Abstract
Background:
Raised incidences of respiratory tract infections due to fungal agents in immunocompetent individuals are a cause of concern due to the unavailability of rapid diagnostic methods.
Materials and Methods:
Sputum and serum samples were collected from patients having lower respiratory tract infections (LRTIs), serum samples were screened for the presence of anti Aspergillus antibodies and sputum samples were homogenized and processed for identification of Aspergillus by conventional methods and further subjected to polymerase chain reaction (PCR) using genus-specific ITS 4-5 primers.
Results:
PCR identified Aspergillus in 28% sputum samples, which was high as compared to conventional methods.
Conclusion:
Simple conventional PCR technique proves to be useful screening in for early identification of Aspergillus colonization in patients with LRTI, which can prevent irreversible damage in their lungs by fungal invasion.
Keywords: Aspergillus flavus, Aspergillus fumigatus, lower respiratory tract infections, polymerase chain reaction
Introduction
Respiratory tract infections are globally responsible for one-third of the infectious diseases of which, fungal agents remain largely unrecognized. Most commonly Aspergillus, Candida, and Mucorales and rarely Fusarium, Scedosporium, Penicillium, and Basidiomycetes have been reported to be responsible for invasive fungal infections.[1] Among these Aspergillus spores due to its ubiquitous distribution gets suspended in air and sediment in distal airways and alveolar spaces.[2]
Respiratory samples such as sputum samples are easy to obtain and do not require any invasive procedure. Sputum of lower respiratory tract infected patients is routinely not sent for fungal culture. Furthermore, culture isolation for invasive infection has a variable sensitivity from 5% to 75% and poor specificity hence, repeated isolation is needed for diagnosing invasive aspergillosis.[3]
Detection of Aspergillus spp., implementing molecular methods have been documented in immunocompromised individuals, but not in immunocompetent individuals.[4] As there are rising incidences of invasive pulmonary aspergillosis (IPA) in immunocompetent individuals without traditional risk factors, rapid diagnostic tests such as polymerase chain reaction (PCR) are warranted along with other conventional methods, for early diagnosis of invasion by Aspergillus spp.[5]
Sensitivity and specificity of PCR in bronchoalveolar lavage fluid have been estimated to be 67%–100% and 55%–95%, respectively.[5] Few studies conducted in India emphasize on Aspergillus isolation from patients with complaints of lower respiratory tract infection (LRTI). Hence, the present study was undertaken to assess the ability of PCR for Aspergillus DNA detection in a sputum sample of patients suffering from LRTI and to evaluate the sensitivity and specificity of PCR comparing it to conventional culture methods.
Materials and Methods
The study was conducted in the Department of Microbiology and TB-Chest Clinic of Santosh Medical College and Hospital Ghaziabad in collaboration with the Department of Microbiology, University College of Medical Sciences, GTB Hospital, New Delhi.
A total number of 150 patients (EpiInfo 4 Software - Centers for Disease Control and Prevention - www.cdc.gov) in the age group 18 years and above visiting Department of TB and Chest, having acute episode of cough for 21 days, sputum production, dyspnoea, wheeze, chest discomfort/pain with chest radiography showing symptoms of LRTIs, were selected for the study as defined in guidelines of the European Respiratory Society and the European Society for Clinical Microbiology and Infectious Diseases on the management of LRTI in adults.[1,6] Our study group included patients of all socioeconomic backgrounds. Institutional ethical clearance was obtained. Patients with active tuberculosis, atypical mycobacterial infections, malignancies, HIV reactive, and immunocompromised were excluded from the study.
Early morning sputum and whole blood samples were collected from patients, and direct microscopy in 10% potassium hydroxide (KOH) was done to observe the presence of fungal elements. Sputum samples were then homogenized by adding N-acetyl L-cystine in M/50 Trisodium citrate and diluting double the amount with phosphate buffer and divided into two parts.[7] A part of the sputum was used to culture on Sabouraud's dextrose agar supplemented with cycloheximide and gentamycin and incubated for 3–4 days at 25°C–26°C for isolation of Aspergillus species. Isolates were identified and confirmed on the basis of microscopic and macroscopic morphological characteristics following standard mycological procedures.[8] The second part of the sputum was used for DNA extraction and PCR. Serum separated from whole blood was used for detecting anti –Aspergillus IgG, IgM and IgE antibodies using commercially available kit (Omega Diagnostics, Calbiotech)-based ELISA method of identification.
DNA was extracted using Qiagen Q Amp mini kit following manufacture's guidelines with modifications in the initial few steps and stored at −20°C. PCR was performed using Taq DNA Polymerase and Taq PCR core kit-(Qiagen) with a set of ITS 5-4 primers having following oligonucleotide sequence for Aspergillus genus detection (Qiagen. DNeasy® Blood and Tissue kit). ITS 5-5® GGAAGTAAAAGTCGTAACAAGG-3® and ITS 4®TCCTCCGCTTATTGATATGC-3®.[9]
PCR was standardized and concentrations of various components of PCR were optimized along with the cycling conditions. The reaction mixture consisted of 2.5 μl 10X PCR buffer, 0.5 μl dNTP mixture, 1 μl forward and reverse primers, 0.15 μlTaq DNA polymerase, 5 μl 5X Q buffer, 3 μl of sample DNA, and total volume of reaction mixture was achieved 25 μl by adding 11.85 μl nuclease-free water. Thermocycler was programmed for 40 cycles with initial denaturation at 95°C for 5 min followed by denaturation at 95°C for 30 min, annealing at 50°C for 30 s, extension at 72°C for 45 s and final extension at 72°C for 10 min. Amplified PCR products were visualized under ultra-violet transilluminator following 1.5% agrose gel electrophoresis at 100V for 30 min. The presence of a unique band around 600bp in positive samples was indicative of the presence of Aspergillus spp. DNA in sputum samples of patients. As can be depicted in Figure 1.
Figure 1.
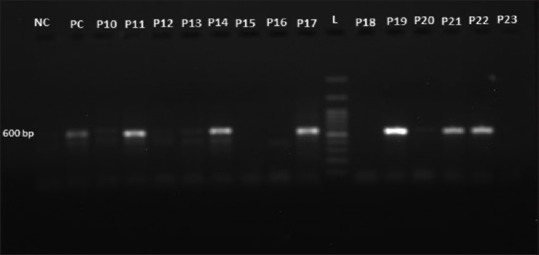
Visualization of amplified polymerase chain reaction products under ultra-violet transilluminator on gel electrophoresis showing unique band around 600bp for Aspergillus genus, NC: Negative control; PC: Positive control; L: 100 base pair ladder, P10, P11, P12, P13, P14, P15, P16, P17, P18, P19, P20, P21, P22, P23: Patient's sample
Conventional methods were compared with PCR, and sensitivity and specificity of PCR were calculated taking culture as a gold standard. The statistical analysis was done using Statistical Package for Social Sciences software (Statistical Package for social Sciences - www.ibm.com).
Results
Of the 150 patients suffering from LRTI, 30% (45) were clinically categorized as a chronic obstructive pulmonary disease (COPD), 22% (33) bronchial asthma (BA), and 4% (6) chronic bronchitis (CB). The remaining 44% (66) patients had exaggerated symptoms of LRTI but could not be categorized in any specific clinical group. None of the patients were immunocompromised or HIV reactive or suspected to have pulmonary tuberculosis. Among the study population, male patients were more affected than females with majority having COPD. Graph 1 Shows Clinical and gender wise distribution of LRTI patients.
Graph 1.
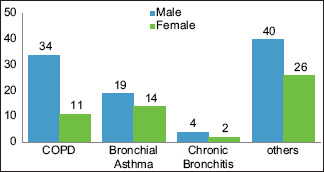
Clinical and gender-wise distribution of lower respiratory tract infection patients. Others: Rest of the patients with exaggerated symptoms of lower respiratory tract infection
Of 150 sputum samples, 19% (29/150) grew Aspergillus species, of which 11% (17/150) were Aspergillus fumigatus, 7% (11/150) Aspergillus flavus, and 0.6% (1/150) Aspergillus niger.
Table 1 shows 13% sputum samples were positive for the presence of septate fungal hyphae of which 11% were culture positive. Two percent of sputum samples could not yield any growth on culture though were positive for KOH and 87% sputum samples were KOH negative of which 8% was culture positive.
Table 1.
Microscopy and culture positivity in sputum samples of lower respiratory tract infection patients (n=150)
| Microscopy (KOH mount) | Culture | Total | P | |
|---|---|---|---|---|
| Positive | Negative | |||
| Positive, n (%) | 17 (11) | 3 (2) | 20 (13) | <0.00001 |
| Negative, n (%) | 12 (8) | 118 (78) | 130 (87) | |
| Total, n (%) | 29 (19) | 121 (80) | 150 | |
KOH: Potassium hydroxide
On screening serum samples of all LRTI patients, raised absolute eosinophil counts was observed in 13% (20/150) patients. 14% patients had raised total serum IgE, majority of which were patients with BA, 3% had raised IgE specific for A. fumigatus and 9% with raised IgG specific for A. fumigatus.
PCR detected 28% (43/150) of LRTI patients as positive for Aspergillus. Graph 2 shows the clinical distribution of PCR-positive patients.
Graph 2.
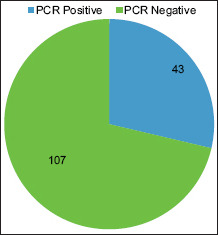
Molecular detection of Aspergillus by polymerase chain reaction among lower respiratory tract infection patients
Of the 28% (43/150) PCR-positive sputum samples only 17% (26/150) were also culture positive for Aspergillus species. While 2% (3/150) were culture positive and PCR negative and 69% (104/150) were culture as well as PCR negative. As shown in Table 2.
Table 2.
Comparison of culture and polymerase chain reaction positivity for Aspergillus detection among lower respiratory tract infection patients (n=150)
| PCR | Culture | Total | P | |
|---|---|---|---|---|
| Positive | Negative | |||
| Positive, n (%) | 26 (17) | 17 (11) | 43 (28) | <0.00001 |
| Negative | 3 | 104 | 107 | |
| Total, n (%) | 29 (19) | 121 (80) | 150 | |
PCR: Polymerase chain reaction
Sensitivity and specificity of PCR were found to be 89.66% and 85.96% with culture as gold standard. The positive predictive value was 60.47% and a higher negative predictive value 97.21% was obtained (P < 0.00001).
Discussion
IPA is a severe disease that can be found not only in severely immunocompromised patients but also in immunocompetent patients such as COPD.[5,10] A strong clinical suspicion to identify patients at risk for invasive aspergillosis is the first step in evaluating for aspergillosis since Aspergillus conidia are constantly inhaled.[11] With raised incidences and difficulty to diagnose IPA early, sensitive assays for accurate diagnosis of the fungal infections are warranted.[12]
In the present study, among LRTI patients majority presented with COPD followed by BA and CB. A male predominance is suggestive of the involvement in outdoor activity hence exposure to airborne pathogens. Patients mostly presented complaints of cough and chest pain, followed by difficulty in breathing. Only a few patients had history of pulmonary tuberculosis.
In our study, radiological findings of most of the patients had nonspecific signs however, in elderly patients and patients showing worsened COPD, presence of abnormal radiological findings proved to be helpful for its correlation with other diagnostic tests to diagnose IPA.
Colonization of Aspergillus spp., in LRTI patients, can be a predisposing condition for the development of IPA, as observed in a study by Dakshina et al., among stable COPD patients who had exacerbations but yielded Aspergillus spp., were followed to assess their stability and it was found that exacerbations increased in COPD patients along with severity of clinical features and isolation Aspergillus spp.[13]
Seropositivity for A. fumigatus was not significant but sputum culture was positive for A. flavus, Due to lack of other species-specific antibody detection, IgG A. fumigatus may not be a useful tool to detect the prevalence of IPA in LRTI patients.
On correlating various radiological, clinical, and serological findings with culture positivity of sputum samples of LRTI patients for Aspergillus spp., they were diagnosed to be suffering from aspergilloma and allergic bronchopulmonary aspergillosis (ABPA). On the basis of criteria proposed by the European Organization for Research on Treatment of Cancer/Mycoses Study Group, patients were categorized as probable and possible invasive aspergillosis, but due to inability to perform histopathological investigations, we could not categorize our patients into proven invasive aspergillosis.
Among the species A. fumigatus and A. flavus were the most common pathogen, A. fumigatus was the major isolates among patients with possible IPA unlike other studies which observe A. flavus as the predominant species. In a study by Kurhade et al. observed 16.2% of Aspergillus spp., from cases with chronic pulmonary infections.[14] A retrospective study of 8 years by Barberan et al. found 56.6% LRTI patients with single culture positive for Aspergillus spp., which was higher as compared to other studies by Vandewoude et al., Bouza et al., and Soubani et al.[10,15,16,17]
In contrast to our study, Tashiro et al. isolated 42% Aspergillus spp., in cases representing colonization, and the most common colonizing species was A. niger, followed by Aspergillus versicolor, A. fumigatus, Aspergillus terreus, A. flavus, Aspergillus sydowii, and Aspergillus nidulans.[18] Due to geographical diversity though predominance of A. flavus as the causative agent other spp. may predominate locally hence, leading to exposure and infection.
Examination of the sputum by Mwaura et al., revealed that 37.8% (65/172) samples were positive for fungal elements, while 62.2% (107/172) samples were negative.[19] We also observed the presence of septate fungal hyphae on direct microscopy of sputum samples of 13% of LRTI patients. These observations demonstrate the diagnostic importance of direct microscopy of sputum samples. However, the presence of septate fungal hyphae in direct microscopy demonstrates invasion or colonization remained questionable.
A study by Kurhade et al., revealed that 16% of respiratory specimens were culture and KOH positive for Aspergillus spp.[14] Similar observations were found in our study too. Of the 150 sputum samples, 20 (13%) samples were KOH positive of which 17 (11%) were also culture positive, while 3 sputum samples were KOH positive but culture negative. Remaining 130 (87%) sputum samples were KOH negative of which 12 (8%) samples were culture positive. Correlating direct microscopy along with culture aided in the identification of Aspergillus spp., as an etiological agent.
The high degree of genetic variations of nucleotide sequences of the ITS1-5.8S-ITS2 region makes the comparison among Aspergillus species very useful for strain classification and phylogenetic studies.[9] The sensitivity of PCR for detecting Aspergillus DNA in sputum is unknown.[12] The ability of PCR for the detection of Aspergillus in sputum is sensitive but it does not provide quantitative information on the fungal burden.[11,20]
In a study by Singh et al., Aspergillus DNA was detected in 25% of sputum samples of suspected aspergillosis patients using nested PCR using specific primer sets (Nig, fmi, and Fla). In our study, PCR detected Aspergillus in sputum of 28% (43) LRTI patients.[21]
Among (28%) PCR-positive sputum samples, (17%) was also found to be culture positive for Aspergillus species. 3 (2%) sputum samples were found to be culture positive and PCR negative while 104 (69%) were culture as well as PCR negative. In the present study, false-positive results of PCR may be attributed to the inability of PCR to differentiate between colonization and infection, also due to high susceptibility of contamination of PCR in laboratory. Patients at the risk of IPA are often prescribed antibiotics and fluids which may act as inhibitors and may hamper PCR which may result in false-negative results.
Overall sensitivity (89.66%) and specificity (85.95%) of PCR were found higher than culture and statistically significant with P < 0.00001. Singh et al. also found sensitivity of nested PCR to be higher than culture and microscopy. However, their sensitivity of PCR was observed to be 25% (50/200). PCR and culture positivity of LRTI patients for Aspergillus spp., in our study, indicates the importance of screening of respiratory samples for Aspergillus infection so that antifungal therapy can be initiated at an appropriate time, to improve patient's outcome.[21]
Kawamura et al. also found high sensitivity of the nested PCR for the detection of Aspergillus DNA in serum samples of patients with pulmonary aspergillosis with a different set of primers M5c and M6b.[22] However, we detected Aspergillus DNA in sputum samples using ITS 5-4 primers. In our study PCR was found to be helpful for detecting Aspergillus DNA in sputum samples of LRTI patients especially in aspergilloma and ABPA which are noninvasive forms of aspergillosis.
According to Knutsen and Slavin PCR for detecting Aspergillus spp., in sputum is more sensitive than culture in ABPA but needs to be interpreted with other clinical laboratory features.[23] Positive culture of Aspergillus species although indicative but does not prove infection. Gold standard of myological/or histological evidence of invasion is not always feasible to obtain. Therefore, in the present study, we performed radiological and serological investigations of each LRTI patient along with culture and PCR for increasing sensitivity of interpretation of Aspergillus infection in these patients.
Conclusion
PCR helped in the early detection of Aspergillus DNA in LRTI patients where sputum cultures were negative. Therefore, as a point of care, routine screening of their sputum samples for the presence of Aspergillus by a simple conventional PCR technique along with culture can be done. There was a growing realization during the conduct of the study that Aspergillus species may also play a role in worsening the pulmonary conditions and exacerbations in patients without classic risk factors. Therefore, appropriate early diagnostic strategies are needed to aid in prompt management, prevent invasion and subsequent serious complications by the fungi.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Moore M, Little P, Rumsby K, Kelly J, Watson L, Warner G, et al. Predicting the duration of symptoms in lower respiratory tract infection. Br J Gen Pract. 2008;58:88–92. doi: 10.3399/bjgp08X264045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Naaraayan A, Kavian R, Lederman J, Basak P, Jesmajian S. Invasive pulmonary aspergillosis – Case report and review of literature. J Community Hosp Intern Med Perspect. 2015;5:26322. doi: 10.3402/jchimp.v5.26322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barton RC. Laboratory diagnosis of invasive aspergillosis: From diagnosis to prediction of outcome. Scientifica (Cairo) 2013;2013:459405. doi: 10.1155/2013/459405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Latgé JP. Aspergillus fumigatus and aspergillosis. Clin Microbiol Rev. 1999;12:310–50. doi: 10.1128/cmr.12.2.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zmeili OS, Soubani AO. Pulmonary aspergillosis: A clinical update. QJM. 2007;100:317–34. doi: 10.1093/qjmed/hcm035. [DOI] [PubMed] [Google Scholar]
- 6.Woodhead M, Blasi F, Ewig S, Garau J, Huchon G, Ieven M, et al. Guidelines for the management of adult lower respiratory tract infections full version. Clin Microbiol Infect. 2011;17(Suppl 6):E1–59. doi: 10.1111/j.1469-0691.2011.03672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collee J, Marmion PB, Fraser GA, Simmons A. Mackie and Mc Cartney Practical Medical Microbiology. Fourteenth edition. NewYork: Churchill livingstone Elsevier; 2011. pp. 695–717. [Google Scholar]
- 8.Chander J. Textbook of Medical Mycology. 3rd ed. New Delhi: Mehta Publishers; 2013. pp. 343–60. [Google Scholar]
- 9.Batista PP, Santos JF, Oliveira NT, Pires AP, Motta CM, Luna-Alves Lima EA. Genetic characterization of Brazilian strains of Aspergillus flavus using DNA markers. Genet Mol Res. 2008;7:706–17. doi: 10.4238/vol7-3gmr422. [DOI] [PubMed] [Google Scholar]
- 10.Soubani AO, Khanchandani G, Ahmed HP. Clinical significance of lower respiratory tract Aspergillus culture in elderly hospitalized patients. Eur J Clin Microbiol Infect Dis. 2004;23:491–4. doi: 10.1007/s10096-004-1137-1. [DOI] [PubMed] [Google Scholar]
- 11.Vuong FM, Waymack RJ. Aspergillosis Treasure Island National institute of health. Statpearls publishing; 2018. [Google Scholar]
- 12.Shuzen Z, Wang S, Wan Z, Li R, Yu J. The diagnosis of invasive and non invasive pulmonary aspergillosis by serum and bronchoalveolar lavage fluid galactomannanassay. Biomed Res Int. 2015;943691:1–5. doi: 10.1155/2015/943691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma A, et al. Repeated Aspergillus isolation in respiratory samples from non-immunocompromised patients not selected based on clinical diagnoses: Colonisation or infection? BMC Infect Dis. 2012;12:295. doi: 10.1186/1471-2334-12-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khurade AM, et al. Invasive aspergillosis in critically ill patients: Attributable mortality and excesses in length of ICU stay and ventilator dependence. J Hosp Infect. 2004;56:269–76. doi: 10.1016/j.jhin.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 15.Barberan J, et al. Workload due to Aspergillus fumigatus and significance of the organism in the microbiology laboratory of a general hospital. J Clin Microbiol. 2005;43:2075–9. doi: 10.1128/JCM.43.5.2075-2079.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vandewoude KH, et al. Aspergillus colonization: A marker of exacerbation in chronic obstructive pulmonary disease. Euro J Biomed Pharma Sci. 2017;4:330–4. [Google Scholar]
- 17.Bouza, et al. Mycological and serological study of pulmonary aspergillosis in central India. Indian J Med Microbiol. 2002;20:141–4. [PubMed] [Google Scholar]
- 18.Tashiro T, Izumikawa K, Tashiro M, Takazono T, Morinaga Y, Yamamoto K, et al. Diagnostic significance of Aspergillus species isolated from respiratory samples in an adult pneumology ward. Med Mycol. 2011;49:581–7. doi: 10.3109/13693786.2010.548084. [DOI] [PubMed] [Google Scholar]
- 19.Mwaura EN, Matiru V, Christine BI. Mycological findings of sputum samples from pulmonary tuberculosis patients attending Tb clinic in Nairobi, Kenya. Virol Mycol. 2013;2:3. [Google Scholar]
- 20.Tang CM, Holden DW, Aufauvre-Brown A, Cohen J. The detection of Aspergillus spp. by the polymerase chain reaction and its evaluation in bronchoalveolar lavage fluid. Am Rev Respir Dis. 1993;148:1313–7. doi: 10.1164/ajrccm/148.5.1313. [DOI] [PubMed] [Google Scholar]
- 21.Singh R, Singh G, Urhekar DA. Detection of Aspergillus species by polymerase chain reaction. Int J Curr Microbiol App Sci. 2016;5:254–60. [Google Scholar]
- 22.Kawamura S, Maesaki S, Tomono K, Tashiro T, Kohno S. Clinical evaluation of 61 patients with pulmonary aspergilloma. Intern Med. 2000;39:209–12. doi: 10.2169/internalmedicine.39.209. [DOI] [PubMed] [Google Scholar]
- 23.Knutsen AP, Slavin RG. Allergic bronchopulmonary aspergillosis in asthma and cystic fibrosis. Clin Dev Immunol. 2011;2011:843763. doi: 10.1155/2011/843763. [DOI] [PMC free article] [PubMed] [Google Scholar]


