Abstract
Background:
Inorganic arsenic (iAs) is an environmental toxicant associated with an increased risk of prostate cancer in chronically exposed populations worldwide. However, the biological mechanisms underlying iAs-induced prostate carcinogenesis remain unclear.
Objectives:
We studied how iAs affects normal human prostate stem-progenitor cells (PrSPCs) and drives transformation and interrogated the molecular mechanisms involved.
Methods:
PrSPCs were enriched by spheroid culture from normal human primary or immortalized prostate epithelial cells, and their differentiation capability was evaluated by organoid culture. Microarray analysis was conducted to identify iAs-dysregulated genes, and lentiviral infection was used for stable manipulation of identified genes. Soft agar colony growth assays were applied to examine iAs-induced transformation. For in vivo study, PrSPCs mixed with rat urogenital sinus mesenchyme were grafted under the renal capsule of nude mice to generate prostatelike tissues, and mice were exposed to () iAs in drinking water for 3 months.
Results:
Low-dose iAs () disturbed PrSPC homeostasis in vitro, leading to increased self-renewal and suppressed differentiation. Transcriptomic analysis indicated that iAs activated oncogenic pathways in PrSPCs, including the KEAP1-NRF2 pathway. Further, iAs-exposed proliferative progenitor cells exhibited NRF2 pathway activation that was sustained in their progeny cells. Knockdown of NRF2 inhibited spheroid formation by driving PrSPC differentiation, whereas its activation enhanced spheroid growth. Importantly, iAs-induced transformation was suppressed by NRF2 knockdown. Mechanistically, iAs suppressed Vacuolar ATPase subunit VMA5 expression, impairing lysosome acidification and inhibiting autophagic protein degradation including p62, which further activated NRF2. In vivo, chronic iAs exposure activated NRF2 in both epithelial and stroma cells of chimeric human prostate grafts and induced premalignant events.
Conclusions:
Low-dose iAs increased self-renewal and decreased differentiation of human PrSPCs by activating the p62-NRF2 axis, resulting in epithelial cell transformation. NRF2 is activated by iAs through specific autophagic flux blockade in progenitor cells, which may have potential therapeutic implications. https://doi.org/10.1289/EHP6471
Introduction
Inorganic arsenic (iAs) is a ubiquitously distributed environmental and industrial toxicant, classified as a class I carcinogen by the International Agency for Research on Cancer (IARC 2004). More than 137 million people in more than 70 countries are exposed to iAs at levels of greater than () (Ravenscroft 2007), which is the drinking water standard mandated by the U.S. Environmental Protection Agency (U.S. EPA) (Carlin et al. 2016; Polya and Charlet 2009). Of particular concern, growing epidemiological evidence reveals adverse health effects at lower iAs levels (iAs level of well water ) than previously realized (Carlin et al. 2016; Schmidt 2014). Epidemiological studies and experimental evidence link chronic iAs exposure with increased risk of certain cancers (Tokar et al. 2011) and multiple noncancerous adverse health conditions (Carlin et al. 2016; Polya and Charlet 2009; Tokar et al. 2011). Although not conclusive, emerging epidemiological data suggest a direct causal relationship between iAs exposure and prostate cancer incidence and mortality (Benbrahim-Tallaa and Waalkes 2008; Chen and Wang 1990; Tokar et al. 2011); however, the precise mechanisms underlying this iAs-induced tumorigenesis remain unclear.
Properties of adult stem-progenitor cells make them compelling targets of tumorigenesis (Blanpain 2013; White and Lowry 2015). Indeed, transcriptomic analysis has shown that basal stem cell signatures are enriched in metastatic prostate cancer (Smith et al. 2015). Environmental factors play an important role in cancer etiology (Tokar et al. 2011; White and Lowry 2015), and prostate stem-progenitor cells (PrSPCs) are hormone (e.g., estrogen and bisphenol A) targets associated with prostate cancer susceptibility (Prins et al. 2015). Epidemiological studies indicate prenatal arsenic exposure is linked with increased mortality risks related to cancer and noncancer diseases (Bailey and Fry 2014), suggesting that arsenic may target the long-lived stem cell population. Further, experimental findings revealed that iAs can directly alter neural, skin, and prostate stem cells, shifting their differentiation capability and augmenting transformation (Carlin et al. 2016; Tokar et al. 2011). These observations are highly significant because recent evidence suggests that cancer may originate from transformation of resident tissue stem cells that self-renew and differentiate into abnormal progeny that continuously seed tumor growth (Blanpain 2013; White and Lowry 2015). Previous studies by Waalkes et al. interrogated iAs-driven prostate carcinogenesis using an immortalized benign human prostate epithelial cell line (RWPE-1 cells) and its stem-progenitor cell-line derivative, WPE cells, finding that extended in vitro exposure to iAs transformed the cells which, upon transplantation in mice, formed malignant, poorly differentiated tumors (Tokar et al. 2010b). Although these studies yield significant mechanistic insights, the HPV-18 immortalized RWPE-1 cells are primed for transformation and can form low-grade tumors in vivo without secondary hits (Zhang et al. 2010). As such, it remains to be determined whether a range of iAs doses can transform the normal human prostate stem and progenitor cell populations.
Autophagy is a conserved, tightly regulated process with an essential role in protein and organelle quality control through capture, degradation, and recycling of intracellular proteins and organelles (Mizushima and Komatsu 2011). It is noteworthy that stem cells, including PrSPCs (Hu et al. 2017), have elevated autophagic activity relative to daughter progenitor cells and differentiated cells, and this heightened autophagic capability was critical for long-term survival (Guan et al. 2013; Maycotte et al. 2015). Although autophagy suppresses initiation of tumors, it can also facilitate progression of established tumors (Amaravadi et al. 2016; Kenific and Debnath 2015; White 2015). Of significance, arsenic exposures have been shown to affect autophagy in a cell- and dose-dependent manner (Qi et al. 2014). Although the mechanistic underpinnings remain elusive, many consider autophagy to be a cellular protective mechanism against arsenic-related tumorigenesis (Lau et al. 2013; Qi et al. 2014). Recently, arsenic was shown to block autophagic flux in a variety of mouse and human cell lines, leading to persistent Kelch-like ECH-associated protein 1 (KEAP1)-NRF2 pathway activation in vitro (Lau et al. 2013; Qi et al. 2014). However, the same has not been determined for stem-progenitor cells in any system or with prostate carcinogenesis. It is interesting that, similar to autophagy, NRF2 is a tumor suppressor in normal cells but also plays an oncogenic role in aggressive tumor cells (Jaramillo and Zhang 2013; Son et al. 2015).
The present study sought to address previously unresolved issues regarding iAs-induced prostate carcinogenesis by using primary prostate epithelial cells (PrECs) from young organ donors to first determine whether iAs exposure at environmentally relevant levels disturbs normal stem-progenitor cell homeostasis and whether this may be capable of transformation of this long-lived population. Current U.S. EPA guidelines indicate a maximum allowable level for arsenic in drinking water of (; ) (https://www.epa.gov/dwreginfo/chemical-contaminant-rules) (Carlin et al. 2016; Polya and Charlet 2009), although levels may reach (; ) in contaminated groundwater (Ravenscroft et al. 2009). As such, we first assessed responses to iAs to embody a full range of chronic arsenic exposure levels. Next, we sought to determine the molecular underpinnings of observed alterations in the PrSPCs by combining transcriptomic analysis with functional assays including an in vivo chimeric prostate model. The data led to the interrogation of the KEAP1-NRF2 pathway and the autophagy status within the PrSPC population as a function of iAs exposure.
Materials and Methods
Reagents and Antibodies
Sodium arsenate solution (VWR, 3; Cat. No. 5000-1L) was used as the source of iAs for all the studies. The following reagents were used: LysoHunt Red DND-99 (Setareh Biotech; Cat. No. 7522), Oltipraz (Sigma, Cat. No. O9389), N-acetyl-L-cysteine (Sigma, Cat. No. A9165), Sea Plague low-melt agarose (Lonza; Cat. No. 50101), BafA1 (Sigma, Cat. No. B1793), and chloroquine (Sigma, Cat. No. C6628). The information of the antibodies is included in Table S2, including source, identifier, and dilution for immunoblotting (IB) and immunofluorescence (IF).
Cell Culture and Treatment
Primary human PrECs were obtained from 4 young (19–21 y old) disease-free donors (Lifeline® Cell Technology) and cultured in Prostalife™ Epithelial Medium (Lifeline® Cell Technology; Cat. No. LL-0041), as previously described (Hu et al. 2011). Cells from passages 3–8 were used. RWPE1 cells were obtained from ATCC and cultured in Keratinocyte Serum-Free Medium (KSFM; ThermoFisher Scientific; Cat. No. 17005042). Cells from passages 5–20 were used. Embedded prostasphere (PS) culture was conducted as previously described with Prostalife™ medium (Hu et al. 2011, 2019). Briefly, for each well of a 12-well plate, 50,000–100,000 cells were suspended in ice-cold 1:1 mixture of culture medium and Matrigel® (Corning®; Cat. No. 356231) on ice. They were then seeded around the bottom rim of the well using pipette transfer, and the mixture was solidified by incubating at 37°C for 30 mins. Warm medium ( at 37°C) was added over the Matrigel® slurry and changed every 2–3 d over a 5- to14-d culture period. For ultralow-attachment PS cultures, 1,000–2,000 dissociated cells were resuspended in Prostalife™ medium with 5% Matrigel®, seeded to wells of 96-well plate (Corning®; Cat. No. 3474), and cultured for 1 wk without change of medium.
Prostate organoid (PO) culture derived from primary human PrECs is similar to embedded PS culture but uses a different medium to drive epithelial differentiation. PO medium used was a modification of previously described (Chua et al. 2014) human PO medium, a medium using FBS and testosterone to drive differentiation in vitro. The final PO medium was KSFM with 5% charcoal-stripped FBS (ThermoFisher Scientific; Cat. No. 12676), Y-27632 dihydrochloride (Abmole Bioscience; Cat. No. M1817), and DHT (Sigma; Cat. No. A-8380).
To retrieve PS and PO from embedded cultures, the Matrigel® mixture was digested with Dispase® (Stemcell Technologies; Cat. No. 07913). Briefly, Dispase® was added to the Matrigel® mixture containing PS or PO (), mixed thoroughly by pipetting up and down several times, incubated at 37°C for 30 mins, and centrifuged at for 5 min to collect a PS or PO pellet. For iAs exposure, commercially available sodium arsenate solution (VWR, ; ) was diluted with water of cell culture grade to make 1000× stock solution, which was stored at 4°C before adding to the cell culture medium directly at the stated final concentration.
Bromodeoxyuridine (BrdU)-Label Retention Assay
Parental PrEC were 2D cultured with BrdU (Sigma-Aldrich) for 10 d for labeling of dividing cells as previously described (Hu et al. 2017, 2019). Cells were transferred to 3D Matrigel® culture for 7 d to permit BrdU wash-out during spheroid growth (). PS were harvested by Dispase® digestion and attached to chamber slides during overnight culture in Prostalife™ culture medium. Spheres were fixed in ice-cold methanol at for 20 min; BrdU epitopes were exposed by incubation with HCl at room temperature (RT) for 30 min and then washed 3 times with PBS before immunostaining using mouse anti-BrdU antibody.
Immunoblotting
Cell lysates were prepared in cell lysis buffer (Cell Signaling Technology; Cat. No. 9803). Protein loading was standardized with a BCA assay (ThermoFisher Scientific; Cat. No. 23225). Total protein () from each sample was resolved on 10% or 15% SDS-PAGE gels and transferred to PVDF membranes (Bio-Rad; Cat. No. 162-0177). Membranes were blocked with 5% nonfat dry milk in TBST buffer ( Tris base, NaCl, EDTA, 0.1% Tween-20, pH 7.5) for 30 min at RT and then incubated with primary antibodies diluted in 5% nonfat dry milk in TBST at 4°C overnight. After washing with TBST ( min, RT), membranes were incubated with appropriate horseradish peroxidase-conjugated secondary antibodies (Cell Signaling Technology; Cat. Nos. 7074 and 7076) and developed using ECL or ECL-Plus on Biomax MR film (Kodak). Integrated band densities were quantified using ImageJ software (https://imagej.nih.gov/ij/).
Immunostaining
Immunostaining was conducted as previously described (Hu et al. 2011, 2017, 2019; Rangel-Huerta and Maldonado 2017). Briefly, PS or PO were attached overnight on chamber slides (Millipore Sigma; Cat. No. PE2GS0416) to permit limited cell outgrowth and improve reagent penetration. Cell culture samples on slides were washed with PBS, then fixed with 5% paraformaldehyde (PFA) at RT for 20 min, permeabilized by incubating with 0.5% Triton X-100 at RT for 5 min. Sample slides were blocked with 5% normal goat serum in PBS with 0.25% Triton X-100 for 30 min at RT, then incubated with primary antibody diluted with incubating buffer (PBS with 2% normal goat serum and 0.125% Triton X-100) overnight at 4°C. After washing (PBS, at RT), slides were incubated in the dark with secondary antibody diluted with incubating buffer at RT for 1 h. After washing, sample slides were mounted for imaging. For animal tissues, samples were fixed in acetone and methanol (1:1) overnight at 4°C, and then incubated in 70% ethanol overnight at 4°C, followed by a regular immunohistochemistry procedure of processing, embedding, and section. For antigen retrieval, tissue sections of were heat-treated for antigen retrieval in a Decloaker pressure cooker (Biocare Medical) in Tris-EDTA, pH 9.0 buffer for 3 min. Then, tissue slides were immunostained as described above. The dilution factor of antibodies is included in Table S2. Stained slides were imaged with a Zeiss Axioskop 20 fluorescent microscope and Axio camera.
Hoechst Exclusion Assay
The Hoechst exclusion assay was performed as previously described (Hu et al. 2012; Prins et al. 2015) to assess the stem-like cells in 2D-cultured cells. PrEC cells were preincubated for 10 min with or without verapamil hydrochloride (Sigma-Aldrich; Cat. No. V4620-1G), which inhibits ABCG2 transporter protein expressed at high levels in stem cells, blocking their Hoechst exclusion ability. Cells were next incubated in Hoechst 33342 (Sigma-Aldrich; Cat. No. B2261) in Hank’s balance salt solution, 10% FBS, 1% D-glucose, and HEPES for 30 min at 37°C, washed in PBS and incubated with Propidium iodide (PI) (Sigma-Aldrich; Cat. No. P4170-10MG) for dead cell exclusion. Hoechst-stained cells were separated by single-channel FACS (CyAnTM ADP Analyzer). All results were confirmed by side-population double-channel FACS analysis (Beckman Coulter; MoFloTM XDP analyzer) using Hoechst dye. The percent prostate stem-like cells were calculated as the difference in Hoechst excluding cells incubated (with or without) verapamil.
Transcriptome Analysis
Total RNA was extracted from PS and PO cultured iAs for 2 wk ( for each group; total 12 samples) with the same RNA isolation method described below for real-time PCR analysis. Microarray analysis with the Illumina HumanHT12 V4 Gene Expression Microarray were performed according to the standard protocol at the University of Chicago Genomics Facility. Raw data are available at Gene Expression Omnibus (GSE137357). Differentially expressed genes (DEG) with iAs exposures were identified by Limma R package (Limma v3.28.14; R v3.2.4) with a cutoff of fold change and -value (Ritchie et al. 2015), which is provided as Table S3. Enriched pathways for DEG were identified by Wikipathway Analysis on WEB-based GEne SeT AnaLysis Toolkit (WebGestalt 2017; http://www.webgestalt.org/2017/option.php) (J Wang et al. 2013) and GSEA software (v3.0) (Subramanian et al. 2005).
Quantitative Real-Time PCR Analysis
For total RNA isolation, PS/PO samples from wells of 12-well plates were collected from Matrigel® cultures using Dispase® as described above. PS and PO pellets were dissolved in Trizol reagent before processing by Direct-Zol RNA miniprep kit according to the manufacturer’s instructions (Zymo Research; Cat. No. 11-331). cDNA synthesis was conducted by iScript cDNA synthesis kit (Bio-Rad, Cat. No. 1708891). Quantitative real-time PCR was conducted using SYBR green master mix reagent (Bio-Rad; Cat. No. 1725271) with CFX96 Real-Time system (Bio-Rad). Primer sequences are provided in Table S1. Data were analyzed with method and normalized to the housekeeping gene RPL13.
Plasmids and Lentiviral Delivery
For knockdown, shRNA for the gene of interest was selected from Genetic Perturbation Platform (GPP) at Broad Institute (https://portals.broadinstitute.org/gpp/public/), where sequences are available through searching clone ID (TRCN #). shRNA targeting p62 (TRCN0000007235), NRF2 (TRCN0000007558), ATG7 (TRCN0000007587), VMA5 (TRCN0000101541), and luciferase (TRCN0000072259) were cloned to pLKO.1-TRC (a gift from David Root, Addgene plasmid 10878) as described (Moffat et al. 2006) or following cloning protocol on the GPP website (https://portals.broadinstitute.org/gpp/public/resources/protocols).
For overexpression, the protein coding sequences (CDS) of NRF2 and VMA5 were cloned from cDNA of human primary prostate epithelial cells by high-fidelity PCR using Phusion enzyme (New England Biolabs; Cat. No. M0530S). This was subcloned to a Tet-On overexpression lentiviral vector pLIX_402 (a gift from David Root, Addgene plasmid 41393) with a C-terminal HA tag. mCherry-EGFP-LC3B CDS were subcloned from pBabe-puro mCherry-EGFP-LC3B (a gift from Jayanta Debnath, Addgene plasmid 22418) to pLVX-puro (Clontech; Cat. No. 632164). Cloning was conducted with a Gibson Assembly Cloning Kit (New England Biolabs; Cat. No. E5510S). Lentiviral packaging and infection were conducted as described (Moffat et al. 2006). Briefly, lentivirus was produced with a second packaging system with pMD2.G and psPAX2 plasmids (a gift from Didier Trono; Addgene plasmid 12259 and 12260) using HEK-293FT cells (ThermoFisher Scientific; Cat. No. R70007). Lentivirus-containing medium was collected 48 h post-transfection and stored at . For infection using 6-well plates, prewarmed lentivirus-containing medium was added to cells at 70%–80% confluence in the presence of polybrene, incubated at 37°C overnight and changed to fresh culture medium. Infected cells were selected with puromycin () 24 h after infection for 1 wk, and then used for experiments.
Intracellular Reactive Oxygen Species (ROS) Assay
To assess intracellular ROS, day-7 PS were collected from embedded culture by Dispase® digestion, dispersed to single cells by trypsin digestion for 5 min at 37°C and attached to chamber slides by overnight culture in Prostalife™ culture medium. Cells were incubated with 2′,7′-dichlorofluorescin diacetate (Setareh Biotech; Cat. No. 7543) for 10 min at 37°C before washing, fixing, and assessing with a fluorescent microscope.
Soft-Agar Colony Formation Assay
RWPE1 cells (10,000 cells/well) were mixed with 0.4% agarose in growth medium and plated on top of a solidified layer of 0.8% agarose in 6-well plates. After 3 wk, colonies were dyed with crystal violet ( PBS containing 4% formaldehyde and 0.005% crystal violet per well), imaged with a dissecting microscope or EVOS FL Auto2 Cell Imaging System (Invitrogen), and quantified with ImageJ (https://imagej.nih.gov/ij/).
NRF2 Luciferase Reporter Assay
To long-term monitor real-time NRF2 pathway activation in spheres, we created a lentiviral reporter with Gaussia luciferase (GLuc) expression under an artificial promoter containing NRF2 binding motifs. The lentiviral reporter was based on FUGW plasmid backbone (Lois et al. 2002), which was a gift from David Baltimore (Addgene plasmid 14883). First, we inserted a DNA fragment at BamHI restriction enzyme site, a fragment containing hPGK promoter, puromycin resistant gene, and T2A peptide from pLIX402 plasmid (a gift from David Root, Addgene plasmid 41393), which enabled a puromycin screening for lentivirus infected cells. Next, we replaced original hUBC promoter with a DNA fragment of NRF2 reporter using PcaI and BamH1 restriction enzyme sites. The NRF2 reporter fragment was synthesized by Integrated DNA Technologies (IDT) (gBlock gene fragments), which consisted of 3 NRF2 binding motifs (TGACTCAGCACTGACTCAGCAGTGACTCAGCA) followed by a mini-TK promoter and the coding sequence of a modified GLuc enzyme. The humanized coding sequence of wild-type Gluce was acquired from pGLuc-Basic 2 plasmid (New England Biolabs; Cat. No. N0802). The modified GLuc contains M43I and M110I mutations to yield an enzyme with glow-like properties (Welsh et al. 2009) and thus more suitable for future high-throughput screening. In addition, we attached a synthetic fragment to GLuc that encoded the proteolytic “PEST” signal from mouse ornithine decarboxylase (Ghoda et al. 1990) to reduce the half-life of the GLuc protein dramatically, thus the NRF2 activation was monitored in real time. The NRF2 activation was quantified by measuring the activity of secreted GLuc. Cell culture medium containing GLuc was collected at appropriate time points, and was mixed with assay buffer containing GLuc substrate ( coelenterazine in PBS). Within 30 min, the bioluminescent signal (maximal emission at ) produced by GLuc from the oxidation of coelenterazine was detected using a Glomax Multi Plus detection system (Promega; Cat. No. E7081) with Luminescence Module (Promega; Cat. No. E8041).
LysoHunt Assay
All treatments were performed at 37°C. Live cells were incubated with culture medium containing LysoHunt for 60 min and imaged () by fluorescent microscopy (Zeiss Axioskop). Images were quantified with ImageJ (https://imagej.nih.gov/ij/).
Recombinants of Chimeric Prostatelike Structures
In vivo tissue recombinant experiments used PrSPCs mixed with rat embryonic urogenital sinus mesenchyme (UGM) and grafted under the renal capsule of 6-wk-old male nude mice as described previously (Hu et al. 2011, 2017). All animals were handled according to Principles for Care and Use of Animal Research and Studies approved by the Institutional Animal Care Committee. Timed pregnant female Sprague-Dawley rats were purchased from Harlan, as well as male nude mice. Animals were housed in polysulfone solid-bottom cages at 21°C with 14-h light/10-h dark cycle with water supplied from glass bottles and standard Purina® rodent chow ad libitum.
Rat embryonic day-17 fetuses were collected by Caesarian section under ketamine/xylazine (5:2, University of Illinois at Chicago pharmacy) anesthesia; the urogenital sinus (UGS) was removed and digested; and the UGM was separated from the urogenital epithelium (UGE). After microscopically confirming absence of UGE, the UGM was used for tissue recombination. For each graft, PrSPCs from dispersed PS were mixed with UGM from 1 embryo and resuspended in Matrigel®. Recombinants were incubated overnight on 1% agar in DMEM in 5% CO2 at 37°C prior to grafting under the renal capsule of 7- to 8-wk-old male nude mice as previously described (Huang et al. 2009) with each kidney receiving 1 graft. Grafts with UGM or UGE alone were used for quality controls. Recipient mice were supplemented with testosterone-containing containing SILASTIC (Dow Corning Corp.) capsules () that produced serum testosterone levels of at 1 month.
Due to the differing iAs pharmacokinetics in rodents and humans, higher iAs must be provided orally to mice to reach similar blood levels in humans exposed to lower doses (Tokar et al. 2010a; Waalkes et al. 2007). The doses aim to mimic typical human exposure levels in geographic hotspots and in occupational settings. For iAs exposure, mice bearing renal grafts were exposed to iAs in the drinking water, which mimics iAs in geographical hotspots (Tokar et al. 2010a), and grafts were harvested after 3 months for histological analysis. A total of 9 mice (18 grafts) were treated with () iAs in the drinking water, and 5 control mice (10 grafts) had no exposure in the water. Animals were anesthetized with isoflurane prior to tissue collections, and animals were euthanized by cervical dislocation.
Statistical Analysis
Data were analyzed using GraphPad Prism version 6.0 (GraphPad Software Inc.) and presented as . Statistically significant differences were assessed by Student’s t-test or, with multiple groups, analysis of variance (ANOVA), followed by Tukey-Kramer or Student-Newman-Keuls post hoc tests. was considered significant.
Results
Self-Renewal and Differentiation of PrSPCs Treated with or without 0.5 to Arsenic
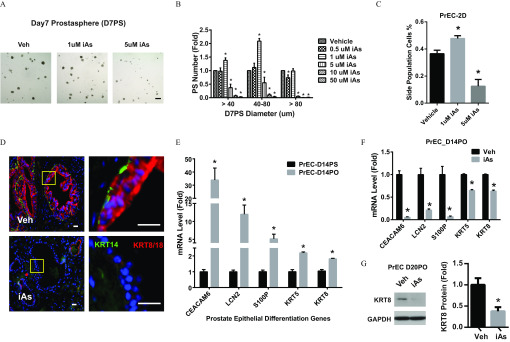
To determine the effects of iAs on primary PrSPCs grown from disease-free human primary PrEC cultures, we studied two unique properties of stem-progenitor cells: self-renewal and differentiation. The sphere formation assay enriches stem-progenitor cells based on their unique capability to survive and expand in 3D serum-free culture, whereas differentiated primary cells undergo apoptosis or senescence. The number and size of prostate spheres (PS) represents stem cell self-renewal and progenitor cell proliferation capacity, respectively (Hu et al. 2011, 2012; Prins et al. 2014). Low-dose iAs () increased, whereas higher-dose iAs () decreased total PS numbers (Figure 1A; PS ). Size analysis revealed that iAs markedly increased the formation of medium-sized () PS, whereas iAs at higher doses suppressed formation of medium and large () PS (Figure 1B). A Hoechst exclusion assay was next used to quantitate the stem-like cell population in 2D primary PrEC cultures upon iAs exposure. Consistent with PS formation assay results, iAs increased and iAs decreased the percentage of stem-like cells (Figure 1C). Of note, iAs significantly suppressed proliferation of 2D-cultured PrECs at all iAs doses including , suggesting that differentiated primary cells are highly sensitive to iAs exposure (Figure S1A). Further, the higher-dose iAs () induced early apoptosis and G1 cell cycle arrest, whereas lower-dose iAs () had no significant effect on cell cycle progression (Figure S1B, S1C). Thus, was used for further experiments, because this dose was nontoxic to PrECs and mimics environmentally relevant iAs exposures.
Figure 1.
Effects of arsenic on self-renewal and differentiation of prostate stem-progenitor cells. (A,B) Prostate spheres (PS) derived from primary human prostate epithelial cells (PrEC) were cultured with inorganic arsenic (iAs) at indicated doses for 7 d (PrEC-D7PS) (A) and quantified according to size (B). . Data shown are (); * vs. vehicle. (C) Percentage of stem-like cell side population in 2-dimensional cultured PrEC (PrEC-2D) using a Hoechst exclusion side-population assay. Data shown are (); * vs. vehicle. (D) Immunostaining of KRT14 and KRT8/18 in recombinant human rat prostate gland structures in nude mice renal grafts exposed to vehicle or iAs in drinking water for 3 months. Red: KRT14, basal epithelial cells. Green: KRT8/18, luminal epithelial cells. (left: low power; right: high power). (E) mRNA level of prostate epithelial differentiation genes in 2-wk cultured prostate organoid (D14PO) and prostate spheres (D14PS) derived from PrEC. Data shown are (); * vs. D14PS. (F) mRNA levels of prostate epithelial differentiation genes in D14PO treated arsenic. Data shown are (); * vs. vehicle. (G) KRT18 protein level of D20PO treated arsenic. Data shown are (); * vs. vehicle.
Lineage commitment and cell differentiation capability are essential features of stem-progenitor cells that permit tissue homeostasis. In the human prostate, PrSPCs give rise to basal cells, luminal PrECs, and rare neuroendocrine cells (Strand and Goldstein 2015). Because iAs alters differentiation in a variety of stem-progenitor cells (Bain et al. 2016; McCoy et al. 2015; Rebuzzini et al. 2015; Yen et al. 2010), we tested whether iAs influences this process in PrSPCs. Primary PrSPCs cultured for 7 d from disease-free PrECs were isolated and mixed with embryonic rat urogenital sinus mesenchyme (UGM) and grafted under renal capsules of nude mice. The hosts were exposed to 0 or () iAs in drinking water for 3 months. The PrSPCs formed normal prostate gland structures with basal () and luminal () epithelial cells in control hosts (Figure 1D). In contrast, expression of KRT14 and KRT8/18 in the graft epithelial cells was markedly lower in iAs-treated mice vs. controls, indicating suppressed prostate epithelial differentiation. As an alternative approach, 3D prostate organoid (PO) culture, which drives epithelial differentiation, was used to assess iAs effects on this process. Compared with PS cultures, PO cultured from primary PrECs expressed prostate epithelial differentiation genes (CEACAM6, LCN2, and S100P) (Rane et al. 2014) as well as marker genes for basal (KRT5) and luminal (KRT8) cells (Figure 1E). Consistent with in vivo data, iAs significantly inhibited expression of these differentiation marker genes (Figure 1F). KRT8 suppression was further confirmed at the protein level (Figure 1G).
Microarray Analysis and Subsequent Evaluation of the NRF2 Pathway in PrSPCs Treated with or without Arsenic
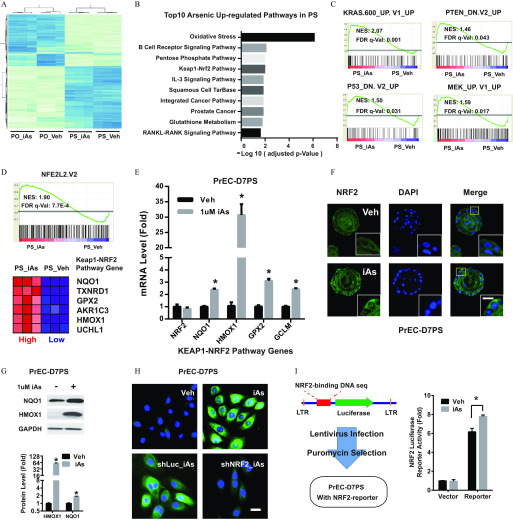
To investigate the underlying mechanisms of PrSPC perturbation, we analyzed the transcriptomes of iAs-treated PS and PO using microarrays. Although nonsupervised clustering revealed that the majority of differences were between PO and PS (Figure 2A), iAs-regulated genes were consistently observed in both PS and PO groups. Because the focus of this study was stem and progenitor cells, we concentrated on iAs-regulated genes in the PS groups. At the mechanistic level, revealed by Wikipathway analysis (J Wang et al. 2013), oxidative stress and KEAP1-NRF2, among others, were top enriched pathways in iAs-treated PS (Figure 2B) as was prostate cancer, supporting PrSPCs as compelling oncogenic targets of iAs. Using gene sets of oncogenic signatures, gene set enrichment analysis (GSEA) revealed that iAs up-regulated well-known oncogenic molecular pathways in PrSPCs, including KRAS, PTEN-deficiency, P53-deficiency, and ERK pathways (Figure 2C).
Figure 2.
Effects of arsenic on the NRF2 pathway in prostate stem-progenitor cells. (A) Heatmap showing the top 1,000 differentially expressed genes across four groups: prostate spheres arsenic (PS_Veh, PS_iAs), prostate organoids arsenic (PO-Veh, PO-iAs). Prostate spheres and organoids were derived from primary human prostate epithelial cells (PrEC), and cultured arsenic for 2 wk. (B) Top 10 iAs up-regulated pathways in prostate spheres, iAs up-regulated genes (, adjusted ) were sorted for pathway analysis with Wikipathway. (C) Enriched oncogenic pathways in PS_iAs group vs. PS_Veh group. Raw microarray data were subject to GSEA analysis using gene sets of oncogenic signatures. (D) GSEA data showing NRF2 pathway enrichment between PS-iAs and PS-Veh groups (top) and most regulated genes of NRF2 pathway (bottom). (E) mRNA level of NRF2 pathway genes in PS_Veh and PS_iAs group. Data shown are (); * vs. vehicle. (F) Representative NRF2 immunostaining showing nuclear translocation in PS upon iAs treatment for 7 d (PrEC-D7PS). . (G) Immunoblot of HMOX1 in PrEC-D7PS iAs treatment. Data shown are (); * vs. vehicle. (H) Representative HMOX1 immunostaining in day-7 spheres derived from primary human prostate epithelial cells (PrEC-D7PS) with indicated treatment, . iAs, arsenic; shLuc, negative control shRNA targeting luciferase gene; shNRF2, shRNA targeting NRF2 gene; Veh, vehicle control. . (I) NRF2-luciferase reporter assay showing luciferase activity in PrEC-D7PS iAs. Data shown are (); * vs. vehicle.
As a master antioxidative transcription factor, NRF2 is also involved in tumorigenesis and stem cell function regulation, but its role in PrSPCs remains unclear. To examine this further, microarray results were validated with qPCR, revealing that iAs significantly increased expression of NRF2 marker genes NQO1, HMOX1, GPX2, and GLCM (Figure 2D,E). Although NRF2 mRNA was not affected by iAs, cytoplasmic-to-nuclear translocation of NRF2 indicated that iAs activated NRF2 at the post-translational level (Figure 2F). Induction of HMOX1 (the top up-regulated NRF2 pathway gene) and NQO1 was further confirmed by immunoblotting (Figure 2G). NRF2 knockdown by shRNA (shNRF2) (Figure S2) attenuated the elevated HMOX1 immunofluorescence induced by iAs (Figure 2H), indicating that HMOX1 was downstream of NRF2 activation. A luciferase reporter with an artificial NRF2 binding motif that was introduced into PS by lentivirus revealed that iAs exposure significantly increased luciferase activity, confirming direct NRF2 activation by iAs (Figure 2I).
Effects of NRF2 Loss and Gain of Function on PrSPC Cells Treated with Arsenic
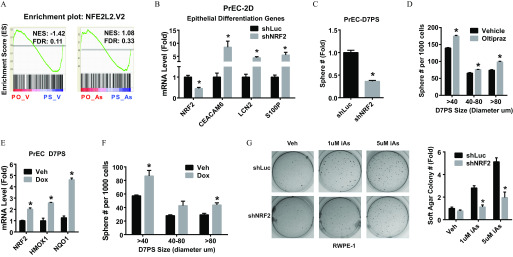
Because studies of hematopoietic stem cells demonstrate that their quiescence and capacity for bone marrow reconstitution are compromised by NRF2 activation (Tsai et al. 2013), we tested whether NRF2 may similarly regulate PrSPC homeostasis. GSEA revealed enrichment of the NRF2 pathway genes in PS relative to differentiated PO culture, whereas iAs mitigated this difference (Figure 3A). PrSPC responses to NRF2 loss- and gain-of-function were next examined. shNRF2 markedly induced expression of differentiation genes in 2D-cultured PrECs (Figure 3B) and reduced PS formation upon transfer to 3D culture (Figure 3C), suggesting a critical role for NRF2 in maintaining stemness. In contrast, the NRF2 inducer Oltipraz significantly increased PS formation and size (Figure 3D). Further, overexpression of NRF2 in PS using a Tet-On expression system showed that forced NRF2 expression similarly significantly increased NRF2 target gene expression (Figure 3E) as well as PS number and size (Figure 3F), suggesting that NRF2 enhances self-renewal of PrSPCs.
Figure 3.
The role of NRF2 plays in iAs-induced transformation. (A) GSEA data of NRF2 pathway genes comparing prostate organoids (PO) and spheres (PS) derived from primary human prostate epithelial cells (PrEC) and treated with vehicle (PO-V vs. PS-V) or arsenic (PO-As vs. PS-As). (B) Expression of prostate epithelial differentiation genes in 2-dimensional culture PrEC (PrEC-2D) with NRF2 knockdown (shNRF2). Data shown are (); * vs. shRNA control (shLuc). (C) Sphere formation capability of PrEC with NRF2-knockdown, D7PS: day-7 spheres. Data shown are (); * vs. vehicle. (D) Quantification of sphere formation capability of PrEC treated with NRF2-inducer Oltipraz () for 7 d. D7PS: day-7 prostate spheres. Data shown are (); * vs. vehicle. (E) mRNA level of NRF2 gene and NRF2 pathway marker genes. Tet-On NRF2 expression was induced in PrEC spheres by lentivirus. Spheres were (Dox, ) for 7 d (Tet-ON-NRF2 PrEC-D7PS) to induce NRF2 overexpression. Data shown are (); * vs. vehicle. (F) Quantification of sphere formation capability of PrEC with Tet-On overexpression of NRF2. Data shown are (); * vs. vehicle. (G) Soft-agar colony formation assay of RWPE1 with indicated treatment. Note: iAs, inorganic arsenic; shLuc, RWPE1 with shRNA targeting luciferase gene, negative control; shNRF2, RWPE1 with NRF2 knockdown. Data shown are (); * vs. shRNA control (shLuc).
NRF2 plays both pro-oncogenic and antioncogenic roles in tumor progression (Jaramillo and Zhang 2013; Son et al. 2015). To determine whether it plays a role in iAs-induced transformation, the effects of NRF2 knockdown on iAs-induced tumorigenesis were investigated using a soft-agar colony formation assay. Because this assay was not possible using primary cells, we used RWPE1 cells that are known to be transformed by extended iAs exposure (Benbrahim-Tallaa and Waalkes 2008; Tokar et al. 2010c). As expected, 3-wk exposure to or iAs significantly increased soft-agar colony formation in a dose-dependent manner in comparison with vehicle controls (Figure 3G). Of significance, shNRF2 abrogated this iAs-induced colony formation activity.
Evaluation of ROS and p62 Regulation of NRF2 Activity in Arsenic Exposed PrSPCs
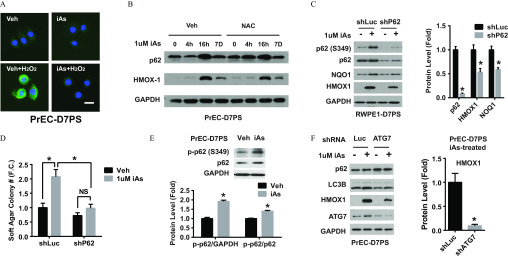
NRF2 activity is primarily regulated in a negative fashion by KEAP1, which targets NRF2 for proteasome degradation. During canonical NRF2 activation, electrophilic or oxidative stress modifies cysteine residues of KEAP1, which disrupts KEAP1-mediated degradation and stabilizes NRF2 (Jaramillo and Zhang 2013). Although high-dose iAs has been shown to induce production of reactive oxygen species (ROS) (Carlin et al. 2016), we did not observe increased ROS production in PrSPCs after treatment with low-dose iAs (Fig. 4A). In fact, low-dose iAs attenuated hydroperoxide-induced ROS production in these cells. Further, iAs-induced HMOX1 expression was not suppressed by the ROS scavenger N-acetyl cysteine (Figure 4B), suggesting that low-dose iAs-induced NRF2 activation is not mediated through ROS in PrSPCs.
Figure 4.
Mechanism of arsenic activation of the NRF2 pathway and autophagy dependence. (A) DCFDA assay showing intracellular levels of reactive oxygen species (ROS) in day-7 prostate spheres derived from primary human prostate epithelial cells (PrEC-D7PS) upon indicated treatment. iAs, treated with inorganic arsenic (iAs) for 7 d; , treated with hydroperoxide for 30 min; Veh, vehicle. . (B) Immunoblotting of p62 and HMOX1 protein from day-7 spheres of primary human prostate epithelial cells (PrEC-D7PS) with indicated treatment: iAs treated for 0, 4, 16 h (16h) and 7 d (7D) without (Veh) or with N-acetyl cysteine (NAC) 30 min prior to iAs exposure. (C) Immunoblot of p-p62 (S349), p62, NQO1 and HMOX1 protein from day-7 spheres of RWPE1 cells (RWPE1-D7PS) with indicated treatment. iAs, inorganic arsenic; shLuc, negative control shRNA targeting luciferase gene; shP62, shRNA targeting p62 gene. Quantification data shown are (); * vs. shRNA control (shLuc). (D) Soft-agar colony formation capability of RWPE1 with indicated treatment: iAs; shLuc, negative shRNA control targeting luciferase gene; shP62, shRNA targeting p62 gene. Quantification data are (); * vs. shRNA control (shLuc). (E) Immunoblot of phosphorylation at p62-serine349 from day-7 spheres of primary human prostate epithelial cells (PrEC-D7PS) treated with iAs for 7 d. Quantification data are (); * vs. vehicle control. (F) Immunoblot of indicated proteins from day-7 spheres derived from primary human prostate epithelial cells (PrEC-D7PS) treated with iAs for 7 d with indicated treatment. shATG7, shRNA targeting ATG7 gene; shLuc, negative control shRNA targeting luciferase gene. Quantification data are (); * vs. shRNA control (shLuc).
Recently, noncanonical p62-dependent NRF2 activation was identified in which p62 competitively binds KEAP1, thus preventing NRF2 degradation (Jaramillo and Zhang 2013; Lau et al. 2013). Given that iAs induces noncanonical NRF2 activation in a variety of 2D-cultured cells (Dodson et al. 2018), we examined whether iAs activated NRF2 in PrSPCs through the p62-KEAP1-NRF2 pathway. Indeed, p62 levels increased in PS within 16 h of iAs exposure, concomitant with HMOX1 induction (Figure 4B). In PrSPCs derived from RWPE1 cells, iAs increased p-p62 (S349), NQO1, and HMOX1 levels, whereas knockdown of p62 attenuated these responses (Figure 4C) and decreased their soft-agar colony formation capability (Figure 4D). Phosphorylation of p62-S349 (S351 for mouse) increases its KEAP1-binding capability (Ichimura et al. 2013) and iAs increased p62-S349 phosphorylation in PrSPCs from normal prostate epithelial cells as well (Figure 4C,E), supporting iAs activation of p62-KEAP1-NRF2. Because p62-S349 modification depends on autophagy initiation, the role of autophagy in iAs-induced NRF2 activation was examined. Significantly, blockade of autophagy initiation by ATG7 knockdown suppressed iAs-induced HMOX1 (Fig. 4F), suggesting that iAs actions may involve autophagy in PrSPCs.
Autophagic Flux Activity in PrSPCs Exposed to Arsenic
Previous studies have reported that iAs can either activate or inhibit autophagy, depending on the context of dosage and cell type (Qi et al. 2014). To examine a potential role of autophagy in iAs-induced NRF2 activation, we examined the effects of iAs on autophagy of PrSPCs in greater detail.
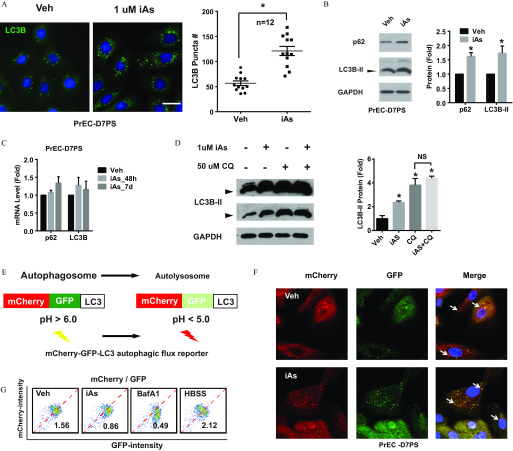
Microtubule-associated protein 1A/1B-light chain 3 (LC3) is a marker of autophagy and is ubiquitously distributed in cells. After autophagy initiation, the cytosolic form of LC3 (LC3-I) is converted to LC3-II by lipidation and recruited to autophagosome membranes. SQSTM/p62 (p62) is an autophagy protein that delivers cargo to the autophagosome for degradation and is also degraded with cargo in the lysosome. Thus, autophagic flux, a measure of autophagic degradation activity, can be assessed by the status of LC3 and p62. Exposure to iAs induced autophagosome accumulation in PrSPCs (Figure 5A). Further, iAs significantly increased both LC3B-II and p62 protein levels (Figure 5B), whereas mRNA levels were not significantly affected (Figure 5C), suggesting that autophagosome accumulation may be a result of impaired degradation. Indeed, cotreatment with the lysosome inhibitor chloroquine did not significantly increase LC3B-II accumulation after iAs treatment (Figure 5D), which suggests autophagic flux blockade.
Figure 5.
Arsenic effects on autophagic flux in prostate stem-progenitor cells. (A) Spheres of primary human prostate epithelial cells (PrEC-D7PS) were treated with inorganic arsenic ( iAs) for 7 d, and single cells of spheres were attached to chamber slides overnight for immunostaining. Autophagosomes were visualized with LC3B staining as green puncta. Puncta in single cells were counted and quantified data shown are ( cells from wells of same slide); * vs. vehicle control. . (B) Immunoblots of p62 and LC3B protein from PrEC-D7PS treated with iAs. Quantified data are (); * vs. vehicle control. (C) mRNA level of p62 and LC3B from PrEC-PS treated with arsenic for 48 h (iAs_48h) or 7 d (iAs_7d). Data are (); no significant difference vs. vehicle. (D) Immunoblots of LC3B from PrEC-D7PS with indicated treatment. Spheres were cultured with iAs, and chloroquine (CQ, ) was added 4 h before sample collection. Arrowheads indicate lipidified LC3B-II blots at two exposure lengths. The top blot of a long exposure time shows the unmodified LC3B-I, which migrates more slowly than the lipidified LC3B-II. The bottom blot shows a shorter exposure time where relative levels were quantified. Quantification data are (); * vs. vehicle control; NS, between two groups. (E) Schematic of the autophagic flux reporter EGFP-mCherry-LC3. (F) Confocal images showing GFP and mCherry in autophagosomes/autolysosomes in PrEC-D7PS overexpressing flux reporter. Arrowheads indicate GFP-mCherry-LC3 positive puncta of autophagosomes/autolysosomes. (G) Flow cytometry data quantification of GFP and mCherry intensity of PrEC-D7PS cells, over-expressing flux reporter, after indicated treatment. BafA1, treatment for 4 hrs; HBSS, cells treated with Hank’s Balanced Salt Solution (HBSS) overnight; iAs, treated for 7 d; Veh, vehicle. Number indicates the ratio of mCherry/GFP for the inset.
To directly assess iAs-induced blockade, an autophagic flux reporter of tandem fluorescent-tagged LC3 (mCherry-GFP-LC3) was used. By integrating to the autophagosome membrane, mCherry-GFP-LC3 fusion protein is delivered to the lysosome, where acidic conditions () quench GFP, whereas the mCherry signal remains (Figure 5E). The ratio of mCherry and GFP fluorescence (mCherry/GFP) reflects autophagic flux. Foci with high mCherry and low GFP signal were observed in vehicle-treated PrSPCs (Figure 5F). In contrast, iAs induced more foci with colocalization of mCherry and GFP, indicating flux blockade. Quantification by flow cytometry (Figure 5G) showed that iAs exposure decreased the mCherry/GFP ratio by ; Hank’s Balanced Salt Solution (HBSS), an autophagy inducer, increased the mCherry/GFP ratio; whereas BafA1, a lysosome inhibitor, decreased the mCherry/GFP ratio. Together the findings show that iAs induces autophagic flux blockade, resulting in p62 accumulation in PrSPCs.
Effects of Arsenic on Lysosome Acidification V-ATPase Expression
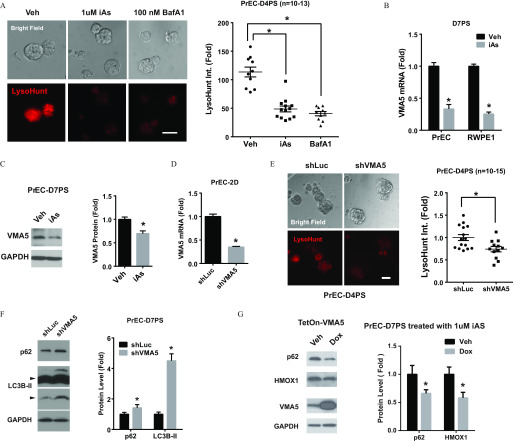
Autophagy uses a lysosomal degradation pathway, and inhibitors of lysosome acidification have been shown to block autophagic flux. As such, the effects of iAs on lysosome acidification were next interrogated. Use of LysoHunt Red, a fluorescent dye that localizes to acidic organelles of living cells, revealed that iAs suppressed organelle acidification of PrSPCs (Figure 6A). Vacuolar ATPase (V-ATPase) plays a central role in organelle acidification (Maxson and Grinstein 2014), and the specific V-ATPase inhibitor BafA1 similarly inhibited organelle acidification in PrSPCs (Figure 6A). We thus evaluated whether iAs inhibited lysosome acidification by suppressing V-ATPase function. V-ATPase is a multisubunit enzyme that is highly conserved among eukaryotes, consisting of a cytosolic ATP-hydrolytic V1 domain and a transmembrane proton-translocation V0 domain (Maxson and Grinstein 2014). Transcriptome analysis revealed that iAs markedly suppressed ATP6V1C2 (VMA5) in PrSPCs, which was further confirmed by qPCR (Figure 6B) and immunoblotting (Figure 6C). This was not a function of reduced lysosome levels because LAMP1, a lysosome marker, was not altered by iAs treatment (Figure S3A).
Figure 6.
Arsenic effects on lysosome acidification and expression of V-ATPase subunit VMA5. (A) LysoHunt staining showing acidification of organelles from day-4 spheres of primary human prostate epithelial cells (PrEC-D4PS) cultured on low-attached plate with indicated treatment. Higher intensity of red signal from LysoHunt indicates higher acidification. BafA1, bafilomycin A1, V-ATPase inhibitor, treatment for 2 h; iAs, iAs treatment for 4 d; Veh, vehicle. Quantification data are ( spheres from wells of same low-attach plate); * vs. vehicle. (B) mRNA level of VMA5 gene in D7PS of RWPE1 and PrEC treated with iAs for 7 d. (); * vs. vehicle. (C) Immunoblot of VMA5 protein in PrEC-D7PS treated with iAs for 7 d. (); * vs. vehicle. (D) mRNA level of VMA5 in PrEC with shRNA targeting VMA5 (shVMA5). (); * vs. shRNA control (shLuc). (E) LysoHunt staining showing acidification of organelles from day-4 spheres of primary human prostate epithelial cells (PrEC-D4PS) with VMA5 gene knockdown (shVMA5). Quantitative data are (); * vs. shRNA control (shLuc). (F) Immunoblotting of p62 and LC3B from PrEC-D7PS with VMA5 gene knockdown (shVMA5). (); * vs. shRNA control (shLuc). (G) Immunoblot of indicated protein in PrEC-D7PS with Tet-On VMA5 overexpression treated with iAs for 7 d. Doxycycline (Dox, ) was added to induce VMA5 overexpression for 7 d. Quantification data are (); * vs. vehicle control.
VMA5 encodes the subunit C of the V-ATPase V1 domain, and studies with yeast and fruit flies indicate that VAM5 is essential for V-ATPase assembly (Maxson and Grinstein 2014). To determine whether VMA5 plays a functional role in iAs-induced autophagic flux blockade, we knocked-down VMA5 in PrECs (Figure 6D) and generated 3D PS cultures. VMA5 knockdown significantly suppressed acidification of intracellular organelles (Figure 6E). An important finding is that VMA5 knockdown induced autophagic flux blockade, as shown by significant accumulation of LC3B-II and p62 (Figure 6F). Although VMA5 loss did not elevate HMOX1 expression (Figure S3B), VMA5 overexpression in PS using a Tet-On system significantly attenuated iAs-induced HMOX1 and p62 accumulation (Figure 6G) but had no effect on basal HMOX1 expression in vehicle control (Figure S3C).
Evaluation of NRF2 Pathway Activation in Separate Prostate Stem, Progenitor, and Differentiated Cells
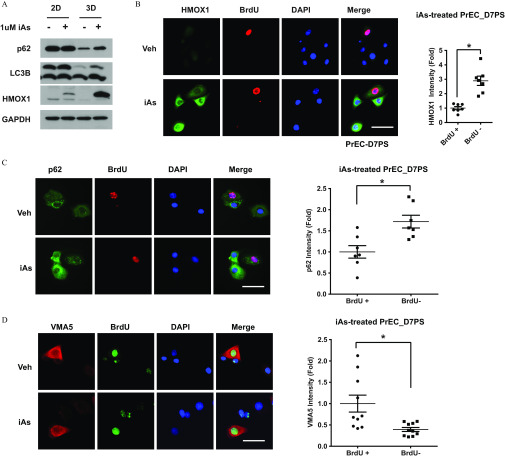
Upon asymmetric cell division, prostate stem cells give rise to daughter progenitor cells that proliferate and give rise to differentiated cell types. To better understand the primary targets of iAs exposures, we next assessed NRF2 activation as a function of treatment with iAs in these three cell populations. Compared with the 3D PS cells, the parenteral differentiated 2D primary PrECs had higher p62 and lower LC3B-II/LC3-I ratio levels, indicating lower basal autophagy activity (Figure 7A). Despite subtle HMOX1 induction, iAs treatment had no obvious effect on p62 and LC3B-II levels in 2D PrECs (Figure 7A) indicating that these pathways are not arsenic targets in differentiated prostate epithelial cells.
Figure 7.
Assessment of arsenic effects on the NRF2 pathway in progenitor cells and their progeny. (A) Representative immunoblotting () of indicated protein in primary human prostate epithelial cells (PrEC) under indicated culture condition with inorganic arsenic (iAs). 2D, 2-dimensional cultured PrEC represent differentiated prostate epithelial cells; 3D, 3-dimensional cultured PrEC represent prostate stem-progenitor cells. (B–D) Representative immunostaining of BrdU and indicated protein in day-7 sphere derived from PrEC (PrEC-D7PS) treated with iAs for 7 d. BrdU-positive cells represent quiescent stem cells. BrdU-negative cells represent transient amplifying progenitor cells. Quantification data of iAs-induced HMOX1(b)/p62(c)/VMA5(d) expression in stem cells () and progenitor cells (). Data shown are ( paired cells from same staining); * between two groups.
Using a long-term bromodeoxyuridine (BrdU)-retaining assay in PS (Hu et al. 2017, 2019), we next studied NRF2 activation in the separate stem and progenitor cell populations. In the prostate spheres, relative quiescent stem cells retain 2D-labeled BrdU, whereas the rapidly proliferating progenitor cells dilute BrdU-label through cell division (Hu et al. 2017), permitting identification of the two cell populations. Surprisingly, iAs induced much higher HMOX1, a marker of NRF2 activation, in BrdU-negative progenitor cells than in BrdU-positive stem cells (Fig. 7B). Because prostate stem cells have higher autophagic flux in comparison with progenitor cells (Hu et al. 2017), it is possible that the stem cells have a higher capacity to overcome iAs-induced flux blockade. Indeed, p62 protein levels were lower in stem cells in comparison with daughter progenitor cells under basal conditions as well as upon iAs-exposure (Figure 7C). Because iAs blocked autophagic flux by VMA5 suppression, we next examined VMA5 expression in the PS stem and progenitor cell types. Markedly higher VMA5 expression was observed in the BrdU-retaining stem cells and remained high expression even upon iAs exposure (Figure 7D). These results indicate that the progenitor cells may be the primary iAs targets in the prostate and further confirm the essential role of VMA5 in iAs-induced NRF2 action as well.
Ability of Arsenic Exposure to Induce Chronic NRF2 Activation and Prostate Cell Transformation
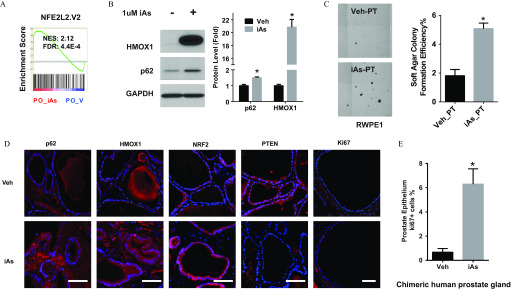
Transcriptomic analysis indicated that iAs activated the NRF2 pathway in day-14 PO cultures (Figure 8A), which was confirmed by elevated protein levels of p62 and HMOX1 in iAs-exposed organoids (Figure 8B). Because differentiated PrECs are largely resistant to iAs-induced NRF2 activation (Figure 7A), we hypothesized that NRF2 activation in differentiating PO epithelial cells might be a result of iAs initially targeting the progenitor cell population that forms the organoids over a 14-d time span. To test whether iAs-induced effects on progenitor cells could be carried forward to their unexposed progeny, we examined the transformation status of unexposed daughter cells following iAs-pretreatment of progenitor cells. RWPE1 cells were transferred to spheroid culture w/wo iAs for 1 wk. The PS were collected, dispersed to single cells, and transferred to soft-agar culture for 3 wk in the absence of iAs to assess transformation. Compared with vehicle control cells, progeny cells of iAs-pretreated PS formed significantly more colonies (Figure 8C), suggesting that iAs-induced transformation initiates in progenitor cells and is passed forward to daughter cells.
Figure 8.
Assessment of arsenic effects on the NRF2 pathway in the progeny of progenitor cells. (A) GSEA NRF2 pathway enrichment analysis of prostate organoid (PO) treated with iAs for 14 d. NES, normalized enrichment score. indicates that NRF2 pathway is enriched in PO_iAs groups. (B) Representative immunoblotting of HMOX1 and p62 in day-14 prostate organoid derived from PrEC (PrEC-D14PO) treated with iAs for 14 d. Quantification data shown are (); * vs. vehicle. (C) Representative image of soft-agar transformation assay of prostate stem-progenitor cells pretreated with iAs for 7 d. Quantification data of soft-agar colony formation efficiency are (); * between two groups. (D) Representative immunostaining () of indicated protein in chimeric human prostate gland structure derived from human PrEC and rat UGM grafted under renal capsule exposed to iAs in drinking water for 3 months. . (E) Quantitative data of Ki67 positive cell percent in chimeric human prostate gland. Data shown are ( chimeric gland); * between two groups.
Consistent with the in vitro studies, immunostaining of in vivo renal grafts of prostate cell-tissue recombinants revealed that 3 months of iAs exposure led to higher protein levels of p62, NRF2, and HMOX1 in the human-derived epithelial cells and p62 and HMOX1 in the rat-derived stromal cells compared with controls, suggesting chronic activation of the NRF2 pathway (Figure 8D). Although no overt pathologic changes were observed in comparison with vehicle grafts, chronic iAs exposure decreased tumor suppressor PTEN levels, consistent with previous in vitro studies in PrECs (Tokar et al. 2010b) and increased the number of Ki67-positive proliferating epithelial cells (Figure 8E), hallmarks of a precancerous state.
Discussion
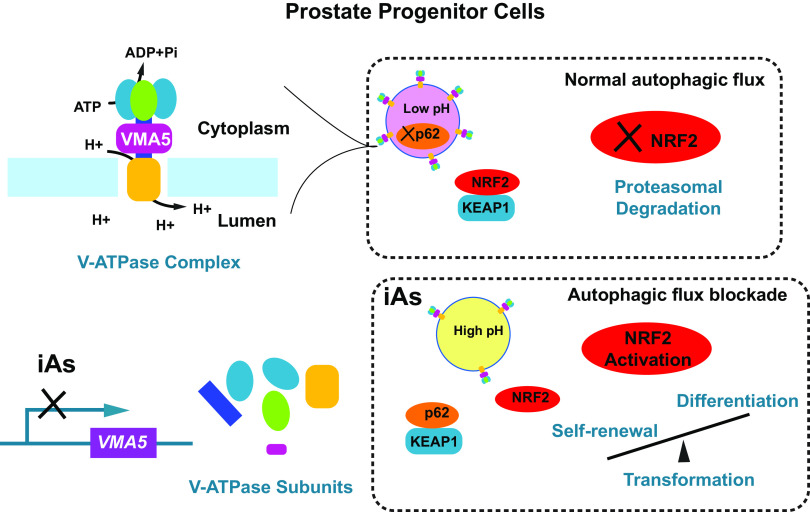
Using normal human primary cells, we herein demonstrate that iAs, at doses relevant to human exposures, disturbs homeostasis of PrSPCs through constitutive NRF2 activation in prostate progenitor cells, leading to their accumulation due to enhanced proliferation and suppressed differentiation. Cancer may arise from maturation arrest during progression of stem cells to transient amplifying progenitor cells and on to differentiated cells (Clarke and Fuller 2006). Perturbations of this formative process could have long-term effects that drive increased prostate cancer risk susceptibility. Indeed, continuous iAs exposures led to transformation of PrSPCs in vitro, which was mitigated by NRF2 knockdown. Similarly, in vivo iAs exposure activated NRF2 in prostate chimeric grafts, which initiated a pre-malignant molecular phenotype in the human prostate epithelium. Mechanistically, iAs activated NRF2 through a noncanonical p62-dependent pathway in which p62 accumulated from iAs-induced autophagic flux blockade, a result of suppressed lysosomal protein degradation due to attenuated lysosome acidification. Specifically, we found that iAs suppressed VMA5 expression, leading to impaired V-ATPase and subsequent inhibition of acidification. This is summarized schematically in Figure 9.
Figure 9.
Model of how inorganic arsenic (iAs) transforms prostate progenitor cells through NRF2 activation via autophagic flux blockade.
Our transcriptome analysis and functional studies identified a novel mechanism underlying iAs-induced autophagic flux blockade. Upon iAs exposure, suppression of VMA5 gene led to compromised V-ATPase function and lysosome acidification, inhibiting the lysosomal degradation step of autophagic flux. Conversely, overexpression of VMA5 partially attenuated iAs-induced p62 accumulation and NRF2 activation. Although VMA5 knockdown has been linked with attenuated Wnt signaling by impairing V-ATPase mediated acidification (Cruciat et al. 2010), the present study provides direct evidence for an essential role of VMA5 in V-ATPase function and autophagic flux regulation in mammal cells. V-ATPase has recently been implicated in various additional roles independent of its proton pump ability (Maxson and Grinstein 2014). Whether these noncanonical functions, which include endocytic traffic, metabolic sensing, and cell signaling, are affected by iAs are areas for potential exploration.
As a multidomain protein, p62 is a signaling hub linking autophagy and tumorigenesis. Downstream effectors of p62 determine its oncogenic or antioncogenic role in a context-dependent way (Katsuragi et al. 2015; Moscat et al. 2016; Shah et al. 2017). In this study, we demonstrated that NRF2 is an oncogenic effector of p62 during iAs-induced transformation. More important, we find that the status of the autophagic machinery of cells dictates the NRF2 responses upon iAs exposure, which may help to explain the controversial roles of autophagy in iAs-induced tumorigenesis (Qi et al. 2014). With high autophagy initiation and low VMA5 expression, progenitor cells have robust NRF2 activation upon iAs exposure, whereas it is limited in stem cells and differentiated epithelial cells due to lower autophagy initiation. Of significance is that iAs-induced NRF2 activation of progenitor cells is sustained in their differentiated progeny cells, which sets the stage for propagation of transformed cells and predisposition to a carcinogenic state.
Additionally, the phosphorylation status of p62 determines its downstream pathways. Phosphorylation of S349 (S351 for mouse) on the KIR domain activates the KEAP1-NRF2 pathway (Ichimura et al. 2013), whereas phosphorylation of S403 on the UBA domain regulates clearance of ubiquitinated proteins through autophagy (Matsumoto et al. 2011). A recent study shows that T269/S272 phosphorylation is critical for amino acid activation of mTORC1 by p62 (Linares et al. 2015). Herein, we demonstrated that iAs-induced S349 phosphorylation of p62 leads to constitutive NRF2 activation in PrSPCs. Whether other modifications of p62 occur in these cells upon iAs exposure is currently unclear. NF-KB, beta-catenin, and ERK1 pathways are known downstream effectors of p62 and are also dysregulated by arsenic in other systems (Huang et al. 1999; Katsuragi et al. 2015; Z Wang et al. 2013; Wei et al. 2016), and these effectors will be interrogated in future studies.
NRF2 and ROS have been shown to involved in iAs-induced tumorigenesis. Acute high-dose iAs () activates NRF2 and produces ROS (Pi et al. 2003; Wang et al. 2008) in mammary epithelial cells and keratinocytes (Pi et al. 2003; Shah et al. 2017; Wang et al. 2008), whereas chronic low-dose iAs () inhibits autophagic flux in differentiated cells (such as fibroblast, kidney, and lung epithelial cells), leading to noncanonical NRF2 activation and reduced ROS levels (Lau et al. 2013; Son et al. 2015; Yang et al. 2015). In the current study, we demonstrated for the first time in prostate progenitor cells that low-dose iAs induces constant NRF2 activation through autophagic flux blockade. As a master transcription factor for cell defense and survival, NRF2 activates antioxidative and antiapoptotic pathways, facilitates iAs-transformed cell survival of microenvironmental stress, and increases tumor growth and metastasis (Jaramillo and Zhang 2013; Son et al. 2015; Yang et al. 2015). Indeed, we demonstrated that iAs-induced NRF2 activation attenuated inducive ROS generation, which may grant a survival advantage to iAs-transformed PrSPCs, allowing accumulation of tumorigenic events.
Recently, the role of NRF2 in iAs-induced malignant transformation has been linked with stem cells through reprogramming lung epithelial cells in vitro to form cancer stem-like cells (Bi et al. 2020). In our study, we extended the role for NRF2 in iAs tumorigenesis to dysregulation of human PrSPCs homeostasis though progenitor cells. iAs has been shown to target the stem cell populations during malignant transformation of WPE-stem cells, which is derived from immortalized RWPE1 cells (Tokar et al. 2010c). However, given the properties of rapid growth and high KRT14 expression (Tokar et al. 2005), we propose that the majority of WPE-stem cells are progenitor rather than quiescent tissue stem cells. In this regard, observations from both cell lines and primary cells indicate that progenitor cells are major targets of iAs tumorigenesis in the prostate. This finding raises the possibility that iAs-induced prostate cancer may arise from progenitor cell dysregulation.
Mounting evidence suggests that dysregulation of progenitor cells is associated with tumorigenesis. In a classical stem cell–progenitor cell–committed cell hierarchy model, progenitor cells serve as a fast lane linking stem cells with differentiated progeny, as they rapidly proliferate, lineage commit, and form the bulk of tissue (Rangel-Huerta and Maldonado 2017; Zhang and Hsu 2017). These unique properties of progenitor cells make them vulnerable targets during tumorigenesis. Multipotent progenitor cells generated from infrequent divisions of hematopoietic stem cells can be transformed, leading to acute myeloid lymphoma (Lavau et al. 2000). In addition, progenitor cells are physically close to stem cells, regulating both the stem cells and their niche. Accumulation of progenitor cells from intestinal stem cells by blocking their differentiation is sufficient to drive tumorigenesis in adult Drosophila. During this process, progenitor cells generate a feed-forward loop promoting stem cell proliferation and remodeling the stem cell environment (Chen et al. 2016; Zhai et al. 2015). In the present study, suppression of NRF2 activation in RWPE-derived progenitor cells by NRF2 shRNA knockdown attenuated their transformation upon iAs exposure, which is in agreement with previous studies showing that blockade of persistent NRF2 activation mitigates iAs-induced transformation in lung epithelial cells (Son et al. 2015; Yang et al. 2015). Further, we demonstrate that NRF2 activation leads to both enhanced progenitor self-renewal and decreased differentiation. Together this supports the central role of prostate progenitor cells in iAs tumorigenesis.
Although prostate cancer arises from epithelial cells, the stroma has been increasingly recognized as an important contributor for prostate tumorigenesis and progression (Barron and Rowley 2012). In the current in vivo study using a chimeric gland regeneration model, iAs activated the p62-NRF2 pathway in both epithelial and stromal cells. Recently, in a prostate cancer cell–conditioned media model, conditioned media from iAs-treated adipose-derived mesenchymal stem/stromal cells increased prostate tumor cell viability (Shearer et al. 2016), and cytokine array analysis revealed that iAs increased HMOX1 expression but suppressed pathway activation in mesenchymal stem/stromal cells. Interestingly, in contrast to a tumorigenic role in epithelial cells, stromal p62 plays a protective role in prostate cancer progression by modulating metabolism in the tumor stroma (Linares et al. 2017; Valencia et al. 2014). Thus, iAs may also affect PrSPCs indirectly through stromal microenvironment.
Although iAs induced transformation in vitro, we observed only a pre-cancerous phenotype of PTEN suppression, high proliferation, and de-differentiation, but not transformation associated with iAs exposure in vivo. There are a few possible explanations for this discrepancy. First, compared with lifelong iAs exposure, 3 months of iAs exposure may not be sufficient for full transformation of normal human PrSPCs derived from young donors. Second, elevated p62 expression in stroma by iAs may suppress tumorigenesis of epithelial cells as previously revealed (Linares et al. 2017; Valencia et al. 2014). Further, the iAs metabolism in humans is different from that in mouse. Monomethylarsonic acid (MMA) is significantly more toxic than either iAs or the other metabolites, and humans have significantly higher MMA metabolite levels than most mammals have, including mice (States et al. 2011).
Just as iAs affects multiple organs, multiple signaling pathways are dysregulated by iAs as demonstrated in Figure 2B,C and previous studies (Benbrahim-Tallaa and Waalkes 2008; Carlin et al. 2016; Tokar et al. 2011). Although we focused on NRF2 activation through autophagy dysregulation, other iAs-affected pathways may crosstalk with NRF2 pathway as well. Oncogenic KRAS, which was also up-regulated in iAs-treated PrSPCs (Figure 2C), has been reported to directly increase NRF2 gene transcription in lung cancer cells through a TPA response element located in the NRF2 promoter (Tao et al. 2014). Although NRF2 mRNA was not affected by iAs in day-7 PrSPCs (Figure 2E), KRAS may activate NRF2 in daughter cells of PrSPCS over time. It has been shown that p53 counteracts NRF2-induced transcription through direct interaction with NRF2-binding sites of promoters (Faraonio et al. 2006). As shown in Figure 2C, iAs suppressed p53 in PrSPCs as well, which may contribute to NRF2 activation in the context of p62 knockdown (Figure 4C). How iAs affects oncogenic pathways (Figure 2B,C) cross-talk with each other will be actively explored in future studies.
Conclusion
In summary, we outlined a novel mechanism by which iAs disturbs homeostasis of PrSPCs and predisposes to tumorigenesis. This work adds to knowledge of both iAs toxicology and prostate cancer, linking a novel role for NRF2 in PrSPC regulation. In addition to demonstrating that iAs activates NRF2 through autophagic flux blockade in prostate progenitor cells, this work reveals a unique molecular mechanism by which iAs inhibits lysosomal degradation via suppression of V-ATPase function. Further, we demonstrate for the first time, to our knowledge, the dynamics of autophagy in prostate epithelial cells during stem cell differentiation and find the autophagic status of these cell populations determines their NRF2 responses to iAs exposure. Our findings support the hypothesis that transient amplifying prostate progenitors are primary iAs targets and that iAs exposure may lead to their accumulation and transformation by increasing self-renewal and inhibiting differentiation through NRF2 activation. As such, this work sheds light on the potential for rational development of novel strategies targeting autophagy to mitigate adverse effects of iAs and for therapeutic approaches of prostate cancer.
Supplementary Material
Acknowledgments
This study was supported by National Institutes of Health grants R01-ES02207 (G.S.P. and L.X.) and P30-ES027792 (G.S.P.), and by The Michael Reese Research and Education Foundation (G.S.P).
References
- Amaravadi R, Kimmelman AC, White E. 2016. Recent insights into the function of autophagy in cancer. Genes Dev 30(17):1913–1930, PMID: 27664235, 10.1101/gad.287524.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey K, Fry RC. 2014. Long-term health consequences of prenatal arsenic exposure: links to the genome and the epigenome. Rev Environ Health 29(1–2):9–12, PMID: 24552957, 10.1515/reveh-2014-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain LJ, Liu JT, League RE. 2016. Arsenic inhibits stem cell differentiation by altering the interplay between the Wnt3a and notch signaling pathways. Toxicol Rep 3:405–413, PMID: 27158593, 10.1016/j.toxrep.2016.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barron DA, Rowley DR. 2012. The reactive stroma microenvironment and prostate cancer progression. Endocr Relat Cancer 19(6):R187–R204, PMID: 22930558, 10.1530/ERC-12-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benbrahim-Tallaa L, Waalkes MP. 2008. Inorganic arsenic and human prostate cancer. Environ Health Perspect 116(2):158–164, PMID: 18288312, 10.1289/ehp.10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi Z, Zhang Q, Fu Y, Wadgaonkar P, Zhang W, Almutairy B, et al. 2020. Nrf2 and HIF1α converge to arsenic-induced metabolic reprogramming and the formation of the cancer stem-like cells. Theranostics 10(9):4134–4149, PMID: 32226544, 10.7150/thno.42903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C. 2013. Tracing the cellular origin of cancer. Nat Cell Biol 15(2):126–134, PMID: 23334500, 10.1038/ncb2657. [DOI] [PubMed] [Google Scholar]
- Carlin DJ, Naujokas MF, Bradham KD, Cowden J, Heacock M, Henry HF, et al. 2016. Arsenic and environmental health: state of the science and future research opportunities. Environ Health Perspect 124(7):890–899, PMID: 26587579, 10.1289/ehp.1510209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CJ, Wang CJ. 1990. Ecological correlation between arsenic level in well water and age-adjusted mortality from malignant neoplasms. Cancer Res 50(17):5470–5474, PMID: 2386951. [PubMed] [Google Scholar]
- Chen J, Xu N, Huang H, Cai T, Xi R. 2016. A feedback amplification loop between stem cells and their progeny promotes tissue regeneration and tumorigenesis. eLife 5:e14330, PMID: 27187149, 10.7554/eLife.14330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua CW, Shibata M, Lei M, Toivanen R, Barlow LJ, Bergren SK, et al. 2014. Single luminal epithelial progenitors can generate prostate organoids in culture. Nat Cell Biol 16(10):951–961, PMID: 25241035, 10.1038/ncb3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke MF, Fuller M. 2006. Stem cells and cancer: two faces of Eve. Cell 124(6):1111–1115, PMID: 16564000, 10.1016/j.cell.2006.03.011. [DOI] [PubMed] [Google Scholar]
- Cruciat CM, Ohkawara B, Acebron SP, Karaulanov E, Reinhard C, Ingelfinger D, et al. 2010. Requirement of prorenin receptor and vacuolar H+ATPase-mediated acidification for Wnt signaling. Science 327(5964):459–463, PMID: 20093472, 10.1126/science.1179802. [DOI] [PubMed] [Google Scholar]
- Dodson M, de la Vega MR, Harder B, Castro-Portuguez R, Rodrigues SD, Wong PK, et al. 2018. Low-level arsenic causes proteotoxic stress and not oxidative stress. Toxicol Appl Pharmacol 341:106–113, PMID: 29408041, 10.1016/j.taap.2018.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraonio R, Vergara P, Di Marzo D, Pierantoni MG, Napolitano M, Russo T, et al. 2006. p53 suppresses the Nrf2-dependent transcription of antioxidant response genes. J Biol Chem 281(52):39776–39784, PMID: 17077087, 10.1074/jbc.M605707200. [DOI] [PubMed] [Google Scholar]
- Ghoda L, Phillips MA, Bass KE, Wang CC, Coffino P. 1990. Trypanosome ornithine decarboxylase is stable because it lacks sequences found in the carboxyl terminus of the mouse enzyme which target the latter for intracellular degradation. J Biol Chem 265(20):11823–11826, PMID: 2365702. [PubMed] [Google Scholar]
- Guan JL, Simon AK, Prescott M, Menendez JA, Liu F, Wang F, et al. 2013. Autophagy in stem cells. Autophagy 9(6):830–849, PMID: 23486312, 10.4161/auto.24132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu WY, Hu DP, Xie L, Birch LA, Prins GS. 2019. Isolation of stem-like cells from 3-dimensional spheroid cultures. J Vis Exp (154):e60357, PMID: 31885380, 10.3791/60357. [DOI] [PubMed] [Google Scholar]
- Hu WY, Hu DP, Xie L, Li Y, Majumdar S, Nonn L, et al. 2017. Isolation and functional interrogation of adult human prostate epithelial stem cells at single cell resolution. Stem Cell Res 23:1–12, PMID: 28651114, 10.1016/j.scr.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu WY, Shi GB, Hu DP, Nelles JL, Prins GS. 2012. Actions of estrogens and endocrine disrupting chemicals on human prostate stem/progenitor cells and prostate cancer risk. Mol Cell Endocrinol 354(1–2):63–73, PMID: 21914459, 10.1016/j.mce.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu WY, Shi GB, Lam HM, Hu DP, Ho SM, Madueke IC, et al. 2011. Estrogen-initiated transformation of prostate epithelium derived from normal human prostate stem-progenitor cells. Endocrinology 152(6):2150–2163, PMID: 21427218, 10.1210/en.2010-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Ma WY, Li J, Goranson A, Dong Z. 1999. Requirement of Erk, but not JNK, for arsenite-induced cell transformation. J Biol Chem 274(21):14595–14601, PMID: 10329651, 10.1074/jbc.274.21.14595. [DOI] [PubMed] [Google Scholar]
- Huang L, Pu Y, Hu WY, Birch L, Luccio-Camelo D, Yamaguchi T, et al. 2009. The role of Wnt5a in prostate gland development. Dev Biol 328(2):188–199, PMID: 19389372, 10.1016/j.ydbio.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IARC (International Agency for Research on Cancer Working Group on the Evaluation of Carcinogenic Risk to Humans). 2004. Some Drinking-Water Disinfectants and Contaminants, Including Arsenic. IARC Monogr Eval Carcinog Risks Hum, No. 84. https://www.ncbi.nlm.nih.gov/books/NBK402251/ [accessed 2 June 2020]. [Google Scholar]
- Ichimura Y, Waguri S, Sou YS, Kageyama S, Hasegawa J, Ishimura R, et al. 2013. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol Cell 51(5):618–631, PMID: 24011591, 10.1016/j.molcel.2013.08.003. [DOI] [PubMed] [Google Scholar]
- Jaramillo MC, Zhang DD. 2013. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev 27(20):2179–2191, PMID: 24142871, 10.1101/gad.225680.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuragi Y, Ichimura Y, Komatsu M. 2015. P62/SQSTM1 functions as a signaling hub and an autophagy adaptor. FEBS J 282(24):4672–4678, PMID: 26432171, 10.1111/febs.13540. [DOI] [PubMed] [Google Scholar]
- Kenific CM, Debnath J. 2015. Cellular and metabolic functions for autophagy in cancer cells. Trends Cell Biol 25(1):37–45, PMID: 25278333, 10.1016/j.tcb.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau A, Zheng Y, Tao S, Wang H, Whitman SA, White E, et al. 2013. Arsenic inhibits autophagic flux, activating the Nrf2-Keap1 pathway in a p62-dependent manner. Mol Cell Biol 33(12):2436–2446, PMID: 23589329, 10.1128/MCB.01748-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavau C, Luo RT, Du C, Thirman MJ. 2000. Retrovirus-mediated gene transfer of MLL-ELL transforms primary myeloid progenitors and causes acute myeloid leukemias in mice. Proc Natl Acad Sci USA 97(20):10984–10989, PMID: 10995463, 10.1073/pnas.190167297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares JF, Cordes T, Duran A, Reina-Campos M, Valencia T, Ahn CS, et al. 2017. ATF4-induced metabolic reprograming is a synthetic vulnerability of the p62-deficient tumor stroma. Cell Metab 26(6):817–829, PMID: 28988820, 10.1016/j.cmet.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares JF, Duran A, Reina-Campos M, Aza-Blanc P, Campos A, Moscat J, et al. 2015. Amino acid activation of mTORC1 by a PB1-domain-driven kinase complex cascade. Cell Rep 12(8):1339–1352, PMID: 26279575, 10.1016/j.celrep.2015.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. 2002. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science 295(5556):868–872, PMID: 11786607, 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- Matsumoto G, Wada K, Okuno M, Kurosawa M, Nukina N. 2011. Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol Cell 44(2):279–289, PMID: 22017874, 10.1016/j.molcel.2011.07.039. [DOI] [PubMed] [Google Scholar]
- Maxson ME, Grinstein S. 2014. The vacuolar-type H(+)-ATPase at a glance - more than a proton pump. J Cell Sci 127(pt 23):4987–4993, PMID: 25453113, 10.1242/jcs.158550. [DOI] [PubMed] [Google Scholar]
- Maycotte P, Jones KL, Goodall ML, Thorburn J, Thorburn A. 2015. Autophagy supports breast cancer stem cell maintenance by regulating IL6 secretion. Mol Cancer Res 13(4):651–658, PMID: 25573951, 10.1158/1541-7786.MCR-14-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy CR, Stadelman BS, Brumaghim JL, Liu JT, Bain LJ. 2015. Arsenic and its methylated metabolites inhibit the differentiation of neural plate border specifier cells. Chem Res Toxicol 28(7):1409–1421, PMID: 26024302, 10.1021/acs.chemrestox.5b00036. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Komatsu M. 2011. Autophagy: renovation of cells and tissues. Cell 147(4):728–741, PMID: 22078875, 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, et al. 2006. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124(6):1283–1298, PMID: 16564017, 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- Moscat J, Karin M, Diaz-Meco MT. 2016. p62 in cancer: signaling adaptor beyond autophagy. Cell 167(3):606–609, PMID: 27768885, 10.1016/j.cell.2016.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi J, Qu W, Reece JM, Kumagai Y, Waalkes MP. 2003. Transcription factor Nrf2 activation by inorganic arsenic in cultured keratinocytes: involvement of hydrogen peroxide. Exp Cell Res 290(2):234–245, PMID: 14567983, 10.1016/s0014-4827(03)00341-0. [DOI] [PubMed] [Google Scholar]
- Polya D, Charlet L. 2009. Environmental science: Rising arsenic risk? Nature Geosci 2(6):383–384, 10.1038/ngeo537. [DOI] [Google Scholar]
- Prins GS, Calderon-Gierszal EL, Hu WY. 2015. Stem cells as hormone targets that lead to increased cancer susceptibility. Endocrinology 156(10):3451–3457, PMID: 26241068, 10.1210/en.2015-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prins GS, Hu WY, Shi GB, Hu DP, Majumdar S, Li G, et al. 2014. Bisphenol a promotes human prostate stem-progenitor cell self-renewal and increases in vivo carcinogenesis in human prostate epithelium. Endocrinology 155(3):805–817, PMID: 24424067, 10.1210/en.2013-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Li H, Zhang M, Zhang T, Frank J, Chen G. 2014. Autophagy in arsenic carcinogenesis. Exp Toxicol Pathol 66(4):163–168, PMID: 24560536, 10.1016/j.etp.2014.01.004. [DOI] [PubMed] [Google Scholar]
- Rane JK, Droop AP, Pellacani D, Polson ES, Simms MS, Collins AT, et al. 2014. Conserved two-step regulatory mechanism of human epithelial differentiation. Stem Cell Reports 2(2):180–188, PMID: 24527392, 10.1016/j.stemcr.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangel-Huerta E, Maldonado E. 2017. Transit-amplifying cells in the fast lane from stem cells towards differentiation. Stem Cells Int 2017:7602951, PMID: 28835754, 10.1155/2017/7602951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravenscroft P. 2007. Predicting the global distribution of natural arsenic contamination of ground water. In: Symposium on Arsenic: The Geography of a Global Problem. London: Royal Geographical Society. [Google Scholar]
- Ravenscroft P, Brammer H, Richards K. 2009. Arsenic Pollution: A Global Synthesis. Malden, MA: Wiley. [Google Scholar]
- Rebuzzini P, Cebral E, Fassina L, Alberto Redi C, Zuccotti M, Garagna S. 2015. Arsenic trioxide alters the differentiation of mouse embryonic stem cell into cardiomyocytes. Sci Rep 5:14993, PMID: 26447599, 10.1038/srep14993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. 2015. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43(7):e47, PMID: 25605792, 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt CW. 2014. Low-dose arsenic: in search of a risk threshold. Environ Health Perspect 122(5):A130–A134, PMID: 24784018, 10.1289/ehp.122-A130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P, Trinh E, Qiang L, Xie L, Hu WY, Prins GS, et al. 2017. Arsenic induces p62 expression to form a positive feedback loop with Nrf2 in human epidermal keratinocytes: implications for preventing arsenic-induced skin cancer. Molecules 22(2):194, PMID: 28125038, 10.3390/molecules22020194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer JJ, Wold EA, Umbaugh CS, Lichti CF, Nilsson CL, Figueiredo ML. 2016. Inorganic arsenic-related changes in the stromal tumor microenvironment in a prostate cancer cell-conditioned media model. Environ Health Perspect 124(7):1009–1015, PMID: 26588813, 10.1289/ehp.1510090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BA, Sokolov A, Uzunangelov V, Baertsch R, Newton Y, Graim K, et al. 2015. A basal stem cell signature identifies aggressive prostate cancer phenotypes. Proc Natl Acad Sci USA 112(47):E6544–E6552, PMID: 26460041, 10.1073/pnas.1518007112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son YO, Pratheeshkumar P, Roy RV, Hitron JA, Wang L, Divya SP, et al. 2015. Antioncogenic and oncogenic properties of Nrf2 in arsenic-induced carcinogenesis. J Biol Chem 290(45):27090–27100, PMID: 26385919, 10.1074/jbc.M115.675371. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- States JC, Barchowsky A, Cartwright IL, Reichard JF, Futscher BW, Lantz RC. 2011. Arsenic toxicology: translating between experimental models and human pathology. Environ Health Perspect 119(10):1356–1363, PMID: 21684831, 10.1289/ehp.1103441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand DW, Goldstein AS. 2015. The many ways to make a luminal cell and a prostate cancer cell. Endocr Relat Cancer 22(6):T187–T197, PMID: 26307022, 10.1530/ERC-15-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102(43):15545–15550, PMID: 16199517, 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao S, Wang S, Moghaddam SJ, Ooi A, Chapman E, Wong PK, et al. 2014. Oncogenic KRAS confers chemoresistance by upregulating NRF2. Cancer Res 74(24):7430–7441, PMID: 25339352, 10.1158/0008-5472.CAN-14-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokar EJ, Ancrile BB, Cunha GR, Webber MM. 2005. Stem/progenitor and intermediate cell types and the origin of human prostate cancer. Differentiation 73(9–10):463–473, PMID: 16351690, 10.1111/j.1432-0436.2005.00047.x. [DOI] [PubMed] [Google Scholar]
- Tokar EJ, Benbrahim-Tallaa L, Ward JM, Lunn R, Sams RL 2nd, Waalkes MP. 2010a. Cancer in experimental animals exposed to arsenic and arsenic compounds. Crit Rev Toxicol 40(10):912–927, PMID: 20812815, 10.3109/10408444.2010.506641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokar EJ, Diwan BA, Waalkes MP. 2010b. Arsenic exposure transforms human epithelial stem/progenitor cells into a cancer stem-like phenotype. Environ Health Perspect 118(1):108–115, PMID: 20056578, 10.1289/ehp.0901059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokar EJ, Qu W, Liu J, Liu W, Webber MM, Phang JM, et al. 2010c. Arsenic-specific stem cell selection during malignant transformation. J Natl Cancer Inst 102(9):638–649, PMID: 20339138, 10.1093/jnci/djq093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokar EJ, Qu W, Waalkes MP. 2011. Arsenic, stem cells, and the developmental basis of adult cancer. Toxicological Sci 120(suppl 1):S192–S203, PMID: 21071725, 10.1093/toxsci/kfq342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JJ, Dudakov JA, Takahashi K, Shieh JH, Velardi E, Holland AM, et al. 2013. Nrf2 regulates haematopoietic stem cell function. Nat Cell Biol 15(3):309–316, PMID: 23434824, 10.1038/ncb2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia T, Kim JY, Abu-Baker S, Moscat-Pardos J, Ahn CS, Reina-Campos M, et al. 2014. Metabolic reprogramming of stromal fibroblasts through p62-mTORC1 signaling promotes inflammation and tumorigenesis. Cancer Cell 26(1):121–135, PMID: 25002027, 10.1016/j.ccr.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waalkes MP, Liu J, Diwan BA. 2007. Transplacental arsenic carcinogenesis in mice. Toxicol Appl Pharmacol 222(3):271–280, PMID: 17306315, 10.1016/j.taap.2006.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Duncan D, Shi Z, Zhang B. 2013. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res 41 (Web Server issue):W77–W83, PMID: 23703215, 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Humphries B, Xiao H, Jiang Y, Yang C. 2013. Epithelial to mesenchymal transition in arsenic-transformed cells promotes angiogenesis through activating β-catenin-vascular endothelial growth factor pathway. Toxicol Appl Pharmacol 271(1):20–29, PMID: 23643801, 10.1016/j.taap.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XJ, Sun Z, Chen W, Li Y, Villeneuve NF, Zhang DD. 2008. Activation of Nrf2 by arsenite and monomethylarsonous acid is independent of Keap1-C151: enhanced Keap1-Cul3 interaction. Toxicol Appl Pharmacol 230(3):383–389, PMID: 18417180, 10.1016/j.taap.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei M, Liu J, Xu M, Rui D, Xu S, Feng G, et al. 2016. Divergent effects of arsenic on NF-κΒ signaling in different cells or tissues: a systematic review and meta-analysis. IJERPH 13(2):163, PMID: 26821040, 10.3390/ijerph13020163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh JP, Patel KG, Manthiram K, Swartz JR. 2009. Multiply mutated Gaussia luciferases provide prolonged and intense bioluminescence. Biochem Biophys Res Commun 389(4):563–568, PMID: 19825431, 10.1016/j.bbrc.2009.09.006. [DOI] [PubMed] [Google Scholar]
- White E. 2015. The role for autophagy in cancer. J Clin Invest 125(1):42–46, PMID: 25654549, 10.1172/JCI73941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AC, Lowry WE. 2015. Refining the role for adult stem cells as cancer cells of origin. Trends Cell Biol 25(1):11–20, PMID: 25242116, 10.1016/j.tcb.2014.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Wang D, Ma Y, Xu X, Zhu Z, Wang X, et al. 2015. Continuous activation of Nrf2 and its target antioxidant enzymes leads to arsenite-induced malignant transformation of human bronchial epithelial cells. Toxicol Appl Pharmacol 289(2):231–239, PMID: 26420645, 10.1016/j.taap.2015.09.020. [DOI] [PubMed] [Google Scholar]
- Yen YP, Tsai KS, Chen YW, Huang CF, Yang RS, Liu SH. 2010. Arsenic inhibits myogenic differentiation and muscle regeneration. Environ Health Perspect 118(7):949–956, PMID: 20299303, 10.1289/ehp.0901525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Z, Kondo S, Ha N, Boquete JP, Brunner M, Ueda R, et al. 2015. Accumulation of differentiating intestinal stem cell progenies drives tumorigenesis. Nat Commun 6:10219, PMID: 26690827, 10.1038/ncomms10219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Hsu YC. 2017. Emerging roles of transit-amplifying cells in tissue regeneration and cancer. Wiley Interdiscip Rev Dev Biol 6(5), PMID: 28670819, 10.1002/wdev.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Zhang H, Wang Q, Lal P, Carroll AM, de la Llera-Moya M, et al. 2010. Suppression of human prostate tumor growth by a unique prostate-specific monoclonal antibody F77 targeting a glycolipid marker. Proc Natl Acad Sci USA 107(2):732–737, PMID: 20080743, 10.1073/pnas.0911397107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.