SARS-CoV-2 RNA in patients with confirmed COVID-19
WHAT IS NEW HERE
-
•
We performed a comprehensive systematic review and meta-analysis in MEDLINE, Embase and the Chinese biomedical database up to 25, March 2020.
-
•
The overall detection rate of fecal SARS-CoV-2 RNA was 43.7% (95% CI 32.6%−55.0%) and 33.7% (95% C.I. 33.7%, 95% C.I. 20.1%−48.8%) by patient and number of specimens as a unit count, respectively.
-
•
Subgroup analysis showed that female individuals, those who presented with gastrointestinal symptoms, and patients with more severe disease tended to have a higher detection rate.
-
•
Publication bias was not detected, and sensitivity analysis by consecutively removing each study from the overall analysis showed that the results were reliable and robust.
-
•
Specific infection control strategies focusing on community and hospital spread via fecal contamination and faulty toilet drainage are urgently needed.
Introduction
The novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was first identified in Wuhan city of China in December 2019, and since then its spread has been observed in various countries.1 Owing to an increasing number of confirmed cases worldwide, the WHO has announced the emerging outbreak of coronavirus disease 2019 (COVID-19) as an International Public Health Emergency of Concern (PHEIC).2 As of 02 April 2020, there were 827,419 confirmed cases, 40,777 deaths among patients with confirmed diagnosis, and affected 206 countries or regions.3 Given its high infectivity and morbidity, the pandemic poses a substantial global burden of disease and represents a serious threat to population health.4 , 5
Real-time reverse transcription-polymerase chain reaction (rRT-PCR) of nasopharyngeal swabs has been typically used to confirm clinical diagnosis.6 Affected patients usually present with respiratory tract symptoms such as cough, fever, fatigue and shortness of breath, which reflects the infection of respiratory epithelial cells and the human-to-human transmission via the airways.7 Chest computed tomography of affected patients is typically ground-glass opacity. However, some patients with confirmed infection reported gastrointestinal symptoms like diarrhea and vomiting,7 and recent studies showed that the SARS-CoV-2 RNA could be detected in stool specimens.8 , 9 These observations supported involvement of the gastrointestinal tract in SARS-CoV-2 infection, and raised the potential of using fecal specimens for case detection and monitoring clearance. More importantly, there are concerns about faecal contamination as source of spread. However, the sample size of individual reported studies has been too small to make a definitive conclusion. Besides, there has been no studies that examined the characteristics of infected patients who shed SARS-CoV-2 in faeces.
The primary objective of this systematic review and meta-analysis is to examine the detection rate of SARS-CoV-2 RNA in faecal specimens of patients with confirmed COVID-19. Second, we attempted to evaluate the prevalence of faecal SARS-CoV-2 RNA in populations of different sociodemographic features, clinical characteristics including gastrointestinal and respiratory symptoms, disease severity, and timing of the disease course. In addition, we explored if published literature contains sufficient information that allowed pooling of data on the duration of positive fecal specimens from illness onset, respiratory clearance and disease recovery.
Methods
Search strategy and selection criteria
This systematic review was registered in PROSPERO of the National Institute for Health Research. We searched MEDLINE, Embase, Scopus, CNKI (China National Knowledge Infrastructure), WANFANG (China Online Journals), VIP (Chinese) up to 25 March 2020 for observational studies reporting the prevalence of SARS-CoV-2 in feces of patients with confirmed diagnosis of SARS-CoV-2 infection. We included literature published in Chinese as many datasets were generated from China, the first epicenter of this outbreak. A pre-determined search strategy (eTable 1) was used to search literature according to the quality of reporting of the MOOSE (Meta-analyses Of Observational Studies in Epidemiology) guidelines (eTable 2).10
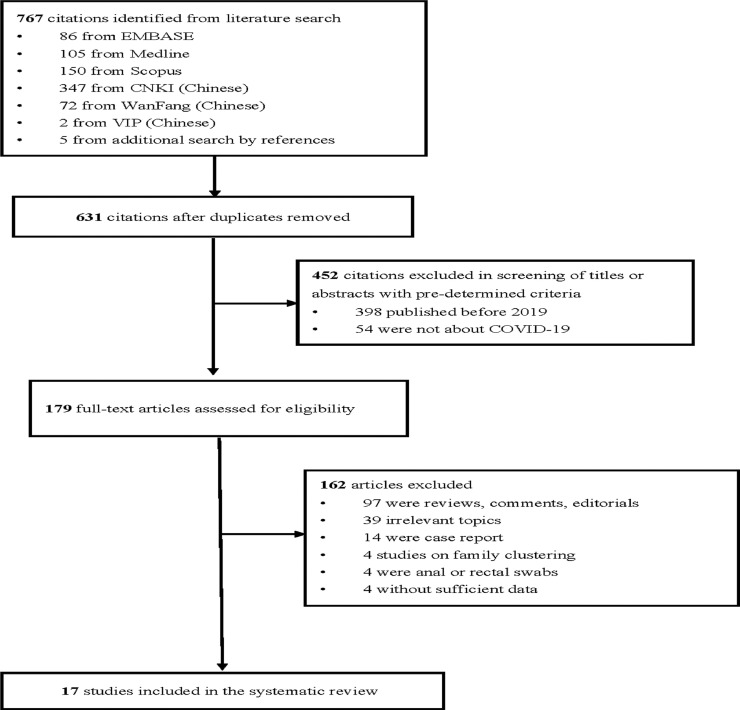
A multidisciplinary group conducted the meta-analysis led by MCSW with JH as reviewers (Fig. 1 ). Consensus was reached by referral to a third reviewer when there was disagreement. All returned citations were screened by title and abstract first, followed by full texts if relevant. The citations in the initial screening stage were excluded if they were published before 2019 or did not investigate the disease of SARS-CoV-2. Citations remaining were eligible for full-text screening. After reading the full texts, we excluded reviews, comments, editorials, case reports/series that focused on the clinical characteristics of several individuals (sample size less than five), and those that reported data on family clustering. We also excluded those studies that did not investigate the detection rate of SARS-CoV-2 RNA in fecal specimens; used anal or rectal swabs; or did not report enough data to compute the positivity rates. Both population-based and clinical studies were reviewed, with the former being defined as those that involved all residents in a specific region as the sampling frame based on a sampling method that was representative of that region. If there were citations based on the same study, the one reporting the most detailed information was used.
Fig. 1.
Selection of articles for systematic review.
Data extraction and quality assessment
Basic information collected from the individual studies consists of the name of first author; country and city of recruitment; age range; sex ratio; as well as sample size of the study participants. Relevant information was extracted to estimate the detection rate of SARS-CoV-2 RNA in fecal specimens, and the data retrieved included symptoms, disease severity, time of testing, methods of detection, duration of positive test results, number of samples tested, number of samples tested positive, number of patients tested and number of patients tested positive. Two reviewers (MCSW, JH) independently evaluated the quality of each included citation using the Appraisal tool for Cross-Sectional Studies (AXIS).11 It is a 20-point questionnaire that addressed study quality and reporting. The key areas addressed in the AXIS include study design, sample size justification, target population, sampling frame, sample selection, measurement validity and reliability, and overall methods.
Statistical analysis
A systematic, analytical method was used to compute the pooled prevalence rate of fecal SARS-CoV-2 from all included studies. The command “metaprop” was adopted to conduct meta-analysis of rates to generate pooled estimates with exact binomial and score test-based confidence intervals (CIs).12 The method provided appropriate ways of combining rates close to the margins by using the Freeman-Tukey Double Arcsine Transformation to stabilize the variances. A random-effects model was used to pool the prevalence of fecal SARS-CoV-2 detection with proportions and 95% CIs. Heterogeneity across different studies was computed using Cochran's Q test and chi square statistics. Subgroup analysis was conducted to explore the observed heterogeneity according to (1). age, (2). sex, (3). the presence of respiratory symptoms at the time of detection of SARS-CoV-2 in feces, (4). period of positive respiratory SARS-CoV-2, (5). timing of detection of SARS-CoV-2 in feces in relation to illness onset as well as respiratory and gastrointestinal symptoms, (6). timing of detection of SARS-CoV-2 in feces in relation to clearance of respiratory viruses. (7). disease severity (mild with no pneumonia, moderate with pneumonia, severe with pneumonia and desaturation, critical requiring mechanical ventilation), and (8). duration of positive fecal SARS-CoV-2 from illness onset, respiratory clearance, symptoms subside, and patient recovery. I2 > 50% referred to substantial heterogeneity. Publication bias was evaluated by Begg's and Egger's funnel plot with a significant p value of 0.05. Sensitivity analysis was conducted to evaluate the stability and robustness of the estimation of meta-analysis. All statistical analysis and graphic compositions were performed by Stata version 14.0 (College Station, Texas) and R version 3.3.2 (R Core Team).
Role of the funding source
There is no funding for this study. All investigators had full access to all the study data and had final responsibility for the decision to submit for publication.
Results
A total of 17 publications fulfilling the inclusion criteria were included in the analysis.7 , 9 , 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27 In the literature search, 767 citations were identified, of which 86 were from Embase,105 from MEDLINE, 150 from Scopus, 347 from CNKI (Chinese), 72 from WanFang (Chinese), 2 from VIP (Chinese), and 5 from additional search in reference sections (Fig. 1). There were 179 citations after removal of duplicates, those published before 2019, or literature that did not report SARS-CoV-2. We retrieved 179 full-text articles assessed for eligibility after 588 citations were excluded during title or abstract screening with pre-determined criteria. We excluded 162 articles as they were review, comments, editorials, case reports or case series with a sample size less than five; covered irrelevant topics; focused on family clustering; used anal or rectal swabs which were regarded as not a reliable quality of fecal sample; or did not present with sufficient data to evaluate the crude prevalence. Finally, there were 17 articles included in the present meta-analysis.
Study characteristics and quality assessment
The study characteristics of the selected articles were summarized in Table 1 . Among 17 studies, 15 were conducted in Mainland China and two others were performed in the United States and Singapore, respectively. Apart from the study by Cai et al. which evaluated fecal specimens among children,15 the average age of the study participants ranged from 32 to 53 years. Male patients consisted of 40% to 70% of all subjects. Only seven studies reported whether the patients presented with respiratory or gastrointestinal symptoms. Four studies reported the disease severity. The time from symptoms onset to SARS-CoV-2 testing ranged from 0 to 26 days, and patients in all studies received rRT-PCR as the diagnostic test. The duration of positive fecal test results ranged from 1 to 30 days. The positive rate of SARS-CoV-2 RNA with respect to the total number of specimens tested in the study ranged from 6.5% to 58.1%, whereas the fecal positive rate respect to patients with one or more positive samples ranged from 16.7% to 88.9%. The average number of samples tested per patient ranged from 1.024, 26 to 10.9.13
Table 1.
Characteristics of studies in the systematic review.
| Author | City | Country | Sample size | Age | Sex (male) | Respiratory symptoms | Digestive symptoms | Severity^ |
|---|---|---|---|---|---|---|---|---|
| Wang WL | Wuhan, Qingdao Beijing | China | 14/205* | 44 (5–67)* | 0.68* | NA | NA | C:14/14 |
| Xiao F | Zhuhai | China | 73 | 43 (0.83–78) | 0.56 | 53/73 | 26/73 | NA |
| Young BE | Singapore | Singapore | 8/18* | 47 (31–73)* | 0.5* | 12/18* | 3/18* | C: 6/18*; D: 2/18* |
| Cai JH | Shanghai, Hainan, Hefei, Qingdao | China | 6/10* | 6(0.25–11) | 0.40 | 5/6 | 0/6 | NA |
| Pan XF | Beijing | China | 17/82* | NA | NA | NA | NA | NA |
| Kujawski SA | NA | USA | 10/12* | 53 (21–68)* | 0.67* | 8/12* | 3/12* | C: 4/12*; D: 1/12* |
| Deng LS | Zhuhai | China | 56 | ≥18 y | NA | NA | NA | NA |
| Guan WJ | Wuhan | China | NA | NA | NA | NA | NA | NA |
| Ling Y | Shanghai | China | 66 | 44 (16–78) | 0.58 | NA | NA | NA |
| Xie CB | Chengdu | China | 9/19* | 34 (18–62) | 0.44 | 6/9 | 1/9 | NA |
| Zhang JC | Jinhua | China | 14 | 41 (18–87) | 0.50 | 10/14 | 0/14 | NA |
| Shi SR | Yibin | China | 7 | 32 (20–44) | 0.71 | NA | NA | mild |
| Chen Y | Ningbo | China | 11 | 41 (12–70) | 0.55 | 7/11 | 3/11 | NA |
| Wu BS | Xiamen | China | 36 | 49 (17–86) | 0.61 | NA | 4/36 | A: 8/36; B: 16/36; C: 9/36; D: 3/36 |
| Tang A | Zhoushan | China | 10 | 51 (28–67) | 0.70 | 7/10 | NA | NA |
| Li BL | Luzhou | China | 15 | NA | NA | NA | NA | NA |
| Zou JB | Chongqing | China | 27 | NA | NA | NA | NA | NA |
| Author | Test time | Method | Duration+ | Samples+ | Samples tested | Positivity (sample) | Patients+ | Patients tested | Positivity (patient) | Samples/patient# |
|---|---|---|---|---|---|---|---|---|---|---|
| Wang WL | NA | rRT-PCR | NA | 44 | 153 | 28.8% | 6 | 14 | 42.9% | 10.9 |
| Xiao F | 3–26 d | rRT-PCR | 1–12 d | NA | NA | NA | 39 | 73 | 53.4% | NA |
| Young BE | 5–15 d | rRT-PCR | 1–3 d | 8 | 22 | 36.4% | 4 | 8 | 50.0% | 2.8 |
| Cai JH | 3–13 d | rRT-PCR | 10–30+ d | NA | NA | NA | 5 | 6 | 83.3% | NA |
| Pan XF | 0–13 d | rRT-PCR | NA | NA | NA | NA | 9 | 17 | 52.9% | NA |
| Kujawski SA | 3–25 d | rRT-PCR | 25 d | 21 | 47 | 44.7% | 7 | 10 | 70.0% | 4.7 |
| Deng LS | NA | rRT-PCR | NA | NA | NA | NA | 25 | 56 | 44.6% | NA |
| Guan WJ | NA | rRT-PCR | NA | 4 | 62 | 6.5% | NA | NA | NA | NA |
| Ling Y | NA | rRT-PCR | NA | NA | NA | NA | 11 | 66 | 16.7% | NA |
| Xie CB | NA | rRT-PCR | NA | NA | NA | NA | 8 | 9 | 88.9% | NA |
| Zhang JC | 3–14 d | rRT-PCR | 1–3 d | 8 | 22 | 36.4% | 5 | 14 | 35.7% | 1.6 |
| Shi SR | 7–15 d | rRT-PCR | NA | NA | NA | NA | 0 | 7 | 0.0% | NA |
| Chen Y | 4–10 d | rRT-PCR | NA | NA | NA | NA | 4 | 11 | 36.4% | NA |
| Wu BS | 3–17 d | rRT-PCR | NA | 18 | 31 | 58.1% | 18 | 31 | 58.1% | 1.0 |
| Tang A | NA | rRT-PCR | NA | NA | NA | NA | 4 | 10 | 40.0% | NA |
| Li BL | NA | rRT-PCR | NA | 6 | 15 | 40.0% | 6 | 15 | 40.0% | 1.0 |
| Zou JB | NA | rRT-PCR | NA | NA | NA | NA | 6 | 27 | 22.2% | NA |
rRT-PCR, real-time reverse transcription polymerase chain reaction; NA, not available.
Test positive.
Average number of samples tested per patient.
Data from total patient group; NA, not available.
A: mild (no pneumonia), B: moderate (pneumonia), C: severe (desaturation needing Oxygen), D: critical (mechanical ventilation).
From quality assessment of all the included studies (eTable 3), All studies presented with: clear objectives (domain 1); appropriate study design (domains 2); clearly defined target and reference population (domain 4); sampling frame taken from an appropriate population base which represented the target and reference population under investigation (domain 5); robust sampling where subjects were representative of the target population (domain 6); measures undertaken to address non-responders (domain 7); statistical methods that were sufficiently described to enable repeatability of the study (domain 11); basic data that were adequately described (domain 12); response rates that did not raise concerns about bias (domain 13); and results for the analyses were described in the methods (domain 16). For other domains, the quality of reporting varied between the studies.
The prevalence of SARS-CoV-2 RNA in fecal specimens of confirmed COVID patients
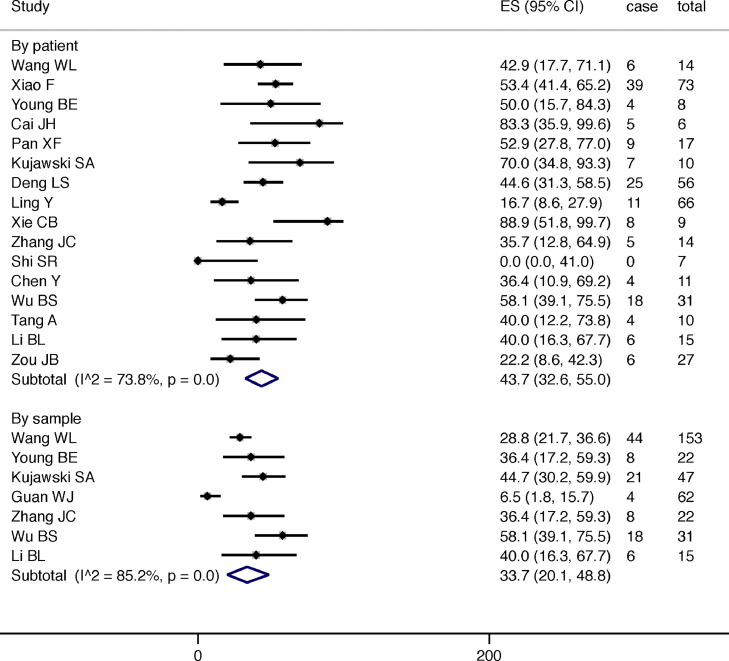
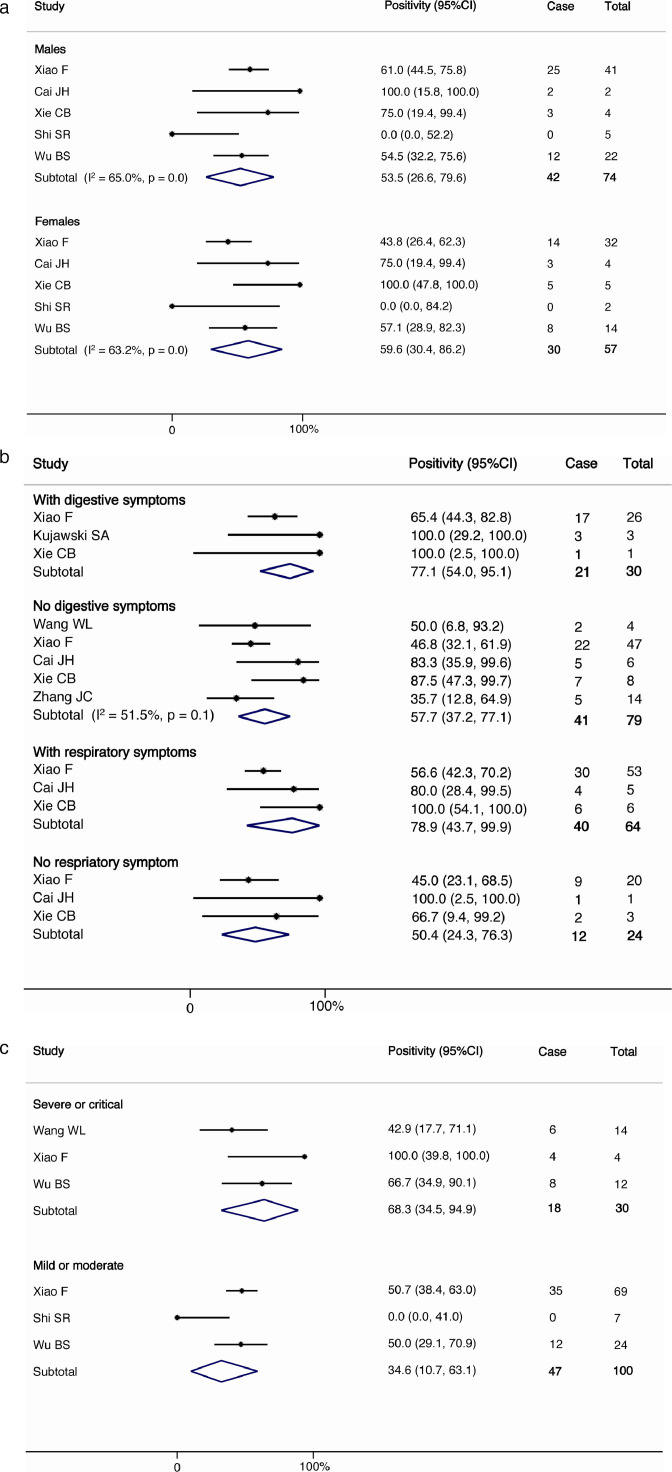
The pooled detection rate of SARS-CoV-2 RNA in fecal specimens among patients with confirmed diagnosis was 43.7% (95% C.I. 32.6%−55.0%, I2=73.8%, p<0.001) and 33.7% (95% C.I. 20.1%−48.8%, I2=85.2%, p<0.001) by patients enrolled (16 studies) and by total number of specimens tested in the whole study (7 studies), respectively (Fig. 2 ). The proportion of patients ever positive for fecal viral RNA was significantly correlated with the number of samples tested per patient in the study (r = 0.704) (eFig. 1). Female individuals (59.6%, 95% C.I. 30.4%−86.2%, I2=63.2%, p<0.001 vs. 53.5%, 95% C.I. 26.6%−79.6%, I2=65.0%, p<0.001 in male; Fig. 3a ), those who presented with gastrointestinal symptoms (77.1%, 95% C.I. 54.0%−95.1% vs. 57.7%, 95% C.I. 37.2%−77.1%, I2=51.5%, p<0.001) or respiratory symptoms (78.9%, 95% C.I. 43.7%−99.9% vs. 50.4%, 95% C.I. 24.3%−76.3%; Fig. 3b ), and patients with higher disease severity (68.3%, 95% C.I. 34.5–94.9% vs. 34.6%, 95% C.I. 10.7%−63.1%; Fig. 3c) tended to have a higher proportion with fecal SARS-CoV-2 RNA detected, although the between group proportions did not attain statistical significance. There is inadequate information on patients’ age, timing of the disease course, and the duration of positive fecal specimens at different occasions for meta-analysis.
Fig. 2.
The overall prevalence of SARS-CoV-2 in faecal samples of patients with confirmed diagnosis.
Fig. 3.
(a) The prevalence of SARS-CoV-2 according to gender (b)The prevalence of SARS-CoV-2 according to digestive and respiratory symptoms. (c) The prevalence of SARS-CoV-2 according to disease severity.
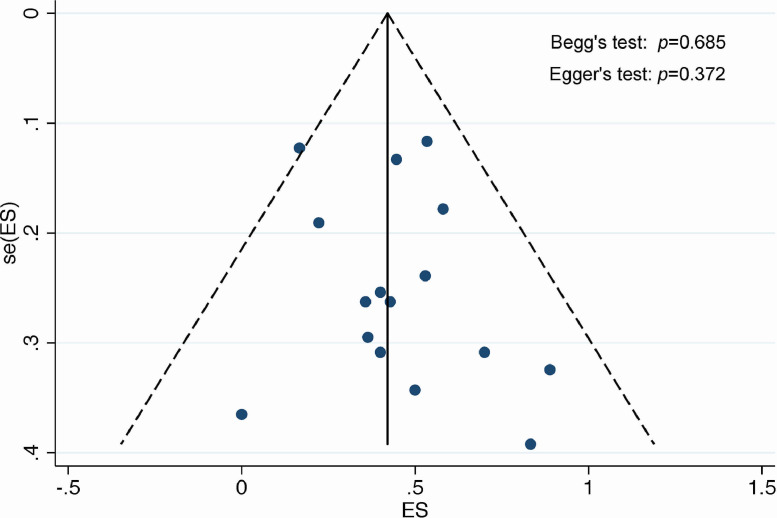
Fig. 4.
Funnel plot for assessment of publication bias.
Publication bias and sensitivity analysis
Based on funnel plot with pseudo 95% confidence intervals, we did not detect the presence of publication bias (Begg's test: p = 0.685; Egger's test: p = 0.372) (Fig. 4). Sensitivity analysis by consecutively removing each study from the overall analysis showed that the results were reliable and robust (eFig. 2).
Discussion
This systematic review and meta-analysis evaluated the pooled prevalence of fecal SARS-CoV-2 RNA in patients with COVID-19, and found that a significant proportion, pooled prevalence of 43.7%, had positive fecal test results. The prevalence was dependent on number of fecal samples tested per patient in the study. We also found that higher proportion of fecal SARS-CoV-2 RNA were detected in female subjects, patients with gastrointestinal symptoms or respiratory symptoms, and those with more severe disease, although the between-group difference did not attain statistical significance. There was no publication bias detected.
It has previously been shown that SARS-CoV-2 used Angiotensin Converting Enzyme (ACE) 2 as a viral receptor to gain access and infect cells.28 It was found that in the gastrointestinal system, the expression of ACE2 mRNA was high and was stabilized by a neutral amino acid transporter B0AT1, which provides a prerequisite for SARS-CoV-2 infection via the surface spike glycoprotein of the coronavirus.29 , 30 This is particularly true for the glandular epithelial cells of the ileum, jejunum, caecum and colon.29 The transmission of the virus via the extra-respiratory route such as the gastrointestinal system is compatible with the rapid spread of COVID-19. Transmission via faulty toilet drainage systems has been suspected in Hong Kong. To achieve stringent case detection in the community, a specimen type that can be self-collected at home is necessary. Fecal specimen fulfills such criteria, and therefore testing stool samples could be an option for community wide screening, especially for young children for whom it is difficult to collect a reliable respiratory sample by parents. Furthermore, fecal test for SARS-CoV-2 RNA also bear a potential implication for physicians to decide the appropriateness of discharge from the hospital, and subsequent quarantine or isolation strategies for those with positive fecal samples. While testing fecal specimen has potential clinical applications, it should be noted that it often requires multiple samples to identify infected persons as revealed in the current study. The existing literature, however, remains inadequate to fully understand the gastrointestinal involvement of SARS-CoV-2, in terms of the pathology, prognosis and treatment strategy.31
This is the largest systematic review and meta-analysis that evaluated the detection rate of SARS-CoV-2 RNA in fecal specimens of patients with confirmed diagnosis, including a total of 436 patients which is significantly higher than individual reports that often consist of data from less than 20 patients. In addition, we performed a comprehensive literature search which was extended to the gray literature extracted from the Chinese biomedical databases. The use of these databases has been recommended for meta-analysis that aimed for incorporation of these Chinese resources,32 potentially allowing more accurate evaluation of summary estimates with higher precision. This is especially true as a vast majority of these studies were performed in Mainland China, the first epicenter of COVID-19 outbreak. In addition, the quality of the studies was assessed as being high as they met most of the criteria from AXIS. However, there are several limitations that should be addressed. Firstly, the degree of heterogeneity is high, and this may be due to different study settings, patient characteristics, and the involvement of patients from various centers that might have their own testing tools for fecal specimens. Besides, the sample size is not large as the number of published studies on fecal SARS-CoV-2 RNA might be limited. Also, some studies had incomplete documentation of the exposure history, symptom onset, timing of clearance from respiratory infection, and patient discharge. Finally, quality assessment of the included studies showed that there was inadequate description of study non-responders.
In summary, this study found that a significant proportion of infected patients had positive SARS-CoV-2 RNA detected in their fecal specimens. Although, the method of detection does not differentiate between live and dead viruses, fecal contamination of environment objects as a means of transmission should not be neglected. Furthermore, possibility of fecal-oral route of transmission of SARS-CoV-2 should be investigated. The higher fecal virus detection rate in patients with gastrointestinal or respiratory symptoms and more critical illness indicated its potential predictive value as well as a pathogenic role. Future research should be performed with a multi-center design, involving a larger number of COVID patients with regular serial sampling and correlated with clinical and pathological characteristics in a systematic manner.
Guarantor of the article
Junjie Huang.
Financial support
None.
Potential competing interests
None declared.
Funding
This study was not funded.
CRediT authorship contribution statement
Martin CS Wong: Conceptualization, Writing - original draft, Data curation, Formal analysis. Junjie Huang: Data curation, Formal analysis. Christopher Lai: Formal analysis. Rita Ng: Formal analysis. Francis K.L. Chan: Conceptualization, Writing - original draft, Formal analysis. Paul K.S. Chan: Conceptualization, Writing - original draft, Formal analysis.
Declaration of Competing Interest
None declared.
Acknowledgments
We express our gratitude to Mr Peter Choi and Ms Xiao Chen for technical assistance in the research process.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jinf.2020.06.012.
Appendix. Supplementary materials
References
- 1.Huang C., Wang Y., Li X. Clinical features of patients with 2019 novel coronavirus in Wuhan, China. Lancet. 2020 doi: 10.1016/S0104-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO. COVID-19. Available athttps://www.who.int/zh/news-room/detail/30-01-2020-statement-on-the-second-meeting-of-the-international-health-regulations-(2005)-emergency-committee-regarding-the-outbreak-of-novel-coronavirus-(2019-ncov). Accessed 31 March 2020.
- 3.WHO. Coronavirus disease (COVID-19) outbreak situation. Available at:https://www.who.int/emergencies/diseases/novel-coronavirus-2019. Accessed 31 March 2020.
- 4.Chan J.F.W., Yuan S., Kok K.H. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen N., Zhou M., Dong X. Epidemiological and clinical characteristics of 99 cases of 2019 novel pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang D., Hu B., Hu C. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020 doi: 10.1001/jama.2020.1585. Published February 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guan W.J., Ni Z., Hu Y. Clinical characteristics of 2019 novel coronavirus infection in China. medRxiv. 2020 doi: 10.1101/2020.02.06.20020974. [DOI] [Google Scholar]
- 8.Holshue M.L., DeBolt C., Lindquist S. First Case of 2019 Novel Coronavirus in the United States. N Engl J Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stroup D.F., Berlin J.A., Morton S.C. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA. 2000;283:2008–2012. doi: 10.1001/jama.283.15.2008. [DOI] [PubMed] [Google Scholar]
- 11.Downes M.J., Brennan M.L., Williams H.C. Development of a critical appraisal tool to assess the quality of cross-sectional studies (AXIS) BMJ Open. 2020;6 doi: 10.1136/bmjopen-2016-011458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nyaga V.N., Arbyn M., Aerts M. Metaprop: a Stata command to perform meta-analysis of binomial data. Arch Public Health. 2014;72:39. doi: 10.1186/2049-3258-72-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang W., Xu Y., Gao R. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. March 11, 2020 doi: 10.1001/jama.2020.3786. Published online on. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Young B.E., Ong S.W., Kalimuddin S. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA. March 3, 2020 doi: 10.1001/jama.2020.3204. Published online on. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai J., Xu J., Lin D. A case series of children with 2019 novel coronavirus infection: clinical and epidemiological features. Clin Infect Dis. 28 February 2020 doi: 10.1093/cid/ciaa198. ciaa198Published. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pan Y., Zhang D., Yang P. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis. 2020 Feb 24 doi: 10.1016/S1473-3099(20)30113-4. pii: S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kujawski S.A., Wong K.K., Collins J.P. First 12 patients with coronavirus disease 2019 (COVID-19) in the United States. MedRxiv. 2020 doi: 10.1101/2020.03.09.20032896. [DOI] [PubMed] [Google Scholar]
- 18.Deng L., Li C., Zeng Q. Arbidol combined with LPV/r versus LPV/r alone against corona virus disease 2019: a retrospective cohort study. J Infect. 2020 doi: 10.1016/j.jinf.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ling Y., Xu S., Lin Y. The persistence and clearance of viral RNA in 2019 novel coronavirus disease survivors. Chin Med J. 2020 doi: 10.1097/CM9.0000000000000774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie C., Jianga L., Huang G. Comparison of different samples for 2019 novel coronavirus detection by nucleic acid amplification tests. Int J Infect Dis. 2020;93:264–267. doi: 10.1016/j.ijid.2020.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J.C., Wang, S.B., Xue Y.D. Fecal specimen diagnosis 2019 novel coronavirus–infected pneumonia. J Med Virol DOI: 10.1002/jmv.25742 [DOI] [PMC free article] [PubMed]
- 22.Shi S., Nie B., Yu Guo. Detection of 2019 novel coronavirus in various biological specimens of novel coronavirus pneumonia. West China Med J. 2020;35:132–136. [Google Scholar]
- 23.Chen Y., Li X., Jiang Y. Clinical features and treatment of 11 cases of COVID-19. Modern Pract Med. 2020;32:150–202. [Google Scholar]
- 24.Wu B., Yu T., Huang Z. Nucleic acid detection of fecal samples from confirmed cases of COVID-19. Chin J Zoonoses. 2020 doi: 10.3969/j.issn.1002-2694.2020.00.027. [DOI] [Google Scholar]
- 25.Tang A., Tong Z., Li K. et al. Epidemiological characteristics of coronavirus disease 2019 (COVID-19) cases in Zhoushan. Prev Med DOI: 10.19485/j.cnki.issn 2096-5087.2020.02.002.
- 26.Li B., Li Q., Wu G. Comparison of novel coronavirus test results of sputum and fecal specimens of 15 patients with COVID-19 after treatment. Clin J Infect Control. 2020;19:1–6. [Google Scholar]
- 27.Zou J., Zhou Y., Qiao J. et al. The report on the cured novel coronavirus-infected pneumonia patients with viral nucleic acid test positive in fecal specimens in Chongqing, China. Chin J Virol. DOI:10.13242/j.cnki.bingduxuebao.003653
- 28.Zhou P., Yang X., Wang X. et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harmer D., Gilbert M., Borman R. Quantitative mRNA expression profiling of ACE 2, a novel homologue of angiotensin converting enzyme. FEBS Lett. 2002;532:107–110. doi: 10.1016/s0014-5793(02)03640-2. [DOI] [PubMed] [Google Scholar]
- 30.Yan R. et al. Structural basis for the recognition of the 2019-nCoV by human ACE2. BioRxiv. 2020 doi: 10.1101/2020.02.19.956946. [DOI] [Google Scholar]
- 31.Chen C., Gao G., Xu Y. SARS-CoV-2-positive sputum and feces after conversion of pharyngeal samples in patients with COVID-19. Ann Intern Med. 2020 doi: 10.7326/M20-0991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen J.F., Korevaar D.A., Wang J. Should we search Chinese biomedical databases when performing systematic reviews. Syst Rev. 2015;4:23. doi: 10.1186/s13643-015-0017-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.