Abstract
Viruses, as obligate intracellular parasites, exploit cellular pathways and resources in a variety of fascinating ways. A striking example of this is the remodelling of intracellular membranes into specialized structures that support the replication of positive-sense ssRNA (+RNA) viruses infecting eukaryotes. These distinct forms of virus-induced structures include double-membrane vesicles (DMVs), found during viral infections as diverse and notorious as those of coronaviruses, enteroviruses, noroviruses, or hepatitis C virus. Our understanding of these DMVs has evolved over the past 15 years thanks to advances in imaging techniques and modern molecular biology tools. In this article, we review contemporary understanding of the biogenesis, structure, and function of virus-induced DMVs as well as the open questions posed by these intriguing structures.
Keywords: viral replication organelles, DMV, replication membrane, endomembranes, positive-sense RNA viruses, membrane remodelling
Highlights
Positive-sense RNA viruses utilize host membranes to generate viral replication organelles (ROs), inducing either invaginated spherules or double-membrane vesicles (DMVs) to support viral RNA synthesis.
DMVs have emerged as a recurrent RO motif among important human pathogenic viruses. These include picornaviruses such as enteroviruses and poliovirus, hepatitis C virus, noroviruses, and coronaviruses such as Middle East respiratory syndrome (MERS)- and severe acute respiratory syndrome (SARS)-coronaviruses.
Virus-induced DMVs appear to derive from membranes of the secretory pathway and form via several membrane remodelling steps coordinated by specific viral nonstructural proteins and host factors.
Recent evidence demonstrates that DMVs are a central hub for the synthesis of viral RNA (vRNA), making them attractive targets for novel antiviral interventions.
Viral Replication Organelles and DMVs
The replication of all +RNA viruses infecting eukaryotes occurs in the cytoplasm of the host cell in association with intracellular membranes that are transformed into distinctive and captivating structures. These virus-induced membrane modifications are increasingly referred to as viral replication organelles (ROs), a term that captures the idea of a subcellular structure that serves the invading virus rather than the host cell. Viral ROs may provide optimal platforms for viral RNA (vRNA) synthesis by concentrating viral replicative proteins and relevant host factors, and may hide replication intermediates, contributing to the evasion of innate immune sensors that can detect vRNA [1]. Additionally, they may help to spatially coordinate different steps in the viral replication cycle. Interestingly, while viral ROs are a hallmark of +RNA virus infections, the confinement of viral replication to a specific altered region of the cytoplasm appears to be an advantageous strategy that is also employed by other viruses [2., 3., 4.]. Due to their conserved role in the replication cycle of +RNA viruses, viral ROs are attractive targets for novel antiviral strategies.
Different families of +RNA viruses target diverse intracellular membranes to generate complex and unique membrane rearrangements. However, +RNA viruses seem to segregate into two main groups regarding the ROs they induce. The first group includes viruses that generate spherules via the formation of invaginations in specific host organelles, such as the endoplasmic reticulum (ER), mitochondria, or endolysosomes, (reviewed in [5]; see also [6,7]). These invaginated spherules remain connected to the cytosol by a narrow channel that can mediate import of metabolites and export of newly-synthesized positive-sense vRNAs to the cytosol for translation and packaging into new virions. The second class of +RNA viruses induces vesiculotubular membrane clusters containing different structural elements, among which DMVs have emerged as a recurrent motif. Double-membrane organelles are somewhat atypical structures, limited in animal cells to the nucleus, mitochondria, and autophagosomes. Yet DMVs are induced in cells infected with a number of +RNA viruses, including picornaviruses (e.g., poliovirus) [8., 9., 10.], noroviruses [11], hepatitis C virus (HCV) [12], arteriviruses [13,14], and coronaviruses such as the severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) coronaviruses [15., 16., 17.]. Moreover, DMV-like structures have also been reported in infections with a number of plant +RNA viruses [18,19]. In general, our understanding of the ROs associated with DMV-inducing viruses is relatively poorly developed due to the higher complexity and morphological variations in these membrane arrangements. This complexity implies an elaborate biogenesis process and makes straightforward structure–function interpretations challenging. Here, we review recent studies that have offered significant progress in this regard, with a special focus on human and animal viruses. We present emerging views on the replication of DMV-inducing viruses and highlight the conceptual and practical challenges that these structures continue to pose.
Common Structural Themes and Variations among DMV-Inducing Viruses
In the past decade, advances in electron microscopy (EM) imaging and EM sample preparation have greatly contributed to our knowledge of the ROs of DMV-inducing viruses (Box 1 ). In particular, electron tomography has provided exquisitely detailed 3D images of these ROs and revealed that, while broadly speaking, all virus-induced DMVs are ~100–300 nm vesicles delimited by two paired membranes, there are significant variations in their architecture (Figure 1 ). At one extreme, the DMVs induced by nidoviruses appear to define closed compartments whose outer membranes are frequently connected to other virus-induced membrane structures or to the ER, thus establishing large reticulovesicular membrane networks [13., 14., 15.,17,20]. At the other extreme, picornavirus-induced DMVs are disconnected structures that often appear in an open vase-like configuration [8., 9., 10.]. The DMVs induced by HCV and noroviruses seem to represent intermediate cases [11,12].
Box 1. Advances in EM for the Study of Viral ROs.
During the past 15 years, electron tomography (ET) has revealed the detailed 3D architecture of viral ROs [8., 9., 10., 11., 12., 13., 14., 15.,17,20]. To generate a 3D volume from the projection images generated in a transmission EM by ET (reviewed in [94]), a series of projection images is recorded while the sample (usually a 100–300 nm thick section through the infected cell) is gradually tilted in the microscope along one or two orthogonal axes. Subsequently, these tilt-series are computationally aligned and reconstructed into a 3D volume (tomogram) whose virtual slices are only a few nanometres thick, thus much thinner than the slices achievable by manual sectioning. In this way fine details of the RO architecture (e.g., small neck-like membrane connections), which may be obscured in the 2D projection images, can be resolved.
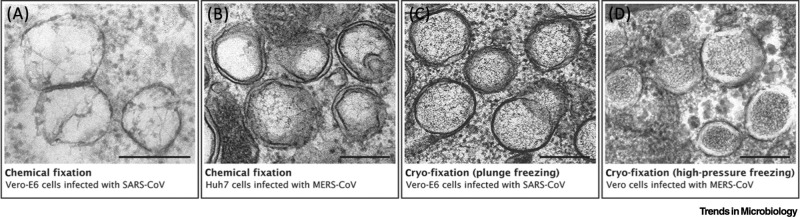
Prior to transmission electron microscopy (TEM) imaging, samples are typically fixed, stained and sectioned. The classical approach is chemical fixation, using cross-linkers such as glutaraldehyde or formaldehyde to stabilize the cellular ultrastructure for further processing. This method, however, is prone to structural artefacts which can be minimized by the use of cryo-fixation followed by freeze substitution [95]. With cryo-fixation, the sample is quickly frozen by immersion in a cryogen (plunge-freezing) or, more commonly, under pressure (high-pressure freezing) to hamper ice-crystal formation. Cryo-fixation allows better preservation of membranous structures; this can be particularly beneficial for the visualization of viral ROs [13,96]. Depending on the approach to fixation and the protocol used for subsequent staining steps, the appearance of different subcellular elements can vary significantly. Virus-induced DMVs, which appear to be particularly fragile, are perhaps one of the best examples to illustrate this point (Figure I) and how the interpretation of structural details should always be guided by the technical limitations in EM sample preparation.
Correlative light and electron microscopy (CLEM) has also become a valuable imaging approach to study viral infection [11,12]. CLEM methods facilitate the identification of specific target areas for sequential light and electron microscopy, after which data from the two modalities can be overlaid and interpreted together. Furthermore, the ability to monitor live cells ahead of CLEM adds temporal information to intrinsically static EM images, meaning rare or transient events, such as RO biogenesis, can be pinpointed ahead of fixation [26,76,97].
Figure I.
The Appearance of Double-Membrane Vesicles (DMVs) Can Drastically Change with Different Preparation Protocols for Electron Microscopy (EM).
The changing appearance of DMVs in different preparation protocols illustrated by images of coronavirus-induced DMVs. (A,B) Examples of DMVs found in cells infected with severe acute respiratory syndrome (SARS)-coronavirus (A) and Middle East respiratory syndrome (MERS)-coronavirus (B), both prepared using chemical fixation but with different protocols. The protocol used in (A) results in poor membrane preservation, and the DMV morphology is difficult to interpret. In (B), both DMV membranes are readily visible, though large artefactual gaps between the inner and outer membrane are present. In general, chemically fixed DMVs are deformed and only roughly spherical. (C,D) Examples of cryo-fixed DMVs induced by SARS-coronavirus (C, fixed by plunge freezing), and MERS-coronavirus (D, fixed by high-pressure freezing), respectively. DMV membranes appear tightly apposed and with minimal deformation, illustrating the superior structural preservation of this approach. (D) High-pressure freezing is considered to be the optimal fixation method for room temperature EM [95] and reveals a core of dense material inside the DMVs, which are surrounded by a denser cytosol. (A) was adapted from [96], copyright © American Society for Microbiology, and (C) was adapted from [15]. (B) and (D) are unpublished images from samples described in [17,16], respectively. Scale bars, 250 nm.
Alt-text: Box 1
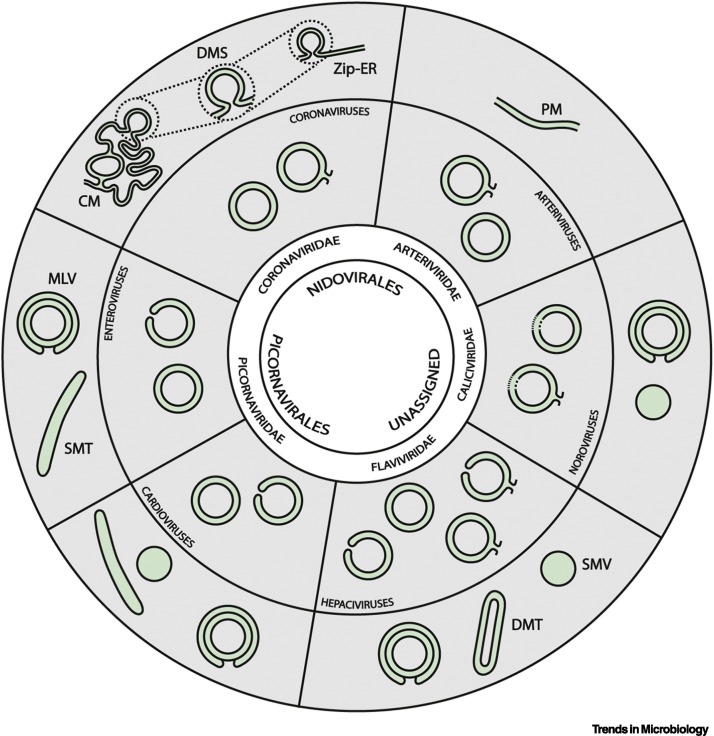
Figure 1.
Schematic Representing the Membrane Modifications Induced by Double-Membrane Vesicle (DMV)-Inducing Viruses.
The viral order and family are indicated in the core and inner ring respectively. The next ring depicts the types of DMV architecture that have been described for different virus families, which include open and/or closed DMVs that may be disconnected or continuous with other structures as shown. In most cases, these connections are established with the endoplasmic reticulum (ER), but nidoviral DMVs are also interconnected and connected to other virus-induced membrane structures. For noroviruses the open/closed state of DMVs is unreported, indicated by broken lines. The outer ring depicts additional virus-induced membrane structures. Single-membrane vesicles (SMVs) have been documented for HCV and noroviruses [11,12,25,27]. Single-membrane tubules (SMTs), sometimes together with SMVs, appear early in picornavirus infections and are DMV precursors [8., 9., 10.,98]. Picornaviruses, HCV, and noroviruses also generate multilamellar vesicles (MLVs), which arise late in infection from massive enwrapping of certain replication organelle (RO) elements by others [8., 9., 10., 11., 12.]. Double-membrane tubules (DMTs) are also late structures that may arise from DMVs [12]. Nidoviruses induce additional double-membrane structures, including paired membranes (PMs) [13,29] which, in the case of coronaviruses, adopt different configurations, from a nonbranching form, termed zippered ER (Zip-ER) [20,99], to a highly labyrinthine structure known as convoluted membranes (CMs) [15., 16., 17.,100]. Small open double-membrane spherules (DMSs), highly reminiscent of the invaginated spherules induced by other +RNA viruses, are formed as invaginations in Zip-ER or CM [17,20,99]. Colours indicate cytosolic (grey) or luminal (green) space.
Further confounding a universal view, these viruses induce different additional membrane structures, which often arise at particular stages in infection (Figure 1). When considered collectively, these structures define divergent and complex ROs that can transform over the course of infection. This raises questions about their potentially differentiated roles that remain largely unanswered. A particularly pertinent question is which of these elements serve as platforms for vRNA synthesis. Localization of key viral components (e.g., by immuno-EM) is insufficient to shed light on this matter as only a small proportion of these may be engaged in active replication complexes, as elegantly demonstrated using biochemical approaches [21]. Many viral proteins are known or suspected to have additional functions in the virus-directed manipulation of the infected cell. In recent years, the more reliable method of metabolic labelling of newly synthesized vRNA has produced abundant experimental evidence that points to DMVs as active sites of vRNA synthesis [9,10,17,22., 23., 24.]. However, intriguing differences seem to exist among virus groups. While the single-membrane structures of picornaviruses are also active sites of vRNA replication and, as predominant structures at the peak of viral replication, may be the most relevant RO elements [9,10,24], coronavirus-induced DMVs appear to be the primary – if not exclusive – sites of vRNA synthesis [17].
The Biogenesis of DMVs
While virus-induced invaginations can form on membranes of very diverse organelles [5], all DMV-inducing viruses studied so far appear to hijack membranes of the secretory pathway. In recent years, 3D ultrastructural analyses have provided key insights by revealing membranous connections between viral ROs and their putative membrane donor organelle (Figure 1). The detection of direct membranous continuities between DMVs and the ER points towards a primary role of ER membranes in the formation of ROs induced by nidoviruses [13., 14., 15.,17,20], HCV [12], noroviruses [11,25], and cardioviruses [24]. Enterovirus ROs, however, have typically been found as separate compartments lacking connections [8,9], which complicates the identification of the donor organelle. Only recently, a study combining live-cell imaging and whole-cell 3D-EM to capture early stages in RO formation revealed that enterovirus ROs originate from both ER and Golgi membranes, which seem to be used in a sequential manner [26].
From a mechanistic point of view, DMV biogenesis is a complex process that requires several membrane-remodelling steps. These include the induction of positive and negative curvature, membrane paring, and, depending on the DMV topology, several membrane fission events. These steps can be combined in numerous ways to form a DMV. Consequently, finding structural intermediates is key to elucidating the exact biogenesis path and has been accomplished in some cases. It should be noted, however, that short-lived intermediates may not be captured in EM images and, therefore, the existence of additional rapid DMV biogenesis routes cannot be entirely discarded. Possible intermediates in DMV formation have been clearly identified for several picornaviruses [8., 9., 10.], supporting a mechanism in which single-membrane structures transform into membrane-paired cisternae that subsequently curve and seal to form DMVs (Figure 2 ). The formation of abundant single-membrane structures early in infection is also a characteristic of HCV and norovirus infections [11,12,25,27], and DMV biogenesis paths similar to that of picornaviruses have been suggested [11,27,28]. Different lines of evidence point towards an alternative DMV biogenesis route for nidoviruses, where segments of paired ER membranes progressively bend to form a DMV (Figure 2). Putative intermediates in this transformation have been observed in arterivirus-infected cells [29], and, both for arteriviruses and coronaviruses, in conditions where DMV formation appeared to be impaired [30,31], suggesting a common mechanism for nidovirus DMV biogenesis.
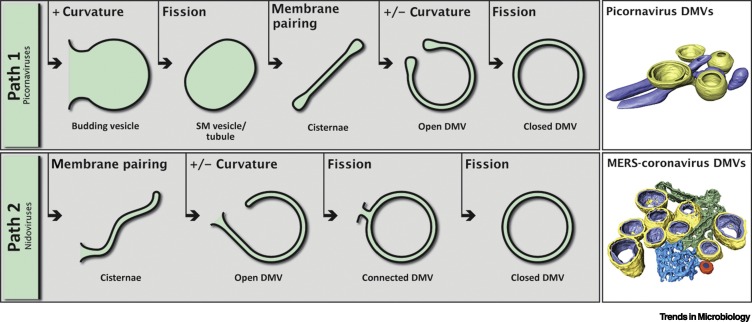
Figure 2.
Pathways of Double-Membrane Vesicle (DMV) Biogenesis and Representative 3D Models.
The biogenesis of DMVs induced by different +RNA viruses appear to occur through two main pathways. (Path 1, top) Initially, by the induction of positive membrane curvature, single-membrane (SM) vesicles or tubules bud out from the donor organelle. Through membrane pairing, these structures form cisternae that curve (inducing positive and negative curvature at the outer and inner membrane, respectively) to finally seal and transform into a closed DMV. This mechanism is supported by intermediate structures found in picornavirus infections [8., 9., 10.] and it has been speculated that norovirus- and Hepatitis C (HCV)-induced DMVs may be formed in a similar way [11,27,28]. (Top right) A 3D model of DMVs (yellow) next to virus-induced SM tubules (blue) in cells infected with encephalomyocarditis virus (family Picornaviridae, genus Cardiovirus), adapted from [10]. (Path 2, bottom) A different pathway of DMV biogenesis appears to take place in nidovirus-infected cells [29., 30., 31.]. Here, endoplasmic reticulum (ER) membranes would pair to form cisternae; these would subsequently curve and undergo one or two fission events to result in connected or free-floating DMVs. (Bottom right) A 3D model of Middle East respiratory syndrome (MERS)-coronavirus-induced DMVs (yellow and lilac, outer and inner membrane, respectively), connected to the ER (green) and convoluted membranes (blue) that include a double-membrane spherule (orange). Model adapted from [17].
The Role of Viral Proteins
+RNA viruses express polyproteins that are cleaved into numerous subunits, including nonstructural proteins that perform the enzymatic functions driving vRNA synthesis. Some nonstructural proteins contain transmembrane or membrane-tethering domains that play critical roles in DMV biogenesis. Upon ectopic expression in noninfected cells, some of these nonstructural proteins have been shown to be sufficient to induce the formation of DMVs. By contrast, the biogenesis of invagination-type ROs appears in some cases to be critically dependent on the presence of vRNA [32,33].
Using these systems, the nonstructural proteins nsp3 and nsp4, which contain predicted transmembrane domains, were identified as critical for coronavirus DMV biogenesis [31,34]. However, additional nonstructural proteins may be required in some coronavirus genera [35]. Their functional equivalents in arteriviruses [36], namely nsp2 and nsp3, have also been shown to be necessary and sufficient for DMV formation [37]. In the case of poliovirus, the most extensively studied of the picornaviruses, a combination of the membrane-associated nonstructural proteins 2BC and 3A seems to be sufficient for the formation of DMVs [38]. Strikingly, protein 3AB even induced the formation of structures resembling DMVs that appeared to form by invagination of liposomes in vitro [39]. For some viruses, expression of a single nonstructural protein is sufficient for the formation of DMVs. This is the case for norovirus NS4 and HCV NS5A, both of which contain an N-terminal amphipathic helix [11,12].
Nevertheless, the membrane phenotype observed after expression of the (minimal set of) proteins capable of inducing DMVs does not fully resemble that of infection. In addition to the lack of vRNA synthesis, differences between these structures and native DMVs include their abundance, their morphology, and the absence (or unusual abundance) of other membrane alterations. This indicates that additional factors may have a strong influence on DMV biogenesis. Some studies using expression of polyprotein fragments suggest that expression in the context of a complete viral polyprotein, polyprotein processing, and even the speed of this process can affect DMV formation [31,40]. Processing intermediates produced in a time-controlled fashion could be important, for example, in favouring interactions between nonstructural proteins that facilitate membrane remodelling. Intriguingly, while HCV DMV formation can be induced by NS5A alone [12], there is an additional requirement for a functional NS3 helicase domain when expression occurs in the context of an NS3-5B polyprotein [40]. Additional nonstructural proteins frequently assist or modulate DMV formation, as documented for HCV NS4B [28,41] and NS5B [12] (a highly hydrophobic protein and the RNA-dependent RNA polymerase, respectively), as well as for the arterivirus nsp5 transmembrane protein [29]. This may also be true of coronavirus nsp6 or norovirus NS1-2 and NS3, which have putative transmembrane helices and have been found to induce proliferation of altered membranes when expressed alone [11,34].
Despite the knowledge accumulated in recent years, the role of viral proteins in DMV biogenesis is only starting to unfold. Studies identifying critical domains and residues for DMV formation are still relatively scarce [28,40., 41., 42., 43., 44.]. Gaining insight into this aspect, and dissecting specific roles in the complex sequence of membrane remodelling steps required for DMV formation, will be essential to understand this process from a molecular perspective.
The Role of Host Factors
While viral factors are evidently critical for replication, their success is dependent on host factors that are recruited and subverted by +RNA viruses throughout the infection cycle [5,45]. In addition to supporting viral replication more broadly, host factors are also likely essential for RO formation, for example, as lipids or (recruited) protein complexes that act as scaffolds or introduce local asymmetry. While much of the available evidence is suggestive rather than mechanistic, this section discusses those host factors thought to play a role in DMV biogenesis.
Of particular interest are factors with canonical membrane-remodelling functions that are associated with the membranes from which DMVs are derived. In this regard, canonical membrane-scaffolding proteins, like the ER-associated reticulons, which have been shown to have an important role in the formation of virus-induced invaginations [46], are intuitive host-factor candidates that may contribute to DMV formation. Although the role of reticulons in DMV formation has not been specifically addressed, the association of reticulon 3 with enterovirus 71 (EV-71) protein 2C appears to promote enterovirus infection [47]. Intriguingly, both pro- and anti-replicative effects of reticulon 3 during HCV infection, presumably mediated by its interaction with HCV NS4B, have been reported [48,49]. Another host protein with canonical roles in membrane remodelling is the F-BAR domain-containing proline-serine-threonine phosphatase-interacting protein 2 (PSTPIP2). While abolishing the interaction between HCV NS4B/NS5A and PSTPIP2 limited DMV formation, viral replication reduced correspondingly [50]. In many cases it is difficult to establish whether reduced levels of DMV formation are a cause or consequence of reduced levels of replication under intervention. Indeed, given the multifaceted function of many host factors, and their requirement for viral replication, untangling specific roles across infection is often challenging and may require the use of systems where protein expression is uncoupled from viral replication.
Two other major cellular systems relying on ER membrane remodelling are the secretory and autophagy pathways, and both have been implicated in DMV formation. In particular, the autophagy pathway, which produces morphologically analogous structures to viral DMVs, is an intuitive candidate. Interestingly, the autophagy marker LC3 has been found to colocalize with viral replicase proteins for a number of DMV-inducing viruses [51., 52., 53., 54.]. Nevertheless, the significance of this association and, more broadly, the specific involvement of (canonical) autophagy in DMV formation, remains unclear. For HCV, the early steps of autophagy have been suggested to be involved in DMV biogenesis. ATG5 has been strongly implicated in HCV replication [54,55], and limiting the formation of ATG5-12 conjugates during infection resulted in the formation of fewer and smaller viral DMVs [55]. A role has also been suggested for the early-acting autophagy protein DFCP1 [54,56], while inhibiting the recruitment or components of the ATG14L autophagosome initiation complex was found to reduce DMV numbers [57].
While COPI- or COPII-coated secretory vesicles, or proteins promoting their formation, have been implicated in the replication of many DMV-inducing +RNA viruses, DMV biogenesis via the subversion of machineries ostensibly used for the production of single-membrane vesicles is conceptually challenging. In the case of coronaviruses, inhibition of the early secretory protein GBF1 has been shown to reduce DMV numbers, albeit with a corresponding drop in replication [58]. However, while GBF1 appears to play an important role in the replication of enteroviruses and HCV, its inhibition was not found to affect DMV formation [59,60].
Lipids are also important determinants of membrane asymmetry, curvature, and DMV formation [45,61]. Both phosphatidylinositol 4-phopshate (PI4P) and cholesterol are potent modifiers of membrane properties [62,63]. In the case of picornaviruses, PI4P is an essential host factor for replication and has been shown to have a direct role in expediting enterovirus RO formation [24,64]. PI4P is also critical in HCV infection, and it modulates the morphology of the HCV ROs, often referred to as the membranous web [65]. Inhibition of oxysterol-binding protein (OSBP), which mediates a PI4P-cholesterol counter current during enterovirus and HCV infections [66., 67., 68.], has also been shown to alter HCV membranous web formation [68]. Together with data demonstrating that endosomal cholesterol homeostasis can influence DMV size, this suggests a role for cholesterol in HCV RO formation [69].
Viruses must also stimulate the import or synthesis of lipid constituents to support the rapid proliferation of membranes for DMV biogenesis. While they are relatively minor membrane constituents, lysophospholipids (LPLs) have nonetheless the potential to modify membrane curvature and have been shown to be important for coronavirus infections, as reduced LPL levels resulted in a dramatic reduction in replication and loss of DMVs [70]. Unlike LPL, phosphatidylcholine (PC) is a major structural component of secretory pathway membranes. Its biosynthesis and regulators thereof have been shown to be crucial for the replication of enteroviruses, cardioviruses, and HCV [71., 72., 73.], where it may serve to support the expansion of suitable membranes for DMV formation. Indeed, enterovirus DMV formation was severely impaired under conditions where PC biosynthesis was limited [73]. Access to the PC required for enterovirus RO formation and replication is now thought to depend upon membrane contact sites between viral ROs and lipid droplets (LDs) [26,73,74], mediated by the membrane-associated viral 2C protein [74]. In the case of HCV, the association between the HCV core protein and LDs, and their importance for virus particle assembly, is long established [75]. More recently, a striking physical association has been found between apparently depleted LDs enwrapped in ER and viral DMVs [76]. This could suggest tightly co-ordinated roles for LDs both in sustaining HCV DMV biogenesis for replication and in supporting virion assembly.
Connecting DMV Structure and Function
Despite the accumulated knowledge on the structure, function, and biogenesis of DMVs, a fundamental question remains unanswered: what are the specific advantages of forming these atypical structures for viral replication? Strictly speaking, this question could be extended to other types of viral ROs, as many of the proposed RO functions are based on rational speculation rather than on direct evidence. Answering this question may ultimately require comparison with systems in which virus replication can occur without (typical) ROs, which are understandably rare. A remarkable example is that of a mutant CVB3 enterovirus that, in the exponential phase of vRNA synthesis, can replicate its genome in the absence of ROs, using instead morphologically intact Golgi membranes [24]. Together with evidence that replication can be redirected to alternative platforms for some +RNA viruses that induce invaginations [77,78], this underlines that the core requirements for replication are not always tied to specific membrane compartments. Interestingly, this delay in enterovirus RO formation did not seem to induce accelerated sensing of vRNA or an enhanced innate immune response during one cycle of viral replication, challenging the idea that ROs may be critical in shielding vRNA from cellular sensors of the innate immune system. A subsequent study on poliovirus showed that, under conditions where RO formation may be more severely impaired, recognition of vRNA by a cellular sensor can be enhanced, though it may take several replication cycles to generate an antiviral response that translates to reduced virus propagation [73].
The implication that vRNA in ROs could be shielded from detection by the host is also supported by the observation that immune sensors of double-stranded RNA (dsRNA, present in vRNA replication intermediates), such as RIG-I and MDA5, have restricted access to ROs in HCV-infected cells, which benefits viral replication [79]. The mechanistic details involved, and even the specific contribution of the ROs to this phenomenon, are unclear; however, components of the nuclear transport machinery, some of which are found in HCV RO regions [80], appear to play key roles in the observed segregation of immune sensors and ROs. Along similar lines, several studies with different DMV-inducing viruses indicate that (intact) ROs have a vital function in shielding vRNA from RNase access or from immunodetection [21,23,81., 82., 83.] making it plausible that this protective function could also serve to conceal viral replication intermediates from cellular sensors. Nevertheless, it remains unknown whether this effect depends on specific RO morphologies. Unlike the spherules induced by other +RNA viruses, which define compartments that are relatively isolated from the cytosol, all the membrane structures induced by DMV-inducing viruses represent large exposed surfaces that could be vulnerable to sensing. For these viruses, the concept that the morphology of their ROs is adapted to shield vRNA is less intuitive. In this respect, picornavirus-induced single-membrane structures, which are proven active sites of vRNA synthesis and predominate at the peak of replication [8., 9., 10.], probably represent the most challenging example. Here, the cytosol-facing replication complex must associate with an exposed RO surface, which defies the idea of compartmentalization as a general RO mechanism to conceal viral replication intermediates and suggests alternative evasion strategies, for example, through shielding by the viral replication machinery.
Although DMVs also have large exposed outer surfaces, replication may instead occur in association with the inner membrane facing the cytosolic interior of the DMV, which could clearly provide the kind of secluded environment ideal for viral replication. Such a compartment, however, should remain capable of exchanging material with the cytosol in order to import metabolites and export the positive-sense vRNA products for translation and encapsidation into progeny virions. In a virus-induced spherule, the neck-like channel that connects it with the cytosol appears to serve this purpose. By contrast, closed DMVs without apparent openings to the surrounding cytosol have regularly been observed in infected cells (Figure 1). For HCV and picornaviruses, these coexist with open DMVs, which are likely their precursors [8., 9., 10.,12]. It is thus conceivable that productive vRNA synthesis occurs inside open DMVs that eventually become sealed, perhaps to hide an excess of vRNA accumulated throughout infection. Alternative functions of these DMVs have also been proposed. Recent studies suggest that some viruses may be able to transfer the DMV content to other cells, using mechanisms such as extracellular vesicles or nanotubes [84., 85., 86.]. This content potentially includes genomic RNA, which would be sufficient to initiate an infection if delivered into a naïve cell. In the case of enteroviruses, where some DMVs appear to contain progeny virions, DMVs have been linked to en bloc transmission of mature virions [84], which could increase the likelihood of a viable infection if complementary viral quasispecies are exported together. These mechanisms could confer an additional benefit by facilitating viral propagation in a manner that eludes antibody neutralization.
The specific case of nidoviruses challenges the idea that only open DMVs are involved in effective RNA synthesis. Nidovirus-induced DMVs have been systematically documented as closed structures [13,15,17,20] yet they have been shown to be the central hub for vRNA synthesis in coronavirus infection [17] and they accumulate the bulk of immunodetected dsRNA [13,15,17,87]. Despite the closed appearance of many virus-induced DMVs, mechanisms for material exchange with the cytosol are still conceivable, for example, in the form of molecular pores that may be undetectable in conventional EM samples. Furthermore, it should be noted that it is still unclear whether DMV-associated vRNA synthesis occurs on the inner, outer, or both membranes.
Concluding Remarks
The nature of viral DMVs, as effective sites for replication and/or shields against cellular defences, has been extensively explored over previous decades. DMVs are sites of vRNA replication for HCV [23], appear to be the major replication platform for coronaviruses [17,22], and a substantial support for vRNA replication in picornaviruses [9,10,24], demonstrating that these structures possess the minimum requirements to serve as replication membranes. While it is currently unclear whether the roles of DMVs are conserved across these viruses, and how critical DMVs are for effective viral propagation, these remain vital questions as demand for broad-acting antivirals remains high and viral ROs constitute a largely unexplored target [88., 89., 90.]. Our understanding of the additional membrane structures commonly found alongside DMVs also remains incomplete. If and how these diverse morphologies arise from a single membrane origin, and what their functional significance might be, remain fascinating open questions. An important conceptual distinction exists between virus-induced membrane structures that support viral genome replication and those that do not. Indeed, our definition of the viral RO may evolve as our understanding of replication membranes versus alternative membrane structures and functions advances (see Outstanding Questions). Further identifying and investigating host factors will play an important part in determining the roles of these structures and the compositional divergences that allow different morphologies to arise. To this end, effective methods for host factor discovery, including proximity labelling [91] and lipidomic approaches [92], will be increasingly valuable. These investigations can be complemented with powerful imaging approaches such as cryoelectron tomography and sub-tomogram averaging, which can be used to reveal details of RO complexes at the molecular level [93], representing a holistic approach for elucidating how viral and host factors come together to confer replication membrane form and function.
Outstanding Questions.
What are the morphological constraints and minimal components required to create a membrane platform suitable for viral replication?
Do the specific morphologies of membrane modifications induced by +RNA viruses, such as DMVs, confer a selective advantage for viral replication? Is the DMV morphology critical to the success of those invading viruses that induce their formation?
Can DMVs be effectively targeted for antiviral drug development?
What are the molecular details of the interplay between viral proteins and host factors that generate DMVs? How conserved are the host factor requirements for DMV biogenesis among the viruses that utilize them?
How do we overcome the challenge of identifying essential host and viral factors with specific roles in DMV biogenesis? What are the roadblocks to developing broadly applicable replication-independent assays that uncouple protein expression from the need for viral replication?
Does viral RNA synthesis occur at the outer or inner surface of DMVs? In the latter case, how is access between the cytosol and replication complex maintained during the peak of replication?
What additional functions might DMVs perform beyond serving as effective platforms for viral RNA synthesis? How critical are these functions?
Could some of the membrane modifications found alongside DMVs be by-products of viral infection (e.g., of viral protein overexpression)? Or do they serve specific functions in the viral replication cycle?
Alt-text: Outstanding Questions
Acknowledgments
The authors would like to thank A.J. Koster [Leiden University Medical Center (LUMC)] for his suggestions regarding the manuscript, and the many other colleagues at LUMC and elsewhere who have contributed over many years to our virus research.
References
- 1.Scutigliani E.M., Kikkert M. Interaction of the innate immune system with positive-strand RNA virus replication organelles. Cytokine Growth Factor Rev. 2017;37:17–27. doi: 10.1016/j.cytogfr.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Castro I.F. Virus factories: biogenesis and structural design. Cell. Microbiol. 2013;15:24–34. doi: 10.1111/cmi.12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu L. From crescent to mature virion: vaccinia virus assembly and maturation. Viruses. 2014;6:3787–3808. doi: 10.3390/v6103787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tenorio R. Function, architecture, and biogenesis of reovirus replication neoorganelles. Viruses. 2019;11:288. doi: 10.3390/v11030288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harak C., Lohmann V. Ultrastructure of the replication sites of positive-strand RNA viruses. Virology. 2015;479:418–433. doi: 10.1016/j.virol.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cortese M. Ultrastructural characterization of Zika virus replication factories. Cell Rep. 2017;18:2113–2123. doi: 10.1016/j.celrep.2017.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernandez de Castro I. Three-dimensional imaging of the intracellular assembly of a functional viral RNA replicase complex. J. Cell Sci. 2017;130:260–268. doi: 10.1242/jcs.181586. [DOI] [PubMed] [Google Scholar]
- 8.Limpens R.W. The transformation of enterovirus replication structures: a three-dimensional study of single- and double-membrane compartments. mBio. 2011;2 doi: 10.1128/mBio.00166-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belov G.A. Complex dynamic development of poliovirus membranous replication complexes. J. Virol. 2012;86:302–312. doi: 10.1128/JVI.05937-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Melia C.E. The origin, dynamic morphology, and PI4P-independent formation of encephalomyocarditis virus replication organelles. mBio. 2018;9 doi: 10.1128/mBio.00420-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doerflinger S.Y. Membrane alterations induced by nonstructural proteins of human norovirus. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Romero-Brey I. Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knoops K. Ultrastructural characterization of arterivirus replication structures: reshaping the endoplasmic reticulum to accommodate viral RNA synthesis. J. Virol. 2012;86:2474–2487. doi: 10.1128/JVI.06677-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang W. An integrated analysis of membrane remodeling during porcine reproductive and respiratory syndrome virus replication and assembly. PLoS One. 2018;13 doi: 10.1371/journal.pone.0200919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knoops K. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6 doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Wilde A.H. MERS-coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin A or interferon-alpha treatment. J. Gen. Virol. 2013;94:1749–1760. doi: 10.1099/vir.0.052910-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.E.J. Snijder, et al., A unifying structural and functional model of the coronavirus replication organelle: tracking down RNA synthesis, PLoS Biol. 18, e3000715. 10.1371/journal.pbio.3000715 [DOI] [PMC free article] [PubMed]
- 18.Gushchin V.A. Beet yellows virus replicase and replicative compartments: parallels with other RNA viruses. Front. Microbiol. 2013;4:38. doi: 10.3389/fmicb.2013.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin X. Three-dimensional architecture and biogenesis of membrane structures associated with plant virus replication. Front. Plant Sci. 2018;9:57. doi: 10.3389/fpls.2018.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maier H.J. Infectious bronchitis virus generates spherules from zippered endoplasmic reticulum membranes. mBio. 2013;4 doi: 10.1128/mBio.00801-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quinkert D. Quantitative analysis of the hepatitis C virus replication complex. J. Virol. 2005;79:13594–13605. doi: 10.1128/JVI.79.21.13594-13605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gosert R. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 2002;76:3697–3708. doi: 10.1128/JVI.76.8.3697-3708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paul D. Morphological and biochemical characterization of the membranous hepatitis C virus replication compartment. J. Virol. 2013;87:10612–10627. doi: 10.1128/JVI.01370-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Melia C.E. Escaping host factor PI4KB inhibition: enterovirus genomic RNA replication in the absence of replication organelles. Cell Rep. 2017;21:587–599. doi: 10.1016/j.celrep.2017.09.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wobus C.E. Replication of Norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol. 2004;2 doi: 10.1371/journal.pbio.0020432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Melia C.E. Origins of enterovirus replication organelles established by whole-cell electron microscopy. mBio. 2019;10 doi: 10.1128/mBio.00951-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferraris P. Sequential biogenesis of host cell membrane rearrangements induced by hepatitis C virus infection. Cell. Mol. Life Sci. 2013;70:1297–1306. doi: 10.1007/s00018-012-1213-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paul D. Glycine zipper motifs in hepatitis c virus nonstructural protein 4B are required for the establishment of viral replication organelles. J. Virol. 2018;92 doi: 10.1128/JVI.01890-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Hoeven B. Biogenesis and architecture of arterivirus replication organelles. Virus Res. 2016;220:70–90. doi: 10.1016/j.virusres.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oudshoorn D. Antiviral innate immune response interferes with the formation of replication-associated membrane structures induced by a positive-strand RNA virus. mBio. 2016;7 doi: 10.1128/mBio.01991-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oudshoorn D. Expression and cleavage of middle east respiratory syndrome coronavirus nsp3-4 polyprotein induce the formation of double-membrane vesicles that mimic those associated with coronaviral RNA replication. mBio. 2017;8 doi: 10.1128/mBio.01658-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kopek B.G. Nodavirus-induced membrane rearrangement in replication complex assembly requires replicase protein A, RNA templates, and polymerase activity. J. Virol. 2010;84:12492–12503. doi: 10.1128/JVI.01495-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spuul P. Assembly of alphavirus replication complexes from RNA and protein components in a novel trans-replication system in mammalian cells. J. Virol. 2011;85:4739–4751. doi: 10.1128/JVI.00085-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Angelini M.M. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. mBio. 2013;4 doi: 10.1128/mBio.00524-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doyle N. Infectious bronchitis virus nonstructural protein 4 alone induces membrane pairing. Viruses. 2018;10:477. doi: 10.3390/v10090477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gorbalenya A.E. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117:17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Snijder E.J. Non-structural proteins 2 and 3 interact to modify host cell membranes during the formation of the arterivirus replication complex. J. Gen. Virol. 2001;82:985–994. doi: 10.1099/0022-1317-82-5-985. [DOI] [PubMed] [Google Scholar]
- 38.Suhy D.A. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 2000;74:8953–8965. doi: 10.1128/jvi.74.19.8953-8965.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J. Double-membraned liposomes sculpted by poliovirus 3AB protein. J. Biol. Chem. 2013;288:27287–27298. doi: 10.1074/jbc.M113.498899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Romero-Brey I. NS5A domain 1 and polyprotein cleavage kinetics are critical for induction of double-membrane vesicles associated with hepatitis C virus replication. mBio. 2015;6 doi: 10.1128/mBio.00759-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paul D. NS4B self-interaction through conserved C-terminal elements is required for the establishment of functional hepatitis C virus replication complexes. J. Virol. 2011;85:6963–6976. doi: 10.1128/JVI.00502-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gouttenoire J. Aminoterminal amphipathic alpha-helix AH1 of hepatitis C virus nonstructural protein 4B possesses a dual role in RNA replication and virus production. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hagemeijer M.C. Membrane rearrangements mediated by coronavirus nonstructural proteins 3 and 4. Virology. 2014;458–459:125–135. doi: 10.1016/j.virol.2014.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Posthuma C.C. Formation of the arterivirus replication/transcription complex: a key role for nonstructural protein 3 in the remodeling of intracellular membranes. J. Virol. 2008;82:4480–4491. doi: 10.1128/JVI.02756-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Z. Host lipids in positive-strand RNA virus genome replication. Front. Microbiol. 2019;10:286. doi: 10.3389/fmicb.2019.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diaz A., Ahlquist P. Role of host reticulon proteins in rearranging membranes for positive-strand RNA virus replication. Curr. Opin. Microbiol. 2012;15:519–524. doi: 10.1016/j.mib.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang W.F. Reticulon 3 binds the 2C protein of enterovirus 71 and is required for viral replication. J. Biol. Chem. 2007;282:5888–5898. doi: 10.1074/jbc.M611145200. [DOI] [PubMed] [Google Scholar]
- 48.Wu M.J. Reticulon 3 interacts with NS4B of the hepatitis C virus and negatively regulates viral replication by disrupting NS4B self-interaction. Cell. Microbiol. 2014;16:1603–1618. doi: 10.1111/cmi.12318. [DOI] [PubMed] [Google Scholar]
- 49.Lin C.L. Intrahepatic HCV RNA level and genotype 1 independently associate with hepatic reticulon 3 expression. Anticancer Res. 2017;37:2885–2891. doi: 10.21873/anticanres.11641. [DOI] [PubMed] [Google Scholar]
- 50.Chao T.C. Proline-serine-threonine phosphatase-interacting protein 2 (PSTPIP2), a host membrane-deforming protein, is critical for membranous web formation in hepatitis C virus replication. J. Virol. 2012;86:1739–1749. doi: 10.1128/JVI.06001-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jackson W.T. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3 doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reggiori F. Coronaviruses hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe. 2010;7:500–508. doi: 10.1016/j.chom.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Monastyrska I. An autophagy-independent role for LC3 in equine arteritis virus replication. Autophagy. 2013;9:164–174. doi: 10.4161/auto.22743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang L. HCV-induced autophagosomes are generated via homotypic fusion of phagophores that mediate HCV RNA replication. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fahmy A.M., Labonte P. The autophagy elongation complex (ATG5-12/16L1) positively regulates HCV replication and is required for wild-type membranous web formation. Sci. Rep. 2017;7:40351. doi: 10.1038/srep40351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mohl B.P. Early events in the generation of autophagosomes are required for the formation of membrane structures involved in hepatitis C virus genome replication. J. Gen. Virol. 2016;97:680–693. doi: 10.1099/jgv.0.000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee J.S. RACK1 mediates rewiring of intracellular networks induced by hepatitis C virus infection. PLoS Pathog. 2019;15 doi: 10.1371/journal.ppat.1008021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Verheije M.H. Mouse hepatitis coronavirus RNA replication depends on GBF1-mediated ARF1 activation. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Belov G.A. A critical role of a cellular membrane traffic protein in poliovirus RNA replication. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goueslain L. Identification of GBF1 as a cellular factor required for hepatitis C virus RNA replication. J. Virol. 2010;84:773–787. doi: 10.1128/JVI.01190-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Knorr R.L. Curvature of double-membrane organelles generated by changes in membrane size and composition. PLoS One. 2012;7 doi: 10.1371/journal.pone.0032753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Furse S. Lipid membrane curvature induced by distearoyl phosphatidylinositol 4-phosphate. Soft Matter. 2012;8:3090–3093. [Google Scholar]
- 63.Yang S.T. The role of cholesterol in membrane fusion. Chem. Phys. Lipids. 2016;199:136–143. doi: 10.1016/j.chemphyslip.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hsu N.-Y. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell. 2010;141:799–811. doi: 10.1016/j.cell.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reiss S. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe. 2011;9:32–45. doi: 10.1016/j.chom.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ilnytska O. Enteroviruses harness the cellular endocytic machinery to remodel the host cell cholesterol landscape for effective viral replication. Cell Host Microbe. 2013;14:281–293. doi: 10.1016/j.chom.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Roulin P.S. Rhinovirus uses a phosphatidylinositol 4-phosphate/cholesterol counter-current for the formation of replication compartments at the ER-Golgi interface. Cell Host Microbe. 2014;16:677–690. doi: 10.1016/j.chom.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 68.Wang H.L. Oxysterol-binding protein is a phosphatidylinositol 4-kinase effector required for HCV replication membrane integrity and cholesterol trafficking. Gastroenterology. 2014;146:1373. doi: 10.1053/j.gastro.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stoeck I.K. Hepatitis C virus replication depends on endosomal cholesterol homeostasis. J. Virol. 2017;92 doi: 10.1128/JVI.01196-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Muller C. Inhibition of cytosolic phospholipase A2α impairs an early step of coronavirus replication in cell culture. J. Virol. 2018;92 doi: 10.1128/JVI.01463-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Plagemann P.G. Effect of mengovirus replication on choline metabolism and membrane formation in novikoff hepatoma cells. J. Virol. 1970;6:800–812. doi: 10.1128/jvi.6.6.800-812.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang J. Positive-strand RNA viruses stimulate host phosphatidylcholine synthesis at viral replication sites. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E1064–E1073. doi: 10.1073/pnas.1519730113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Viktorova E.G. Phospholipid synthesis fueled by lipid droplets drives the structural development of poliovirus replication organelles. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Laufman O. Viral generated inter-organelle contacts redirect lipid flux for genome replication. Cell. 2019;178:275–289.e16. doi: 10.1016/j.cell.2019.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Miyanari Y. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 2007;9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- 76.Lee J.Y. Spatiotemporal coupling of the hepatitis C virus replication cycle by creating a lipid droplet- proximal membranous replication compartment. Cell Rep. 2019;27:3602–3617.e5. doi: 10.1016/j.celrep.2019.05.063. [DOI] [PubMed] [Google Scholar]
- 77.Schwartz M. Alternate, virus-induced membrane rearrangements support positive-strand RNA virus genome replication. Proc. Natl. Acad. Sci. U. S. A. 2004;101:11263–11268. doi: 10.1073/pnas.0404157101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xu K., Nagy P.D. Expanding use of multi-origin subcellular membranes by positive-strand RNA viruses during replication. Curr. Opin. Virol. 2014;9:119–126. doi: 10.1016/j.coviro.2014.09.015. [DOI] [PubMed] [Google Scholar]
- 79.Neufeldt C.J. The hepatitis C virus-induced membranous web and associated nuclear transport machinery limit access of pattern recognition receptors to viral replication sites. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Neufeldt C.J. Hepatitis C virus-induced cytoplasmic organelles use the nuclear transport machinery to establish an environment conducive to virus replication. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bienz K. Structural and functional characterization of the poliovirus replication complex. J. Virol. 1992;66:2740–2747. doi: 10.1128/jvi.66.5.2740-2747.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Du X. Replicase-mediated shielding of the poliovirus replicative double-stranded RNA to avoid recognition by MDA5. J. Gen. Virol. 2018;99:1199–1209. doi: 10.1099/jgv.0.001111. [DOI] [PubMed] [Google Scholar]
- 83.van Hemert M.J. SARS-coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen Y.H. Phosphatidylserine vesicles enable efficient en bloc transmission of enteroviruses. Cell. 2015;160:619–630. doi: 10.1016/j.cell.2015.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guo R. Porcine reproductive and respiratory syndrome virus utilizes nanotubes for intercellular spread. J. Virol. 2016;90:5163–5175. doi: 10.1128/JVI.00036-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Grunvogel O. Secretion of hepatitis C virus replication intermediates reduces activation of Toll-like receptor 3 in hepatocytes. Gastroenterology. 2018;154:2237–2251.e16. doi: 10.1053/j.gastro.2018.03.020. [DOI] [PubMed] [Google Scholar]
- 87.Avila-Perez G. Ultrastructural characterization of membranous torovirus replication factories. Cell. Microbiol. 2016;18:1691–1708. doi: 10.1111/cmi.12620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Berger C. Daclatasvir-like inhibitors of NS5A block early biogenesis of hepatitis C virus-induced membranous replication factories, independent of RNA replication. Gastroenterology. 2014;147:1094–1105.e25. doi: 10.1053/j.gastro.2014.07.019. [DOI] [PubMed] [Google Scholar]
- 89.Garcia-Nicolas O. The small-compound inhibitor K22 displays broad antiviral activity against different members of the family Flaviviridae and offers potential as a panviral inhibitor. Antimicrob. Agents Chemother. 2018;62 doi: 10.1128/AAC.01206-18. pii: e01206-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lundin A. Targeting membrane-bound viral RNA synthesis reveals potent inhibition of diverse coronaviruses including the middle East respiratory syndrome virus. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.V'Kovski P. Determination of host proteins composing the microenvironment of coronavirus replicase complexes by proximity-labeling. eLife. 2019;8 doi: 10.7554/eLife.42037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yuan S. SREBP-dependent lipidomic reprogramming as a broad-spectrum antiviral target. Nat. Commun. 2019;10:120. doi: 10.1038/s41467-018-08015-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ertel K.J. Cryo-electron tomography reveals novel features of a viral RNA replication compartment. eLife. 2017;6 doi: 10.7554/eLife.25940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Barcena M., Koster A.J. Electron tomography in life science. Semin. Cell Dev. Biol. 2009;20:920–930. doi: 10.1016/j.semcdb.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.McDonald K.L. Out with the old and in with the new: rapid specimen preparation procedures for electron microscopy of sectioned biological material. Protoplasma. 2014;251:429–448. doi: 10.1007/s00709-013-0575-y. [DOI] [PubMed] [Google Scholar]
- 96.Snijder E.J. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 2006;80:5927–5940. doi: 10.1128/JVI.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bykov Y.S. Correlative light and electron microscopy methods for the study of virus–cell interactions. FEBS Lett. 2016;590:1877–1895. doi: 10.1002/1873-3468.12153. [DOI] [PubMed] [Google Scholar]
- 98.Monaghan P. The ultrastructure of the developing replication site in foot-and-mouth disease virus-infected BHK-38 cells. J. Gen. Virol. 2004;85:933–946. doi: 10.1099/vir.0.19408-0. [DOI] [PubMed] [Google Scholar]
- 99.Doyle N. The porcine Deltacoronavirus replication organelle comprises double-membrane vesicles and zippered endoplasmic reticulum with double-membrane spherules. Viruses. 2019;11:1030. doi: 10.3390/v11111030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ulasli M. Qualitative and quantitative ultrastructural analysis of the membrane rearrangements induced by coronavirus. Cell. Microbiol. 2010;12:844–861. doi: 10.1111/j.1462-5822.2010.01437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]