Abstract
The Anaphase Promoting Complex/Cyclosome (APC/C) is a ubiquitin ligase that initiates anaphase and mitotic exit. The APC/C is activated by Cdc20 and inhibited by the mitotic checkpoint complex (MCC), which delays mitotic exit when the spindle assembly checkpoint (SAC) is activated. We previously identified apcin as a small molecule ligand of Cdc20 that inhibits APC/CCdc20 and prolongs mitosis. Here we find that apcin paradoxically shortens mitosis when SAC activity is high. These opposing effects of apcin arise from targeting a common binding site in Cdc20 required for both substrate ubiquitination and MCC-dependent APC/C inhibition. Furthermore, we found that apcin cooperates with p31comet to relieve MCC-dependent inhibition of APC/C. Apcin therefore causes either net APC/C inhibition, prolonging mitosis when SAC activity is low, or net APC/C activation, shortening mitosis when SAC activity is high, demonstrating that a small molecule can produce opposing biological effects depending on regulatory context.
Introduction
The Anaphase Promoting Complex/Cyclosome (APC/C) is a multi-subunit ubiquitin ligase (E3) that catalyzes ubiquitin transfer from associated E2s (Ube2C and Ube2S) to substrates, targeting them for degradation via the 26S proteasome1-3. The APC/C initiates anaphase by targeting securin for degradation and triggers mitotic exit by inducing degradation of the Cdk1 activator cyclin B1. APC/C activity in mitosis depends on binding of a co-activator, Cdc20, which recruits substrates and stimulates catalysis. Distinct surfaces on Cdc20 recognize specific sequence motifs in substrates, including the destruction box (D-box), KEN box, and ABBA motif1-3. The D-box receptor (DBR) of Cdc20 binds to the RxxL sequence of the D-box, using an acidic patch to recognize the basic arginine side chain and an adjacent hydrophobic pocket to accommodate the leucine side chain1-3.
Accurate chromosome segregation requires that APC/C not be activated until all chromosomes have become properly attached to the mitotic spindle. The mitotic checkpoint complex (MCC) is the effector of the spindle assembly checkpoint (SAC), which is triggered by insufficient kinetochore-microtubule attachments that arise during early stages of mitosis. MCC binds and inhibits APC/CCdc20 to ensure sufficient time for proper chromosome attachment prior to anaphase onset4,5. The MCC consists of BubR1, Mad2, Bub3, and Cdc20 itself, and thus the inhibited APC/CCdc20-MCC complex contains of two molecules of Cdc206, designated Cdc20-A (the co-activator) and Cdc20-M (in MCC). The MCC makes multiple contacts with APC/CCdc20 to inhibit its activity7,8, including the binding of D-box sequences in BubR1 to the Cdc20 DBR6,9-11. In addition, KEN-box and ABBA motifs in BubR1 interact with other sites on Cdc20 to efficiently inhibit APC/CCdc20 6,9. The formation of MCC is dynamic and regulated by a network of protein kinases and phosphatases, including the kinase Mps14,5. SAC inactivation and mitotic exit are promoted by disassembly of free MCC, mediated by p31comet and TRIP13, as well as dissociation of MCC from APC/CCdc20, which requires ubiquitination of Cdc20-M5. How these dynamic processes are integrated to determine the overall level of APC/CCdc20 activity in mitosis is not fully understood.
Through an unbiased screen in Xenopus extract, we previously identified two small molecule inhibitors of APC/C : TAME (tosyl-L-arginine methyl ester) and apcin (APC Inhibitor)12. Subsequent studies revealed that these compounds also inhibit human APC/C, and work by distinct mechanisms13-15. TAME binds Cdc27 and Apc8, subunits of APC/C, to block Cdc20 binding13,14,16. Apcin binds the leucine pocket of the Cdc20 DBR, interfering with association, ubiquitination and proteolysis of D-box-containing substrates15. TAME and apcin synergize to inhibit APC/CCdc20-dependent ubiquitination and proteolysis in mitotic Xenopus extract and block mitotic exit in human cells15.
Mitotic exit can also be inhibited by microtubule-targeting agents (MTAs), which cause defects in microtubule-kinetochore attachment, triggering MCC production, MCC-dependent APC/CCdc20 inhibition and a SAC-induced mitotic arrest. However, cells can prematurely exit from mitosis through a process known as mitotic slippage17-19. The rate of slippage varies across cell lines20 and blocking slippage by inhibiting APC/CCdc20 may potentiate the apoptotic effect of MTA-based cancer therapies 21,22. It has been shown that proTAME, the cell permeable form of TAME13, in combination with MTAs stabilizes cyclin B123, increases apoptosis23,24 and reduces mitotic slippage in cancer cells23. We hypothesized that apcin might also cooperate with MCC to inhibit APC/CCdc20 more robustly, preventing mitotic slippage. However, we found that apcin paradoxically induces mitotic slippage during a SAC-induced mitotic arrest. Using a reconstituted biochemical system and experiments in engineered cells, we provide a mechanism by which apcin causes this paradoxical effect.
Results
Apcin promotes slippage from a SAC-induced mitotic arrest
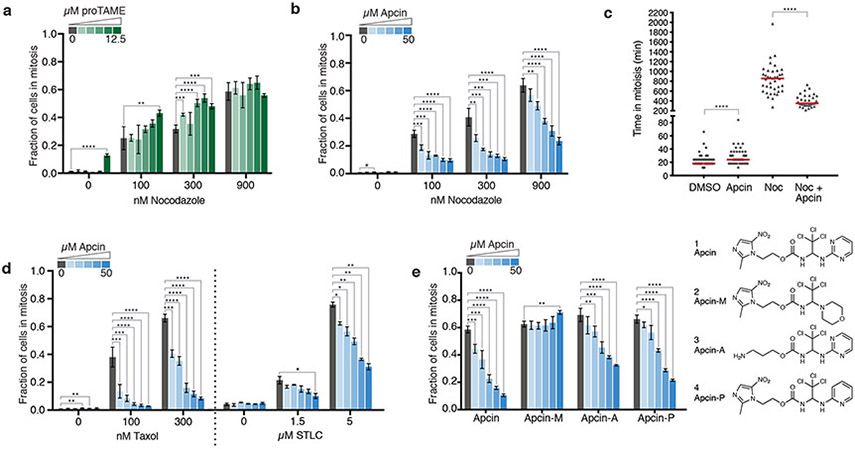
To determine whether apcin can cooperate with MTAs to strengthen mitotic arrest, we treated cells with nocodazole in combination with apcin or proTAME. We measured the fraction of cells in mitosis using a previously validated high-throughput fixed cell assay15. At low nocodazole concentrations, proTAME increased the fraction of cells in mitosis, but at high nocodazole concentrations, proTAME had no effect on mitotic fraction (Fig. 1a). Therefore, consistent with previous studies23, APC/C inhibition by proTAME enhances a SAC-dependent mitotic arrest. Surprisingly, apcin, in a dose-dependent manner, decreased the fraction of cells in mitosis across multiple nocodazole concentrations (Fig. 1b) in multiple cell lines (Supplementary Fig. 1). Time-lapse microscopy confirmed that apcin shortened mitotic duration in nocodazole-treated cells, from a median of 858 minutes to 351 minutes (Fig. 1c). Premature mitotic exit occurred in the absence of chromosome segregation and cell division, consistent with mitotic slippage17,18. In contrast, in the absence of nocodazole, apcin lengthened mitotic duration from 18 to 24 minutes (Fig. 1c), consistent with apcin’s ability to antagonize APC/CCdc20 15. Apcin also reduced mitotic fraction in cells treated with taxol or the kinesin-5 inhibitor S-trityl-L-cysteine (STLC) (Fig. 1d; Supplementary Fig. 1), indicating that apcin promotes mitotic slippage regardless of the mechanism of SAC activation. The ability of apcin derivatives to induce mitotic slippage paralleled their ability to bind Cdc20, as defined by previous structure-activity relationship analysis15. Specifically, apcin-M, which neither binds Cdc20 nor inhibits APC/CCdc20-dependent degradation, was unable to reduce mitotic fraction (Fig. 1e). In contrast, apcin-A and apcin-P, which retain the ability to bind Cdc20 and inhibit APC/CCdc20, though somewhat less effectively15, also reduced mitotic fraction, but not to the same extent as apcin (Fig. 1e). Together these results suggest that apcin binding to the leucine-pocket of the Cdc20 DBR is required to promote mitotic slippage. Because proTAME does not induce mitotic slippage, this effect seems to be a specific consequence of perturbing the Cdc20 DBR rather than a general consequence of APC/C inhibition.
Fig. 1. Apcin promotes slippage from a SAC-induced mitotic arrest.
a, Fraction of hTERT-RPE1 H2B-GFP cells in mitosis after 24-hour treatment with proTAME (0, 0.8, 1.5, 3.1, 6.2, 12.5 μM) +/− nocodazole. Mitotic fraction was determined using a high-throughput fixed cell assay. The mean of four biological replicates is shown; error bars represent standard deviation. Statistical significance was calculated using a two-tailed unpaired t test (* p-value < 0.05, ** p-value < 0.01, *** p-value < 0.001, **** p-value < 0.0001). b, Fraction of hTERT-RPE1 H2B-GFP cells in mitosis after 24-hour treatment with apcin (0, 3.1, 6.2, 12.5, 25, 50 μM) +/− nocodazole. Cells were analyzed as in a. c, hTERT-RPE1 H2B-GFP cells were treated with apcin (50 μM) and/or nocodazole (300 nM) and imaged via time-lapse microscopy for 24 hours. Each dot represents mitotic duration of a single cell (n = 30) quantified manually from nuclear envelope breakdown (NEB) to anaphase, slippage or death. Horizontal bars (red) represent median time in mitosis. **** p-value < 0.0001 calculated using a two-sided Mann-Whitney test. d, Fraction of hTERT-RPE1 H2B-GFP cells in mitosis after 24-hour treatment with apcin (concentrations as in b) +/− taxol or STLC. Cells were analyzed as in panel a. e, Fraction of hTERT-RPE1 H2B-GFP cells in mitosis after 24-hour treatment with 300 nM nocodazole and apcin derivatives (concentrations as in b). Cells were analyzed as in a. Structure of apcin derivatives is shown on the right. Exact p-values for a-e are shown in Supplementary Table 2. Experiments in a,b,d,e were repeated independently with similar results, with the exception of STLC in d, which was not repeated. The experiment in c was repeated twice independently and repeated once in three other cell lines showing similar results.
Apcin hastens cyclin B1 degradation prior to slippage
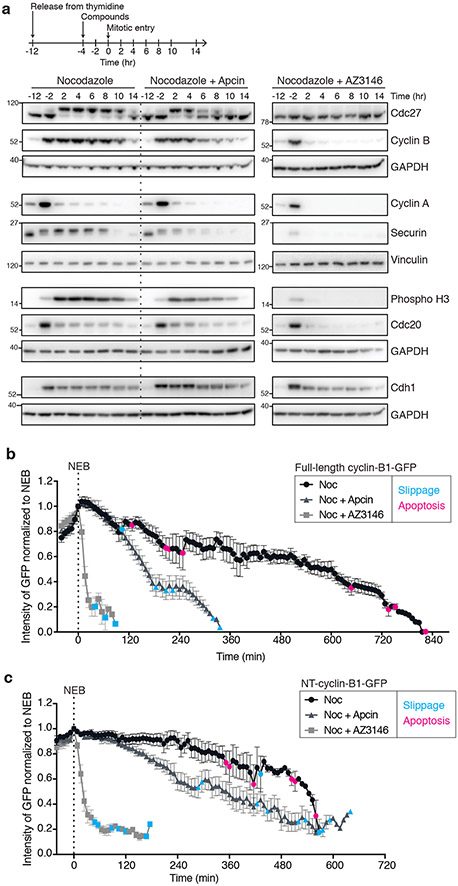
To characterize the mechanism by which apcin induces mitotic slippage, we first measured how apcin influences the timing of APC/C substrate degradation during slippage. Thymidine-synchronized cells were released into nocodazole in the presence of apcin or AZ314625, an Mps1 inhibitor that inhibits SAC activity by slowing MCC formation, and substrate levels were measured by immunoblotting. Nocodazole-treated cells retained persistent Cdc27 phosphorylation, phospho-H3, securin and cyclin B1 levels for up to 10 hours before exiting from mitosis (Fig. 2a, Supplementary Fig. 2a), whereas cyclin A levels declined within 2 hours of mitotic entry, consistent with the known insensitivity of cyclin A to SAC-dependent APC/C inhibition26,27. As expected, Mps1 inhibition caused an almost immediate reduction in cyclin B1 and securin levels with no observable Cdc27 phosphorylation (Fig. 2a). Apcin induced slippage more slowly, as indicated by Cdc27 dephosphorylation and a reduction in cyclin B1, securin, and phospho-H3 levels beginning 4–6 hours after mitotic entry (Fig. 2a). Apcin had little effect on Cdc20 levels, but increased Cdh1 abundance (Fig. 2a). However, Cdh1 knockdown did not alter apcin’s ability to decrease mitotic fraction (Supplementary Fig. 2b), indicating that Cdh1 is not required for apcin-induced mitotic slippage.
Fig. 2. Apcin hastens cyclin B1 degradation prior to slippage.
a, HeLa cells were thymidine synchronized and treated with nocodazole (100 nM) +/− apcin (25 μM) or AZ3146 (4 μM). Cells were collected at indicated times and extracts analyzed by immunoblotting. The same samples were analyzed on separate immunoblots with separate loading controls. Timing of mitotic entry was determined by examining Cdc27 phosphorylation at earlier time points (Supplementary Fig. 2a). Uncropped blots are shown in Supplementary Fig. 8. Measurement of Cdc27, cyclin B1, Cdc20, and Cdh1 levels were repeated twice across different time points with similar results. b, HCT116 cells were thymidine synchronized and treated with adenovirus expressing cyclin B1-GFP, followed by treatment with nocodazole (333 nM) +/− apcin (50 μM) or AZ3146 (4 μM). Cells were imaged for 24 hours by time-lapse fluorescence microscopy. The fluorescence of individual cells was measured, the intensity value at nuclear envelope breakdown (NEB) set to 1 and the mean and SEM for all cells were plotted (n = 10). Colored dots indicate time points at which at least one cell underwent apoptosis (pink) or mitotic slippage (blue). A two-sided F-test comparing the slopes of the transformed log (fluorescence intensity) values indicated that the rate of cyclin B1 degradation differed in a statistically significant manner between all paired conditions (p-value < 0.0001). The experiment was independently repeated twice with similar results. c, The same experiment was performed as in b, however cells were infected with an adenovirus expressing an N-terminal fragment of cyclin B1-GFP (NT-cyclin B1-GFP). A two-sided F-test comparing the slopes of the transformed log (fluorescence intensity) values indicated that the rate of NT-cyclin B1-GFP degradation differed in a statistically significant manner between all paired conditions (p-value < 0.0001). The experiment was not repeated. Exact p-values for b,c are shown in Supplementary Table 2.
To more carefully define the kinetics of cyclin B1 degradation during apcin-induced mitotic slippage, we monitored fluorescence intensity in cells expressing near-endogenous levels of cyclin B1-GFP (Supplementary Fig. 2c). Consistent with earlier studies19,28, we observed slow cyclin B1 degradation in cells treated with nocodazole alone (Fig. 2b), resulting primarily in apoptosis rather than mitotic exit. In apcin-treated cells, the rate of cyclin B1 degradation was accelerated, leading to mitotic exit rather than apoptosis (Fig. 2b). Importantly, apcin promoted a decrease in cyclin B1 levels before cells exited mitosis in multiple cell lines (Fig. 2b, Supplementary Fig. 2d,e), consistent with the idea that accelerated cyclin B1 degradation drives mitotic exit. Mps1 inhibition led to more rapid cyclin B1 degradation and exit from mitosis (Fig. 2b, Supplementary Fig. 2d). These experiments are consistent with a model in which apcin induces mitotic slippage by promoting premature cyclin B1 degradation in the face of an activated SAC.
Given that apcin blocks the Cdc20 DBR15, we were curious if apcin-induced cyclin B1 degradation relied upon its D-box-independent recruitment to APC/CCdc20. Cyclin B1-Cdk1 can be recruited to MCC-inhibited APC/CCdc20 independent of the D-box through binding of Cks proteins29. We therefore analyzed the degradation kinetics of an N-terminal fragment of cyclin B1 fused to GFP (NT-cyclin-B1-GFP) as this protein lacks cyclin boxes and therefore cannot bind Cdk1 or Cks proteins. Apcin enhanced degradation of NT-cyclin B1-GFP and promoted mitotic slippage in NT-cyclin B1-GFP expressing cells (Fig. 2c, Supplementary Fig. 2c), indicating that Cks-dependent recruitment to APC/C is not required for acceleration of cyclin B1 degradation by apcin. The cyclin B1 cyclin box domain is also required for its degradation mediated by CRL2ZYG−11, a ubiquitin ligase that can function redundantly with APC/CCdc20 to target cyclin B1 for degradation, particularly during mitotic slippage30. Since apcin promotes NT-cyclin B1-GFP degradation (Fig. 2c), apcin-induced mitotic slippage is unlikely to depend on CRL2ZYG−11-dependent degradation. Instead, these results suggest that apcin accelerates cyclin B1 degradation by somehow perturbing MCC-dependent APC/CCdc20 inhibition.
Apcin-induced mitotic slippage depends on APC/CCdc20
We suspected that apcin caused mitotic slippage by activating APC/C-dependent cyclin B1 degradation, despite the fact that apcin is a known inhibitor of APC/CCdc20 15. Therefore, we tested whether proTAME was sufficient to block slippage, since proTAME did not cause slippage on its own (Fig. 1a). In a fixed cell assay, in the absence of nocodazole, apcin and proTAME synergized to increase mitotic fraction, as previously reported15 (Fig. 3a). In the presence of nocodazole, proTAME blocked apcin’s ability to decrease mitotic fraction in a dose-dependent manner (Fig. 3a), indicating that apcin-induced mitotic slippage depends on APC/C activity. These findings suggest that apcin may antagonize the ability of the SAC to effectively inhibit APC/CCdc20.
Fig. 3. Apcin-induced mitotic slippage depends on APC/CCdc20.
a, Fraction of HCT116 cells in mitosis after 24-hour treatment with combinations of proTAME, apcin (0, 6.2, 12.5, 25, 50 μM) and nocodazole. Mitotic fraction was determined using a high-throughput fixed cell assay. The mean of four biological replicates is shown; error bars represent standard deviation. The experiment was not repeated. Statistical significance was calculated using a two-tailed unpaired t test (* p-value < 0.05, ** p-value < 0.01, *** p-value < 0.001, **** p-value < 0.0001). Exact p-values shown in Supplementary Table 2. b, hTERT-RPE1 H2B-GFP cells were treated with 10 nM control or Cdc20 siRNA for 24 hours, followed by combinations of nocodazole, apcin (0, 6.2, 12.5, 25 μM), and AZ3146 (4 μM) for 24 hours. The fraction of mitotic cells was determined using a high-throughput fixed cell assay. The mean of four biological replicates from two independent experiments is shown; error bars represent standard deviation. Statistical significance was calculated using a two-tailed unpaired t test (* p-value < 0.05, ** p-value < 0.01, *** p-value < 0.001, **** p-value < 0.0001). Exact p-values are shown in Supplementary Table 2.
Since Cdc20 is a co-activator of APC/C and apcin binds the Cdc20 DBR, we tested whether the effects of apcin were dependent on Cdc20 levels. We first examined the effects of apcin and Cdc20 knockdown in the absence of nocodazole. As previously reported31,32, Cdc20 knockdown in cycling cells was not sufficient to increase mitotic fraction (Fig. 3b). However, this level of Cdc20 knockdown (Supplementary Fig. 3) strongly sensitized cells to apcin’s ability to increase mitotic fraction (Fig. 3b). The ability of apcin to increase mitotic fraction was independent of the SAC since Mps1 inhibition did not affect the mitotic fraction (Fig. 3b). This finding suggests that high levels of Cdc20 may explain why apcin treatment alone is insufficient to strongly prolong mitotic duration in cycling cells.
We next tested how Cdc20 knockdown influences the ability of apcin to overcome a SAC-induced mitotic arrest. We found that Cdc20 knockdown abrogated apcin’s ability to reduce mitotic fraction (Fig. 3b), suggesting that apcin-induced mitotic slippage depends on Cdc20. In contrast, Cdc20 knockdown did not interfere with the ability of an Mps1 inhibitor to induce slippage (Fig. 3b), suggesting that apcin and Mps1 inhibition induce slippage by different mechanisms.
Apcin treatment does not affect MCC generation
In principle, apcin might induce mitotic slippage by affecting MCC formation or reducing MCC binding to APC/C. To test whether apcin affects MCC generation, we immunoprecipitated BubR1 from nocodazole-treated cells and measured the effect of apcin on the amount of associated MCC components Mad2, Cdc20 and Bub3. As expected, inhibition of MCC generation with combined use of Mps1 and Aurora B inhibitors, AZ3146 and Hesperidin, caused a strong reduction of Mad2 associated with BubR1. However, apcin had no effect (Fig. 4a). We also tested whether apcin influences BubR1 localization to unattached kinetochores, a necessary step in MCC generation4,5. In nocodazole-treated cells, BubR1 localized to kinetochores in a manner that required Mps1 activity (Fig. 4b, Supplementary Fig. 4a). In contrast, apcin had no effect on BubR1 localization (Fig. 4b). Thus, apcin does not seem to interfere with MCC generation.
Fig. 4. Apcin treatment does not affect MCC generation.
a, HeLa cells were thymidine synchronized and mitotic cells were collected after treatment with nocodazole (800 nM) and MG132 (15 μM), +/− apcin (50 μM) or AZ3146 (4 μM) + hesperidin (300 nM). BubR1 was immunoprecipitated from cellular extracts and binding of MCC components analyzed by immunoblot. The experiment was repeated independently with similar results. b, Immunofluorescence of BubR1 and centromeres (Crest) in HeLa cells treated with nocodazole (333 nM) and MG132 (10 μM), +/− apcin (50 μM) or AZ3146 (4 μM). Cells treated with siRNA against BubR1 (20 nM) were used as a positive control. Graph represents the mean quantification of BubR1 signal intensities co-localized with Crest. Error bars represent standard deviation (n = 5 cells with an average of 300 kinetochores per condition). **** p-value < 0.0001, ns p-value 0.385 calculated using a two-sided Mann-Whitney test. Exact p-values are shown in Supplementary Table 2. Representative images are shown. Scale bar, 10 μm. The experiment was not repeated. c, HCT116 cells were thymidine synchronized and mitotic cells were collected after treatment with nocodazole (800 nM) and MG132 (15 μM), +/− apcin (50 μM) or AZ3146 (4 μM) + hesperidin (300 nM). Cdc27 was immunoprecipitated and binding of MCC components analyzed by immunoblot. The experiment was repeated independently in HCT116 cells with similar results and repeated twice in HeLa cells with similar results. d, HCT116 cells were thymidine synchronized and mitotic cells were collected after treatment with nocodazole (300 nM) and MG132 (15 μM), +/− apcin (50 μM) or AZ3146 (4 μM). Cdc27 was immunoprecipitated and bound proteins were analyzed by TMT-based quantitative mass spectrometry. Graph represents mean quantification of APC/C subunits and MCC components, detected with more than 10 peptides, in three biological replicates; error bars represent standard deviation. Ratio of protein bound to Cdc27 beads was normalized to the average amount of protein detected in the sample treated with nocodazole alone. Empty bead control indicates the relative amount of protein detected when beads without Cdc27 antibody were used for immunoprecipitation. Asterisk (*) represents proteins that exceeded the threshold for significance and fold-change in volcano plots shown in Supplementary Fig. 4b. The experiment was not repeated. Uncropped images for a and c are shown in Supplementary Fig. 8.
To test whether apcin affects the amount of MCC bound to APC/C, we isolated APC/C by immunoprecipitating Cdc27 from nocodazole-treated cells and analyzed the effect of apcin or Mps1 inhibition on the pattern of associated MCC components. As predicted, the combined inhibition of Mps1 and Aurora B led to a reduction in the amount of associated MCC. However, apcin treatment had no measurable effect (Fig. 4c). We next used quantitative tandem-mass tag mass spectrometry to assess the amount of MCC associated with APC/C in apcin-treated cells, as well as identify any other changes in APC/C-associated proteins that might occur as a consequence of apcin treatment. As expected, Mps1 inhibition alone reduced the level of MCC bound to APC/C (Fig. 4d), but apcin did not, consistent with the results from the immunoblotting experiment. Furthermore, we did not identify other interpretable changes in APC/C-associated proteins that might explain the ability of apcin to induce mitotic slippage (Supplementary Fig. 4b; Supplementary Table 1). Together these findings suggest that apcin can induce mitotic slippage even in the absence of bulk changes in the amount of MCC bound to APC/C.
p31comet and apcin cooperate to reactivate APC/C
Because apcin induces a relatively slow rate of cyclin B1 degradation and mitotic slippage without affecting MCC generation or the bulk population of APC/C-MCC complexes, we suspected that an additional, limiting factor might be required for apcin-induced mitotic slippage. We therefore investigated a role for p31comet, a Mad2-binding protein implicated in MCC disassembly5. Previous work has shown that p31comet depletion slows mitotic slippage, whereas p31comet overexpression accelerates mitotic slippage, consistent with the possibility that limiting levels of p31comet can influence the duration of SAC-induced mitotic arrest33,34. Additionally, p31comet associates with APC/C-MCC complexes35-37, although the role of this interaction is not understood. We therefore depleted cells of p31comet using RNAi (Supplementary Fig. 5a), treated cells with apcin and nocodazole and quantified mitotic fraction. We found that p31comet was required for apcin to reduce mitotic fraction of nocodazole-treated cells (Fig. 5a). In contrast, mitotic exit induced by Mps1 inhibition did not require p31comet. Apcin did not affect p31comet levels during mitotic slippage (Supplementary Fig. 5b), suggesting that apcin-induced mitotic slippage is not due to elevated p31comet levels. Together these findings suggest that apcin-induced mitotic slippage depends on p31comet.
Fig. 5. p31comet and apcin cooperate to reactivate APC/C.
a, HCT116 cells were treated with 5 nM control or p31comet siRNA, followed by combinations of nocodazole, apcin (0, 6.2, 12.5, 25, 50 μM) or AZ3146 (0, 0.25, 0.5, 1, 2 μM) for 24 hours. The fraction of mitotic cells was determined using a high-throughput fixed cell assay. The mean of four biological replicates from two independent experiments is shown; error bars represent standard deviation. Statistical significance was calculated using a two-tailed unpaired t test (* p-value < 0.05, ** p-value < 0.01, *** p-value < 0.001, **** p-value < 0.0001). Exact p-values are shown in Supplementary Table 2. b, Reactions were carried out in the presence of APC/C, MCC containing 3Flag-Cdc20-M, 3Myc-His-Cdc20-A, Ube2S, Ube2C, E1, WT ubiquitin, ATP and/or apcin (500 μM) and/or p31comet (1 μM). All components (- substrate) were pre-incubated for 5 minutes followed by addition of fluorescently labeled cyclin A (CycA*). Ubiquitination was analyzed by fluorescence imaging. The experiment was repeated independently with similar results. c, APC/C carrying a Strep tag on APC4 was incubated with strep-tactin resin, 3Myc-His-Cdc20-A, MCC containing 3Flag-Cdc20-M, p31comet (1 μM), apcin (500 μM) or apcin-M (500 μM) for 1 hour and eluates were analyzed by immunoblotting. d, Quantification of experiment in c. Protein abundance was normalized to Cdc27. The fraction of bound protein was set to 1 for the minus compound condition. The mean of five independent experiments is shown; error bars represent SEM. Statistical significance was calculated using a two-tailed unpaired t test (* p-value < 0.05, ** p-value < 0.01, *** p-value < 0.001, **** p-value < 0.0001). e, MCC containing 3Flag-Cdc20-M was incubated with anti-FLAG-beads, 3Myc-His-Cdc20-A, p31comet (1 μM), apcin (500 μM) or apcin-M (500 μM) for 1 hour and eluates were analyzed by immunoblotting. f. Quantification of experiment in e. The mean of 5 independent experiments is shown. Quantification and representation is identical to d, except protein abundance was normalized to 3FLAG-Cdc20-M. Uncropped images for b, c, e are shown in Supplementary Fig. 8. Exact p-values for d,f are shown in Supplementary Table 2.
To understand the mechanism of apcin-induced mitotic slippage, we turned to a reconstituted ubiquitination system containing recombinant human APC/C, Cdc20 and MCC8,38. In this system, distinct epitope tags are used to distinguish free Cdc20 (Cdc20-A) from that which is present in MCC (Cdc20-M). We first demonstrated that apcin inhibits APC/CCdc20-dependent ubiquitination of cyclin A, securin, and an N-terminal fragment of cyclin B1 (cyclin B1-NTD), with strongest effects on cyclin B1-NTD (Supplementary Fig. 5c). Apcin-M, an inactive derivative, did not inhibit substrate ubiquitination (Supplementary Fig. 5d). Together these results suggest that this reconstituted system is sensitive to apcin in a manner that is consistent with our previous findings15.
We next tested how MCC, p31comet, and apcin influence substrate ubiquitination in this system. As expected, addition of MCC alone inhibited the ubiquitination of cyclin A (Fig. 5b), cyclin B1-NTD (Supplementary Fig. 5e) and securin (Supplementary Fig. 5f). Interestingly, inclusion of apcin led to a slight relief of MCC-dependent inhibition of cyclin A ubiquitination, which was further stimulated by addition of p31comet (Fig. 5b). Addition of p31comet alone had little effect (Fig. 5b). Similar results were observed with securin (Supplementary Fig. 5f), but effects on cyclin B1-NTD ubiquitination (Supplementary Fig. 5e) were more difficult to interpret due to the fact that apcin inhibits ubiquitination of cyclin B1-NTD more strongly than that of cyclin A or securin in the absence of MCC (Supplementary Fig. 5c). We also investigated the effect of apcin and p31comet on ubiquitination of Cdc20-M by APC/C, since this is an important mechanism for stimulating dissociation of MCC from APC/C5. We found that apcin strongly suppressed Cdc20-M ubiquitination with no added effect of p31comet (Supplementary Fig. 5g). Together these findings suggest that apcin and p31comet can cooperate to reduce MCC-dependent inhibition of APC/CCdc20, and that this effect occurs independent of MCC ubiquitination.
We next tested the possibility that apcin and p31comet promote APC/C-dependent substrate ubiquitination by reducing the amount of MCC bound to APC/C. The presence of apcin alone reduced the amount of MCC bound to APC/C (Fig. 5c,d), whereas the inactive derivative apcin-M had little effect. The addition of p31comet alone also reduced the amount of MCC bound to APC/C, though to a lesser extent than apcin. However, apcin in combination with p31comet strongly reduced the amount of MCC and p31comet bound to APC/C, whereas the inactive derivative apcin-M had little effect (Fig. 5c,d). Together these findings suggest that p31comet and apcin can cooperate to reduce MCC binding to the APC/C in this reconstituted system. Limiting availability of p31comet in cells may explain why apcin does not produce measurable decreases in the amount of MCC associated with APC/C in this context.
To determine whether apcin and p31comet cause dissociation of MCC before binding to APC/C, we assessed the integrity of MCC in the presence of Cdc20-A, apcin or p31comet (Fig. 5e,f). Apcin and/or p31comet had no significant effect on Mad2 binding to Cdc20-M, whereas p31comet slightly reduced BubR1 association (Fig. 5e,f). Furthermore, apcin did not affect the ability of p31comet to bind MCC. However, apcin and p31comet were each sufficient to strongly reduce MCC association with a second molecule of Cdc20 (Cdc20-A) (Fig. 5e,f; Supplementary Fig. 5h,i), with the combination of apcin and p31comet showing an even greater effect (Fig. 5e,f). Based on these findings, we conclude that the ability of apcin and p31comet to reduce MCC binding to APC/C does not arise from disassembly of MCC prior to APC/C binding. Instead, apcin and p31comet appear to alter the ability of MCC to bind Cdc20-A as well as the ability of MCC to bind APC/C.
Apcin-induced slippage requires a functional Cdc20 DBR
Our biochemical experiments support a model in which apcin induces mitotic slippage by interfering with MCC binding or inhibition of APC/CCdc20, rather than by inhibiting MCC generation. To test this model in cells, we took advantage of a mutation in the Cdc20 DBR (Cdc20D177A) that interferes with MCC-dependent APC/C inhibition, without affecting MCC generation6. Using our in vitro reconstituted system, we confirmed that this mutant abrogates MCC-dependent APC/C inhibition when present in Cdc20-A, but does not inhibit MCC assembly when present in Cdc20-M (Supplementary Fig. 6a,b). If apcin works by a similar mechanism, we predicted that apcin should fail to further accelerate slippage in cells expressing mutant Cdc20.
We generated a CRISPR knock-in cell line homozygous for Cdc20D177A and verified the knock-in via mass spectrometric analysis of Cdc20 (Supplementary Fig. 6c) and sequencing (Supplementary Fig. 6d). The knock-in cell line expressed Cdc20 at levels similar to wild-type cells (Supplementary Fig. 6e). Consistent with the fact that D177 lies outside the apcin binding pocket15 (Fig. 6a), apcin bound Cdc20D177A as efficiently as Cdc20WT (Supplementary Fig. 6f). During unperturbed division, Cdc20D177A cells spent less time in mitosis than wild-type cells, consistent with defective SAC-dependent APC/C inhibition (Supplementary Fig. 6g). Addition of apcin to asynchronous cells lengthened mitotic duration in the mutant background, indicating that apcin retains its ability to bind and inhibit Cdc20D177A (Supplementary Fig. 6g).
Fig. 6. Apcin-induced slippage requires a functional Cdc20 DBR.
a, Structure of apcin (blue, PDB 4N14)15, the substrate Hsl1 (yellow, PDB 5G04)16 or BubR1 (orange, PDB 5LCW)7 bound to Cdc20 (grey). Acidic residues of the Cdc20 DBR (red) and the RxxL D-box motif of substrate and BubR1 are noted. b, Fraction of Cdc20D177A knock-in cells in mitosis after 24-hour treatment with nocodazole and/or apcin (0, 6.2, 12.5, 25, 50 μM). Mitotic fraction was determined using a high-throughput fixed cell assay. Three separate Cdc20D177A knock-in clones were analyzed (2G2, 2B6 and 2B7). The mean of three biological replicates from two independent experiments is shown; error bars represent standard deviation. Statistical significance was calculated using a two-tailed unpaired t test (* p-value < 0.05, ** p-value < 0.01, *** p-value < 0.001, **** p-value < 0.0001). Exact p-values are shown in Supplementary Table 2. c, Cdc20WT and Cdc20D177A knock-in cells were treated with nocodazole +/− apcin and imaged via time-lapse microscopy for 24 hours. Each dot represents a single cell time in mitosis quantified from nuclear envelope breakdown (NEB) to anaphase, slippage or death. Horizontal bars (red) represent median time in mitosis (n = 30). **** p-value < 0.0001 calculated using a two-sided Mann-Whitney test. The experiment was repeated independently with a different clone with similar results. d, Cdc20D177A knock-in cells were treated with combinations of nocodazole (333 nM), apcin (50 μM), and/or AZ3146 (4 μM) and imaged via live-cell time-lapse microscopy for 24 hours. Experiment was analyzed as described in c. The experiment was not repeated. e, Cdc20WT and Cdc20D177A knock-in cells were thymidine synchronized and treated with adenovirus expressing cyclin B1-GFP and nocodazole (333 nM) +/− apcin (50 μM), and imaged for 24 hours by time-lapse fluorescence microscopy. The fluorescence of individual cells was measured and the intensity value at nuclear envelope breakdown (NEB) set to 1. The mean and SEM for all cells were plotted (n = 10). A two-sided F-test comparing the slopes of the transformed log (fluorescence intensity) values indicated that the rate of cyclin B1-GFP degradation differed in a statistically significant manner between all paired conditions (p-value ≤ 0.0001). The experiment was not repeated. f, Model summarizing the effect of apcin on APC/CCdc20 activity in an unperturbed mitosis. Apcin inhibits APC/CCdc20 activity in a dose-dependent manner, leading to increased mitotic duration. When Cdc20 levels are reduced by knock-down (KD), apcin further lowers APC/CCdc20 activity. g, Model summarizing the effect of apcin and Cdc20D177A mutation on APC/CCdc20 activity when the SAC is stimulated. When the SAC is active due to nocodazole treatment, APC/CCdc20 activity is strongly reduced (red). Addition of apcin at low concentrations increases APC/CCdc20 activity by antagonizing the SAC; however, at high concentrations, apcin reduces net APC/C activity by direct inhibition of APC/CCdc20. In contrast, the Cdc20D177A mutant (gold) is much less sensitive to the SAC and shows increased APC/CCdc20 activity compared to Cdc20WT. Since Cdc20D177A is already defective in MCC-dependent inhibition of APC/C, binding of apcin no longer increases APC/CCdc20 activity, but instead inhibits APC/CCdc20 activity. Exact p-values for c-e are shown in Supplementary Table 2.
We next examined how the presence of the Cdc20D177A mutation influenced the ability of cells to respond to nocodazole. The Cdc20D177A mutant strongly reduced mitotic fraction across a range of nocodazole doses (Fig. 6b), consistent with previous results of knockdown-replacement experiments6. Importantly, the addition of apcin did not further reduce mitotic fraction. Similar behaviors were observed in three independent Cdc20D177A knock-in clones (Fig. 6b). Additionally, the ability of Cdc20D177A to reduce mitotic fraction was independent of p31comet (Supplementary Fig. 6h). In time-lapse imaging experiments, the Cdc20D177A mutation induced rapid slippage in nocodazole, shortening mitotic duration from a median of 908 minutes in Cdc20WT cells to 128 minutes in Cdc20D177A cells (Fig. 6c). In nocodazole-treated Cdc20WT cells, apcin induced mitotic slippage, shortening mitotic duration to about 600 minutes. However, in nocodazole-treated Cdc20D177A cells, apcin did not accelerate slippage, but instead extended mitotic duration from a median of 128 minutes to 256 minutes (Fig. 6c). MCC continued to inhibit APC/CCdc20 activity in Cdc20D177A cells, as Mps1 inhibition further accelerated mitotic exit (Fig. 6d). We further characterized the ability of apcin to promote cyclin B1 degradation in nocodazole-treated cells expressing Cdc20WT or Cdc20D177A. Consistent with earlier knockdown replacement experiments6, we found that the rate of cyclin B1 degradation was strongly accelerated in Cdc20D177A cells compared to Cdc20WT (Fig. 6e). Whereas apcin accelerated cyclin B1 degradation in nocodazole-treated Cdc20WT cells, as described earlier, we found that apcin instead inhibited the rate of cyclin B1 degradation in nocodazole-treated Cdc20D177A cells (Fig. 6e). In contrast, Mps1 inhibition further accelerated the rate of cyclin B1 degradation in Cdc20D177A cells (Supplementary Fig. 6i). In summary, our findings suggest that in nocodazole-treated Cdc20WT cells, apcin relieves MCC-dependent inhibition of APC/CCdc20, accelerating cyclin B1 degradation and promoting mitotic slippage. However, when APC/CCdc20 is rendered less sensitive to MCC inhibition by the Cdc20D177A mutant, apcin instead slows the rate of cyclin B1 degradation and mitotic exit. Together these findings are consistent with a model in which apcin induces mitotic slippage by perturbing the DBR of Cdc20, reducing the ability of MCC to bind or inhibit APC/CCdc20, rather than by altering MCC levels.
Discussion
In this study, we have discovered that the direction of the biological effect of a small molecule can be inverted depending on the cellular regulatory context, resulting in paradoxical net activation of a pathway that would otherwise be inhibited. We show that apcin binding to Cdc20 can result in either net inhibition of APC/CCdc20, delaying mitotic exit when SAC activity is low, or net activation of APC/CCdc20, promoting mitotic exit when SAC activity is high. This context-dependent effect is specific to apcin’s ability to bind the DBR of Cdc20, as other mechanisms of inhibiting APC/CCdc20 such as treatment with proTAME or knockdown of Cdc20, do not exhibit context-dependent behavior.
When the SAC is not strongly engaged, apcin delays mitotic exit, exhibiting the predicted effect of an APC/CCdc20 inhibitor (Fig. 6f). However, the delay is brief, raising the question as to what limits apcin’s APC/C inhibitory effect on mitotic exit. We found that reducing Cdc20 levels strongly sensitizes cells to apcin’s ability to prolong mitosis. In a pharmacological context, Cdc20 therefore behaves like a “spare receptor”39, in which only a fraction of the total receptors is required for a full biological effect. Our results are consistent with previous RNAi-based experiments demonstrating that Cdc20 must be reduced to less than 5% of normal levels to significantly delay mitotic exit31. The ability of a recently-reported apcin-based proTAC to induce mitotic arrest and cell death may thus arise from its ability to both reduce Cdc20 levels and antagonize Cdc20 function40. Furthermore, reducing APC/CCdc20 activity with proTAME also strongly sensitizes cells to the ability of apcin to block mitotic exit15. Therefore, partially inhibiting APC/CCdc20 activity by treatment with proTAME or reducing Cdc20 levels is sufficient to enhance apcin’s ability to inhibit mitotic exit (Fig. 6f).
Because apcin efficiently blocks mitotic exit when APC/CCdc20 activity is partially inhibited by other means, we expected that apcin would also cooperate with MCC, an endogenous APC/CCdc20 inhibitor, to further delay mitotic exit. Instead we observed the paradoxical result that apcin promotes mitotic slippage by relieving MCC-dependent APC/C inhibition, resulting in premature APC/CCdc20 -dependent cyclin B1 degradation. Apcin does not fully restore APC/CCdc20 activity when the SAC is active, either because apcin does not fully relieve MCC-dependent APC/CCdc20 inhibition or because apcin also acts as an inhibitor of APC/CCdc20 activity (Fig. 6g). However, the fact that apcin accelerates cyclin B1 degradation and promotes mitotic slippage in SAC-arrested cells indicates that apcin must relieve MCC-dependent APC/CCdc20 inhibition to a greater extent than it inhibits APC/CCdc20 ubiquitin-ligase activity.
The ability of apcin to promote mitotic slippage depends on p31comet. We recapitulated apcin’s ability to cooperate with p31comet to relieve MCC-dependent APC/C inhibition in a reconstituted system, where we saw a reduction in MCC binding to APC/C. We also found that p31comet and apcin were each sufficient to reduce MCC binding to Cdc20-A. The ability of apcin to reduce binding of Cdc20-A to MCC is consistent with previous studies showing that the DBR in Cdc20-A is essential for binding a D-box in BubR16. In contrast, the ability of p31comet to hamper Cdc20-A association with MCC has not been reported and thus represents a new mechanism by which p31comet may antagonize SAC-dependent signaling. Analysis of reported structures7,41 suggests that MCC would have to undergo dramatic structural reorganization to accommodate binding of p31comet to Mad2, explaining why binding of p31comet to MCC may perturb binding of Cdc20-A.
The ability of apcin and p31comet to reduce MCC binding to Cdc20-A may generate a free pool of Cdc20-A that can bind and activate APC/C. However, the liberation of Cdc20-A is unlikely to fully account for rescue of substrate ubiquitination, since p31comet alone does not efficiently rescue substrate ubiquitination, despite its ability to strongly block the MCC-Cdc20-A interaction. Therefore, apcin may also bind the DBR of Cdc20-M, perhaps interfering with the binding of a second D-box of BubR1 to this site. Together our results suggest a model for cooperation between apcin and p31comet (Supplementary Fig. 7). Apcin and p31comet can each reduce the binding of MCC to Cdc20-A, making more Cdc20-A available to activate APC/C. Furthermore, the resulting formation of an APC/C-MCC-p31comet complex, instead of an APC/C-MCC-Cdc20-A complex, renders the system vulnerable to apcin, as binding of apcin to Cdc20-M may destabilize the APC/C-MCC-p31comet complex. This would generate free APC/C to make it available to bind free Cdc20-A, leading to net activation of APC/CCdc20.
In addition to these in vitro results, our study provides genetic evidence that apcin induces mitotic slippage by weakening MCC-dependent APC/CCdc20 inhibition due to binding the Cdc20 DBR. Apcin produces the same phenotype as a DBR mutant (Cdc20D177A)6, in which the SAC is weakened without affecting MCC formation. Importantly, this mutant inverts the direction of the biological effect of apcin in MTA-treated cells, as apcin inhibits rather than promotes mitotic slippage in this genetic background. In contrast, an inhibitor of MCC assembly (Mps1 inhibitor) still promotes mitotic exit in the mutant, indicating that the SAC remains partially functional and dependent on MCC formation. Since Cdc20D177A produces a strong SAC defect independent of p31comet, we propose that the Cdc20D177A mutation strongly destabilizes MCC-APC/CCdc20 to the point that apcin has no further effect on MCC-dependent inhibition of APC/CCdc20. This model explains why the primary effect of apcin is to prolong mitotic duration in cells expressing the Cdc20D177A mutation compared to wild-type cells (Fig. 6g).
Paradoxical activation of a cellular pathway by an inhibitor is a relatively rare phenomenon. Some opioids act as mixed agonist-antagonists, which bind to distinct receptors in different tissues to produce opposite effects42. Additionally, selective estrogen response modulators can act as antagonists in some tissues but agonists in others43. Alternatively, small molecule inhibitors can produce paradoxical effects as a consequence of mutations in a receptor. In the context of cancer, B-Raf inhibitors block the MAP kinase signaling pathway in cells that contain activating B-Raf mutations, but paradoxically activate the pathway in wild-type cells44. In contrast, apcin’s paradoxical effects on mitotic exit arise not from differences in receptor expression or mutation, but rather from the complexity of APC/CCdc20 regulation by the spindle checkpoint. Our findings therefore establish a new mechanism through which a small molecule, by targeting a single site on a dynamic protein interface, can lead to opposite biological effects depending on the regulatory context.
Online Methods
Cell Culture and Synchronization
HeLa, HCT116, hTERT-RPE1, A549 and U2OS cells (American Type Culture Collection, Manassas, VA) were cultured in a humidified incubator at 37 °C in the presence of 5% CO2. For hTERT-RPE1 and U2OS cells, stable cell lines expressing H2B–GFP were derived using described methods45. HeLa, HCT116 and A549 cells were grown in DMEM (10–013-CV, Corning) with 10% FBS. hTERT-RPE1 H2B-GFP cells were grown in DMEM/F12 (10–090-CV, Corning) with 10% FBS and 0.01 mg/ml hygromycin B. U2OS H2B-GFP cells were grown in McCoy’s 5A (10–050-CV, Corning) with 10% FBS.
Unless otherwise noted, HeLa and HCT116 cells were synchronized by treating with 2 mM thymidine for 18 hours, releasing for 8 hours and retreating for 18 hours. In all cellular experiments involving chemicals dissolved in DMSO, the final DMSO concentration was kept below 0.5%.
Immunoblot Analysis
Cell extracts were prepared in RIPA lysis buffer (150 mM NaCl, 50 mM Tris pH 8, 1 mM EDTA, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 0.1 mM Na3VO4, 10 mM NaF, and Pierce protease inhibitor tablet) or directly in NuPAGE LDS sample buffer (Thermo Fisher Scientific) with 200 mM DTT. Cell pellets lysed in LDS sample buffer were vortexed and boiled at 100 °C for 10 minutes. Extracts lysed in RIPA buffer were incubated on ice for 30 minutes and centrifuged at 20,000xg for 10 minutes. Protein concentrations were determined using the bicinchoninic acid (BCA) assay (Thermo Fisher Scientific). Supernatants were resuspended in NuPAGE SDS sample buffer + 200 mM DTT and heated at 100 °C for 5 minutes. Cell lysates (10–20 μg) were separated via SDS-PAGE using 4–12% Bis-Tris gels (Bio-Rad) and transferred to polyvinylidene difluoride (PVDF) membranes followed by immunoblotting as indicated.
Antibodies and Chemicals
Commercial antibodies used for western blotting were as follows: anti-Cyclin A (H-432: sc-751, Santa Cruz Biotechnology, discontinued) 1:200; anti-Bub3 (611730, BD Transduction Laboratories) 1:500; anti-Cdc20 (H-175: sc-8358, Santa Cruz Biotechnology, discontinued) 1:500; anti-Cdc27 (610455, BD Transduction Laboratories) 1:500; anti-Cdh1 (DH01: CC43, EMD Millipore) 1:500; anti-Cyclin B1 (H-433: sc-752, Santa Cruz Biotechnology, discontinued) 1:200; anti-GAPDH (6C5: ab8245, Abcam) 1:1000; anti-GFP (sc-8334, Santa Cruz Biotechnology, discontinued) 1:500; anti-Mad2 (A300–301A, Bethyl Laboratories) 1:1000; anti-p31comet (EPR9584: ab150363, Abcam) 1:1000; anti-phospho H3 (pSer10: 06–570, EMD Millipore) 1:1000; anti-Securin (DCS-280: sc-56207, Santa Cruz Biotechnology) 1:200; anti-Vinculin (7F9: sc-73614, Santa Cruz Biotechnology) 1:1000. Anti-BubR1 (4B12) was a gift from the McKeon Lab46 and used at 1:1000. Secondary antibodies used included anti-rabbit IgG-HRP (NA934; GE Healthcare) and anti-mouse IgG- HRP (NA931V; GE Healthcare).
Antibodies used in recombinant protein in vitro experiments that differ from those used in mammalian cell extracts include: anti-FLAG (OctA-Probe, H-5: sc-166355, Santa Cruz Biotechnology) 1:1000; anti-cMyc (9E10: sc-40, Santa Cruz Biotechnology) 1:500; anti-Mad2 (FL-205: sc-28261, Santa Cruz Biotechnology) 1:500. Fluorescent secondary antibodies used included anti-rabbit IgG DyLite® 488 (35552, Invitrogen) and anti-mouse IgG Alexa Fluor 633 (A-21052, Invitrogen).
For APC/C immunoprecipitation, anti-Cdc27 (AF3.1: sc-9972, Santa Cruz Biotechnology) was used; for BubR1 immunoprecipitation, anti-BubR1 (612502, BD Transduction Laboratories) was used; and for Cdc20 immunoprecipitation, anti-Cdc20 (H-7: sc-5296, Santa Cruz Biotechnology) was used. For immunofluorescence anti-BubR1 (A300–386A, Bethyl Laboratories) 1:1000 and anti-centromere protein (15–234-0001, Antibodies Inc.) 1:50 were used and secondary antibodies conjugated to Alexa Fluor 568 (goat anti-human IgG, A21090, Molecular Probes Invitrogen) or Alexa Fluor 647 (donkey anti-rabbit IgG, A31573, Molecular Probes Invitrogen) were diluted 1:1000.
Chemicals used were as follows: apcin (T0506–3874, Enamine), apcin-P (Amb2237944, Ambinter), apcin-M (Amb1395012, Ambinter), apcin and apcin-A were also synthesized by Sundia Meditech according to the methods previously described15, AZ3146 (sc-361114, Santa Cruz Biotechnology), hesperidin (422513–13-1, Calbiochem), proTAME (I-440, Boston Biochem), MG132 (474790, Calbiochem), nocodazole (31430–18-9, Sigma-Aldrich), STLC (L14384, Alfa Aesar), taxol (33069–62-4, Sigma-Aldrich), thymidine (T9250, Sigma-Aldrich).
Time-Lapse Imaging and Microscopy
For experiments measuring time in mitosis, cells were plated in 24 well coverslip-bottom plates (Greiner BioOne, 662892). After 24 hours cells were either treated directly with compound or subjected to double thymidine synchronization. If cells were synchronized, compound was added 3 hours after release from the second thymidine block. Imaging was initiated immediately after compound treatment.
For cyclin-B1-eGFP imaging experiments, HeLa, HCT116 Cdc20WT and Cdc20D177A cells were plated in 24 well coverslip-bottom plates (Greiner BioOne, 662892) 24 hours before double thymidine synchronization. Cells were treated with adenovirus expressing full length cyclin B1-EGFP or NT1−107-cyclin B1-EGFP 47 after the first thymidine block and maintained in virus until the end of the second thymidine block. One hour after release from the second thymidine block, imaging was initiated. After an additional two hours, imaging was paused and compound was manually added and imaging resumed. To reduce background fluorescence, cells were imaged in DMEM without phenol red (17–205-CV, Corning) with 10% FBS and 584 mg/L of L-glutamine (25–005-CI, Corning).
For all imaging experiments, plates were inserted into a covered OkoLab cage microscope incubator set to 37 °C supplied with humidified 5% CO2 and mounted onto a motorized microscope stage (Prior Proscan III). All images were collected on a Nikon Ti motorized inverted microscope with a 20x/0.75NA Plan Apo objective lens and the Perfect Focus System. EGFP fluorescence was excited with the Lumencor Spectra-X light engine, using a 470/30nm excitation filter and 515/30nm emission filter (Chroma). Images were acquired with Hamamatsu ORCA-ER cooled CCD camera controlled with MetaMorph image acquisition software. Phase contrast and/or fluorescence images were captured at 8-minute intervals for 24 hours.
Videos were manually analyzed using ImageJ. Mitotic duration was defined as the time from nuclear envelop breakdown (NEB) until anaphase, in the case of normal mitosis, or until ‘death’ as indicated by cytoplasmic blebbing or until ‘slippage’ as indicated by the appearance of a single interphase cell resulting after that cell was in a mitotic state for any duration. Cyclin-B1-eGFP abundance was quantified manually by measuring mean intensity of GFP signal starting 40 minutes before NEB and continuing until anaphase, ‘death’, or ‘bypass’. GFP intensity was corrected for background by subtracting background fluorescence intensity in every frame and normalized to the GFP intensity at NEB. In order to determine whether the rate of degradation was statistically significant between treatment conditions, normalized GFP intensities were transformed to log scale, re-plotted (not shown), fit to a linear regression starting at NEB (x = 0) and the slopes were compared using an F-test.
High-throughput image-based assay to measure mitotic fraction
A high-throughput fixed cell assay was adapted15 to measure the fraction of mitotic cells as follows. Asynchronous hTERT-RPE1 H2B-GFP, HeLa, A549 and U2OS H2B-GFP cells were plated using a Multidrop Combi Reagent Dispenser (Thermo Fisher Scientific) onto a black, clear bottom 384 well plate (3712, Corning) with four replicates. HCT116 Cdc20WT and Cdc20D177A cells were plated manually onto a black, clear bottom 96 well plate (3606, Corning) with two replicates. To ensure uniformity of knock-down, experiments involving RNAi treatment were plated onto 96 well plates (3606, Corning) with two replicates. To prevent evaporation, plates were sealed with breathable rayon sealing tape (241205, Nunc) for subsequent incubation. After 24 hours, the cells were treated with indicated concentrations of compound dissolved in DMSO. In the case of RNAi, cells were initially treated with siRNA for 24–30 hours before compound treatment. After 24 hours of compound treatment, cells were fixed and stained directly without wash steps at a final concentration of 10% formalin, 0.1% Triton X-100 and 0.33ug/mL Hoechst 33342 in DPBS. The plates were sealed with aluminum sealing tape (276014, Nunc) and incubated at room temperature overnight before imaging.
All plates were imaged using an ImageXpress Micro (Molecular Devices) high-throughput microscope, with a 10x objective at four positions per well controlled with metaXpress software. ImageJ was used to automatically identify single nuclei, count the number of nuclei and determine the maximum fluorescence intensity of each nucleus. Using the output files from ImageJ, Matlab generated the cumulative frequency curves of maximum fluorescence intensity for the cell population in each treatment condition. To separate mitotic cells from interphase cells, a fluorescence intensity threshold was set on the basis of the mitotic fraction in DMSO treated control wells. The fraction of mitotic cells is the fraction of cells above the set intensity threshold calculated using Matlab. Statistical significance was determined using an unpaired two-tailed t test (* p-value < 0.05, ** p-value < 0.01, *** p-value < 0.001, **** p-value < 0.0001). Every fixed cell assay experiment was repeated on a separate day with similar results, except for STLC in Fig 1d, Fig. 3a and Supplementary Fig 6h.
Small-Interfering RNA (siRNA)
Cells were transfected using RNAiMax (Invitrogen) with the following siRNAs: Non-Targeting Control siRNA #5 5 nM-25 nM (D-001210–05, Dharmacon), Cdc20 10 nM (CGAAAUGACUAUUACCUGAtt, Silencer Select, Life Technologies), Cdh1 10 nM (J-015377–08, Dharmacon), p31comet 5 nM (L-020817–01, Dharmacon), or BubR1 20 nM (GAUGGUGAAUUGUGGAAUAUU, ON-TARGETplus, Dharmacon). Cells were treated for 24 hours with Cdc20 or Cdh1 siRNA, 30 hours with p31comet siRNA and 48 hours with BubR1 siRNA.
Immunofluorescence and Confocal Microscopy
HeLa cells were grown on #1.5 coverslips. After 18 hours cells were transfected with BubR1 siRNA or left untreated. 48 hours later cells were treated with apcin (50 μM) and/or nocodazole (333 nM). MG132 (10 μM) and AZ3146 (4 μM) were added 4 hours after nocodazole treatment. Two hours later cells were pre-permeabilized in PHEM (60 mM piperazine-N,N′-bis(2-ethanesulfonic acid), 25 mM HEPES, 10 mM EGTA, and 4 mM MgSO4) with 0.5% Triton X-100 for 5 minutes and fixed in 4% formaldehyde in phosphate-buffered saline (PBS) for 15 minutes. After fixation, cells were thoroughly rinsed in PBS with 0.05% Tween 20 (PBST) and blocked with 3% BSA in PBS for 30 minutes. Cells were subsequently incubated with primary antibodies diluted in blocking solution for 2 hours at room temperature, washed in PBST and incubated with secondary antibodies for an additional 45 minutes at room temperature. After washing in PBST, DNA was stained using 2 μg/mL of Hoechst 33342 (Invitrogen) in PBS for 2 minutes. Coverslips were mounted in 0.1 M n-propyl gallate (Sigma-Aldrich) in 90% glycerol / 10% PBS. Additional staining controls included: primary antibody alone without secondary; secondary antibody alone without primary; Hoechst DNA staining without primary or secondary antibody; and individual primary/secondary antibody combinations for BubR1 and Crest.
Images were collected the same day as fixation/staining with a Yokogawa spinning disk confocal on an inverted Nikon Ti fluorescence microscope with 100x/1.4NA Plan Apo objective. Images were acquired with a Hamamatsu ORCA-ER cooled CCD camera controlled with MetaMorph image acquisition software. Z-series optical sections were collected with a step-size of 0.25 microns were collected for 10 cells for each condition. AlexaFluor 568 was excited with a 561nm laser and emission was collected with a 620/60nm filter (Chroma). AlexaFluor 647 was excited with a 642nm laser and emission was collected with a 700/75nm filter (Chroma). Laser intensity, exposure and step size were kept constant throughout acquisition. Z-series are displayed as maximum z-projections, and gamma, brightness, and contrast were adjusted (identically for compared image sets) using ImageJ and Imaris software. All immunofluorescence images shown were chosen to represent the mean quantified data.
Image datasets were collected using the same image acquisition settings including magnification, exposure time and laser power. Using Imaris 8.2 (Bitplane, Oxford Instruments), we measured the amount of colocalization between BubR1 and Crest in mitotic cells (identified based on DAPI intensity) (n=5). First, the BubR1 and Crest channels were background subtracted before detecting foci using the Spot Detection method with a spot size of 0.5 μm. Spot quality for Crest was maintained across datasets while all visible BubR1 foci were retained (on average 300 kinetochores as defined by Crest spots were analyzed). Using the colocalize spot function on Imaris, spots from BubR1 and Crest were deemed colocalized if they were within 0.6 μm (center-center) of each other. Parameters for colocalization were set based on nocodazole control where Crest and BubR1 are known to colocalize. In the colocalized spots, intensities of BubR1 spots were plotted. The mean of the BubR1 spots co-localized with Crest is graphed with standard deviation. Mann-Whitney statistical analysis was completed on the collection of co-located spots.
Immunoprecipitation
Cells were synchronized by double thymidine block in 2.5 mM (HCT116) or 2 mM (HeLa) thymidine. Nocodazole (333 nM- 800 nM) was added 6 hours after release from the second thymidine block. Five hours after nocodazole treatment a mitotic shake-off was performed. Mitotic cells were maintained in nocodazole and re-plated with MG132 (10 – 15 μM), apcin (50 μM), and/or AZ3146 (4 μM) and Hesperidin (300 nM). After 2 hours, a second mitotic shake off was performed to collect cells. Proteasome inhibitor (MG132) was used to prevent mitotic exit.
Cells for immunoprecipitation were lysed with HEPES buffer (20 mM HEPES pH 7.6 w/ KOH, 5 mM KCl, 1 mM DTT, 10 mM NaF, 1mM Na3VO4, and Pierce protease inhibitor tablet) with apcin (50 μM) or DMSO for 30 minutes on ice and clarified by a 20,000xg spin for 10 minutes. Protein complexes were immunoprecipitated from 0.5–1 mg of total protein for 1 hour at 4 °C with anti-Cdc27 (AF3.1) or anti-BubR1 antibody covalently coupled to Affi-Prep Protein A Resin (156–0006, BIO-RAD) using HEPES buffer for incubation and washing. For immunoblotting, protein complexes were eluted in NuPAGE LDS sample buffer (Thermo Fisher Scientific) with 50 mM DTT and heated to 100 °C for 5 minutes. For mass spectrometry, protein complexes were eluted in 4% SDS + 120 mM Tris pH 8 heated at 50 °C for 5 minutes.
TMT Mass-Spectrometry
The TMT labeling and subsequent mass spectrometry analysis was based on the SL-TMT sample processing strategy48. Briefly, proteins were precipitated via Trichloroacetic acid (TCA) precipitation. TCA was added to a final concentration of 20% v/v and samples were incubated at 4 °C overnight. Samples were centrifuged at 14,000 RPM for 15 minutes at 4 °C and subsequently washed twice in ice-cold 100% acetone and once in ice-cold 100% methanol. Samples were resuspended in 200 mM EPPS, pH 8.5 and digested at room temperature overnight with Lys-C protease (Wako Chemicals) at a 100:1 protein-to-protease ratio. Trypsin (Pierce Biotechnology) was then added at a 100:1 protein-to-protease ratio, incubated for 6 hours at 37 °C.
Tandem mass tag (TMT) isobaric reagents (Thermo Fisher Scientific) were dissolved in anhydrous acetonitrile to a final concentration of 20 μg/uL of which 7 μL was added to the peptides (~100 μg). Following incubation at room temperature for 1.5 hours, the reaction was quenched with hydroxylamine to a final concentration of 0.3% (v/v). The TMT-labeled samples were pooled at a 1:1 ratio across all samples. The pooled TMT-labeled peptide sample was vacuum centrifuged and fractionated using the Pierce High pH Reversed-Phase Peptide Fractionation Kit (Pierce, 84868). Twelve fractions were collected using: 7.5%, 10%, 12.5%, 15%, 17.5%, 20%, 22.5%, 25%, 27.5%, 30%, 35%, and 60% acetonitrile. Samples were pooled in pairs (every 6th sample was pooled) to generate 6 final fractions, acidified with 1% formic acid and vacuum centrifuged. Each fraction was desalted via StageTip, dried again via vacuum centrifugation, and reconstituted in 5% acetonitrile and 5% formic acid for LC-MS/MS processing.
All samples were analyzed on an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific) coupled to a Proxeon EASY-nLC 1200 liquid chromatography (LC) pump (Thermo Fisher Scientific). Peptides were separated on a 100 μm inner diameter microcapillary column packed with 35 cm of Accucore C18 resin (2.6 μm, 150 Å, Thermo Fisher Scientific). For each analysis, we loaded approximately 2 μg onto the column. Peptides were separated using a 120 minute gradient of 3 to 25% acetonitrile in 0.125% formic acid with a flow rate of 450 nL/min. Each analysis used an MS3-based TMT method49,50, which has been shown to reduce ion interference compared to MS2 quantification51. The scan sequence began with an MS1 spectrum (Orbitrap analysis, resolution 120,000, 400−1400 Th, automatic gain control (AGC) target 5E5, maximum injection time 100 ms). The top ten precursors were then selected for MS2/MS3 analysis. MS2 analysis consisted of: collision-induced dissociation (CID), quadrupole ion trap analysis, automatic gain control (AGC) 2E4, NCE (normalized collision energy) 35, q-value 0.25, maximum injection time 120 ms), and isolation window at 0.7. Following acquisition of each MS2 spectrum, we collected an MS3 spectrum in which multiple MS2 fragment ions are captured in the MS3 precursor population using isolation waveforms with multiple frequency notches. MS3 precursors were fragmented by HCD and analyzed using the Orbitrap (NCE 65, AGC 1.0E5, maximum injection time 150 ms, resolution was 50,000 at 400 Th). For MS3 analysis, we used charge state-dependent isolation windows: For charge state z=2, the isolation window was set at 1.3 Th, for z=3 at 1 Th, for z=4 at 0.8 Th, and for z=5 at 0.7 Th.
CRISPR/Cas9 Mediated Gene Editing
A 20 nt gRNA for CDC20 (GGCGCATCCAGGATACGGTC) was cloned into a pSpCas9(BB)-2A-Puro (PX459) V2.0 CRISPR/Cas9 vector (Addgene 62988) and confirmed by Sanger sequencing. A ssDNA Ultramer oligo (IDT) was used to introduce the D177A (GAT to GCC) mutation into CDC20 (TACAGCCAAAAGGCCACTCCTGGCTCCAGCCGGAAGACCTGCCGTTACATTCCTTCCCTGCCAGACCGTATCCTGgccGCGCCTGAAATCCGAAATGACTATTGTAAGTGCATCCTTATCCTCGCCTCATGCATGGAGAAAGAGGGCCTGGGA).
HCT116 cells (ATCC) were tested for mycoplasma contamination (Lonza, LT07–218) and were found negative. Lipofectamine 3000 (Invitrogen) was used without the p3000 reagent to co-transfect 1 μg of pX459 vector containing gRNA with 2 μL of a 10 μM oligo stock. A 24-hour pulse of puromycin (1 μg/mL) was performed 48 hours post transfection after splitting cells. Single cell clones were plated in 96 well format by limiting dilution 24 hours after puromycin selection. Clones were grown for a week, split and mutant clones were identified via Illumina MiSeq. In brief, two rounds of PCR were performed in a 96 well plate: 1). gene specific PCR with CDC20 primers and 2). indexing PCR with barcoding primers. Three homozygous knock-in Cdc20D177A clones were identified, expanded, and further confirmed via Sanger sequencing. Cdc20D177A CRISPR KI cell line is available upon request from the corresponding author.
CRISPR knock-in clones were also analyzed by mass spectrometry to determine the presence of the D177A peptide. In brief, asynchronous HCT116 Cdc20WT and Cdc20D177A cells were collected and lysed with HEPES buffer (20 mM HEPES pH 7.6 w/ KOH, 5 mM KCl, 1 mM DTT, 10 mM NaF, 1 mM Na3VO4, and Pierce protease inhibitor tablet) for 30 minutes on ice and clarified by a 20,000xg spin for 10 minutes. Protein complexes were immunoprecipitated from ~3 mg of total protein for 1 hour at 4 °C with anti-Cdc20 (H7) antibody covalently coupled to Affi-Prep Protein A Resin (156–0006, BIO-RAD) using HEPES buffer for incubation and washing. Protein complexes were eluted in NuPAGE LDS sample buffer (Thermo Fisher Scientific) with 50 mM DTT and heated to 100 °C for 5 minutes. Bound protein was run on an SDS-PAGE gel and Coomassie stained. A region of the gel containing Cdc20, as estimated by size, was excised for mass spectrometry analysis. Samples were processed using trypsin digestion according to the GeLC-MS/MS strategy as described previously52.
Mass spectrometry data were collected using a Q Exactive mass spectrometer (Thermo Fisher Scientific, San Jose, CA) coupled with an Accela600 liquid chromatography (LC) pump (Thermo Fisher Scientific) and a Famos Autosampler (LC Packings). Peptides were separated on a 100 μm inner diameter microcapillary column packed with ∼30 cm of Accucore C18 resin (2.6 μm, 150 Å, Thermo Fisher Scientific). For each analysis, we loaded ~0.5 μg onto the column. Peptides were separated using a 60 min method of 3 to 25% acetonitrile in 0.125% formic acid with a flow rate of ∼300 nL/min. The scan sequence began with an Orbitrap MS1 spectrum with the following parameters: resolution 70,000, scan range 300−1500 Th, automatic gain control (AGC) target 3 × 106, maximum injection time 250 ms, and centroid spectrum data type. We selected the top 20 precursors for MS2 analysis which consisted of high-energy collision dissociation (HCD) with the following parameters: resolution 17,500, AGC 1 × 105, maximum injection time 60 ms, isolation window 1.6 Th, normalized collision energy (NCE) 30, 2 × 104 intensity threshold, and centroid spectrum data type. Unassigned and singly charged species were excluded from MS2 analysis, while dynamic exclusion was set to automatic. Mass spectra were processed using a Sequest-based in-house software pipeline. Spectra were converted to mzXML using a modified version of ReAdW.exe. Database searching included only the Cdc20 human protein (Q12834). This database was concatenated with one composed of the sequence in the reversed order. Searches were performed using a 3 Da precursor ion tolerance and the product ion tolerance was set to 0.03 Da. Oxidation of methionine residues (+15.995 Da) was set as a variable modification. Sequence-assigned spectra were manually validated after filtering with criteria requiring a mass error within +/− 5 ppm, Xcorr >2, and tryptic cleavage on both ends of the peptide.
Cellular Thermal Shift Assay to Measure Apcin-Cdc20 Interaction
The cellular thermal shift assay (CETSA)53 was adapted to examine binding of apcin to endogenous Cdc20WT and Cdc20D177A in HCT116 cells. In brief, asynchronous HCT116 cells were treated with a dose range of apcin or apcin-M for 1 hour, collected via trypsinization and cell pellets were resuspended in apcin or apcin-M diluted in DPBS with protease inhibitor tablet (Pierce). The resuspended cell pellet was incubated at 59 °C for 3 minutes followed by cooling for 2 minutes at 22–24 °C. These incubation conditions were established in preliminary experiments as the temperature that yielded the greatest degree of Cdc20 precipitation that was rescued by apcin treatment, but not apcin-M treatment (data not shown). The lysates were flash frozen in liquid nitrogen and thawed at 22–24 °C twice. To separate the soluble fractions from precipitates lysates were centrifuged at 20,000 xg for 20 minutes. Supernatants were mixed with SDS sample buffer and analyzed by SDS-PAGE. The fraction of soluble Cdc20, quantified using ImageJ, was normalized to soluble Cdc20 in samples treated with the highest concentration of apcin and statistical analysis was performed using an unpaired, two-tailed t test (** p-value < 0.01, *** p-value < 0.001, **** p-value < 0.0001).
Reconstituted Ubiquitination and Immunoprecipitation
Recombinant APC/CWT and APC/C∆APC15 used in this study contain phospho-mimetic mutations and were purified as previously described38,54. N-terminally 3Myc-His-tagged Cdc20 and MCC containing N-terminally 3Flag-tagged Cdc20 (WT and D177A) were expressed in High Five insect cells, while His6-p31comet was expressed in E. coli. Purification was carried out using nickel affinity, cation exchange, and size exclusion chromatography.
Substrate ubiquitination assays were performed as described 8,38,55 with apcin or apcin-M titrated from 0.05 μM to 500 μM. Briefly, APC/C (90 nM), 3Myc-His-Cdc20 (0.15 μM), +/− MCC containing 3Flag-Cdc20 (0.25 μM), +/− p31comet (1 μM), +/− apcin or apcin-M (0.05 μM – 500 μM), prepared on ice, were mixed with E1 (0.1 μM), Ube2C (1 μM) and/or Ube2S (1 μM), WT ubiquitin (125 μM), and ATP-MgCl2 (5 mM), prepared on ice. After a 5 minute incubation at room temperature, fluorescently labeled substrate was added, including cyclin B1-NTD1-95, cyclin A or securin at indicated concentrations. Ubiquitination of substrate was analyzed at indicated time points by fluorescence imaging using a Typhoon FLA 9500 scanner.
Cdc20 ubiquitination assays were carried out as described8 with p31comet titrated from 1 μM to 10 μM. In brief, two separate mixtures containing 1) APC/CWT (90nM) or APC/C∆APC15 (90nM), 3Myc-His-Cdc20 (0.15 μM), MCC containing 3Flag-Cdc20 (0.25 μM), p31comet (1 μM), and apcin (500 μM) and 2) E1 (0.1 μM), Ube2C (0.5 μM) and/or Ube2S (0.5 μM), ATP-MgCl2 (5 mM), and methylated ubiquitin (125 μM) were mixed on ice and pre-incubated separately at 30 ˚C for 10 minutes, then combined to start the reaction and allowed to react for 10 min at 30 ˚C. Ubiquitination of 3Myc-His-Cdc20 (Cdc20-A) and 3Flag-Cdc20 (Cdc20-M) was detected using two-color immunoblot by primary antibodies conjugated with Alexa 647 and Alexa 488, respectively, and detected using a Typhoon FLA 9500.
For the in vitro APC/C binding assays, 90nM recombinantly expressed APC/C carrying a Strep tag on APC4 was added to 30 μL of strep-tactin resin. 3Myc-His-Cdc20, MCC containing 3Flag-Cdc20, p31comet and apcin were added sequentially at 150 nM, 250 nM, 1 μM, and 500 μM, respectively. For in vitro MCC binding assays, 250 nM recombinantly expressed MCC containing 3Flag-Cdc20 was added to 30 μL of anti-FLAG resin. 3Myc-His-Cdc20, p31comet and/or apcin were added sequentially at 150 nM, 0.125 μM - 4 μM, and 500 μM, respectively. For all binding experiments, binding buffer (20 mM Hepes pH 8.0, 200 mM NaCl, 0.5 % Tween-20) was used to bring the final volume to 60 μL. Samples were incubated on ice for an hour, washed with binding buffer, eluted with SDS PAGE sample buffer, boiled for 5 minutes and analyzed by immunoblot using fluorescent primary or secondary antibodies. Blots were scanned using a Typhoon FLA 9500 scanner. Five independent experiments were quantified in which protein levels were normalized to Cdc27 and the fraction of bound protein set to 1 for the condition in which no was drug was added. Error bars represent SEM and statistical significance was calculated using a two-tailed unpaired t test (* p-value < 0.05, ** p-value < 0.01, *** p-value < 0.001, **** p-value < 0.0001).
Data Availability
Figures with associated raw data include: Fig. 1a-e; Fig. 2b,c; Fig. 3a,b; Fig. 4b,d; Fig. 5a; Fig. 6b,c,d,e. The time-lapse and fixed cell imaging datasets generated during the current study are available from the corresponding author upon reasonable request. All mass spectrometry data generated during this study has been deposited to the ProteomeXchange Consortium via the PRIDE56 partner repository with the dataset identifier PXD013786.
Code Availability
Matlab code used to calculate mitotic fraction is available upon request from the corresponding author.
Supplementary Material
Acknowledgements
We extend our gratitude in memoriam of J. Hoyt, who first observed the phenotype of apcin-induced premature mitotic exit in the presence of nocodazole. We thank X. Zeng for contributing to early investigations on the mechanism of the apcin-induced mitotic slippage and for comments on the manuscript. We thank T. Mitchison and D. Finley for helpful discussions. We thank the Nikon Imaging Facility at Harvard Medical School for assistance with time-lapse microscopy, the Image and Data Analysis Core at Harvard Medical School for assistance in immunofluorescence and time-lapse microscopy analysis, the ICCB-Screening Facility at Harvard Medical School for assistance with high-throughput imaging in fixed cell experiments, and the Harvard Medical School Cell Biology Initiative for Molecular Trafficking and Neurodegeneration for help with MiSeq CRISPR knock-in cell line verification. This work was supported by NIH grant R35GM127032 to R.W.K. T.B. was supported by NSF DGE-1650116 and NIH T32GM00857. N.G.B. was supported by NIH R35GM128855 and UCRF. B.A.S., N.G.B. and M.Y. were supported by ALSAC/St. Jude and NIH 5P30CA021765 (St. Jude Cancer Center).
Footnotes
Competing Financial Interests
There is a patent on this work, filed by Harvard University on behalf of K.L.S. and R.W.K.
References
- 1.Peters J The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol 7, 644–656 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Alfieri C, Zhang S & Barford D Visualizing the complex functions and mechanisms of the anaphase promoting complex/cyclosome (APC/C). Open Biol. 7, 170204 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watson ER, Brown NG, Peters JM, Stark H & Schulman BA Posing the APC/C E3 ubiquitin ligase to orchestrate cell division. Trends Cell Biol. 29, 117–134 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Musacchio A & Salmon ED The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol 8, 379–393 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Liu S-T & Zhang H The mitotic checkpoint complex (MCC): looking back and forth after 15 years. AIMS Mol. Sci 3, 597–634 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Izawa D & Pines J The mitotic checkpoint complex binds a second CDC20 to inhibit active APC/C. Nature 517, 631–634 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alfieri C et al. Molecular basis of APC/C regulation by the spindle assembly checkpoint. Nature 536, 431–436 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamaguchi M et al. Cryo-EM of mitotic checkpoint complex-bound APC/C reveals reciprocal and conformational regulation of ubiquitin ligation. Mol. Cell 63, 593–607 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Di Fiore B, Wurzenberger C, Davey NE & Pines J The mitotic checkpoint complex requires an evolutionary conserved cassette to bind and inhibit active APC/C. Mol. Cell 64, 1144–1153 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burton JL & Solomon MJ Mad3p, a pseudosubstrate inhibitor of APCCdc20 in the spindle assembly checkpoint. Genes Dev. 21, 655–667 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sewart K & Hauf S Different functionality of Cdc20 binding sites within the mitotic checkpoint complex. Curr. Biol 27, 1213–1220 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Verma R Ubistatins inhibit proteasome-dependent degradation by binding the ubiquitin chain. Science 306, 117–120 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Zeng X et al. Pharmacologic inhibition of the anaphase-promoting complex induces a spindle checkpoint-dependent mitotic arrest in the absence of spindle damage. Cancer Cell 18, 382–395 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeng X & King RW An APC/C inhibitor stabilizes cyclin B1 by prematurely terminating ubiquitination. Nat. Chem. Biol 8, 383–392 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sackton KL et al. Synergistic blockade of mitotic exit by two chemical inhibitors of the APC/C. Nature 514, 646–649 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang S et al. Molecular mechanism of APC/C activation by mitotic phosphorylation. Nature 533, 260–264 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kung AL, Sherwood SW & Schimke RT Cell line-specific differences in the control of cell cycle progression in the absence of mitosis. Proc. Natl. Acad. Sci 87, 9553–9557 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andreassen PR & Margolis RL Microtubule dependency of p34cdc2 inactivation and mitotic exit in mammalian cells. J. Cell Biol 127, 789–802 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brito DA & Rieder CL Mitotic checkpoint slippage in humans occurs via cyclin B destruction in the presence of an active checkpoint. Curr. Biol 16, 1194–1200 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gascoigne KE & Taylor SS Cancer cells display profound intra- and interline variation following prolonged exposure to antimitotic drugs. Cancer Cell 14, 111–122 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Huang H, Shi J, Orth JD & Mitchison TJ Evidence that mitotic exit is a better cancer therapeutic target than spindle assembly. Cancer Cell 16, 347–358 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manchado E et al. Targeting mitotic exit leads to tumor regression in vivo: modulation by Cdk1, Mastl, and the PP2A/B55α,δ phosphatase. Cancer Cell 18, 641–654 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Giovinazzi S, Bellapu D, Morozov VM & Ishov AM Targeting mitotic exit with hyperthermia or APC/C inhibition to increase paclitaxel efficacy. Cell Cycle 12, 2598–2607 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crawford LJ, Anderson G, Johnston CK & Irvine AE Identification of the APC/C co-factor FZR1 as a novel therapeutic target for multiple myeloma. Oncotarget 7, 1–5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hewitt L et al. Sustained Mps1 activity is required in mitosis to recruit O-Mad2 to the Mad1–C-Mad2 core complex. J. Cell Biol 190, 25–34 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.den Elzen N & Pines J Cyclin A Is destroyed in prometaphase and can delay chromosome alignment and anaphase. J. Cell Biol 153, 121–136 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geley S et al. Anaphase-promoting complex/cyclosome-dependent proteolysis of human cyclin A starts at the beginning of mitosis and is not subject to the spindle assembly checkpoint. J. Cell Biol 153, 137–147 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clute P & Pines J Temporal and spatial control of cyclin B1 destruction in metaphase. Nat. Cell Biol 1, 82–87 (1999). [DOI] [PubMed] [Google Scholar]
- 29.Van Zon W et al. The APC/C recruits cyclin B1-Cdk1-Cks in prometaphase before D box recognition to control mitotic exit. J. Cell Biol 190, 587–602 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balachandran RS et al. The ubiquitin ligase CRL2ZYG11 targets cyclin B1 for degradation in a conserved pathway that facilitates mitotic slippage. J. Cell Biol 215, 151–166 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wolthuis R et al. Cdc20 and Cks direct the spindle checkpoint-independent destruction of cyclin A. Mol. Cell 30, 290–302 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Baumgarten AJ, Felthaus J & Wäsch R Strong inducible knockdown of APC/CCdc20 does not cause mitotic arrest in human somatic cells. Cell Cycle 8, 643–646 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Ma HT, Chan YY, Chen X, On KF & Poon RYC Depletion of p31comet protein promotes sensitivity to antimitotic drugs. J. Biol. Chem 287, 21561–21569 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Díaz‐Martínez LA et al. Genome‐wide siRNA screen reveals coupling between mitotic apoptosis and adaptation. EMBO J. 33, 1960–1976 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Westhorpe FG, Tighe A, Lara-Gonzalez P & Taylor SS p31comet-mediated extraction of Mad2 from the MCC promotes efficient mitotic exit. J. Cell Sci 124, 3905–3916 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mansfeld J, Collin P, Collins MO, Choudhary JS & Pines J APC15 drives the turnover of MCC-CDC20 to make the spindle assembly checkpoint responsive to kinetochore attachment. Nat. Cell Biol 13, 1234–1243 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Varetti G, Guida C, Santaguida S, Chiroli E & Musacchio A Homeostatic control of mitotic arrest. Mol. Cell 44, 710–720 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Qiao R et al. Mechanism of APC/CCDC20 activation by mitotic phosphorylation. Proc. Natl. Acad. Sci 113, E2570–E2578 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu BT The competitive and noncompetitive antagonism of receptor-mediated drug actions in the presence of spare receptors. J. Pharmacol. Toxicol. Methods 29, 85–91 (1993). [DOI] [PubMed] [Google Scholar]
- 40.Chi J. (Jack) et al. A novel strategy to block mitotic progression for targeted therapy. EBioMedicine 49, 40–54 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang M et al. p31comet blocks Mad2 activation through structural mimicry. Cell 131, 744–755 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoskin PJ & Hanks GW Opioid agonist-antagonist drugs in acute and chronic pain states. Drugs 41, 326–344 (1991). [DOI] [PubMed] [Google Scholar]
- 43.McDonnell DP & Wardell SE The molecular mechanisms underlying the pharmacological actions of ER modulators: implications for new drug discovery in breast cancer. Curr. Opin. Pharmacol 10, 620–628 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karoulia Z, Gavathiotis E & Poulikakos PI New perspectives for targeting RAF kinase in human cancer. Nat. Rev. Cancer 17, 676–691 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-Only References
- 45.Sigoillot FD et al. A time-series method for automated measurement of changes in mitotic and interphase duration from time-lapse movies. PLoS One 6, e25511 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taylor SS & McKeon F Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell 89, 727–735 (1997). [DOI] [PubMed] [Google Scholar]
- 47.Bentley AM, Normand G, Hoyt J & King RW Distinct sequence elements of cyclin B1 promote localization to chromatin, centrosomes, and kinetochores during mitosis. Mol. Biol. Cell 18, 4847–4858 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Navarrete-Perea J, Yu Q, Gygi SP & Paulo JA Streamlined tandem mass tag (SL-TMT) protocol: an efficient strategy for quantitative (phospho)proteome profiling using tandem mass tag-synchronous precursor selection-MS3. J. Proteome Res 17, 2226–2236 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ting L, Rad R, Gygi SP & Haas W MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat. Methods 8, 937–940 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haas W et al. MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal. Chem 86, 7150–7158 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paulo JA, O’Connell JD & Gygi SP A triple knockout (TKO) proteomics standard for diagnosing ion interference in isobaric labeling experiments. J. Am. Soc. Mass Spectrom 27, 1620–1625 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paulo JA Sample preparation for proteomic analysis using a GeLC-MS/MS strategy. J. Biol. Methods 3, 45 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jafari R et al. The cellular thermal shift assay for evaluating drug target interactions in cells. Nat. Protoc 9, 2100–2122 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Weissmann F et al. biGBac enables rapid gene assembly for the expression of large multisubunit protein complexes. Proc. Natl. Acad. Sci 113, E2564–E2569 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brown NG et al. Dual RING E3 architectures regulate multiubiquitination and ubiquitin chain elongation by APC/C. Cell 165, 1440–1453 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Perez-Riverol Y et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Figures with associated raw data include: Fig. 1a-e; Fig. 2b,c; Fig. 3a,b; Fig. 4b,d; Fig. 5a; Fig. 6b,c,d,e. The time-lapse and fixed cell imaging datasets generated during the current study are available from the corresponding author upon reasonable request. All mass spectrometry data generated during this study has been deposited to the ProteomeXchange Consortium via the PRIDE56 partner repository with the dataset identifier PXD013786.