Abstract
In recent years, hyperspectral imaging (HSI) has been shown as a promising imaging modality to assist pathologists in the diagnosis of histological samples. In this work, we present the use of HSI for discriminating between normal and tumor breast cancer cells. Our customized HSI system includes a hyperspectral (HS) push-broom camera, which is attached to a standard microscope, and home-made software system for the control of image acquisition. Our HS microscopic system works in the visible and near-infrared (VNIR) spectral range (400 – 1000 nm). Using this system, 112 HS images were captured from histologic samples of human patients using 20× magnification. Cell-level annotations were made by an expert pathologist in digitized slides and were then registered with the HS images. A deep learning neural network was developed for the HS image classification, which consists of nine 2D convolutional layers. Different experiments were designed to split the data into training, validation and testing sets. In all experiments, the training and the testing set correspond to independent patients. The results show an area under the curve (AUCs) of more than 0.89 for all the experiments. The combination of HSI and deep learning techniques can provide a useful tool to aid pathologists in the automatic detection of cancer cells on digitized pathologic images.
Keywords: Hyperspectral, microscopy, deep learning, histological
1. INTRODUCTION
Traditional diagnosis of histological samples is based on the manual examination of morphological features of samples by skilled pathologists. In recent years, the use of computer-aided technologies for improving these procedures is an emerging trend to reduce the intra and inter-observer variability [1]. Such technologies are intended to improve the diagnosis, make it reproducible and quantitative, and save time in the examination of samples [2] [3].
Hyperspectral imaging (HSI) technology is presented as an interesting alternative to RGB imaging due to its capability to differentiate between different materials by exploiting both morphological and spectral features. The use of this technology is motivated by the fact that spectral information may detect subtle molecular differences between biological samples [4]. This technology is used together with advanced machine learning algorithms to extract useful information about the materials within an HS image. In this context, recent studies have proven the advantages of using deep learning approaches for HSI classification [5], which are able to exploit simultaneously both the spatial and spectral features of HSI. This technology has been successfully applied to aid in the diagnosis of different types of histological samples, such as blood samples [6], brain tumors [7], liver tumors [8], and melanomas [9]. In the field of breast cancer, researchers have explored the HS capabilities of detecting cancers [10–13] or in the differentiation between normal and mitotic cells [14–18]. Nevertheless, these works are characterized for being restricted to the visible spectral range, e.g., 400 – 750 nm, and with only a few spectral bands. Furthermore, their annotations of histologic slides for the subsequent classification usually lacks of details and do not have the information at the cellular level.
In this work, we investigate the use of HSI to differentiate between normal and tumor cells from breast histological samples. The images are acquired using a high spectral resolution system which provides 826 spectral bands from 400 nm to 1000 nm, which are beyond the visible limitation of a naked eye. First, cell-level annotations are performed by a skilled pathologist in digitized whole slides. Then, hyperspectral (HS) images from the annotated areas are captured. We applied an image registration method to translate the annotations from the whole-slide RGB image to the HS domain, thus obtaining the HS data at the annotated cells. Finally, we use a convolutional neural network (CNN) to automatically discriminate between tumor and normal breast cells.
2. METHODS
2.1. Breast cancer tissue samples
In this research work, the samples consist of pathological slides diagnosed with breast tumor. In this study, two different pathological slides from different patients were analyzed. The tissue samples were processed using a conventional histological process, including paraffin embedding, tissue sectioning, and staining using hematoxylin and eosin (H&E). Then, the whole-slides were digitized using a Pannoramic SCAN digital scanner (3D Histech Ltd., Budapest, Hungary). The samples were examined, and some parts of the digitized slides were carefully annotated by pathologists using the Pannoramic Viewer software (3D Histech Ltd., Budapest, Hungary). The annotations were performed at the cellular level, and different types of cells were annotated: tumor cells, mitotic cells, lymphocytes and normal cells. Sample selection, preparation, and annotation was carried out in the Department of Pathology of the Hospital de Tortosa Verge de la Cinta, Spain. Different colors markers were used to highlight different types of cells: red for tumor cells, green for mitotic cells, yellow from lymphocytes, and blue for normal cells. We show an example of the whole digitized slide with and without annotations in Figure 1.a and b, respectively. In Figure 1.c, a magnified region from an annotated area of the slide is presented.
Figure 1.
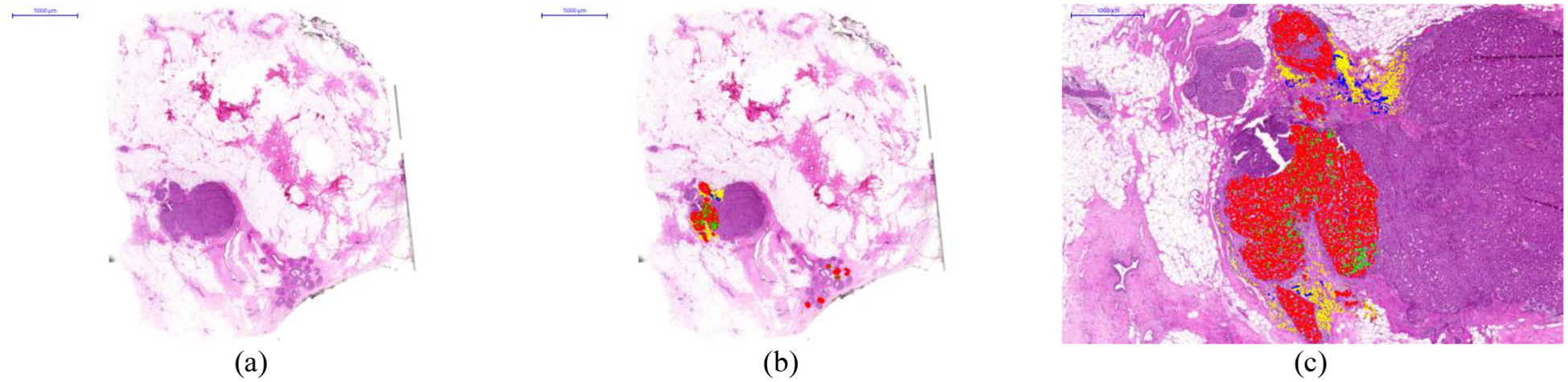
Histological samples used in this study. a) Digitized whole-slide image. b) Digitized whole-slide image including annotations. c) Details of an annotated area in the slide. Different types of cells were annotated: tumor cells (red) mitotic cells (green), lymphocytes (yellow), and normal cells (blue).
2.2. Hyperspectral image acquisition
The instrumentation employed in this study consists of an HS camera coupled to a conventional light microscope. The microscope is an Olympus BX-53 (Olympus, Tokyo, Japan). The HS camera is a Hyperspec VNIR A-Series from HeadWall Photonics (Fitchburg, MA, USA), which is based on an imaging spectrometer coupled to a CCD (Charge-Coupled Device) sensor, the Adimec-1000m (Adimec, Eindhoven, Netherlands). This HS system works in the spectral range from 400 to 1000 nm (VNIR) with a spectral resolution of 2.8 nm, sampling 826 spectral channels and 1004 spatial pixels. The push-broom camera performs spatial scanning to acquire an HS cube with a mechanical stage (SCAN, Märzhäuser) attached to the microscope, which provides accurate movement of the specimens. A software system was developed to synchronize the scanning movement and the camera acquisition. The objective lenses are from the LMPLFLN family (Olympus, Tokyo, Japan), which are optimized for infra-red (IR) observations. The light source is a 12 V - 100 W halogen lamp.
Using the aforementioned instrumentation, most of the areas annotated by the pathologist were captured. We used a 20* magnification for image acquisition, producing an HSI image size of 375 × 299 μm (1004 × 800 pixels). We imaged a total of 112 HS images, 65 from Patient 1 and 47 from Patient 2. Flat-field correction and reduction of the spectral bands by averaging contiguous spectral channels were applied to the HS images. The final goal of this study is to establish a relationship between the outcomes of the HSI detection and the diagnosis provided by the pathologists. For this reason, after capturing each HS cube, the consequent annotations from each area were extracted using the Pannoramic Viewer software. Figure 2 show an example of several HS images and their corresponding digitized counterparts. The annotations for each image are also shown.
Figure 2.
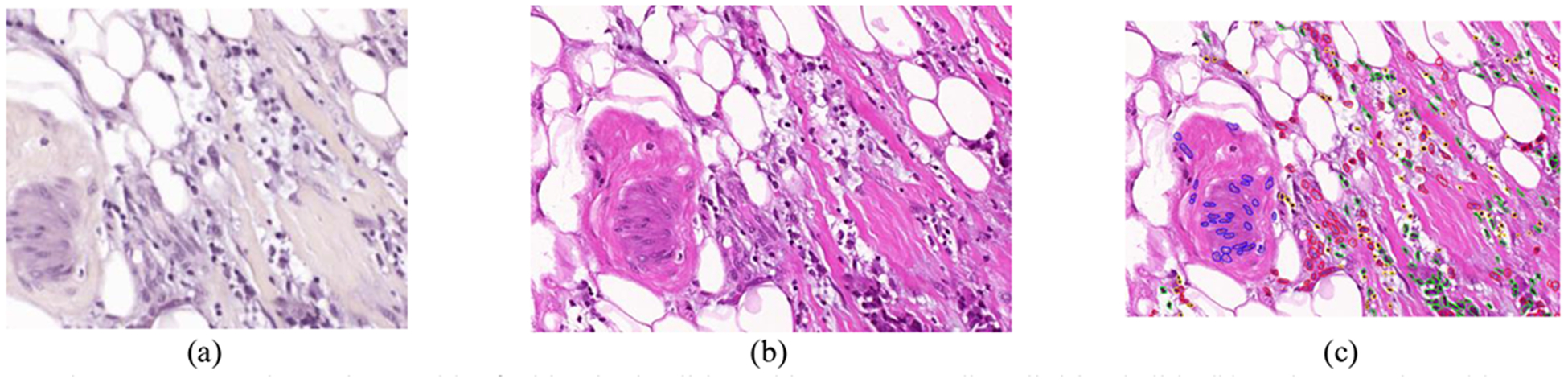
Example HS image (a) of a histologic slide and its corresponding digitized slide (b) and annotations (c).
2.3. Hyperspectral dataset generation
In this study, we processed HS data from different types of cells within breast cancer histological samples. We extracted the information from the areas that had been previously annotated by the pathologist. We had two different types of images: the annotated regions from digitized RGB slides and the corresponding HS images from the same regions. The size and the orientation of both types of images were different. In order to identify the annotated cells in the HS images, image registration between the HS images and the digitized slides was necessary. Our approach for the image registration consisted of searching for a geometric transformation that matched the information from the annotations to our HS images. To this end, we performed an image registration between the digitized RGB slide and an RGB synthetic image extracted from the HS cube.
Our image registration includes the following steps. First, the Speeded-Up Robust Features (SURF) algorithm [19] is applied to both images. The output of the SURF algorithm is a set of relevant points of an image that can be used in subsequent image matching tasks. After applying SURF, feature descriptors of relevant points are extracted. Then, feature matching is applied to the feature descriptors retrieved from each image. This feature matching is based on a nearest neighbors search, using the pixel-wise distance between the different feature descriptors of each image. Finally, using the relevant points of both images that present similar features, we search for an appropriate geometric transformation. This computation was performed using the MATLAB Computer Vision Toolbox (MathWorks Inc., Natick, MA, USA).
Figure 3 shows a flowchart of the workflow for this task, where the inputs of our image registration approach are the digitized slide and an RGB synthetic image extracted from the HS cube. After obtaining the appropriate geometric transformation to map the digitized slides to the HS domain, we apply this geometric transformation exclusively to the annotations. The annotation image is calculated as the subtraction of the digitized image from the pathological slide and its annotated counterpart. As a result, we retrieve the information about the annotations in the HS domain.
Figure 3.
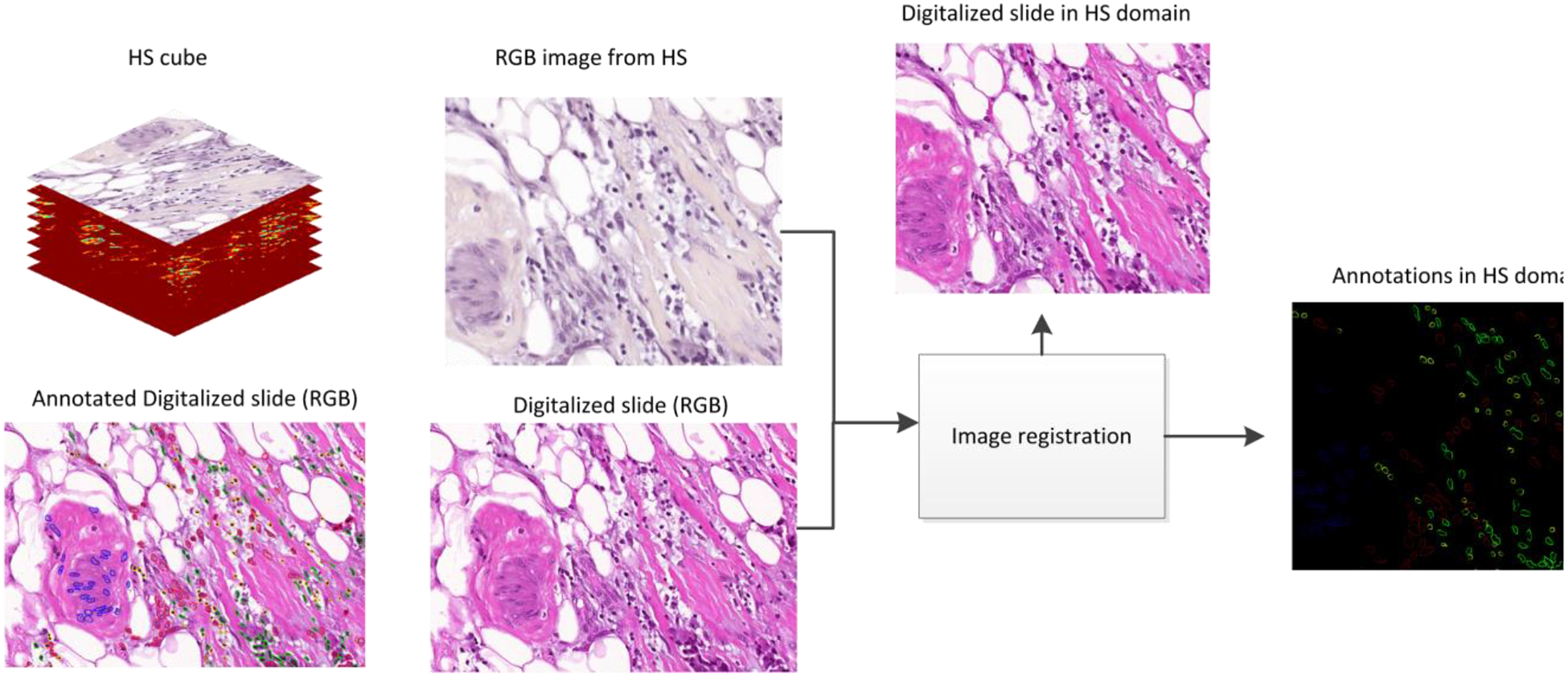
Framework for image registration between the digitized slides and the HS images.
After the image registration process, the annotations are geometrically aligned with the HS image. In order to extract the spectral information of each cell, we perform some image processing over the annotated image. First, as each color used for the cell annotation procedure contains information about a particular cell type, we separate the different colors (red, green, yellow, and blue) for subsequent analysis. These images consist of binary maps containing the contour of the annotations for each cell type. To generate a binary map where the whole cells are identified, we apply morphological operations to retrieve a map containing the location of each cell, i.e. dilation, erosion, opening and closing [20]. Finally, we identify each cell by computing the center of mass (centroid) of each region identified after performing an object search based on 8-connected components in the binary image [21]. Using this information, we can extract the information about the annotated cells within our HS data, generating HS image-patches of 39 × 39 spatial pixels and 159 spectral channels. This procedure is shown in Figure 4.
Figure 4.
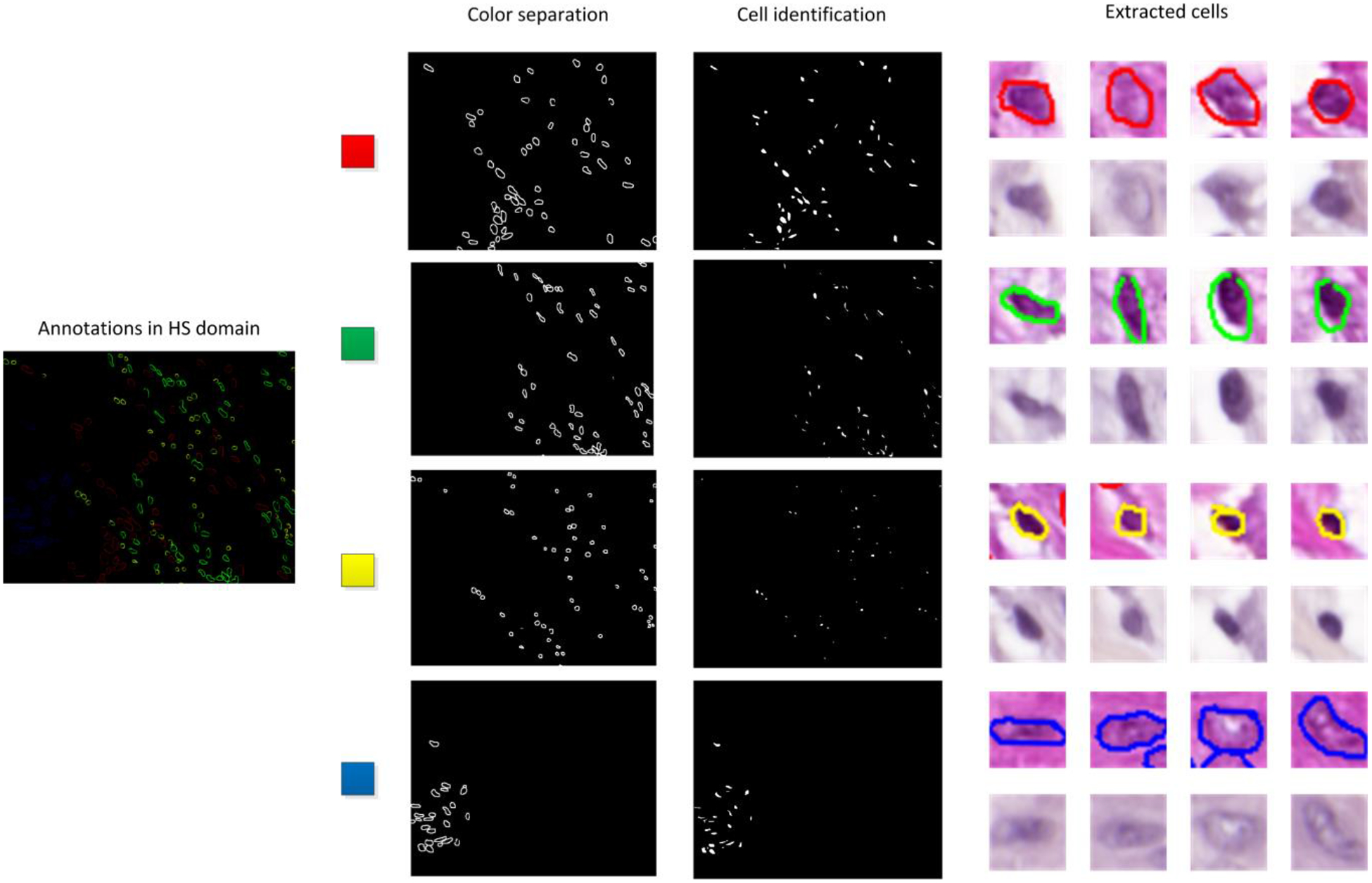
Detection of cells within the spectral image.
Table 1 shows a summary of our annotated dataset of 112 HS images. Figure 5 shows the average spectral signature for each patient and each cell type. In the experiments, we only employed normal and tumor cells to demonstrate the feasibility of the proposed method.
Table 1.
Number of HS image-patches in the dataset.
| Class | Patient 1 (65 images) | Patient 2 (47 images) |
|---|---|---|
| Tumor cells | 12505 | 7607 |
| Mitotic cells | 576 | 1082 |
| Lymphocytes | 2238 | 563 |
| Normal cells | 300 | 365 |
Figure 5.
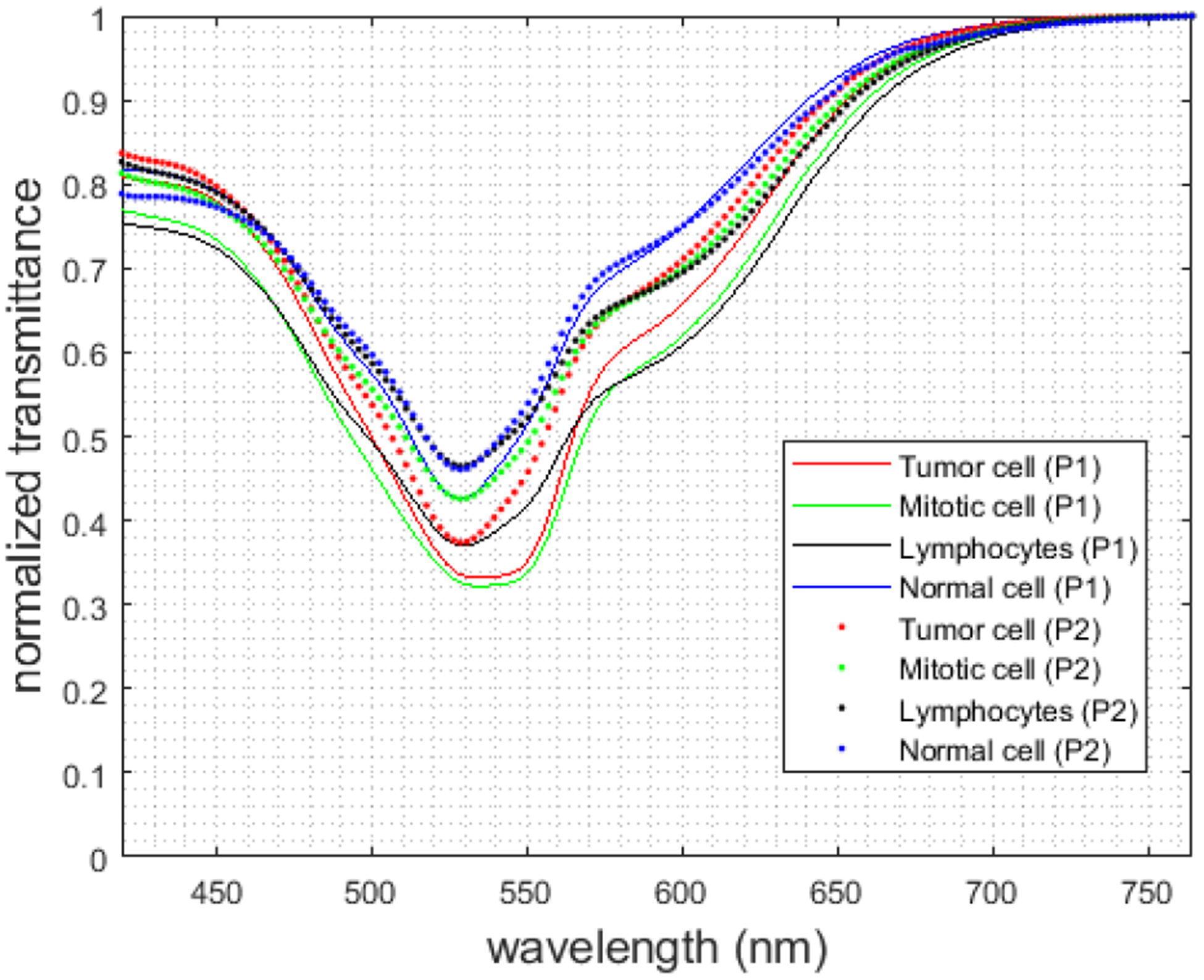
Mean spectral signatures from normal cells (blue), tumor cells (red), mitotic cells (green), and lymphocytes (black) for each patient.
2.4. Convolutional Neural Network
We employed a custom 2D-CNN for the automatic differentiation between normal and tumor cells. The network was developed using the TensorFlow implementation of the Keras Deep Learning API [22–23]. This network is mainly composed by 2D convolutional layers. We detail the description of the network in Table 2, where the input size of each layer is shown in each row, and the output size is the input size of the consequent layer. All convolutional and dense layer were performed with ReLU activation functions with a 10% dropout. The optimizer was the stochastic gradient descend method with a learning rate of 10−4.
Table 2.
Schematic of the CNN method.
| Layer | Kernel size | Input Size |
|---|---|---|
| Conv2D | 3×3 | 39×39×159 |
| Conv2D | 3×3 | 37×37×256 |
| Conv2D | 3×3 | 35×35×256 |
| Conv2D | 3×3 | 33×33×512 |
| Conv2D | 3×3 | 33×33×512 |
| Conv2D | 3×3 | 31×31×1024 |
| Conv2D | 3×3 | 29×29×1024 |
| Conv2D | 3×3 | 27×27×1024 |
| Global Avg. Pool | 25×25 | 25×25×1024 |
| Dense | 256 neurons | 1×1024 |
| Dense | Logits | 1×256 |
| Softmax | Classifier | 1×2 |
In this pilot study, due to the limited number of patients available for this study and due to the extensive work of cell labeling, we decided to train the network using the data from one patient, and using the data from the remaining patient for both validation (25%) and test (75%). A schematic of the data partition is shown in
Figure 6. We refer Experiment 1 and Experiment 2 for the situations where the data from Patient 1 and Patient 2 were used for training, respectively. This preliminary data partition was motivated by the limitation on the number of patients involved in this preliminary study. However, this situation is not realistic for biomedical applications. For this reason, we proposed two additional experiments (namely Experiment 3 and Experiment 4), where the models were trained and validated using the data from one patient, and the test was performed in an independent patient. Additionally, because of the imbalance between classes, we performed data augmentation for the normal cells for the training of the CNN. This data augmentation consisted of spatial flip and rotation of the patches corresponding to normal cells.
Figure 6.
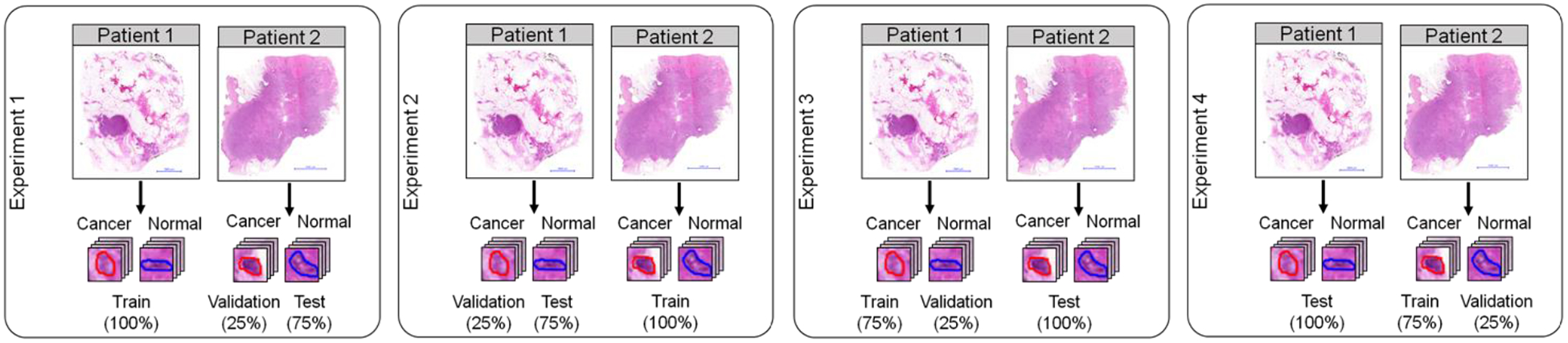
Data partition used for the classification experiments and testing.
3. RESULTS
In order to measure the performance of the classifier for discriminating between normal and tumor cells, we use the receiver operating characteristic (ROC) curve on both the validation and testing datasets. To generate these results, we used different thresholds for each experiment. These thresholds were selected using the validation data, and then were applied to the final classifiers. In Table 3, we show the values for AUC, overall accuracy, sensitivity, and specificity extracted for both the validation and the testing datasets. Additionally, due to the low number of normal tissues in the testing and validation sets, we studied the variations of the classifier performance using bootstrapping, and we reported the 95% confidence intervals for such metrics.
Table 3.
Results on the classification of normal and tumor cells for Experiment 1 and Experiment 2, which performs validation and testing on the same patient (inter-patient validation).
| Group | Experiment | AUC | Accuracy (%) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|
| Validation | Patient 1 | 0.88 (0.83, 0.93) | 80 (74, 85) | 80 (73, 86) | 80 (73, 87) |
| Patient 2 | 0.91 (0.86, 0.95) | 82 (76, 88) | 85 (77, 91) | 80 (73, 87) | |
| Average | 0.89 (0.85, 0.94) | 81 (75,87) | 82 (75, 89) | 80 (73,87 ) | |
| Testing | Patient 1 | 0.87 (0.84, 0.90) | 80 (77, 83) | 80 (76, 84) | 80 (76, 84) |
| Patient 2 | 0.94 (0.91, 0.96) | 87 (84, 90) | 86 (82, 90) | 89 (85, 93) | |
| Average | 0.90 (0.88, 0.93) | 84 (80, 87) | 83 (78, 87) | 85 (81, 89) |
The results of Experiments 1 and 2 are competitive in terms of AUC, reaching an average AUC of 0.89 and 0.90 for both validation and test. The values of accuracy, sensitivity and specificity are greater than 80% in all the experiments, with narrow confidence intervals. Furthermore, specificity and sensitivity are balanced, showing a competitive detection of both normal and tumor cells. The selected threshold values were 50% for Experiment 1, and 10% for Experiment 2.
The results for Experiments 3 and 4 are shown in Table 4. The threshold values were selected during the validation as 25% for both Experiment 3 and Experiment 4. For these experiments, the validation values of all the metrics are high. This is because the data from the same patient was used for training and validation. Nevertheless, the results on an independent patient show competitive results in terms of AUC, with an average of 0.9. In the case of Experiment 4, the accuracy is not worsened significantly compared to Experiment 2. Unlike Experiment 2, however, the specificity and sensitivity values are not as balanced.
Table 4:
Results on the classification of normal and tumor cells for Experiment 3 and Experiment 4, which performs training and validation on the same patient (intra-patient validation).
| Group | Experiment | AUC | Accuracy (%) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|
| Validation | Patient 1 | 0.97 (0.96, 0.98) | 90 (88,91) | 99 (98,100) | 83 (81, 85) |
| Patient 2 | 0.94 (0.93, 0.95) | 87 (85,89) | 84 (82,86) | 90 (88,92) | |
| Average | 0.95 (0.94, 0.96) | 88 (87, 90) | 92 (91, 93) | 87 (85, 89) | |
| Test | Patient 1 | 0.88 (0.86, 0.90) | 78 (75, 81) | 70 (68, 72) | 97 (95, 99) |
| Patient 2 | 0.91 (0.89, 0.93) | 79 (76, 82) | 96 (93, 98) | 71 (68, 74) | |
| Average | 0.90 (0.87, 0.92) | 79 (76, 81) | 83 (80, 86) | 84 (82, 87) |
In Figure 7, we present the ROC curves associated with the intra-patient validation experiments for both patients. The AUCs are similar in both cases. Additionally, the ROC curves suggest the classification for Patient 1 is more specialized in correctly detecting tumor cells (lower false positive rate), while the classification for Patient 2 is more accurate in detecting normal cells (higher true positive rate).
Figure 7.
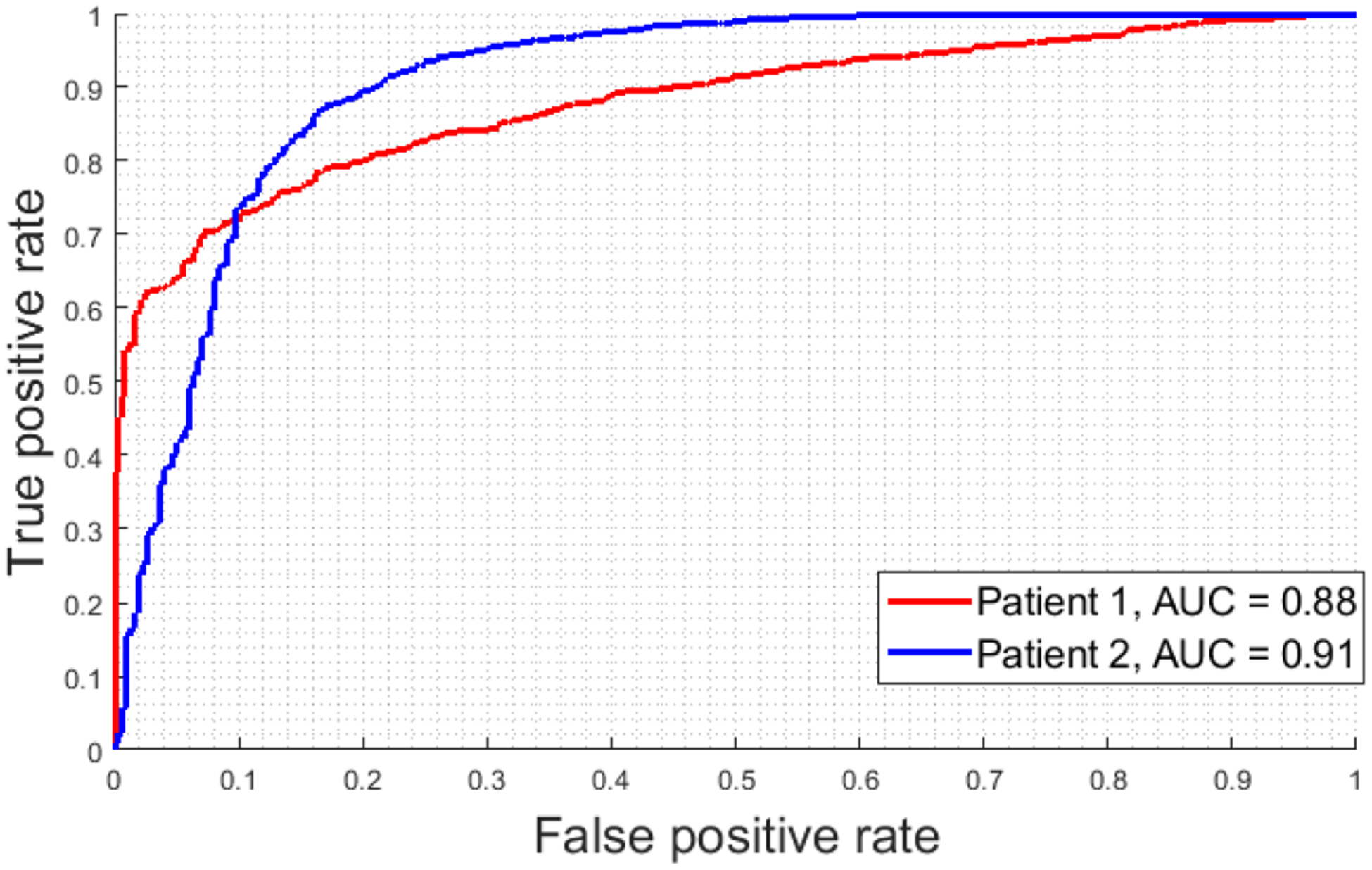
ROC curves from Experiment 3 and Experiment 4
4. RGB Comparison
In order to investigate whether the classification results using HSI boost the classification performance of standard RGB digitized images, we perform a comparison between both types of images. To this end, we generated synthetic RGB images from the HS cubes by using a combination of wavelengths simulating the human eye response. In this way, we ensure a fair comparison between RGB and HSI images as they are perfectly aligned. Then, we generated the patches for only Experiment 3 and Experiment 4 and performed the classification using the same CNN described in Section 2.4. After validation, we selected the best performing models and summarized the results in Table 5. As it can be observed in the table, although the validation results showed models with competitive AUC values, where the specificity and sensitivity values were also high, the results on the test showed unbalanced results. This fact suggests a specialization of the CNN on the prediction of a single class, worsening the overall performance of the classifier for the other classes. This circumstance is more evident in Experiment 3, where the specificity is quite lower than the sensitivity results.
Table 5.
Comparison between HSI and RGB for intra-patient validation in Experiment 3 and Experiment 4.
| Group | Experiment | Technology | AUC | Accuracy (%) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|---|
| Validation | Patient 1 | HSI | 0.97 (0.96, 0.98) | 90 (88, 91) | 99 (98, 100) | 83 (81, 85) |
| RGB | 0.94 (0.93, 0.96) | 85 (83, 86) | 97 (96, 99) | 77 (75, 79) | ||
| Patient 2 | HSI | 0.94 (0.93, 0.95) | 87 (85,89) | 84 (82,86) | 90 (88,92) | |
| RGB | 0.88 (0.86, 0.90) | 76 (74, 79) | 86 (83. 89) | 71 (69, 73) | ||
| Average | HSI | 0.95 (0.94, 0.96) | 88 (87, 90) | 92 (91, 93) | 87 (85, 89) | |
| RGB | 0.91 (0.89, 0.93) | 80 (78, 82) | 91 (89, 94) | 74 (72, 76) | ||
| Test | Patient 1 | HSI | 0.88 (0.86, 0.90) | 78 (75, 81) | 70 (68, 72) | 97 (95, 99) |
| RGB | 0.94 (0.93, 0.95) | 66 (64, 68) | 99 (98, 100) | 60 (59, 61) | ||
| Patient 2 | HSI | 0.91 (0.89, 0.93) | 79 (76, 82) | 96 (93, 98) | 71 (68, 74) | |
| RGB | 0.83 (0.81, 0.86) | 77 (75, 79) | 70 (68, 73) | 89 (86, 92) | ||
| Average | HSI | 0.90 (0.87, 0.92) | 79 (76, 81) | 83 (80, 86) | 84 (82, 87) | |
| RGB | 0.88 (0.87, 0.90) | 71 (69, 73) | 84 (83, 86) | 74 (72, 76) |
5. DISCUSSION & CONCLUSION
The use of machine learning techniques for assisting pathologists in routine examination of samples is an emerging trend. These computer-assisted tools are devoted to provide a quantitative diagnosis of different diseases, and also to decrease the current inter-observer variability in diagnosis. Although most of the approaches are based on conventional RGB image analysis, HSI is presented as a suitable technology that can boost the outcomes of conventional imagery due to its capability of differentiating between materials that present subtle molecular differences. In this work, we present the use of hyperspectral imaging and deep learning for automatic classification of normal and tumor cells in histological samples.
This work is novel due to the use of an annotated cell-level dataset of digitized slides, and the translation of such annotations to the HS domain using image registration techniques. After a cell-based dataset was generated, we used a 2D-CNN for the automatic differentiation of tumor and normal cells. The AUC for all our experiments were above 0.89, and the sensitivity and specificity values were approximately equivalent for Experiments 1, 2 and 3. In the case of Experiment 4, the sensitivity and specificity values of each patient are not as balanced as in the other experiments. These metrics strongly depend on the threshold selected for the final models. Nevertheless, due to the competitive AUC, there is room for improvement for specificity and sensitivity if the threshold is selected using data from an independent patient. Two main challenges must be addressed in the future to improve the performance of the CNN in this application. The number of patients should be increased, and more annotated cells from the normal class should be included in the dataset to balance the classes for the classification problem.
Finally, we compared the classification of using HSI and conventional RGB images. Previous study has proven the feasibility of RGB image analysis for breast cancer applications [24–25]. Our experiments suggest that HSI outperforms over RGB images. Nevertheless, further investigations should be performed to prove this observation.
One of the main challenges in computational pathology is to deal with the variations of the digitized slides among different laboratories. Such variations are mainly caused by the differences both in the instrumentation and the histological processing of samples (e.g. different staining conditions). In this study, we use images acquired with instrumentation from a single institution. However, a future goal in this field is to investigate the capabilities of HSI to reduce the inter-laboratory variability of data compared to conventional RGB imagery. Although the results of this study are promising, the study is limited by the sample size of the current dataset. Future work involves the inclusion of data from additional patients and a study of classifying other types of cells, i.e., mitotic and lymphocyte cells.
In conclusion, we developed a hyperspectral imaging microscope and deep learning software for digital pathology application and demonstrated the feasibility of this new imaging technology for breast cancer cell detection.
ACKNOWLEDGEMENTS
This research was supported in part by the U.S. National Institutes of Health (NIH) grants (R01CA156775, R01CA204254, R01HL140325, and R21CA231911) and by the Cancer Prevention and Research Institute of Texas (CPRIT) grant RP190588. This research was supported in part by the Canary Islands Government through the ACIISI (Canarian Agency for Research, Innovation and the Information Society), ITHACA project under Grant Agreement ProID2017010164 and it has been partially supported also by the Spanish Government and European Union (FEDER funds) as part of support program in the context of Distributed HW/SW Platform for Intelligent Processing of Heterogeneous Sensor Data in Large Open Areas Surveillance Applications (PLATINO) project, under contract TEC2017-86722-C4-1-R. This work was completed while Samuel Ortega was beneficiary of a pre-doctoral grant given by the “Agencia Canaria de Investigación, Innovacion y Sociedad de la Información (ACIISI)” of the “Conserjería de Economía, Industria, Comercio y Conocimiento” of the “Gobierno de Canarias”, which is part-financed by the European Social Fund (FSE) (POC 2014-2020, Eje 3 Tema Prioritario 74 (85%)).
Footnotes
DISCLOSURES
The authors have no relevant financial interests in this article and no potential conflicts of interest to disclose.
REFERENCES
- [1].Van Es SL, “Digital pathology: semper ad meliora,” Pathology, vol. 51, no. 1, pp. 1–10, January 2019. [DOI] [PubMed] [Google Scholar]
- [2].Flotte TJ and Bell DA, “Anatomical pathology is at a crossroads,” Pathology, vol. 50, no. 4, pp. 373–374, June 2018. [DOI] [PubMed] [Google Scholar]
- [3].Madabhushi A and Lee G, “Image analysis and machine learning in digital pathology: Challenges and opportunities,” Med. Image Anal, vol. 33, pp. 170–175, October 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lu G and Fei B, “Medical hyperspectral imaging: a review,” J. Biomed. Opt, vol. 19, no. 1, p. 10901, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Li S, Song W, Fang L, Chen Y, Ghamisi P, and Benediktsson JA, “Deep Learning for Hyperspectral Image Classification: An Overview,” IEEE Trans. Geosci. Remote Sens, pp. 1–20, 2019. [Google Scholar]
- [6].Duan Y et al. , “Leukocyte classification based on spatial and spectral features of microscopic hyperspectral images,” Opt. Laser Technol, vol. 112, pp. 530–538, April 2019. [Google Scholar]
- [7].Ortega S, Fabelo H, Camacho R, de la Luz Plaza M, Callicó GM , and Sarmiento R, “Detecting brain tumor in pathological slides using hyperspectral imaging,” Biomed. Opt. Express, vol. 9, no. 2, p. 818, February 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wang J and Li Q, “Quantitative analysis of liver tumors at different stages using microscopic hyperspectral imaging technology,” J. Biomed. Opt, vol. 23, no. 10, p. 1, October 2018. [DOI] [PubMed] [Google Scholar]
- [9].Wang Q, Li Q, Zhou M, Sun L, Qiu S, and Wang Y, “Melanoma and Melanocyte Identification from Hyperspectral Pathology Images Using Object-Based Multiscale Analysis,” Appl. Spectrosc, p. 000370281878135, June 2018. [DOI] [PubMed] [Google Scholar]
- [10].Boucheron LE, Bi Z, Harvey NR, Manjunath BS, and Rimm DL, “Utility of multispectral imaging for nuclear classification of routine clinical histopathology imagery,” BMC Cell Biol, vol. 8 Suppl 1, no. SUPPL. 1, p. S8, July 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Qi X, Cukierski W, and Foran DJ, “A comparative performance study characterizing breast tissue microarrays using standard RGB and multispectral imaging,” in Proceedings SPIE Multimodal Biomedical Imaging V;, 2010, p. 75570Z. [Google Scholar]
- [12].Qi X, Xing F, Foran DJ, and Yang L, “Comparative performance analysis of stained histopathology specimens using RGB and multispectral imaging,” in Proceedings SPIE Medical Imaging 2011: Computer-Aided Diagnosis, 2011, vol. 7963, p. 79633B. [Google Scholar]
- [13].Khouj Y, Dawson J, Coad J, and Vona-Davis L, “Hyperspectral Imaging and K-Means Classification for Histologic Evaluation of Ductal Carcinoma In Situ,” Front. Oncol, vol. 8, February 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Roux L et al. , “Mitosis detection in breast cancer histological images An ICPR 2012 contest.,” J. Pathol. Inform, vol. 4, p. 8, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Malon CD and Cosatto E, “Classification of mitotic figures with convolutional neural networks and seeded blob features.,” J. Pathol. Inform, vol. 4, p. 9, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Irshad H, Gouaillard A, Roux L, and Racoceanu D, “Multispectral band selection and spatial characterization: Application to mitosis detection in breast cancer histopathology.,” Comput. Med. Imaging Graph, vol. 38, no. 5, pp. 390–402, July 2014. [DOI] [PubMed] [Google Scholar]
- [17].Irshad H, Gouaillard A, Roux L, and Racoceanu D, “Spectral band selection for mitosis detection in histopathology,” in 2014 IEEE 11th International Symposium on Biomedical Imaging (ISBI), 2014, pp. 12791282. [Google Scholar]
- [18].Lu Cheng and Mandal M, “Toward Automatic Mitotic Cell Detection and Segmentation in Multispectral Histopathological Images,” IEEE J. Biomed. Heal. Informatics, vol. 18, no. 2, pp. 594–605, March 2014. [DOI] [PubMed] [Google Scholar]
- [19].Bay H, Ess A, Tuytelaars T, and Van Gool L, “Speeded-Up Robust Features (SURF),” Comput. Vis. Image Underst, vol. 110, no. 3, pp. 346–359, June 2008. [Google Scholar]
- [20].Serra J and Soille P, Eds., Mathematical Morphology and Its Applications to Image Processing, vol. 2 Dordrecht: Springer Netherlands, 1994. [Google Scholar]
- [21].Haralick RM and Shapiro LG, Computer and robot vision, vol. 1 Addison-wesley Reading, 1992. [Google Scholar]
- [22].Abadi M et al. , “TensorFlow: Large-Scale Machine Learning on Heterogeneous Distributed Systems,” March 2016. [Google Scholar]
- [23].Chollet F, “Keras,” GitHub repository. GitHub, 2015. [Google Scholar]
- [24].Ehteshami Bejnordi B et al. , “Diagnostic Assessment of Deep Learning Algorithms for Detection of Lymph Node Metastases in Women With Breast Cancer,” JAMA, vol. 318, no. 22, p. 2199, December 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Pei Z, Cao S, Lu L, and Chen W, “Direct Cellularity Estimation on Breast Cancer Histopathology Images Using Transfer Learning,” Comput. Math. Methods Med, vol. 2019, pp. 1–13, June 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]


