Abstract
Breast parenchymal enhancement (BPE) has shown association with breast cancer risk and response to neoadjuvant treatment. However, BPE quantification is challenging, and there is no standardized segmentation method for measurement. We investigated the use of a fully automated breast fibroglandular tissue segmentation method to calculate BPE from dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) for use as a predictor of pathologic complete response (pCR) following neoadjuvant treatment in the I-SPY 2 TRIAL. In this trial, patients had DCE-MRI at baseline (T0), after 3 weeks of treatment (T1), after 12 weeks of treatment and between drug regimens (T2), and after completion of treatment (T3). A retrospective analysis of 2 cohorts was performed: one with 735 patients and another with a final cohort of 340 patients, meeting a high-quality benchmark for segmentation. We evaluated 3 subvolumes of interest segmented from bilateral T1-weighted axial breast DCE-MRI: full stack (all axial slices), half stack (center 50% of slices), and center 5 slices. The differences between methods were assessed, and a univariate logistic regression model was implemented to determine the predictive performance of each segmentation method. The results showed that the half stack method provided the best compromise between sampling error from too little tissue and inclusion of incorrectly segmented tissues from extreme superior and inferior regions. Our results indicate that BPE calculated using the half stack segmentation approach has potential as an early biomarker for response to treatment in the hormone receptor–negative and human epidermal growth factor receptor 2–positive subtype.
Keywords: Magnetic resonance imaging (MRI), neoadjuvant chemotherapy, background parenchymal enhancement (BPE), breast cancer, segmentation
Introduction
Neoadjuvant chemotherapy (NAC) of breast cancer has shown equivalent effectiveness in comparison to adjuvant chemotherapy in terms of disease-free and overall survival (1, 2). NAC has the advantage of allowing a down-grade of the primary tumor for breast-conserving surgery and providing in vivo information about a patient's response to a specific regimen (3–5). The I-SPY 2 TRIAL (Investigation of Serial Studies to Predict Your Therapeutic Response through Imaging and Molecular Analysis 2) is a multicenter clinical trial for patients with locally advanced breast cancer undergoing NAC with the primary endpoint of pathological complete response (pCR) (6). Patients undergo dynamic-contrast enhanced MRI (DCE-MRI) examinations before, during, and after NAC. DCE-MRI provides additional insight into tumor physiology and may be able to provide better imaging biomarkers to treatment response than anatomical imaging alone (7–10). Results from the ACRIN 6657 trial showed that functional tumor volume measured by magnetic resonance imaging (MRI) was associated with pCR and recurrence-free survival, and functional tumor volume was a stronger indicator of response than clinical assessment (11–14).
Background parenchymal enhancement (BPE) on breast DCE-MRI is a physiological feature describing signal enhancement resulting from the uptake of gadolinium-based intravenous contrast by normal breast tissue (15). BPE observed in breast fibroglandular tissue (FGT) shows an association with breast cancer risk (16–19) and has also been investigated for use as an imaging biomarker to predict NAC response (20–22). Studies have shown BPE to be subtype-dependent with positive association for hormone receptor status (23, 24). Currently, 4 categories of BPE are qualitatively defined in the Breast Imaging Reporting and Data System (BI-RADS) atlas: minimal, mild, moderate, and marked (25). Acceptance of BPE as a biomarker is constrained by limited single-site studies with small sample sizes and varying methods for visual and quantitative BPE assessment (26). A recent review by Liao et al. reported the results of a number of studies using quantitative BPE measurements to evaluate treatment outcomes with varying methods for BPE quantification between studies (19, 22, 27, 28). To address inter-reader variability associated with qualitative BPE assessment, a standardized quantitative method is also needed.
Here, we evaluated 3 segmentation approaches for measuring quantitative contralateral BPE and compared them for prediction of pCR using data from the multicenter I-SPY2 trial. The overall aim was to determine an accurate, fully automatic, and robust segmentation method to quantitatively measure contralateral BPE and optimize its predictive power for assessing treatment response.
Materials and Methods
Patient Population
Women ≥18 years of age diagnosed with stage II/III breast cancer and tumor size measuring ≥2.5 cm were eligible to enroll in the I‐SPY 2 TRIAL (6). Patients with evidence of distant metastasis were excluded from the study. Biomarker assessments based on hormone (estrogen and progesterone) receptors (HR+/−), human epidermal growth factor receptor 2 (HER2+/−) status, and a 70‐gene assay (MammaPrint, Agendia, Amsterdam, The Netherlands) were performed at baseline (T0) and used for treatment randomization (6). In addition to standard immunohistochemical and fluorescence in situ hybridization (FISH) assays, the protocol included a microarray‐based assay of HER2 expression (TargetPrint, Agendia) to assign HR and HER2 statuses. Patients with tumors that were designated as HR+/HER2− and low risk according to the MammaPrint 70‐gene assay were excluded because the potential benefit of receiving investigational drugs along with chemotherapy for patients with less proliferative tumors are low with consideration of the risk of drug side effects (29, 30). All patients provided written informed consent to participate in the trial. A second consent was obtained if the patient was randomized to an experimental treatment.
Pathologic Assessment of Response
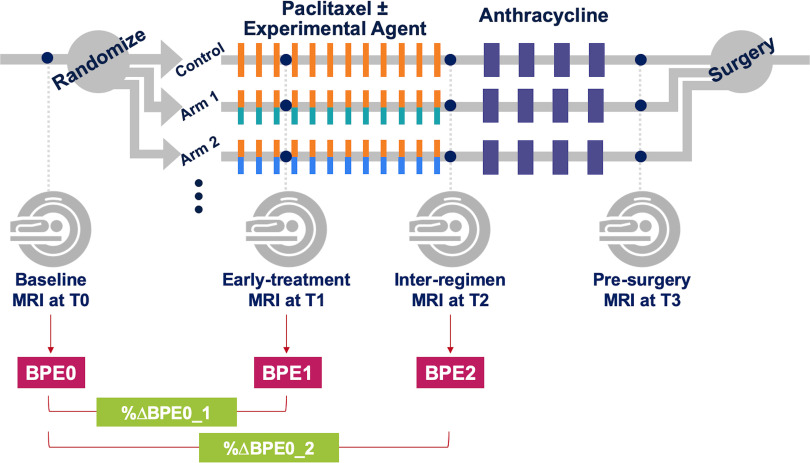
Figure 1 shows the schema of the I‐SPY 2 TRIAL. Pathologic complete response (pCR), defined as the absence of residual cancer in the breast or lymph nodes as evaluated by a trained pathologist at the time of surgery, is the primary endpoint of the trial. All patients were classified as either pCR or non‐pCR. Patients who left the study without completing the entire course of treatment or did not undergo surgery for any reason were labeled as non‐pCR.
Figure 1.
I‐SPY 2 study schema and adaptive randomization. Patients were randomized to the control (paclitaxel for HER2− or paclitaxel + trastuzumab for HER2+) or one of the experimental drug arms. Participants received a weekly dose of paclitaxel alone (control) or in combination with an experimental agent for 12 weekly cycles followed by 4 (every 2–3 weeks) cycles of anthracycline–cyclophosphamide (AC) before surgery.
MRI Acquisition
MRI examinations were performed before the initiation of NAC (baseline, T0), after 3 weeks of treatment (early‐treatment, T1), after 12 weeks and between drug regimens (inter-regimen, T2), and after completion of NAC and before surgery (presurgery, T3). MRI data were acquired with 1.5 T or 3 T scanners with a dedicated breast RF coil, across a variety of vendor platforms and institutions. All MRI examinations for the same patient were performed using the same magnet configuration (manufacturer, field strength, and breast coil model). The standardized image acquisition protocol included T2‐weighted and DCE‐MRI sequences performed bilaterally in the axial orientation.
DCE‐MRI was acquired as a series of 3D fat‐suppressed T1‐weighted images with the following parameters as specified in the I-SPY2 MRI protocol: repetition time = 4–10 milliseconds, minimum echo time, flip angle = 10°–20°, field of view = 260–360 mm to achieve full bilateral coverage, acquisition matrix = 384–512 with in‐plane resolution ≤ 1.4 mm, slice thickness ≤ 2.5 mm, and temporal resolution = 80–100 seconds. Gadolinium contrast agent was administered intravenously at a dose of 0.1 mmol/kg body weight and at a rate of 2 mL/s, followed by a 20‐mL saline flush. The same contrast agent brand was used for all MRI examinations for the same patient. Precontrast and multiple postcontrast images were acquired using identical sequence parameters. Postcontrast imaging continued for at least 8 minutes following contrast agent injection.
Quantitative Image Analysis
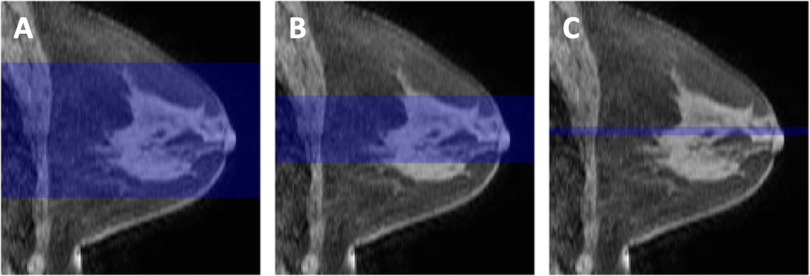
Nonuniformity of low spatial frequency intensity owing to coil sensitivity variations seen in the MRI data is known as bias or inhomogeneity. To correct for image inhomogeneity, all examinations were preprocessed with N4 bias correction, an improvement upon the N3 (nonparametric nonuniformity normalization) method (31). Automatic whole breast segmentation was performed on all examinations on each slice using locally developed software. Both breasts were initially segmented from background for the volumes anterior to the sternal notch using the precontrast image reformatted to the coronal orientation. The FGT volume of only the contralateral breast was then segmented using fuzzy c-means (FCM) clustering (32). Segmentation of 3 different sized subvolumes was investigated: all axial slices containing FGT voxels (full stack), central 50% of included slices (half stack), and the central 5 slices (center 5). A visual representation of the subvolumes is shown in Figure 2. All magnetic resonance examinations were centrally processed at the core I-SPY 2 imaging core laboratory using in‐house software developed in IDL (ITT Visual Information Solutions, Boulder, CO).
Figure 2.
Visual examples of the compared 3 subvolumes: full stack (A), half stack (B), and center 5 subvolumes (C). Each image is a representative sagittal slice of the same breast in which the highlighted region is the segment of axial slices used for background parenchymal enhancement (BPE) quantification.
Within each segmentation mask, mean background parenchymal enhancement (BPE) in the contralateral breast was calculated from DCE-MRI at each treatment time point as:
where S0 is the precontrast signal intensity, S1 is the postcontrast signal intensity of the image volume acquired closest to 2.5 minutes after contrast injection, and N is the number of included voxels.
A subset of 148 patients underwent unilateral manual whole breast segmentation of the contralateral breast followed by automatic FGT segmentation to better encapsulate as much FGT as possible. Manual whole breast segmentation excluded any regions with artifacts such as inhomogeneous fat saturation or coil bias, observed typically in the most superior and inferior axial slices to minimize inclusion of non-FGT voxels in the BPE quantification. Owing to the time-consuming nature of performing manual delineation, the full cohort was not assessed, and this manually delineated subset was used as a reference standard. The Pearson's linear correlation coefficient, r, was calculated to assess the difference in BPE quantification between the fully automated and semimanual methods.
Quality Assessment of BPE Calculation
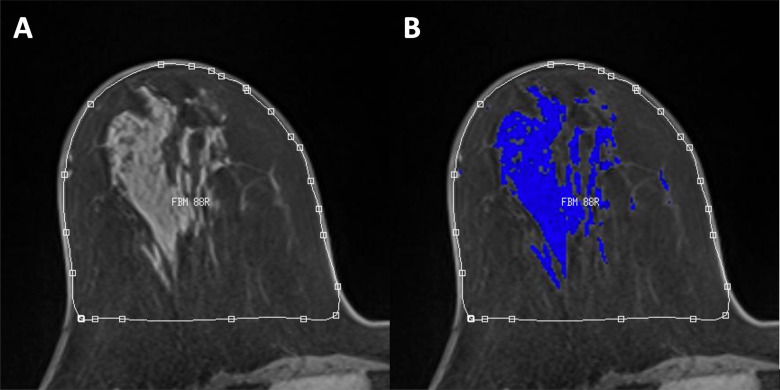
Visual quality of breast tissue segmentation for each examination was examined by a radiologist and was graded on how well the automatic segmentation performed on tissue classification, because image quality (eg, coil artifacts, poor fat suppression) can cause errors in the segmentation process. Automatic segmentation quality was visually graded as good, adequate, poor, or failed quality using representative images chosen at the center slice and at ends of the selected subvolume. Figure 3 shows an example of a typical good tissue segmentation for accurate BPE quantification. The quality assurance grades were used to further stratify the quality of BPE values used for analysis.
Figure 3.
An example of a typical good BPE segmentation on an axial slice: fully-automatic whole breast segmentation (A) and derived FGT mask in blue from fuzzy c-means clustering (B).
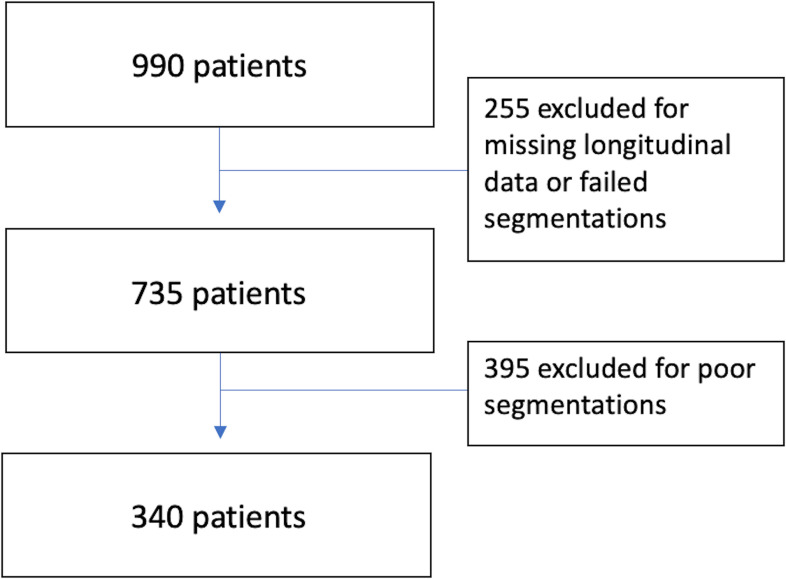
An initial 990 I-SPY2 patients enrolled on drug arms completed by November 2016 were included and considered for analysis. Patients who did not have a DCE-MRI scan at early-treatment (T1) or inter-regimen (T2), had a rejected DCE-MRI scan, or had a failed segmentation quality grade were excluded from analysis and comprised the first cohort for analysis. A final cohort was defined after additional removal of examinations with poor segmentation quality, leaving only good and adequate segmentation-quality examinations.
Statistical Analysis
Statistical analysis was performed to assess the predictive performance of a single magnetic resonance predictor for pCR vs non‐pCR outcomes. All statistical analyses were performed using SciPy 1.3 (https://scipy.org) and Python 3.7 (Python Software Foundation, Wilmington, DE).
The percent change in mean BPE from T0 to T1 (%ΔBPE0_1) was used in a univariate analysis for pCR prediction and is calculated as:
The area under the ROC curve (AUC) of a logistic regression model was used to assess the predictive performance of %ΔBPE0_1 in the full cohort and within subtypes. P-values for AUCs being different from .5 were estimated using the Mann–Whitney U test. Results with P-values < 0.05 were considered statistically significant.
Results
Patient Characteristics
In total, 990 patients with pCR outcome enrolled in the I-SPY2 TRIAL from completed drug arms before November 2016 were included in this study. Patients who did not have a DCE-MRI scan at early-treatment (T1) or inter-regimen (T2) had a failed segmentation quality grade, or had a rejected DCE-MRI scan, because other image quality or protocol adherence issues were excluded. After preliminary exclusion, BPE was calculated in 735 women (median age, 49 years; range, 24–77) and were included in the first cohort analysis, in which 258 (35.1%) patients achieved pCR. An additional 395 patients were excluded owing to strict quality assessment of poor tissue segmentations including undersampling, coil artifacts, poor fat suppression. For the second cohort, 340 women (median age, 49 years; range, 24–77) were included, in which 113 (33.2%) patients achieved pCR. Patients with hormone receptor–negative disease were more likely to achieve pCR than those with hormone receptor–positive disease. Patient characteristics are shown in Table 1 and a flow diagram of patient exclusion is visualized in Figure 4. No statistically significant differences were found in patient characteristics between the enrolled population of 990 patients and final analysis cohort of 340 patient that excluded poor-quality BPE.
Table 1.
Patient Characteristics
| Any Segmentation Quality (N = 990) |
Good or Adequate Segmentation Quality (N = 340) |
P Value | |
|---|---|---|---|
| Age at Screening (Years) | 0.78a | ||
| Missing | 1 | 0 | |
| Mean (SD) | 48.8 (10.6) | 48.9 (10.0) | |
| Range | 23.0–77.0 | 24.0–77.0 | |
| Race/Ethnicity | 0.43b | ||
| Missing | 1 | 0 | |
| American Indian or Alaska Native | 4 (0%) | 2 (1%) | |
| Asian | 68 (7%) | 23 (7%) | |
| Black or African American | 121 (12%) | 28 (8%) | |
| Mixed Race/Ethnicity | 7 (1%) | 4 (1%) | |
| Native Hawaiian or Pacific Islander | 5 (1%) | 2 (1%) | |
| White | 784 (79%) | 281 (83%) | |
| Menopausal Status | 0.52b | ||
| Missing | 202 | 70 | |
| Post/Perimenopausal | 324 (41%) | 117 (43%) | |
| Premenopausal | 464 (59%) | 153 (57%) | |
| Pathologic Complete Response | 0.86b | ||
| pCR | 324 (33%) | 113 (33%) | |
| nPCR | 666 (67%) | 227 (67%) | |
| Receptor Status | 0.70b | ||
| Missing | 2 | 0 | |
| HR+HER2+ | 156 (16%) | 57 (17%) | |
| HR+HER2− | 380 (38%) | 140 (41%) | |
| HR−HER2+ | 89 (9%) | 27 (8%) | |
| HR−HER2− | 363 (37%) | 116 (34%) |
aKruskal–Wallis rank sum test.
bPearson chi-square test.
Figure 4.
Flow diagram of the study database showing the exclusion criteria to obtain the first 735 patient cohort and the second quality-restricted cohort of 340 patients.
Univariate Analysis
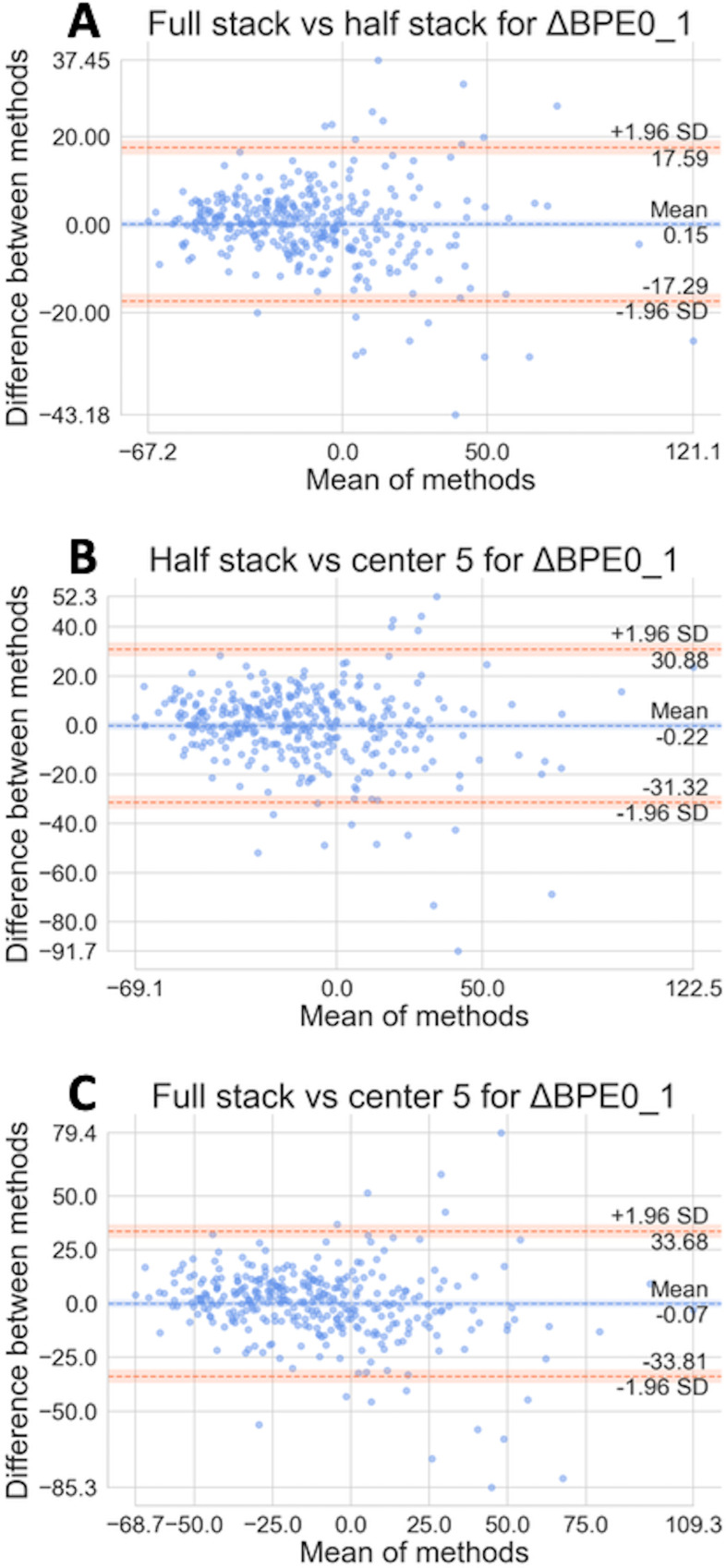
The comparability of 3 segmentation methods, full stack, half stack, and center 5 was assessed in the full cohort and within HR and human epidermal growth factor receptor 2 (HER2) subtypes. To analyze the strength of the linear relationship, the Pearson's linear correlation coefficient (r) was calculated between segmentation methods. The r values for full vs half, half vs center 5, and full vs center 5 were 0.953, 0.867, and 0.840, respectively. However, a high correlation is not necessarily indicative of meaningful results, as it does not provide information about possible bias. To visualize systematic bias versus random variation between the 3 automated segmentation methods, Bland–Altman plots (Figure 5) were calculated in the quality-restricted second cohort to see if our various automated method differed from each other (33). The mean differences for all 3 comparisons are very close to 0, suggesting that the estimated bias is low. All 3 plots also show that there are no apparent variations with mean values, with most of the points within the 95% limits of agreement.
Figure 5.
Bland–Altman plots comparing BPE values from 3 subvolumes of the dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) used for segmentation with our best-quality assessment. Full vs half (A). Half vs center 5 (B). Full vs center 5 (C).
Comparison with Manual BPE Reference Standard.
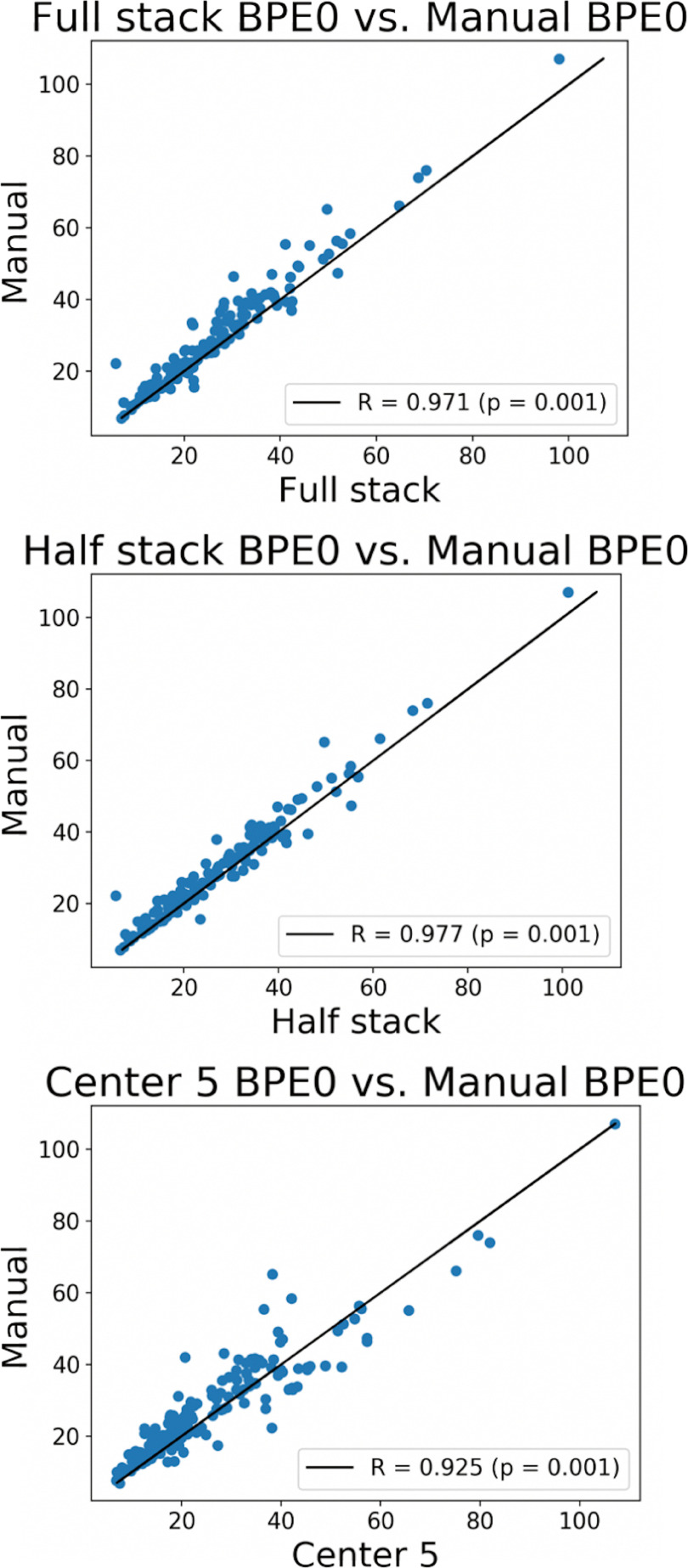
We calculated BPE for a subset of patients that had a manual whole breast segmentation to compare the relationship between automated and manual methods. Figure 6 shows the Pearson's linear correlation. All automated methods showed high agreement with the manual reference method, with best agreement using the half stack method. The r values between manual and full stack, half stack, and center 5 are 0.971, 0.977, and 0.925, respectively, with all P-values = .001.
Figure 6.
Pearson linear correlation plots comparing BPE0 values of the 3 fully automated methods to the semimanual method.
Table 2 shows the pCR rate and the reported AUCs for percent change in mean contralateral BPE from baseline to early treatment (%ΔBPE0_1) and from baseline to inter-regimen (%ΔBPE0_2) for each segmentation method within the full cohort and within subtypes. The data in this table contain all segmentation quality categories including poor, adequate, and good visual segmentations. AUCs in the full cohort ranged from 0.51 to 0.53 and AUCs varied within subtype from 0.56 to 0.58 in HR+/HER2+, 0.52 to 0.53 in HR+/HER2−, 0.56 to 0.59 in HR−/HER2+, and 0.51 to 0.52 in HR−/HER2−. These results reached statistical significance in the HR+/HER2− subtype for the T2 time-point predictor (%ΔBPE0_2).
Table 2.
AUCs from Logistic Regression in the First Cohort of 735 Patients Using Percent Change in Mean cBPE as a Predictor for pCR
| Patient Cohort | N | pCR Rate (%) | Method | %ΔBPE0_1 | %ΔBPE0_2 | ||
|---|---|---|---|---|---|---|---|
| AUC | P-Value | AUC | P-Value | ||||
| Full Cohort | 735 | 35.1 | Full stack | 0.53 | .09* | 0.52 | .14 |
| Half stack | 0.52 | .23 | 0.52 | .18 | |||
| Center 5 | 0.51 | .30 | 0.50 | .50 | |||
| HR+/HER2+ | 112 | 33.9 | Full stack | 0.58 | .08* | 0.53 | .30 |
| Half stack | 0.56 | .14 | 0.53 | .31 | |||
| Center 5 | 0.56 | .15 | 0.52 | .39 | |||
| HR+/HER2− | 299 | 19.1 | Full stack | 0.53 | .23 | 0.61 | .01** |
| Half stack | 0.52 | .32 | 0.60 | .01** | |||
| Center 5 | 0.53 | .24 | 0.59 | .02** | |||
| HR−/HER2+ | 61 | 68.9 | Full stack | 0.57 | .20 | 0.62 | .08* |
| Half stack | 0.59 | .14 | 0.60 | .11 | |||
| Center 5 | 0.56 | .25 | 0.53 | .35 | |||
| HR−/HER2− | 263 | 46.0 | Full stack | 0.52 | .33 | 0.50 | .49 |
| Half stack | 0.51 | .42 | 0.50 | .46 | |||
| Center 5 | 0.52 | .28 | 0.54 | .15 | |||
All measurements were obtained from bias corrected images with poor, adequate, and good segmentation quality. *P < .10; **P < .05.
When patients were restricted to adequate and good visual segmentation quality (Table 3), the associated AUCs for both BPE predictors remained similar between segmentation methods. The pCR rate was higher for HR−/HER2+ patients with adequate/good quality than the patients with any segmentation quality (81.5% versus 68.9%). AUCs in the full cohort ranged from 0.50 to 0.51 and AUCs varied within subtype from 0.44 to 0.57 in HR+/HER2+, 0.54 to 0.57 in HR+/HER2−, 0.78 to 0.87 in HR−/HER2+, and 0.50 to 0.55 in HR−/HER2−. The highest AUC values, which were also statistically significant, were found in the HR−/HER2+ subtype at the early time point (%ΔBPE0_1). Although sample sizes were reduced by ≥50% in every subtype after restricting for segmentation quality, differences in AUCs between subtypes became more apparent, with notably higher AUC values achieved in the HR−/HER2+ subtype at the T1 time point. Comparison of AUC values in the quality-restricted and unrestricted cohorts highlights the small variation between segmentation methods relative to differences between subtypes.
Table 3.
AUCs from Logistic Regression in the Second Quality Restricted Cohort of 340 Patients Using Percent Change in Mean cBPE as a Predictor for pCR
| Patient Cohort | N | pCR Rate (%) | Method | %ΔBPE0_1 | %ΔBPE0_2 | ||
|---|---|---|---|---|---|---|---|
| AUC | P-Value | AUC | P-Value | ||||
| Full Cohort | 340 | 33.2 | Full stack | 0.51 | .40 | 0.55 | .08* |
| Half stack | 0.51 | .35 | 0.54 | .11 | |||
| Center 5 | 0.50 | .49 | 0.50 | .46 | |||
| HR+/HER2+ | 57 | 31.6 | Full stack | 0.44 | .25 | 0.51 | .44 |
| Half stack | 0.45 | .26 | 0.50 | .48 | |||
| Center 5 | 0.57 | .21 | 0.49 | .46 | |||
| HR+/HER2− | 140 | 17.9 | Full stack | 0.54 | .25 | 0.61 | .05** |
| Half stack | 0.53 | .33 | 0.59 | .07* | |||
| Center 5 | 0.57 | .13 | 0.59 | .08* | |||
| HR−/HER2+ | 27 | 81.5 | Full stack | 0.78 | .03** | 0.66 | .14 |
| Half stack | 0.87 | .01** | 0.65 | .15 | |||
| Center 5 | 0.78 | .03** | 0.64 | .18 | |||
| HR−/HER2− | 116 | 41.4 | Full stack | 0.53 | .32 | 0.58 | .08* |
| Half stack | 0.55 | .17 | 0.57 | .09* | |||
| Center 5 | 0.50 | .47 | 0.50 | .47 | |||
All measurements were obtained from bias-corrected images with adequate and good segmentation quality only. *P < .10; **P < .05.
Discussion
BPE observed in breast FGT shows an association with breast cancer risk. We further investigated BPE's use as an imaging biomarker to predict NAC response. For BPE to be used as a robust, clinically meaningful biomarker, an automated quantitative segmentation method is necessary to remove subjectivity and inter-reader variability associated with qualitative methods (34). Although manual segmentation yields promising results, manual delineation of the breast surface and visual confirmation of tissue boundaries are time-consuming and subject to inter-reader variability. The use of automated segmentation may provide reproducible quantitative results required for validation and for ensuring repeatability. This study compared automated quantitative methods for BPE calculation using different levels of tissue sampling to improve segmentation quality and assessed the ability of each method to predict treatment response.
When visual assessment of segmentation quality was used, a large proportion or percentage of cases, 54% of the data set, were excluded from analysis. A limitation of this retrospective study may be the image quality, in which patients up until 2016 were included in the analysis. Since then, we have implemented better equipment and are continually improving our segmentation methods. When the exclusion criteria were relaxed, allowing artifacts or undersampling of tissue, our findings remained consistent within and between subtypes. In Table 1, the Kruskal–Wallis rank sum test and Pearson's chi-square test performed between the second quality-restricted cohort, and the initial 990 patients considered for analysis showed that the difference between cohorts is not statistically significant, suggesting that the included cohort reflects that of the population included in the I-SPY2 trial. However, the results in the HR+/HER2− subtype for %ΔBPE0_2 were reinforced in the quality-limited cohort, indicating that image quality may have different impacts in different subtypes. Results had higher relative AUCs for pCR prediction.
AUC values were similar for each segmentation method with only small differences for the full cohort as well as within subtypes and do not appear substantially meaningful for pCR prediction in the first cohort of 735 patients and when quality was restricted to 340 patients. Interesting AUC results from %ΔBPE0_2 are seen in Table 2 for the HR+ and HER2− subtype group. Our results show that BPE at the later time point may be predictive of HR+/HER2− patient's response to treatment with no clear differences between methods. Variations within subtype were relatively small in comparison to the AUC differences between subtypes. For example, the differences in the full cohort at T0 to T1 only vary by 0.01 in Table 3. Percent change in BPE did not show strong predictive power, which can be generalized to the full cohort. However, in HR− and HER2+ patients where there was a higher percentage of pCR, the change in mean BPE showed statistically significant predictive power toward pCR at the earlier time point in response to taxane-based treatment. Within the HR− and HER2+ subtype, the jump in AUC may signify change in BPE as a good imaging biomarker for early detection of pCR in a neoadjuvant setting. Although the HR−/HER2+ cohort size consists of 27 patients, of whom 22 achieved pCR, additional validation needs to be performed on a larger sample size.
Our results corroborate those of the work of Dong et al., supporting current findings that women with HR− tumors were more likely to achieve pCR than HR+ tumors and indicating that decreased BPE in women with HER2+ breast cancer may predict effective response to NAC treatment (35). BPE is affected by hormonal changes where estrogen can lead to increased contrast uptake in tissue as well as dilation of the blood vessels (36).
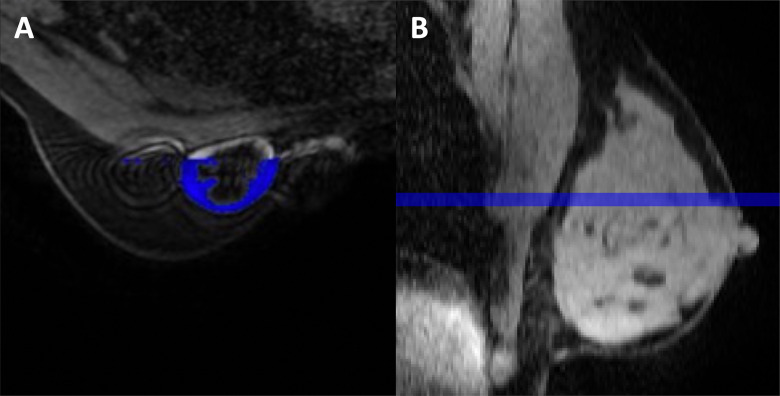
Fully automatic segmentation demonstrated some limitations. Figure 7 visually shows some limitations of the full stack method and the center 5 method. The full stack may pick up noise and false masking in the outermost regions of the DCE-MRI. The example on the left in Figure 7 shows an axial slice that contains a contralateral breast artifact from an implanted venous access port used to deliver chemotherapy. The artifact adversely affected the automatic segmentation, which falsely classified the artifact as tissue. The full stack method is also the most computationally intensive method and does not appear to provide more predictive benefit than the half stack. Another limitation was that the center referenced for the center 5 slice method may not always have been well centered within the breast, and thus, it might not give a representative sample of the tissue; whereas, the half stack method may sample enough of the breast to capture all of the FGT while excluding the other edges that may pick up artifacts. The example on the right in Figure 7 shows the smaller volume of interest using the center 5 slice method.
Figure 7.
Examples showing limitations of the (A) full stack method, which contains an artifact from an implanted venous access port used to deliver chemotherapy, and the (B) center 5 method, which may not always be well-centered within the breast, and thus might not give a representative sample of the tissue.
We showed that using the half stack method was the best compromise to optimize our clinical decision tool through validation. This compromise uses fewer computational resources while still retaining the same predictive performance as the full stack method. Using the half stack method, our study, along with many others, shows the importance of a longitudinal analysis using BPE as a predictor for positive response to treatment. Based on these observations, we recommend using the half-stack volume of interest moving forward. Future plans include comparing our results to a manually segmented reference standard, implementing automatic nipple slice detection, and adding contralateral BPE into a multivariate model to hopefully improve predictive performance for treatment response.
In conclusion, quantitative BPE calculated from DCE-MRI is an emerging imaging biomarker that has shown promise as an indicator of early response to neoadjuvant treatment. We showed that our BPE calculations from different-sized subvolumes of DCE-MRI scans are robust against each other and provide results with close agreement. From our study, we recommend moving forward with the half stack method for a fully automatic segmentation method for repeatable quantitative BPE measurements.
Acknowledgments
This work was supported by NIH U01 CA225427 and NIH R01 CA132870.
Disclosure: N.M.H. reports grants from NIH U01 CA225427 and R01 CA132870, during the conduct of the study; N.M.H, D.C.N., and B.N.J. report research support from Kheiron Medical Technology to institutions outside the submitted work; N.M.H. reports research support from GE Healthcare to institutions outside the submitted work; B.N.J. reports author royalties from UpToDate, outside the submitted work.
Conflict of Interest: None reported.
Footnotes
- NAC
- neoadjuvant chemotherapy
- DCE-MRI
- dynamic contrast-enhanced-Magnetic Resonance Imaging
- BPE
- background parenchymal enhancement
- pCR
- pathological complete response
- FGT
- fibroglandular tissue
- HR
- hormone receptor
- HER2
- human epidermal growth factor receptor 2
- AUC
- area under the curve
References
- 1. Mauri D, Pavlidis N, Ioannidis JP. Neoadjuvant versus adjuvant systemic treatment in breast cancer: a meta-analysis. J Natl Cancer Inst. 2005;97:188–194. [DOI] [PubMed] [Google Scholar]
- 2. Deo SV, Bhutani M, Shukla NK, Raina V, Rath GK, Purkayasth J. Randomized trial comparing neo-adjuvant versus adjuvant chemotherapy in operable locally advanced breast cancer (T4b N0-2 M0). J Surg Oncol. 2003;84:192–197. [DOI] [PubMed] [Google Scholar]
- 3. Clough KB, Acosta-Marín V, Nos C, Alran S, Rouanet P, Garbay J-R, Giard S, Verhaeghe J-L, Houvenaeghel G, Flipo B, Dauplat J, Dorangeon PH, Classe J-M, Rouzier R, Bonnier P. Rates of neoadjuvant chemotherapy and oncoplastic surgery for breast cancer surgery: a French national survey. Ann Surg Oncol. 2015;22:3504–3511. [DOI] [PubMed] [Google Scholar]
- 4. Early Breast Cancer Trialists' Collaborative Group (EBCTCG). Long-term outcomes for neoadjuvant versus adjuvant chemotherapy in early breast cancer: meta-analysis of individual patient data from ten randomised trials. Lancet Oncol. 2018;19:27–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mougalian SS, Soulos PR, Killelea BK, Lannin DR, Abu-Khalaf MM, DiGiovanna MP, Sanft TB, Pusztai L, Gross CP, Chagpar AB. Use of neoadjuvant chemotherapy for patients with stage I to III breast cancer in the United States. Cancer. 2015;121:2544–2552. [DOI] [PubMed] [Google Scholar]
- 6. Barker AD, Sigman CC, Kelloff GJ, Hylton NM, Berry DA, Esserman LJ. I-SPY 2: an adaptive breast cancer trial design in the setting of neoadjuvant chemotherapy. Clin Pharmacol Ther. 2009;86:97–100. [DOI] [PubMed] [Google Scholar]
- 7. Elhalawani H, Ger RB, Mohamed ASR, Awan MJ, Ding Y, Li K, Fave XJ, Beers AL, Driscoll B, H, Ii DA, van Houdt PJ, He R, Zhou S, Mathieu KB, Li H, Coolens C, Chung C, Bankson JA, Huang W, Wang J, Sandulache VC, Lai SY, Howell RM, Stafford RJ, Yankeelov TE, van der Heide UA, Frank SJ, Barboriak DP, Hazle JD, Court LE, Kalpathy-Cramer J, Fuller CD; Joint Head and Neck Radiotherapy MRI Development Cooperative. Dynamic contrast-enhanced magnetic resonance imaging for head and neck cancers. Sci Data. 2018;5:180008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hylton N. Dynamic contrast-enhanced magnetic resonance imaging as an imaging biomarker. J Clin Oncol. 2006;24:3293–3298. [DOI] [PubMed] [Google Scholar]
- 9. Rosen MA, Schnall MD. Dynamic contrast-enhanced magnetic resonance imaging for assessing tumor vascularity and vascular effects of targeted therapies in renal cell carcinoma. Clin Cancer Res. 2007;13:770s–776s. [DOI] [PubMed] [Google Scholar]
- 10. Press RH, Shu HG, Shim H, Mountz JM, Kurland BF, Wahl RL, Jones EF, Hylton NM, Gerstner ER, Nordstrom RJ, Henderson L, Kurdziel KA, Vikram B, Jacobs MA, Holdhoff M, Taylor E, Jaffray DA, Schwartz LH, Mankoff DA, Kinahan PE, Linden HM, Lambin P, Dilling TJ, Rubin DL, Hadjiiski L, Buatti JM. The use of quantitative imaging in radiation oncology: a Quantitative Imaging Network (QIN) perspective. Int J Radiat Oncol Biol Phys. 2018;102:1219–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hylton NM, Blume JD, Bernreuter WK, Pisano ED, Rosen MA, Morris EA, Weatherall PT, Lehman CD, Newstead GM, Polin S, Marques HS, Esserman LJ, Schnall MD; I-SPY TRIAL Investigators. Locally advanced breast cancer: mR imaging for prediction of response to neoadjuvant chemotherapy–results from ACRIN 6657/I-SPY TRIAL. Radiology. 2012;263:663–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hylton NM, Blume JD, Bernreuter WK, Pisano ED, Rosen MA, Morris EA, Weatherall PT, Lehman CD, Polin SM, Newstead G, Marques HS, Schnall MD, Esserman LJ; ACRIN 6657 Trial Team and I-SPY TRIAL Investigators. Comparison of MRI endpoints for assessing breast cancer response to neoadjuvant treatment: preliminary findings of the American College of Radiology Imaging Network (ACRIN) trial 6657. Cancer Res. 2009;69:6043. [Google Scholar]
- 13. Hylton NM, Gatsonis CA, Rosen MA, Lehman CD, Newitt DC, Partridge SC, Bernreuter WK, Pisano ED, Morris EA, Weatherall PT, Polin SM, Newstead GM, Marques HS, Esserman LJ, Schnall MD; ACRIN 6657 Trial Team and I-SPY 1 TRIAL Investigators. Neoadjuvant Chemotherapy for Breast Cancer: functional tumor volume by MR imaging predicts recurrence-free survival-results from the ACRIN 6657/CALGB 150007 I-SPY 1 trial. Radiology. 2016;279:44–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Scheel JR, Kim E, Partridge SC, Lehman CD, Rosen MA, Bernreuter WK, Pisano ED, Marques HS, Morris EA, Weatherall PT, Polin SM, Newstead GM, Esserman LJ, Schnall MD, Hylton NM; ACRIN 6657 Trial Team and I-SPY Investigators Network. MRI, clinical examination, and mammography for preoperative assessment of residual disease and pathologic complete response after neoadjuvant chemotherapy for breast cancer: aCRIN 6657 trial. AJR Am J Roentgenol. 2018;210:1376–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Arslan G, Celik L, Cubuk R, Celik L, Atasoy MM. Background parenchymal enhancement: is it just an innocent effect of estrogen on the breast? Diagn Interv Radiol. 2017;23:414–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Arasu VA, Miglioretti DL, Sprague BL, Alsheik NH, Buist DSM, Henderson LM, Herschorn SD, Lee JM, Onega T, Rauscher GH, Wernli KJ, Lehman CD, Kerlikowske K. Population-based assessment of the association between magnetic resonance imaging background parenchymal enhancement and future primary breast cancer risk. J Clin Oncol. 2019;37:954–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dontchos BN, Rahbar H, Partridge SC, Korde LA, Lam DL, Scheel JR, Peacock S, Lehman CD. Are qualitative assessments of background parenchymal enhancement, amount of fibroglandular tissue on MR images, and mammographic density associated with breast cancer risk? Radiology. 2015;276:371–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. King V, Brooks JD, Bernstein JL, Reiner AS, Pike MC, Morris EA. Background parenchymal enhancement at breast MR imaging and breast cancer risk. Radiology. 2011;260:50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. van der Velden BH, Dmitriev I, Loo CE, Pijnappel RM, Gilhuijs KG. Association between parenchymal enhancement of the contralateral breast in dynamic contrast-enhanced MR imaging and outcome of patients with unilateral invasive breast cancer. Radiology. 2015;276:675–685. [DOI] [PubMed] [Google Scholar]
- 20. Choi JS, Ko ES, Ko EY, Han BK, Nam SJ. Background parenchymal enhancement on preoperative magnetic resonance imaging: association with recurrence-free survival in breast cancer patients treated with neoadjuvant chemotherapy. Medicine. 2016;95:e3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee J, Kim SH, Kang BJ. Pretreatment prediction of pathologic complete response to neoadjuvant chemotherapy in breast cancer: perfusion metrics of dynamic contrast enhanced MRI. Sci Rep. 2018;8:9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. You C, Peng W, Zhi W, He M, Liu G, Xie L, Jiang L, Hu X, Shen X, Gu Y. Association between background parenchymal enhancement and pathologic complete remission throughout the neoadjuvant chemotherapy in breast cancer patients. Transl Oncol. 2017;10:786–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ozturk M, Polat AV, Sullu Y, Tomak L, Polat AK. Background parenchymal enhancement and fibroglandular tissue proportion on breast MRI: correlation with hormone receptor expression and molecular subtypes of breast cancer. J Breast Health. 2017;13:27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vreemann S, Gubern-Merida A, Borelli C, Bult P, Karssemeijer N, Mann RM. The correlation of background parenchymal enhancement in the contralateral breast with patient and tumor characteristics of MRI-screen detected breast cancers. PLoS One. 2018;13:e0191399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. D'Orsi C, Bassett L, Feig S. Breast Imaging Reporting and Data System (BI-RADS). Breast Imaging Atlas. 4th ed Reston, VA: American College of Radiology; 2018 [Google Scholar]
- 26. Liao GJ, Henze Bancroft LC, Strigel RM, Chitalia RD, Kontos D, Moy L, Partridge SC, Rahbar H. Background parenchymal enhancement on breast MRI: a comprehensive review. J Magn Reson Imaging. 2020;51:43–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen JH, Yu HJ, Hsu C, Mehta RS, Carpenter PM, Su MY. Background parenchymal enhancement of the contralateral normal breast: association with tumor response in breast cancer patients receiving neoadjuvant chemotherapy. Transl Oncol. 2015;8:204–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Luo J, Johnston BS, Kitsch AE, Hippe DS, Korde LA, Javid S, Lee JM, Peacock S, Lehman CD, Partridge SC, Rahbar H. Ductal carcinoma in situ: quantitative preoperative breast MR imaging features associated with recurrence after treatment. Radiology. 2017;285:788–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Park JW, Liu MC, Yee D, Yau C, van ‘T Veer LJ, Symmans WF, Paoloni M, Perlmutter J, Hylton NM, Hogarth M, DeMichele A, Buxton MB, Chien AJ, Wallace AM, Boughey JC, Haddad TC, Chui SY, Kemmer KA, Kaplan HG, Isaacs C, Nanda R, Tripathy D, Albain KS, Edmiston KK, Elias AD, Northfelt DW, Pusztai L, Moulder SL, Lang JE, Viscusi RK, Euhus DM, Haley BB, Khan QJ, Wood WC, Melisko M, Schwab R, Helsten T, Lyandres J, Davis SE, Hirst GL, Sanil A, Esserman LJ, Berry DA; I-SPY 2 Investigators. Adaptive randomization of neratinib in early breast cancer. N Engl J Med. 2016;375:11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rugo HS, Olopade OI, DeMichele A, Yau C, van ‘T Veer LJ, Buxton MB, Hogarth M, Hylton NM, Paoloni M, Perlmutter J, Symmans WF, Yee D, Chien AJ, Wallace AM, Kaplan HG, Boughey JC, Haddad TC, Albain KS, Liu MC, Isaacs C, Khan QJ, Lang JE, Viscusi RK, Pusztai L, Moulder SL, Chui SY, Kemmer KA, Elias AD, Edmiston KK, Euhus DM, Haley BB, Nanda R, Northfelt DW, Tripathy D, Wood WC, Ewing C, Schwab R, Lyandres J, Davis SE, Hirst GL, Sanil A, Berry DA, Esserman LJ; I-SPY 2 Investigators. Adaptive randomization of veliparib-carboplatin treatment in breast cancer. N Engl J Med. 2016;375:23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC. N4ITK: improved N3 bias correction. IEEE Trans Med Imaging. 2010;29:1310–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Klifa C, Carballido-Gamio J, Wilmes L, Laprie A, Lobo C, Demicco E, Watkins M, Shepherd J, Gibbs J, Hylton N. Quantification of breast tissue index from MR data using fuzzy clustering. Conf Proc IEEE Eng Med Biol Soc. 2004;3:1667–1670. [DOI] [PubMed] [Google Scholar]
- 33. Altman DG, Bland JM. Measurement in medicine: the analysis of method comparison studies. J R Stat Soc Ser D. 1983;32:307–317. [Google Scholar]
- 34. Farahani K, Tata D, Nordstrom RJ. QIN benchmarks for clinical translation of quantitative imaging tools. Tomography. 2019;5:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dong JM, Wang HX, Zhong XF, Xu K, Bian J, Feng Y, Chen L, Zhang L, Wang X, Ma DJ, Wang B. Changes in background parenchymal enhancement in HER2-positive breast cancer before and after neoadjuvant chemotherapy: association with pathologic complete response. Medicine (Baltimore). 2018;97:e12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kuhl CK, Bieling HB, Gieseke J, Kreft BP, Sommer T, Lutterbey G, Schild HH. premenopausal breast parenchyma in dynamic contrast-enhanced MR imaging of the breast: normal contrast medium enhancement and cyclical-phase dependency. Radiology. 1997;203:137–144. [DOI] [PubMed] [Google Scholar]