Abstract
We investigated the performance of multiple radiomics feature extractors/software on predicting epidermal growth factor receptor mutation status in 228 patients with non–small cell lung cancer from publicly available data sets in The Cancer Imaging Archive. The imaging and clinical data were split into training (n = 105) and validation cohorts (n = 123). Two of the most cited open-source feature extractors, IBEX (1563 features) and Pyradiomics (1319 features), and our in-house software, Columbia Image Feature Extractor (CIFE) (1160 features), were used to extract radiomics features. Univariate and multivariate analyses were performed sequentially to predict EGFR mutation status using each individual feature extractor. Our univariate analysis integrated an unsupervised clustering method to identify nonredundant and informative candidate features for the creation of prediction models by multivariate analyses. In training, unsupervised clustering-based univariate analysis identified 5, 6, and 4 features from IBEX, Pyradiomics, and CIFE as candidate features, respectively. Multivariate prediction models using these features from IBEX, Pyradiomics, and CIFE yielded similar areas under the receiver operating characteristic curve of 0.68, 0.67, and 0.69. However, in validation, areas under the receiver operating characteristic curve of multivariate prediction models from IBEX, Pyradiomics, and CIFE decreased to 0.54, 0.56 and 0.64, respectively. Different feature extractors select different radiomics features, which leads to prediction models with varying performance. However, correlation between those selected features from different extractors may indicate these features measure similar imaging phenotypes associated with similar biological characteristics. Overall, attention should be paid to the generalizability of individual radiomics features and radiomics prediction models.
Keywords: Radiomics, Pyradiomics, IBEX, NSCLC, EGFR, TCIA
Introduction
Radiomics is a rapidly evolving field aiming to link phenotypes characterized from medical images with clinical data, including but not limited to, diagnostic, prognostic, and genomic information (1–7). Quantitative image features (also known as, radiomics features) have been shown, for example, to be associated with distant metastasis in lung adenocarcinoma (8–10), pathological response in a variety of cancer types (11–15), cancer recurrence after radiation therapy (16–19), and disease-free survival (20–23), and even with genotypes in many different cancer types (1, 24–31). Although there are many published prediction models related to both disease and treatment, there is no standardized evaluation of the performance (2), such as, but not limited to, the use of publicly available data and open-source feature extractors. The need for repeatability and reproducibility in radiomics has been increasingly emphasized (32, 33).
Therefore, the National Institutes of Health has encouraged medical imaging researchers to publicly share their data to stimulate open-science collaboration, and The Cancer Imaging Archive (TCIA) has evolved into a leading public database (34). TCIA is a service that hosts a large archive of medical images of cancer accessible for public download. Researchers nationwide are encouraged to submit data sets, and the current collection contains projects sponsored by private institutions and national programs. The Cancer Genome Atlas (TCGA) program is one such project that has generated a huge database of genomic, epigenomic, transcriptomic, and proteomic data from >20,000 samples spanning 33 cancer types (35). Clinical, genetic, and pathological data are stored on the Genomic Data Commons (GDC) data portal, while the radiological data reside in TCIA.
Many research groups have developed and also released open-source software packages with the hopes of establishing standardization to enhance reproducibility and comparability of radiomics results (36–41). The use of textural features for image classification dates back to 1973 (42), and image pattern recognition technologies have been widely deployed in computer-aided detection and diagnosis for the past 3 decades (43). A standard lexicon has been adopted as reference—the Image Biomarker Standardization Initiative (IBSI), version 9, available as of May 19, 2019 (44). Still, differences in image acquisition and preprocessing parameters may impact feature extraction (32, 45–48). Different radiomics software have also been shown to have varied algorithm implementation, which results in different feature values and poor agreement (49–50).
To the best of our knowledge, it is unknown how differences in feature extractor selection and feature calculation may impact the overall classification performance. The purpose of this study was to investigate differences in the overall radiogenomic classification performance on publicly available computed tomography (CT) images of patients with non–small cell lung cancer (NSCLC) owing to the use of different feature extractors. We also reported in detail our experience in the use of public data sets and open-source feature extractors.
Materials and Methods
Study Design
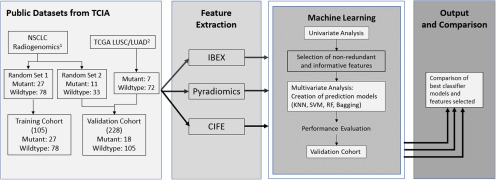
The basic study design diagram is shown in Figure 1. Public imaging data from TCIA relevant to our experiment were collected and split into training and validation cohorts. TCIA data consisted of 3 shared projects, NSCLC-Radiogenomics (51), TCGA-Lung Adenocarcinoma (TCGA-LUAD) (52), and TCGA-Lung Squamous Cell Carcinoma (TCGA-LUSC) (53). The training cohort was created using part of NSCLC-Radiogenomics (data collected from 2 institutions with relatively homogenous CT scanning parameters), while the validation cohort was created using a mix of data from the 3 projects (data collected from 7 institutions with diverse CT scanning parameters). This split aimed to test the generalization ability of radiomics features/models from a relatively homogenous data set to a more heterogeneous data set. Three feature extractors were used for feature extraction. Univariate and multivariate analyses were performed sequentially on predicting epidermal growth factor receptor (EGFR) mutant status by using each individual extractor, and performance was compared between the 3.
Figure 1.
Study design diagram. The design consists of 4 modules. First, projects NSCLC Radiogenomics and The Cancer Genome Atlas-Lung Adenocarcinoma (TGCA-LUSC)/TGCA-Lung Squamous Cell Carcinoma (TCGA/LUAD) were obtained from The Cancer Imaging Archive (TCIA) and split into a homogenous training cohort and a heterogeneous validation cohort. Second, features were extracted from all imaging cases using 3 different feature extractors: IBEX, Pyradiomics, and CIFE. Third, univariate and multivariate analyses are sequentially conducted on features from each extractor to create prediction models for epidermal growth factor receptor (EGFR) mutation status. “x3” means the univariate and multivariate analyses were performed identically 3 times by using the features from IBEX, Pyradiomics, and CIFE. Finally, the best classifier models and optimal features are compared between the 3 individual extractors.
1NSCLC Radiogenomics was produced by Bakr et al. with 211 patients with NSCLC from Stanford University School of Medicine and the Palo Alto Veteran Affairs Healthcare System.
2TCGA-LUSC and -LUAD are projects of TCGA, consisting of lung squamous cell carcinoma and lung adenocarcinoma cases. Imaging is available from 5 centers in the United States (Washington University, University of Pittsburgh, UNC, Roswell Park, and Lahey Health Home).
Patient Imaging and Clinical Data
The 3 data sets, NSCLC-Radiogenomics, TCGA-LUAD and TCGA-LUSC, were obtained through TCIA website. The NSCLC Radiogenomics data set was produced and described in detail by Bakr et al. (51). The NSCLC Radiogenomics data set included 211 cases with 129 EGFR wildtypes, 43 EGFR mutants, and 39 unknowns. TCGA-LUAD (52) and TCGA-LUSC (53) data collections provide clinical images to matched subjects in TCGA. TCGA-LUAD data set included 69 cases with 52 EGFR wildtypes, 11 EGFR mutants, and 6 unknowns. TCGA-LUSC data set included 37 cases with 36 EGFR wildtypes and 1 EGFR mutant. Imaging data for TCGA was collected from many sites worldwide and is very heterogeneous in terms of scanner modalities, manufacturers, and acquisition protocols. We included all patients that had a chest CT scan and a known EGFR mutation status. We excluded cases that had no noticeable lesion, cases with artifacts such as a biopsy needle in the lesion, and cases with multiple lesions and no provided segmentation. In some cases, Pyradiomics and IBEX produced an error during feature extraction, and these cases were excluded as well. Additional details are provided in online supplemental Section S3. In total, 149 cases from NSCLC Radiogenomics and 79 cases from TCGA LUAD and LUSC data sets were ultimately included in the study. Further details regarding all data, including information about scanning parameters, are included in the online supplemental Section S1A.
Training and Validation Set Split
The training and validation cohorts were split by data set and adjusted in order to maintain a balance of EGFR mutants and wildtypes in each cohort. The training cohort consisted of a random subset of the NSCLC Radiogenomics–included cases, totaling 105, with 27 mutant and 78 wildtype cases. The validation cohort included the remaining 44 cases from NSCLC Radiogenomics and the cases from TCGA-LUAD and TGCA-LUSC, totaling 123, with 18 mutant and 105 wildtype cases. The validation cohort had a much more heterogeneous sample owing to contribution from 3 data sets. Figure 1 details this split visually.
The validation cohort was also split into 3 subgroups corresponding to the 3 data sets. The NSCLC Radiogenomics subgroup had 44 cases with 33 EGFR wildtypes and 11 EGFR mutants. The TCGA-LUAD subgroup had 46 cases with 39 EGFR wildtypes and 7 EGFR mutants. The TCGA-LUSC subgroup had 33 EGFR wildtype cases only.
Tumor Segmentation
The NSCLC Radiogenomics data set provided segmentation for only 144 out of 211 cases. The remaining 67 cases and the TCGA-LUAD/-LUSC cases were segmented semiautomatically using a published segmentation algorithm incorporated into an open-source image viewing platform, WEASIS (54–55). The available and newly created segmentations were reviewed by an experienced thoracic radiologist (LE) and manually adjusted if necessary.
Feature Extraction
Three feature extractors were used to extract radiomics features from the segmented tumor volumes. The radiomics feature extractors included 2 open-source software packages, Pyradiomics, developed by Aerts' group (36), and the Imaging Biomarker Explorer (IBEX), developed by Court's group (37), and our in-house extractor, Columbia Image Feature Extractor (CIFE) developed by Zhao's group (32). Conditions between the 3 packages were controlled by using the recommended or if not available, the default settings.
Pyradiomics V2.1.2 (36) is an open-source Python package for the extraction of radiomics features from medical imaging. In total, 1319 features were extracted from each segmented tumor using Pyradiomics.
IBEX version 1.0β (37) is an open-source MATLAB and C/C++ software platform designed to support common radiomics workflow tasks, including but not limited to feature extraction. All available features were extracted without image preprocessing filters. In total, 1767 features were extracted from each segmented tumor using IBEX.
CIFE (32) is our in-house software package based on MATLAB 2016b (The MathWorks, Natick, MA) designed to extract radiomics features from medical imaging. In total, 1126 features were extracted from each segmented tumor using CIFE. (See online supplemental Section S1B for further details about settings of each feature extractor.)
Statistical Analysis
Analysis was run separately and identically on the 3 different feature sets computed from the 3 feature extractors. In this work, the univariate and multivariate analyses were performed sequentially on the feature sets. The univariate analysis was performed on only the training cohort to select features, and the multivariate analysis was performed on the training cohort and validated in the validation cohort.
First, a large number of redundant (ie, highly correlated) and noninformative features were removed using unsupervised clustering and receiver operating characteristic analysis. The unsupervised hierarchical clustering was performed in 3 steps:
Spearman rank correlations were calculated between features.
Features were organized into a hierarchical clustering tree based on these correlations.
Features were separated into groups based on a set correlation threshold.
Within each group containing redundant features, the correlation threshold was set to <0.2, and only features satisfying that criteria were selected as nonredundant (56). Nonredundant features were then examined in the univariate analysis using the area under the receiver operating characteristic curve (AUC) to indicate prediction performance for each feature. Only features with AUC > 0.6 were selected as informative features. Because the data set we used were relatively small, we used an unsupervised clustering–based algorithm instead of other widely used supervised feature selection algorithms (eg, mRMR and Relief) which might result in high risk of overfitting (56–58).
In the multivariate analysis, features attained from the univariate analysis were used to build models on the training set using 4 widely used machine-learning classification algorithms: k-nearest neighbors (KNN), least absolute shrinkage and selection operator (LASSO), support vector machine (SVM), and random forest classifier techniques. Fivefold cross-validation was applied on the training cohort to establish a performance baseline. In the 5-fold cross-validation, the training cohort was randomly separated into 5 subsets. One subset was used as a testing set, whereas the other 4 subsets were used as the training set. The training and testing procedures were repeated 5 times until each sample in the data set was used as a testing sample exactly once. The same 5-fold subsets were used for every model. The final training AUC for the prediction model was estimated using the average of 5 prediction performance.
The performance of model was then evaluated on the independent validation cohort. No samples in the independent validation cohort had ever been seen during training. The input to each model was the selected feature values and the output was the EGFR mutation status. A bootstrap approach reported by Aerts et al. (1) was used to calculate the significance on comparing models attained from each feature extractor. For 100 times, we calculated the AUC from 100 randomly selected samples, and the Wilcoxon test was used to assess significance.
All statistical analysis was performed on MATLAB 2016b platform (The MathWorks). A 2-sided P value of <.05 was regarded as statistically significant.
Results
Patient Characteristics
The clinical characteristics of the 228 patients included in our experiment are presented in Tables 1 and 2. Statistically significant differences were tested using the chi-square test for categorical data and the t test for continuous data. There was no significant difference between the training and validation cohorts in terms of age, sex, or tumor stage (P = .98, .74, and .39, respectively). The histological diagnosis showed a significant difference between the 2 cohorts, likely due to differences in data set origin (detailed in the Materials and Methods section). Although not statistically significant, there is a trend toward a difference between the training and validation cohorts in terms of proportion of EGFR mutants and wildtypes (P = .54) owing to the increased number of EGFR wildtypes in the validation cohort.
Table 1.
Patient Characteristics in Training and Validation Cohorts
| Training Cohort | Validation Cohort | P-Value | |
|---|---|---|---|
| Subjects (N) | 105 | 123 | |
| Age (Years) | 67.96 ± 8.9 | 67.92 ± 10.77 | .98 |
| Sex | .74 | ||
| Female | 40 (38%) | 45 (37%) | |
| Male | 65 (62%) | 55 (45%) | |
| Unknown | 0 (0%) | 23 (19%) | |
| Histology | <.001 | ||
| Adenocarcinoma | 92 (88%) | 84 (68%) | |
| Squamous Cell Carcinoma | 11 (10%) | 38 (31%) | |
| NOS (Not Otherwise Specified) | 2 (2%) | 1 (1%) | |
| Stage | .39 | ||
| Unknown | 22 (21%) | 34 (28%) | |
| 0 | 1 (1%) | 2 (2%) | |
| I | 49 (46%) | 40 (32%) | |
| II | 18 (17%) | 24 (20%) | |
| III | 13 (12%) | 18 (14%) | |
| IV | 2 (2%) | 4 (3%) | |
| EGFR Mutation | .054 | ||
| EGFR-Mutant | 27 (26%) | 18 (15%) | |
| EGFR-Wildtype | 78 (74%) | 105 (85%) |
P-value: chi-square test for categorical data and t test for continuous data.
Univariate Analysis
For the 3 sets of features from each feature extractor, we selected candidate features with a correlation coefficient <0.2 and an AUC > 0.6. These features are presented in Table 2. The definitions of these features are presented in online supplemental Section S1B. From Pyradiomics, IBEX, and CIFE, 6, 5, and 4 features were identified, respectively.
Table 2.
Nonredundant and Informative Features from Each Feature Extractor
| Feature Name | Univariate Analysis (AUC) |
|---|---|
| IBEX | |
| 135-1Correlation | 0.74 |
| LocalRangeStd | 0.72 |
| 1GaussAmplitude | 0.66 |
| VoxelSize | 0.62 |
| -333-4ClusterShade | 0.62 |
| Pyradiomics | |
| log-σ-2-0-mm-3D_firstorder_Minimum | 0.72 |
| log- σ2-0-mm-3D_glszm_SizeZoneNonUniformityNormalized | 0.70 |
| log-σ2-0-mm-3D_glcm_InverseVariance | 0.68 |
| wavelet-LHL_firstorder_Skewness | 0.67 |
| wavelet-LHH_firstorder_Skewness | 0.65 |
| wavelet-HHH_glszm_SmallAreaEmphasis | 0.65 |
| CIFE | |
| DWF_Z_H | 0.72 |
| Intensity_Minimum | 0.71 |
| Gabor_Max_Z | 0.68 |
| Intensity_Skewness | 0.65 |
Optimal features are listed for each individual extractor. These features are then used to build prediction models in the multivariate analysis. Each feature has a correlation coefficient <0.2 and an AUC > 0.6.
Most of the features selected from each extractor were different. Pyradiomics and our CIFE both used forms of intensity minimum and skewness, but owing to the use of image preprocession, Laplacian of Gaussian (LoG) filtering, in Pyradiomics, these values are not interchangeable. The distribution for every feature is included in the online supplemental Section S2B.
Moreover, we performed nonparametric Wilcoxon rank sum test to test the significance of feature distribution between EGFR wildtype and mutant for each individual candidate feature, and the results are shown in the online supplemental Figures S2–S4. All feature values originally had a significant difference in distribution between the wildtype and mutant subsets of the training cohort. However, 5/5 IBEX features, 6/6 Pyradiomics features, and 3/4 CIFE features did not have a significant difference in the wildtype and mutant subsets of the validation cohort. Only CIFE: intensity skewness had a significant difference between wildtype and mutant subsets of both the training and validation cohorts (P = .0072, P = .014) (see online supplemental Figure S4.4).
A correlogram of all features selected is shown in the online supplemental Figure S5 of Section S2C. There is little correlation between features from the same extractor, and there is some correlation between features from different extractors.
Multivariate Model Performance on Differentiating Lung Cancer EGFR Subtypes
The performance of multivariate models built from each feature extractor is summarized by the AUC value and is presented in Table 3. The optimal model from each feature extractor, determined by performance on the validation cohort, was produced using random forest classifier techniques. The performances from IBEX, Pyradiomics, and CIFE random forest models on the validation cohort were AUCs of 0.54, 0.56, and 0.64, respectively.
Table 3.
Performance of Multivariate Models from Each Feature Set in the Training and Validation Cohorts
| IBEX | PY | CIFE | ||||
|---|---|---|---|---|---|---|
| ClassificationAlgorithm | T | V | T | V | T | V |
| KNN | 0.620 | 0.54 | 0.66 | 0.54 | 0.67 | 0.60 |
| SVM | 0.59 | 0.48 | 0.57 | 0.52 | 0.60 | 0.51 |
| Random Forests | 0.68 | 0.54a | 0.67 | 0.56a | 0.68 | 0.64a |
| Bagging | 0.66 | 0.53 | 0.67 | 0.53 | 0.69 | 0.63 |
aOptimal model based on validation performance from each feature set, which was random forest for all extractors. T and V columns represent AUC scores for the indicated model from the training and validation cohorts, respectively. Performance values are presented as AUC values.
A pairwise comparison between each of the best models is shown in the online supplemental Table S2A. Comparisons were done using the bootstrap approach previously reported by Aerts et al. (1). Although the results of IBEX and Pyradiomics were not significantly different (P = .19), CIFE produced results significantly different from those of IBEX (p = 1.54e-14) and Pyradiomics (p = 2.02e-10).
A comparison between the performances of the best models on the training versus the validation data sets is shown in the online supplemental Table S2B. All models had significant differences in performance between the training and validation sets, but the trend for IBEX and Pyradiomics seems to have a greater difference.
Discussion
In this study, we aimed to use different feature extractors on public imaging data to compare classification performance. The radiomics feature extractors included 2 open-source software packages, Pyradiomics (36) and IBEX (37), and our in-house extractor, CIFE (32). These software packages have seen extensive use by researchers worldwide in experiments to predict diagnostic, genomic, prognostic, and response outcomes for a wide range of diseases, and proved ideal candidates for our comparison (8, 13, 15, 59–67).
We initially extracted 1767, 1319, and 1126 features from IBEX, Pyradiomics, and CIFE, respectively. After removing for redundancy and selecting clinical informative features, we ultimately isolated 6, 5, and 4 candidate features for the 3 feature extractors respectively. This result is consistent with that of a previous report that there is a large amount of redundancy within feature extractors (68). Notably, the selected features differed mostly from each group, but there were some similarities. Intensity minimum and skewness features were chosen from Pyradiomics and CIFE, although the implementation of the 2 is not exactly the same. There was some correlation between features from different extractors. This may suggest that similar biological characteristics are described.
Our results match those of existing literature on EGFR radiogenomic classification. Zhang et al. and Li et al. have found skewness to be predictive of EGFR mutation status and subtypes (69–70). Mei et al. also used the Pyradiomics feature extractor and similarly found that Size Zone NonUniformity Normalized was a predictor for EGFR mutation status (71).
We next used these selected nonredundant and informative candidate features to build multivariate prediction models using 4 commonly used machine-learning classification algorithms: k-nearest neighbors, support vector machine, random forest, and bagging. The best models created from IBEX, Pyradiomics, and CIFE features achieved similar training performance with cross-validated AUCs of 0.68, 0.67, and 0.69, respectively.
However, in validation, the performances from IBEX, Pyradiomics, and CIFE were AUCs of 0.54, 0.555, and 0.638 respectively. The validation performances were significantly decreased from the cross-validated training performance for models created from all 3 feature extractors. A pairwise comparison showed that CIFE had a significantly different validation performance than both IBEX and Pyradiomics, whereas the performance between IBEX and Pyradiomics was not significantly different. Our data were split into training and validation cohorts using a single data set for the training cohort and a mix of 3 data sets for the validation cohort. Therefore, the validation cohort will naturally be relatively more heterogeneous in terms of imaging parameters than the training cohort, as the cases come from 7 different institutions. Furthermore, the CT imaging parameters are more lung cancer–specific in the training data than those in the validation data. We believe that the splitting strategy used in our work would allow us to discover better model performance. This may explain the decrease in performance of radiofrequency (RF) models from all groups from the training cohort to the validation cohort. In addition, the trend toward a difference in proportion of EGFR wildtype and mutant cases between the training and validation cohorts may have also affected the performance. Although a decrease in performance from training to validation is commonly seen in machine-learning experiments (72), it is interesting that the performance of RF models built from IBEX and Pyradiomics features decreased more than the performance of the RF model built from CIFE features.
Our study has several limitations. For the open-source feature extractors, features were extracted as suggested by online documentation or by using the default settings of the features' parameters. Other researchers may find different results if they, for example, use different image preprocessing parameters. In addition, although we found 3 data sets with the information for our case example, our data size is still limited. The validation cohort consists of 3 different data sets, which may have affected the performance of our model. Although it would be interesting to see the individual performances of each subgroup within the validation cohort, this analysis was not feasible owing to the limited number of cases and imbalances of mutant and wildtype cases. We did not consider the effect of imaging heterogeneity and segmentation on our results because the purpose of our study was to compare extractors rather than assess the potential effects of segmentation. Although we had a mix of provided segmentation and our own in-house-generated contours, we used the same data set images and tumor segmentation for all different extractors. In addition, the definitions of the implemented features are available, but some are hard for us to fully explain the meanings.
It is important to note that the purpose of the study was to not compare these feature extractors in terms of their capabilities of building prediction models, but to show that differences can exist when applying different feature extractors to the same clinical application.
Future work may include optimization of machine-learning models, larger data sets, and other clinical applications. Generating a combined model from features of all 3 extractors may also potentially increase performance. In addition, although the CIFE feature extractor has been used in several published studies, it has yet to be released to the public.
Overall our experience with public data sets and open-source feature extraction software has been quite smooth. The majority of data cases fulfilled our inclusion criteria for our experiment and are easily accessible and ready for use. We could extract features for the majority of cases with all software packages and had clear documentation to facilitate use by a beginner. Further details regarding our experience are included in the online supplemental Section S3.
Conclusion
Different radiomics features were selected from different feature extractors to predict EGFR mutation status in patients with NSCLC, which resulted in varying prediction performance. Correlation between features from different extractors may indicate similar biological characteristics are measured. However, attention should be paid to the generalizability of both individual radiomics features and radiomics prediction models. In the future, radiomics feature extraction techniques will undoubtedly improve and may further standardize, but for now researchers may find it useful to use multiple packages for their clinical applications.
Supplemental Materials
Acknowledgments
Equal Contribution: L.L. and S.H.S. contributed equally to the study.
This study was in part supported by U01 CA225431 from the National Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the funding sources.
Conflict of Interest: None reported.
Disclosures: No disclosures to report.
Footnotes
- TCIA
- The Cancer Imaging Archive
- TGCA
- The Cancer Genome Atlas
- CT
- computed tomography
- NSCLC
- non–small cell lung cancer
- TCGA-LUAD
- TCGA-Lung Adenocarcinoma
- TCGA-LUSC
- TGCA-Lung Squamous Cell Carcinoma
- EGFR
- epidermal growth factor receptor
- CIFE
- Columbia Image Feature Extractor
- AUC
- area under the receiver operating characteristic curve
- RF
- radiofrequency
References
- 1. Aerts HJWL, Velazquez ER, Leijenaar RTH, Parmar C, Grossmann P, Carvalho S, Bussink J, Monshouwer R, Haibe-Kains B, Rietveld D, Hoebers F, Rietbergen MM, Leemans CR, Dekker A, Quackenbush J, Gillies RJ, Lambin P. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, Sanduleanu S, Larue RTHM, Even AJG, Jochems A, van Wijk Y, Woodruff H, van Soest J, Lustberg T, Roelofs E, van Elmpt W, Dekker A, Mottaghy FM, Wildberger JE, Walsh S. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14:749–762. [DOI] [PubMed] [Google Scholar]
- 3. Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RGPM, Granton P, Zegers CML, Gillies R, Boellard R, Dekker A, Aerts HJWL. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kumar V, Gu Y, Basu S, Berglund A, Eschrich SA, Schabath MB, Forster K, Aerts HJWL, Dekker A, Fenstermacher D, Goldgof DB, Hall LO, Lambin P, Balagurunathan Y, Gatenby RA, Gillies RJ. Radiomics: the process and the challenges. Magn Reson Imaging. 2012;30:1234–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278:563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gatenby RA, Grove O, Gillies RJ. Quantitative imaging in cancer evolution and ecology. Radiology. 2013;269:8–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Aerts HL. The potential of radiomic-based phenotyping in precision medicine: a review. JAMA Oncol. 2016;2:1636–1642. [DOI] [PubMed] [Google Scholar]
- 8. Coroller TP, Grossmann P, Hou Y, Rios Velazquez E, Leijenaar RTH, Hermann G, Lambin P, Haibe-Kains B, Mak RH, Aerts HJWL. CT-based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiother Oncol. 2015;114:345–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dou TH, Coroller TP, van Griethuysen JJM, Mak RH, Aerts H. Peritumoral radiomics features predict distant metastasis in locally advanced NSCLC. PLoS One. 2018;13:e0206108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ferreira-Junior JR, Koenigkam-Santos M, Magalhães Tenório AP, Faleiros MC, Garcia Cipriano FE, Fabro AT, Näppi J, Yoshida H, de Azevedo-Marques PM. CT-based radiomics for prediction of histologic subtype and metastatic disease in primary malignant lung neoplasms. Int J Comput Assist Radiol Surg. 2020;15:163–172. [DOI] [PubMed] [Google Scholar]
- 11. Riyahi S, Choi W, Liu C-J, Zhong H, Wu AJ, Mechalakos JG, Lu W. Quantifying local tumor morphological changes with Jacobian map for prediction of pathologic tumor response to chemo-radiotherapy in locally advanced esophageal cancer. Phys Med Biol. 2018;63:145020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cain EH, Saha A, Harowicz MR, Marks JR, Marcom PK, Mazurowski MA. Multivariate machine learning models for prediction of pathologic response to neoadjuvant therapy in breast cancer using MRI features: a study using an independent validation set. Breast Cancer Res Treat. 2019;173:455–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang Z, He B, Zhuang X, Gao X, Wang D, Li M, Lin Z, Luo R. CT-based radiomic signatures for prediction of pathologic complete response in esophageal squamous cell carcinoma after neoadjuvant chemoradiotherapy. J Radiat Res. 2019;60:538–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lin P, Yang P-F, Chen S, Shao Y-Y, Xu L, Wu Y, Teng W, Zhou X-Z, Li B-H, Luo C, Xu L-M, Huang M, Niu T-Y, Ye Z-M. A Delta-radiomics model for preoperative evaluation of Neoadjuvant chemotherapy response in high-grade osteosarcoma. Cancer Imaging. 2020;20:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Coroller TP, Agrawal V, Narayan V, Hou Y, Grossmann P, Lee SW, Mak RH, Aerts HJWL. Radiomic phenotype features predict pathological response in non-small cell lung cancer. Radiother Oncol. 2016;119:480–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mattonen SA, Palma DA, Johnson C, Louie AV, Landis M, Rodrigues G, Chan I, Etemad-Rezai R, Yeung TPC, Senan S, Ward AD. Detection of local cancer recurrence after stereotactic ablative radiation therapy for lung cancer: physician performance versus radiomic assessment. Int J Radiat Oncol Biol Phys. 2016;94:1121–1128. [DOI] [PubMed] [Google Scholar]
- 17. Meng J, Liu S, Zhu L, Zhu L, Wang H, Xie L, Guan Y, He J, Yang X, Zhou Z. Texture Analysis as Imaging Biomarker for recurrence in advanced cervical cancer treated with CCRT. Sci Rep. 2018;8:11399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li Q, Kim J, Balagurunathan Y, Qi J, Liu Y, Latifi K, Moros EG, Schabath MB, Ye Z, Gillies RJ, Dilling TJ. CT imaging features associated with recurrence in non-small cell lung cancer patients after stereotactic body radiotherapy. Radiat Oncol. 2017;12:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee SL, Ravi A, Morton G, Loblaw A, Tseng C-L, Haider M, Murgic J, Nicolae A, Semple M, Chung HT. Changes in ADC and T2-weighted MRI-derived radiomic features in patients treated with focal salvage HDR prostate brachytherapy for local recurrence after previous external-beam radiotherapy. Brachytherapy. 2019;18:567. [DOI] [PubMed] [Google Scholar]
- 20. Huang Y, Liu Z, He L, Chen X, Pan D, Ma Z, Liang C, Tian J, Liang C. Radiomics signature: a potential biomarker for the prediction of disease-free survival in early-stage (I or II) non-small cell lung cancer. Radiology. 2016;281:947–957. [DOI] [PubMed] [Google Scholar]
- 21. Fang J, Zhang B, Wang S, Jin Y, Wang F, Ding Y, Chen Q, Chen L, Li Y, Li M, Chen Z, Liu L, Liu Z, Tian J, Zhang S. Association of MRI-derived radiomic biomarker with disease-free survival in patients with early-stage cervical cancer. Theranostics. 2020;10:2284–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Franceschini D, Cozzi L, De Rose F, Navarria P, Fogliata A, Franzese C, Pezzulla D, Tomatis S, Reggiori G, Scorsetti M. A radiomic approach to predicting nodal relapse and disease-specific survival in patients treated with stereotactic body radiation therapy for early-stage non-small cell lung cancer. Strahlenther Onkol. 2019. [DOI] [PubMed] [Google Scholar]
- 23. Kirienko M, Cozzi L, Antunovic L, Lozza L, Fogliata A, Voulaz E, Rossi A, Chiti A, Sollini M. Prediction of disease-free survival by the PET/CT radiomic signature in non-small cell lung cancer patients undergoing surgery. Eur J Nucl Med Mol Imaging. 2018;45:207–217. [DOI] [PubMed] [Google Scholar]
- 24. Jansen RW, van Amstel P, Martens RM, Kooi IE, Wesseling P, de Langen AJ, van Oordt C. W M-V D H, Jansen BHE, Moll AC, Dorsman JC, Castelijns JA, de Graaf P, de Jong MC. Non-invasive tumor genotyping using radiogenomic biomarkers, a systematic review and oncology-wide pathway analysis. Oncotarget. 2018;9:20134–20155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nair JKR, Saeed UA, McDougall CC, Sabri A, Kovacina B, Raidu BVS, Khokhar JRA, Probst S, Hirsh V, Chankowsky J, Van Kempen LC, Taylor J. Radiogenomic models using machine learning techniques to predict EGFR mutations in non-small cell lung cancer. Can Assoc Radiol J. 2020. [DOI] [PubMed] [Google Scholar]
- 26. Hoshino I, Yokota H, Ishige F, Iwatate Y, Takeshita N, Nagase H, Uno T, Matsubara H. Radiogenomics predicts the expression of microRNA-1246 in the serum of esophageal cancer patients. Sci Rep. 2020;10:2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lo Gullo R, Daimiel I, Morris EA, Pinker K. Combining molecular and imaging metrics in cancer: radiogenomics. Insights Imaging. 2020;11:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yamamoto S, Maki DD, Korn RL, Kuo MD. Radiogenomic analysis of breast cancer using MRI: a preliminary study to define the landscape. AJR Am J Roentgenol. 2012;199. [DOI] [PubMed] [Google Scholar]
- 29. Karlo CA, Di Paolo PL, Chaim J, Hakimi AA, Ostrovnaya I, Russo P, Hricak H, Motzer R, Hsieh JJ, Akin O. Radiogenomics of clear cell renal cell carcinoma: associations between CT imaging features and mutations. Radiology. 2014;270:464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gevaert O, Mitchell LA, Achrol AS, Xu J, Echegaray S, Steinberg GK, Cheshier SH, Napel S, Zaharchuk G, Plevritis SK. Glioblastoma multiforme: exploratory radiogenomic analysis by using quantitative image features. Radiology. 2014;273:313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zinn PO, Majadan B, Sathyan P, Singh SK, Majumder S, Jolesz FA, Colen RR. Radiogenomic mapping of edema/cellular invasion MRI-phenotypes in glioblastoma multiforme. PLoS One. 2011;6:e25451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhao B, Tan Y, Tsai W-Y, Qi J, Xie C, Lu L, Schwartz LH. Reproducibility of radiomics for deciphering tumor phenotype with imaging. Sci Rep. 2016;6:23428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Traverso A, Wee L, Dekker A, Gillies R. Repeatability and reproducibility of radiomic features: a systematic review. Int J Radiat Oncol Biol Phys. 2018;102:1143–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Prior FW, Clark K, Commean P, Freymann J, Jaffe C, Kirby J, Moore S, Smith K, Tarbox L, Vendt B editors. TCIA: an information resource to enable open science. Engineering in Medicine and Biology Society (EMBC), 2013 35th Annual International Conference of the IEEE; 2013: IEEE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. National Cancer Institute. (2019). The Cancer Genome Atlas Program. [online]. Available at: https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga [Accessed September18, 2019].
- 36. van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, Beets-Tan RGH, Fillion-Robin J-C, Pieper S, Aerts HJWL. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77:e104–e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang L, Fried DV, Fave XJ, Hunter LA, Yang J, Court LE. IBEX: an open infrastructure software platform to facilitate collaborative work in radiomics. Med Phys. 2015;42:1341–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Echegaray S, Bakr S, Rubin DL, Napel S. Quantitative Image Feature Engine (QIFE): an open-source, modular engine for 3D quantitative feature extraction from volumetric medical images. J Digit Imaging. 2018;31:403–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Apte AP, Iyer A, Crispin-Ortuzar M. Technical Note: extension of CERR for computational radiomics: a comprehensive MATLAB platform for reproducible radiomics research. Med Phys. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yuan R, Shi S, Chen J, Cheng G. Radiomics in RayPlus: a web-based tool for texture analysis in medical images. J Digit Imaging. 2019;32:269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pfaehler E, Zwanenburg A, de Jong JR, Boellaard R. RaCaT: an open source and easy to use radiomics calculator tool. PLoS One. 2019;14:e0212223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Haralick RM, Shanmugam K, Dinstein I. Textural features for image classification. IEEE Trans Syst, Man, Cybern. 1973;SMC-3:610–621. [Google Scholar]
- 43. Doi K. Computer-aided diagnosis in medical imaging: historical review, current status and future potential. Comput Med Imaging Graph. 2007;31:198–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zwanenburg A, Vallières M, Abdalah MA, Aerts HJWL, Andrearczyk V, Apte A, Ashrafinia S, Bakas S, Beukinga RJ, Boellaard R, Bogowicz M, Boldrini L, Buvat I, Cook GJR, Davatzikos C, Depeursinge A, Desseroit MC, Dinapoli N, Dinh CV, Echegaray S, El Naqa I, Fedorov AY, Gatta R, Gillies RJ, Goh V, Götz M, Guckenberger M, Ha SM, Hatt M, Isensee F, Lambin P, Leger S, Leijenaar RTH, Lenkowicz J, Lippert F, Losnegård A, Maier-Hein KH, Morin O, Müller H, Napel S, Nioche C, Orlhac F, Pati S, Pfaehler EAG, Rahmim A, Rao AUK, Scherer J, Siddique MM, Sijtsema NM, Fernandez JS, Spezi E, Steenbakkers RJHM, Tanadini-Lang S, Thorwarth D, Troost EGC, Upadhaya T, Valentini V, van Dijk LV, van Griethuysen J, van Velden FHP, Whybra P, Richter C, Löck S. The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology. 2020;295:328–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fave X, Zhang L, Yang J, Mackin D, Balter P, Gomez D, Followill D, Jones AK, Stingo F, Court LE. Impact of image preprocessing on the volume dependence and prognostic potential of radiomics features in non-small cell lung cancer. Transl Cancer Res. 2016;5:349–363. [Google Scholar]
- 46. Shafiq-Ul-Hassan M, Zhang GG, Latifi K, Ullah G, Hunt DC, Balagurunathan Y, Abdalah MA, Schabath MB, Goldgof DG, Mackin D, Court LE, Gillies RJ, Moros EG. Intrinsic dependencies of CT radiomic features on voxel size and number of gray levels. Med Phys. 2017;44:1050–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lu L, Ehmke RC, Schwartz LH, Zhao B. Assessing agreement between radiomic features computed for multiple CT imaging settings. PLoS One. 2016;11:e0166550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lu L, Liang Y, Schwartz LH, Zhao B. Reliability of radiomic features across multiple abdominal CT image acquisition settings: a pilot study using ACR CT phantom. Tomography. 2019;5:226–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Foy JJ, Robinson KR, Li H, Giger ML, Al-Hallaq H, Armato SG. Variation in algorithm implementation across radiomics software. J Med Imag. 2018;5:044505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. McNitt-Gray M, Napel S, Kalpathy-Cramer J, Jaggi A, Emaminejad N, Muzi M, Goldgof D, Yang H, Jones E, Wahi-Anwar M, Balagurunathan Y, Abdalah M, Zhao B, Hadjiiski L, Virkud A, Chan H, Pierce L, Farahani K. Standardization in Quantitative Imaging: A Multi-Center Comparison of Radiomics Feature Values Obtained by Different Software Packages on Digital Reference Objects and Patient Datasets. Radiological Society of North America 2019. Scientific Assembly and Annual Meeting, December 1-December 6, 2019, Chicago IL archive.rsna.org/2019/19018740.html [Accessed December 16, 2019]. [Google Scholar]
- 51. Bakr S, Gevaert O, Echegaray S, Ayers K, Zhou M, Shafiq M, Zheng H, Zhang W, Leung A, Kadoch M, Shrager J, Quon A, Rubin D, Plevritis S, Napel S. Data for NSCLC Radiogenomics Collection. The Cancer Imaging Archive. 2017. [Google Scholar]
- 52. Albertina B, Watson M, Holback C, Jarosz R, Kirk S, Lee Y, Lemmerman J. Radiology Data from The Cancer Genome Atlas Lung Adenocarcinoma [TCGA-LUAD] collection. The Cancer Imaging Archive. 2016. [Google Scholar]
- 53. Kirk S, Lee Y, Kumar P, Filippini J, Albertina B, Watson M, Lemmerman J. Radiology Data from The Cancer Genome Atlas Lung Squamous Cell Carcinoma [TCGA-LUSC] collection. The Cancer Imaging Archive. 2016. [Google Scholar]
- 54. Yang H, Schwartz LH, Zhao B. A response assessment platform for development and validation of imaging biomarkers in oncology. Tomography. 2016;2:406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tan Y, Schwartz LH, Zhao B. Segmentation of lung lesions on CT scans using watershed, active contours, and Markov random field. Med Phys. 2013;40:043502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lu L, Wang D, Wang L, E L, Guo P, Li Z, Xiang J, Yang H, Li H, Yin S, Schwartz LH, Xie C, Zhao B. A quantitative imaging biomarker for predicting disease-free-survival-associated histologic subgroups in lung adenocarcinoma. Eur Radiol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Peng HC, Long F, Ding C. Feature selection based on mutual information: criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans Pattern Anal Mach Intell. 2005;27:1226–1238. [DOI] [PubMed] [Google Scholar]
- 58. Kenji K, Larry R. A Practical Approach to Feature Selection. Proceedings of the Ninth International Workshop on Machine Learning. 1992;p249–256. [Google Scholar]
- 59. Bi WL, Hosny A, Schabath MB, Giger ML, Birkbak NJ, Mehrtash A, Allison T, Arnaout O, Abbosh C, Dunn IF, Mak RH, Tamimi RM, Tempany CM, Swanton C, Hoffmann U, Schwartz LH, Gillies RJ, Huang RY, Aerts H. Artificial intelligence in cancer imaging: clinical challenges and applications. CA Cancer J Clin. 2019;69:127–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. L E, Lu L, Li L, Yang H, Schwartz LH, Zhao B. Radiomics for classification of lung cancer histological subtypes based on nonenhanced computed tomography. Acad Radiol. 2019;26:1245–1252. [DOI] [PubMed] [Google Scholar]
- 61. Chen A, Lu L, Pu X, Yu T, Yang H, Schwartz LH, Zhao B. CT-based radiomics model for predicting brain metastasis in category T1 lung adenocarcinoma. AJR Am J Roentgenol. 2019;213:134–139. [DOI] [PubMed] [Google Scholar]
- 62. Mokrane F-Z, Lu L, Vavasseur A, Otal P, Peron J-M, Luk L, Yang H, Ammari S, Saenger Y, Rousseau H, Zhao B, Schwartz LH, Dercle L. Radiomics machine-learning signature for diagnosis of hepatocellular carcinoma in cirrhotic patients with indeterminate liver nodules. Eur Radiol. 2020;30:558–570. [DOI] [PubMed] [Google Scholar]
- 63. Hawkins S, Wang H, Liu Y, Garcia A, Stringfield O, Krewer H, Li Q, Cherezov D, Gatenby RA, Balagurunathan Y, Goldgof D, Schabath MB, Hall L, Gillies RJ. Predicting malignant nodules from screening CT scans. J Thorac Oncol. 2016;11:2120–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rios Velazquez E, Parmar C, Liu Y, Coroller TP, Cruz G, Stringfield O, Ye Z, Makrigiorgos M, Fennessy F, Mak RH, Gillies R, Quackenbush J, Aerts H. Somatic mutations drive distinct imaging phenotypes in lung cancer. Cancer Res. 2017;77:3922–3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sun R, Limkin EJ, Vakalopoulou M, Dercle L, Champiat S, Han SR, Verlingue L, Brandao D, Lancia A, Ammari S, Hollebecque A, Scoazec JY, Marabelle A, Massard C, Soria JC, Robert C, Paragios N, Deutsch E, Ferte C. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol. 2018;19:1180–1191. [DOI] [PubMed] [Google Scholar]
- 66. Khalvati F, Zhang Y, Baig S, Lobo-Mueller EM, Karanicolas P, Gallinger S, Haider MA. Prognostic value of CT radiomic features in resectable pancreatic ductal adenocarcinoma. Sci Rep. 2019;9:5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Jeong J, Wang L, Ji B, Lei Y, Ali A, Liu T, Curran WJ, Mao H, Yang X. Machine-learning based classification of glioblastoma using delta-radiomic features derived from dynamic susceptibility contrast enhanced magnetic resonance images: introduction. Quant Imaging Med Surg. 2019;9:1201–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Berenguer R, Pastor-Juan M. D R, Canales-Vázquez J, Castro-García M, Villas MV, Mansilla Legorburo F, Sabater S. Radiomics of CT features may be nonreproducible and redundant: influence of CT acquisition parameters. Radiology. 2018;288:407–415. [DOI] [PubMed] [Google Scholar]
- 69. Zhang L, Chen B, Liu X, Song J, Fang M, Hu C, Dong D, Li W, Tian J. Quantitative biomarkers for prediction of epidermal growth factor receptor mutation in non-small cell lung cancer. Transl Oncol. 2018;11:94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Li S, Ding C, Zhang H, Song J, Wu L. Radiomics for the prediction of EGFR mutation subtypes in non-small cell lung cancer. Med Phys. 2019;46. [DOI] [PubMed] [Google Scholar]
- 71. Mei D, Luo Y, Wang Y, Gong J. CT texture analysis of lung adenocarcinoma: can radiomic features be surrogate biomarkers for EGFR mutation statuses. Cancer Imaging. 2018;18:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. González CR, Abu-Mostafa YS. Mismatched training and test distributions can outperform matched ones. Neural Comput. 2015;27:365–387. [DOI] [PubMed] [Google Scholar]
- 73. Zaffino P, Raudaschl P, Fritscher K, Sharp GC, Spadea MF. Technical Note: Plastimatch MABS, an open source tool for automatic image segmentation. Med Phys. 2016;43:5155–5160. [DOI] [PubMed] [Google Scholar]
- 74. Ger RB, Cardenas CE, Anderson BM. Guidelines and experience using Imaging Biomarker Explorer (IBEX) for radiomics. J Vis Exp. 2018:57132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. IBEX Google Forum. 2017. Available from: https://groups.google.com/forum/#!forum/ibex_users.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.