Abstract
We studied admission and dynamic demographic, hematological and biochemical co-variates in 1449 hospitalized subjects with coronavirus infectious disease-2019 (COVID-19) in five hospitals in Wuhan, Hubei province, China. We identified two admission co-variates: age (Odds Ratio [OR] = 1.18, 95% Confidence Interval [CI] [1.02, 1.36]; P = 0.026) and baseline D-dimer (OR = 3.18 [1.48, 6.82]; P = 0.003) correlated with an increased risk of death in persons with COVID-19. We also found dynamic changes in four co-variates, Δ fibrinogen (OR = 6.45 [1.31, 31.69]; P = 0.022), Δ platelets (OR = 0.95 [0.90–0.99]; P = 0.029), Δ C-reactive protein (CRP) (OR = 1.09 [1.01, 1.18]; P = 0.037), and Δ lactate dehydrogenase (LDH) (OR = 1.03 [1.01, 1.06]; P = 0.007) correlated with an increased risk of death. The potential risk factors of old age, high baseline D-dimer, and dynamic co-variates of fibrinogen, platelets, CRP, and LDH could help clinicians to identify and treat subjects with poor prognosis.
Subject terms: Infectious diseases, Risk factors
Introduction
Most people with coronavirus infectious disease-2019 (COVID-19) have mild to moderate symptoms and recover after the appropriate medical intervention(s). However, 15–32 percent develop severe or critical COVID-19 with a case-fatality rate of 1–15% [1–6]. There are few data of hematological abnormalities in persons with COVID-19 [7–13]. We studied hematological co-variates in 1449 hospitalized persons with COVID-19 in five hospitals in Wuhan, China, interrogating correlations of admission parameters with COVID-19 outcomes.
Methods
Study design and subjects
We studied subjects in seven centers of five hospitals of Union Hospital (Central Hospital, Union West Hospital, and Union Tumor Hospital), Wuhan Central Hospital, General Hospital of Central Theater Command, PLA, Wuhan Third Hospital, and Wuhan Jin-Yin-Tan Hospital between January 20, and April 4, 2020. Subjects were studied on admission for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-infection by quantitative real-time reverse transcriptase-polymerase chain reaction (qRT-PCR) of nasal and pharyngeal swabs and/or blood test for anti-SARS-CoV-2 IgG/IgM antibodies using a colloidal-gold-based 2019-nCoV IgG/IgM Detection Kit (Nanjing Vazyme Medical Technology, Nanjing, China). COVID-19 was diagnosed according to World Health Organization interim guidance and the Novel Coronavirus Pneumonia Diagnosis and Treatment Program of the National Health Commission of China [14, 15]. Severity of COVID-19 was graded as follows: (1) mild; mild clinical symptoms, no pneumonia on lung CT; (2) common: fever, cough and lung CT with pneumonia; (3) severe: respiratory distress (respiratory rate > 30 min−1, oxygen saturation (O2Sat) ≤ 93 percent at rest and/or ratio of arterial oxygen partial pressure to fractional inspired oxygen ≤300 mmHg (PaO2/FIO2); and (4) critical: aforementioned criteria of respiratory failure receiving mechanical ventilation, shock, and/or organ failure other than lung and/or intensive care unit (ICU) hospitalization [12, 15]. Subjects were treated as described below or according to Chinese government guidelines [15–18]. The study was approved by the Ethics Committees of Union Hospital (2020-0095) and of Wuhan Central Hospital (2020-007). Written and oral informed consent from subjects was waived by the Ethics Committees.
Data collection
Electronic medical records of subjects including epidemiological, demographic, and laboratory data. Outcomes included: (1) death; (2) alive with two successive negative SARS-Cov-2 qRT-PCR tests; (3) interval from symptoms to hospital admission, discharge, or death; (4) negative qRT-PCT test; and (5) interval from admission to COVID-19 progression. Data collection forms were reviewed and verified independently by two researchers. A third researcher adjudicated discordances.
Statistical analysis
Categorical variables were described as frequency rates and percentages and continuous variables by median and interquartile range (IQR). Significance was tested by Kruskal–Wallis test or the Fisher exact test. A two-sided P < 0.05 was considered significant. Locally weighted regression and smoothing scatterplot was performed to equate a smooth curve about the relationship between time and values of laboratory parameters. Uni- and multivariable logistic regression models were used to interrogate co-variates associated with in-hospital death. Analyses were not been adjusted for multiple comparisons. Data were analyzed as of April 4, 2020.
Results
Baseline co-variates corelated with death from COVID-19
We analyzed data from 3559 consecutive subjects. One thousand seven hundred were excluded because of missing data of SARS-CoV-2 qRT-PCR or anti-SARS-CoV-2 IgG/IgM, 410 with co-morbidities and/or receiving drugs that would potentially affect bone marrow function (Supplementary Fig. 1). The resulting study cohort was 1449 subjects.
Seven hundred and thirty-three subjects (51%) were male. Median age was 57 years (IQR, 42–66 years), 576 (40%) were 60–79 years and 66 (5%), ≥80 years. Common signs and symptoms on admission included dry (485; 43%) or productive cough (551; 38%), fatigue (531; 37%), and shortness of breath (520; 37%). Eighty three subjects (6%) were smokers and 71 (6%), health care providers. Four subjects were exposed in the Hunan Wholesale Seafood Market and 94 (6%), close contact with persons with SARS-CoV-2-infection. Twenty nine subjects (2%) had mild COVID-19, 956 (66%), moderate, 347 (24%), severe and 117 (8%), critical. Lung computed tomography (CT) scan showed bilateral pneumonia in 1215 (87%), ground-glass opacity in 1041 (75%), patchy shadows in 579 (42%), and consolidation in 242 (18%). Eight hundred and twenty-six (57%) subjects had ≥1 abnormal liver function tests. One hundred and forty-four (10%), acute respiratory distress syndrome (ARDS) and 392 (27%), bacterial co-infections. Other complications had <10% frequencies including heart, acute kidney abnormalities, septic shock, and multiple organ failure. One hundred and fifty-six subjects (14%) received high-flow nasal cannula oxygen, 98 (7%), noninvasive ventilation (no intubation), 54 (4%), invasive ventilation (intubation), and four extracorporeal membrane oxygenation (ECMO). Median ICU stay was 12 days (IQR, 5–21 days). Median intervals from symptom onset to a negative SARS-CoV-2 qRT-PCR RNA, progression and death or discharge were 22 days (IQR 17–28 days), 10 days (IQR 7–14 days), and 30 days (IQR 23–37 days).
At the time of data lock 1327 (92%) subjects were alive and discharged; 122 (8%), died (Table 1). Subjects who died were significantly older than survivors (median 69 versus 55 years; P < 0.001) and more likely male (74% versus 48%; P < 0.001), with more frequent fatigue (54% versus 35%; P < 0.001), chills (26% versus 15%; P < 0.001), productive cough (50% versus 37%, P = 0.006), shortness of breath (66% versus 35%; P < 0.001), a higher temperature on admission (median 36.8 °C; IQR 36.5–37.8 °C versus median 36.6 °C; IQR 36.4–37.0 °C; P < 0.001), bilateral pneumonia (95% versus 86%; P = 0.01), and/or consolidation (35% versus 16%, P < 0.001) on lung CT scan but similar dry cough (44% versus 43%, P = 0.824), diarrhea (both 13%; P = 0.897), and myalgia (20% versus 17%; P = 0.502).
Table 1.
Baseline characteristics of survivors and non-survivors with COVID-19.
| Total n = 1449 |
Alive n = 1327 |
Died n = 122 |
P value | |
|---|---|---|---|---|
| Characteristics | ||||
| Age, median (IQR), years | 57 (42, 66) | 55 (41, 65) | 69 (63, 78) | <0.001 |
| Age distribution | ||||
| <40 years | 306 (21) | 305 (23) | 1 (0.8) | |
| 40–59 years | 501 (35) | 481 (36) | 20 (16) | |
| 60–79 years | 576 (40) | 499 (38) | 77 (63) | |
| ≥80 years | 66 (5) | 42 (3) | 24 (20) | |
| Female sex | 716 (49) | 684 (52) | 32 (26) | <0.001 |
| Smoking history | 83 (6) | 66 (5) | 17 (15) | <0.001 |
| Health care provider | 71 (6) | 69 (7) | 2 (2) | 0.038 |
| Exposure history | 0.026 | |||
| Huanan Seafood Market | 4 (0.28) | 2 (0.15) | 2 (2) | |
| Close contact with patients | 94 (6) | 89 (7) | 5 (4) | |
| Signs and symptoms | ||||
| Temperature (°C) | 36.7 (36.4, 37.1) | 36.6 (36.4, 37.0) | 36.8 (36.5, 37.8) | <0.001 |
| Shortness of breath | 520 (37) | 456 (35) | 64 (66) | <0.001 |
| Dry cough | 485 (43) | 437 (43) | 48 (44) | 0.824 |
| Wet cough | 551 (38) | 491 (37) | 60 (50) | 0.006 |
| Fatigue | 531 (37) | 466 (35) | 65 (54) | <0.001 |
| Nausea or vomiting | 88 (6) | 83 (6) | 5 (4) | 0.43 |
| Diarrhea | 186 (13) | 170 (13) | 16 (13) | 0.897 |
| Chill | 226 (16) | 194 (15) | 32 (26) | <0.001 |
| Runny nose | 30 (2) | 28 (2) | 2 (1.6) | 0.999 |
| Myalgia | 255 (18) | 231 (17) | 24 (20) | 0.502 |
| Headache | 89 (6) | 84 (6) | 5 (4) | 0.43 |
| Staging | <0.001 | |||
| Mild | 29 (2) | 29 (2) | 0 (0) | |
| Moderate | 956 (66) | 951 (72) | 5 (4) | |
| Severe | 347 (24) | 332 (25) | 15 (12) | |
| Critical | 117 (8) | 15 (1) | 102 (84) | |
| Imaging features | ||||
| Bilateral pneumonia | 1215 (87) | 1117 (86) | 98 (95) | 0.01 |
| Consolidation | 242 (18) | 207 (16) | 35 (35) | <0.001 |
| Ground-glass opacity | 1041 (75) | 974 (76) | 67 (67) | 0.059 |
| Patchy shadows | 579 (42) | 532 (41) | 47 (47) | 0.218 |
| Complications | ||||
| ARDS | 144 (10) | 37 (3) | 107 (88) | <0.001 |
| Bacterial infections | 392 (27) | 288 (22) | 104 (85) | <0.001 |
| Septic shock | 45 (4) | 1 (0.1) | 44 (40) | <0.001 |
| Acute kidney injury | 49 (3) | 7 (0.5) | 42 (34) | <0.001 |
| Cardiac injury | 125 (9) | 58 (4) | 67 (55) | <0.001 |
| Liver damage | 826 (57) | 721 (54) | 105 (86) | <0.001 |
| Gastrointestinal bleeding | 20 (1) | 4 (0.3) | 16 (13) | <0.001 |
| Coagulopathy | 28 (2) | 0 (0) | 28 (23) | <0.001 |
| Multiple organ failure | 72 (5) | 3 (0.2) | 69 (57) | <0.001 |
| Treatments | ||||
| Antibiotics | 1203 (85) | 1084 (83) | 119 (98) | <0.001 |
| Antimycotics | 44 (3) | 17 (1) | 27 (22) | <0.001 |
| Oseltamivir | 604 (42) | 564 (43) | 40 (33) | 0.037 |
| Umifenovir | 1099 (76) | 1004 (76) | 95 (78) | 0.688 |
| Lopinavir and Ritonavir | 276 (24) | 232 (23) | 44 (40) | <0.001 |
| Interferon | 297 (21) | 269 (20) | 28 (23) | 0.483 |
| Corticosteroids | 576 (40) | 471 (35) | 105 (86) | <0.001 |
| Intravenous immunoglobin | 381 (28) | 314 (25) | 67 (55) | <0.001 |
| High-flow nasal cannula oxygen therapy | 156 (14) | 59 (6) | 97 (88) | <0.001 |
| Noninvasive mechanical ventilation | 98 (7) | 21 (2) | 77 (63) | <0.001 |
| Invasive mechanical ventilation | 54 (4) | 7 (0.5) | 47 (39) | <0.001 |
| ECMO | 4 (0.4) | 1 (0.1) | 3 (3) | 0.003 |
| Outcomes | ||||
| ICU admission | 63 (4) | 23 (2) | 40 (33) | <0.001 |
| Time from illness onset to ICU admission, median (IQR), days | 14 (10, 19) | 10 (8, 18) | 15 (12, 20) | 0.141 |
| ICU length of stay, median (IQR), days | 12 (5, 21) | 11 (8, 21) | 12 (4, 21) | 0.619 |
| Time from illness onset to repeated negatively tests of SARS-CoV-2, median (IQR), days | 22 (17, 28) | 22 (17, 28) | 19 (12, 26) | 0.066 |
| Time from illness onset to admission, median (IQR), days | 10 (7, 15) | 10 (7, 16) | 10 (7, 12) | 0.017 |
| Time from illness onset to progression, median (IQR), days | 10 (7, 14) | 10 (6, 14) | 12 (9, 19) | 0.001 |
| Time from illness onset to outcome, median (IQR), days | 30 (23, 37) | 30 (24, 38) | 21 (15, 29) | <0.001 |
| Time from diagnosis to outcome, median (IQR), days | 19 (13, 27) | 20 (13, 27) | 11 (5, 17) | <0.001 |
| Time from admission to outcome, median (IQR), days | 18 (13, 23) | 18 (13, 23) | 11 (6, 20) | <0.001 |
IQR interquartile ranges, ARDS acute respiratory distress syndrome, ECMO extracorporeal membrane oxygenation, ICU intensive care unit, SARS-CoV-2 severe acute respiratory syndrome coronavirus 2.
Median interval from symptom(s) onset to admission was 10 days in survivors (IQR 7–16 days) versus 10 days (IQR 7–12 days; P = 0.017) in non-survivors. Median interval from admission to progression in survivors was briefer (10 days [IQR 6–14 days] versus 12 days [IQR 9–19; P = 0.001). Median interval from onset to ICU admission was 14 days (IQR 10–19 days) and median ICU stay, 12 days (IQR 5–21 days) with no significate difference between survivors and non-survivors (P = 0.141 and P = 0.619). Subjects who died were more likely to have complications (122 [100%] versus 741 [56%]; P < 0.001) including acute respiratory distress syndrome (ARDS; 107 [88%] versus 37 [3%]; P < 0.001), liver function test abnormalities (105 [86%] versus 721 [54%]; P < 0.001) and bacterial infections (104 [85%] versus 288 [22%]; P < 0.001). More subjects dying received high-flow nasal cannula oxygen therapy (97 [88%] versus 59 [6%]; P < 0.001). Four subjects received ECMO including one survivor.
Blood hematological co-variates of survivors and non-survivors
We first compared admission hematological co-variates between survivors and non-survivors. Subjects who died had a higher median WBC (8 × 10E + 9/L [IQR 6–11 × 10E + 9/L] versus 5 × 10E + 9/L [IQR 4–7 × 10E + 9/L]; P < 0.001), higher median neutrophils (7 × 10E + 9/L [IQR 5–10 × 10E + 9/L] versus 3 × 10E + 9/L [IQR 2–4 ×10E + 9/L]; P < 0.001), lower median lymphocytes (0.5 × 10E + 9/L [IQR 0.4–0.8 × 10E + 9/L] versus 1.2 × 10E + 9/L [IQR 0.9–1.7 × 10E + 9/L]; P < 0.001) and lower median platelets (166 × 10E + 9/L [IQR 109–223 × 10E + 9/L] versus 208 × 10E + 9/L [IQR 164–268 × 10E + 9/L]; P < 0.001) compared with survivors (Table 2).
Table 2.
Blood hematological co-variates of survivors and non-survivors with COVID-19.
| N | Total | Alive | Died | P value | |
|---|---|---|---|---|---|
| Hemoglobin, g/L (115–150) | |||||
| Baseline | 1437 | 129 (119–139) | 128 (119–139) | 131 (122–143) | 0.068 |
| Max | 1101 | 132 (122–142) | 131 (126–146) | 136 (122–142) | 0.049 |
| Min | 1101 | 119 (108–131) | 120 (110–131) | 105 (78–126) | <0.001 |
| White blood cell, ×10E + 9/L (3.5–10) | |||||
| Baseline | 1420 | 5 (4–7) | 5 (4–7) | 8 (6–11) | <0.001 |
| Max | 1421 | 7 (5–9) | 7 (5–8) | 16 (12–21) | <0.001 |
| Min | 1421 | 5 (4–6) | 5 (4–6) | 6 (4–9) | <0.001 |
| Neutrophil, ×10E + 9/L (1.8–6.3) | |||||
| Baseline | 1417 | 3 (2–5) | 3 (2–4) | 7 (5–10) | <0.001 |
| Max | 1421 | 5 (3–7) | 4 (3–6) | 14 (11–20) | <0.001 |
| Min | 1421 | 3 (2–4) | 3 (2–4) | 5 (3–8) | <0.001 |
| Lymphocyte, ×10E + 9/L (1.1–3.2) | |||||
| Baseline | 1440 | 1.2 (0.8–1.6) | 1.2 (0.9–1.7) | 0.5 (0.4–0.8) | <0.001 |
| Min | 1411 | 1.0 (0.7–1.4) | 1.1 (0.8–1.5) | 0.3 (0.2–0.5) | <0.001 |
| Monocyte, ×10E + 9/L (0.1–0.6) | |||||
| Baseline | 1408 | 0.4 (0.3–0.53) | 0.4 (0.3–0.5) | 0.3 (0.2–0.5) | <0.001 |
| Max | 1419 | 0.5 (0.4–0.7) | 0.5 (0.4–0.7) | 0.5 (0.4–0.8) | 0.305 |
| Min | 1419 | 0.3 (0.2–0.4) | 0.3 (0.3–0.4) | 0.2 (0.1–0.3) | <0.001 |
| Platelet, ×109 E + 9/L (125–350) | |||||
| Baseline | 1415 | 206 (159–264) | 208 (164–268) | 166 (109–223) | <0.001 |
| Max | 1420 | 258 (204–325) | 263 (208–331) | 190 (134–255) | <0.001 |
| Min | 1420 | 176 (135–224) | 180 (143–226) | 80 (39–147) | <0.001 |
Data are presented as medians (interquartile ranges, IQR). p values were calculated by Mann–Whitney U test, χ² test, or Fisher’s exact test, as appropriate.
Blood lymphocyte subsets
We next analyzed baseline blood lymphocyte subsets between subjects who died and survivors (Table 3). Subjects who died had lower median CD3-positive cells (140 × 10E + 9/L [IQR 75–190 × 10E + 9/L] versus 381 × 10E + 9/L [IQR 2–918 × 10E + 9/L]; P = 0.023), CD3-positive/CD4-positive cells (71 × 10E + 9/L [IQR 46–107 × 10E + 9/L] versus 227 × 10E + 9/L [IQR 1–487 × 10E + 9/L]; P = 0.036), CD3-positive/CD8-positive cells (49 × 10E + 9/L [IQR 14–64 × 10E + 9/L] versus 141 × 10E + 9/L [IQR, 1–308 × 10E + 9/L]; P = 0.023) and lower median proportions of CD3-positive cells (59% [IQR 50–67%] versus 72% [IQR 63–78%]; P < 0.001), median proportions of CD3-positive/CD8-positive cells (14% [IQR 10–18%] versus 24% [IQR 19–30%]; P < 0.001) and median proportions of B cells (16% [IQR 10–28%] versus 12% [IQR 9–17%]; P = 0.022). There were no significant differences in median proportions of CD3-positive/CD4-positive T- or natural-killer (NK)-cells, nor in concentrations of B cells or NK-cells between survivors and non-survivors.
Table 3.
The lymphocyte subsets of peripheral blood in of survivors and non-survivors with COVID-19.
| Lymphocyte subset | N | Total | Alive | Died | P value |
|---|---|---|---|---|---|
| CD3+ (%) | 579 | 72 (62–78) | 72 (63–78) | 59 (50–67) | <0.001 |
| CD3 concentration × 10E + 9/L | 246 | 359 (2–901) | 381 (2–918) | 140 (75–190) | 0.023 |
| CD3+CD4+ (%) | 579 | 41 (33–48) | 41 (33–48) | 37 (28–48) | 0.443 |
| CD3+CD4+ concentration × 10E + 9/L | 246 | 200 (1–481) | 227 (1–487) | 71 (46–107) | 0.036 |
| CD3+CD8+ (%) | 579 | 23 (18–30) | 24 (19–30) | 14 (10–18) | <0.001 |
| CD3+CD8+ concentration × 10E + 9/L | 246 | 119 (1–302) | 141 (1–308) | 49 (14–64) | 0.023 |
| NK cell (%) | 415 | 10 (6–17) | 10 (6–17) | 10 (5–12) | 0.29 |
| NK cell concentration × 10E + 9/L | 246 | 76 (0.4–185) | 77 (0.4–188) | 41 (19–102) | 0.453 |
| B lymphocyte (%) | 415 | 13 (9–18) | 12 (9–17) | 16 (10–28) | 0.022 |
| B lymphocyte concentration × 10E + 9/L | 246 | 96 (0.3–178) | 98 (0.3–188) | 55 (22–91) | 0.136 |
| CD4+/CD8+ ratio | |||||
| Max | 577 | 2 (1.4–2.6) | 1.9 (1.4–2.6) | 3.0 (1.9–4.5) | <0.001 |
| Min | 489 | 1.6 (1.2–2) | 1.57 (1.2–2) | 1.6 (1.3–2.3) | 0.336 |
Data are median (IQR), n (%), or n/N (%). Cell count at presentation (cells/ul). p values were calculated by Mann–Whitney U test, χ2 test, or Fisher’s exact test, as appropriate.
NK cell natural killer cell
Clotting co-variates
Baseline and maximum values of prothrombin time, activated partial thromboplastin time, and D-dimer concentrations were significantly higher in subjects who died compared with survivors. In contrast, fibrinogen concentration was higher at baseline in subject who died (median 4.3 g/L [IQR, 3.2–5.2 g/L] versus 3.6 g/L [IQR 2.9–4.5 g/L); P < 0.001) but had lower minimum values (2.6 g/L [IQR 1.7–3.9 g/L] versus 3.2 g/L [IQR 2.6–3.9 g/L]; P < 0.001; Table 4).
Table 4.
Clotting factor levels of survivors and non-survivors with COVID-19.
| N | Total | Alive | Died | P value | |
|---|---|---|---|---|---|
| PT, s (11–16) | |||||
| Baseline | 1055 | 13 (12–13) | 13 (12–13) | 14 (13–15) | <0.001 |
| Max | 1035 | 13 (12–14) | 13 (12–14) | 17 (14–20) | <0.001 |
| APTT, s (28–43.5) | |||||
| Baseline | 1055 | 34 (30–38) | 34 (30–37) | 35 (300–40) | 0.019 |
| Max | 1289 | 34 (30–38) | 34 (30–37) | 40 (34–50) | <0.001 |
| D-dimer, mg/L (<0.5) | |||||
| Baseline | 1239 | 0.4 (0.2–0.9) | 0.4 (0.2–0.8) | 3.6 (0.9–8) | <0.001 |
| Max | 1262 | 0.6 (0.2–1.6) | 0.5 (0.2–1) | 8 (6–8) | <0.001 |
| Fibrinogen, g/L (2–4) | |||||
| Baseline | 1304 | 3.7 (2.9–4.6) | 3.6 (2.9–4.5) | 4.3 (3.2–5.2) | <0.001 |
| Min | 976 | 3.2 (2.5–3.9) | 3.2 (2.6–3.9) | 2.6 (1.7–3.9) | <0.001 |
PT prothrombin time, APTT activated partial thromboplastin time.
Inflammatory and biochemical co-variates
Median admission concentrations of C-reactive protein (CRP) (93 mg/L [IQR 58–125 mg/L] versus 9 mg/L [IQR 3–30 mg/L]; P < 0.001), procalcitonin (0.2 ng/ml [IQR 0.12–0.6 ng/ml] versus 0.05 ng/ml [IQR 0.04–0.1 ng/ml]; P < 0.001) and lactate dehydrogenase (LDH) (470 U/L [IQR 359–599 U/L] versus 199 U/L [IQR 161–258 U/L]; P < 0.001) were significantly higher in subjects who died compared with survivors (Table 5). Subjects who died were more likely to have abnormal heart, liver, and/or kidney function and to have higher admission median concentrations of interleukin-6 (IL-6; 71 pg/ml [IQR 29–442 pg/ml] versus 9 pg/ml [IQR 4–30 pg/ml]; P < 0.001) and interleukin-10 (IL-10; 11 pg/ml IQR [6–30 pg/ml] versus 4 pg/ml [IQR 3–5 pg/ml]; P < 0.001). There were no significant differences in concentrations of interleukins-2 (IL-2) or -4 (IL-4), tumor necrosis factor (TNF)-α, or interferon (IFN)-γ between survivors and non-survivors.
Table 5.
Biochemical parameters and inflammatory cytokines of survivors and non-survivors with COVID-19.
| N | Total | Alive | Died | P value | |
|---|---|---|---|---|---|
| CRP, mg/L (<8) | |||||
| Baseline | 1046 | 11 (3, 44) | 9 (3, 30) | 93 (58, 125) | <0.001 |
| Max | 1063 | 14 (4, 55) | 11 (3, 38) | 140 (110, 181) | <0.001 |
| Procalcitonin, ng/ml (<0.5) | |||||
| Baseline | 1273 | 0.05 (0.05, 0.1) | 0.05 (0.04, 0.1) | 0.2 (0.12, 0.6) | <0.001 |
| Max | 1065 | 0.07 (0.04, 0.1) | 0.06 (0.04, 0.1) | 1.2 (0.4, 4) | <0.001 |
| LDH, U/L (109–245) | |||||
| Baseline | 1338 | 207 (165, 283) | 199 (161, 258) | 470 (359, 599) | <0.001 |
| Max | 1354 | 215 (174, 302) | 207 (169, 271) | 707 (509, 1154) | <0.001 |
| Ferritin, ng/ml (4.6–204) | 231 | 542 (226, 1207) | 446 (191, 906) | 1584 (1196, 2000) | <0.001 |
| ALT, U/L (5–35) | 1429 | 39 (22, 68) | 37 (21, 66) | 62 (34, 150) | <0.001 |
| AST, U/L (8–40) | 1428 | 31 (22, 49) | 30 (22, 44) | 68 (45, 143) | <0.001 |
| Total bilirubin, μmol/L (5.1–19) | 1245 | 14 (10, 19) | 13 (10, 17) | 25 (16, 39) | <0.001 |
| Creatine kinase, U/L (26–140) | 1151 | 87 (54, 150) | 81 (52, 130) | 253 (104, 656) | <0.001 |
| BNP, pg/ml (<100) | 627 | 50 (14, 160) | 39 (12, 115) | 454 (116, 1377) | <0.001 |
| Myoglobin, ng/ml (<140) | 718 | 32 (21, 60) | 29 (21, 49) | 476 (147, 1200) | <0.001 |
| Troponin I, ng/L (<26.2) | 830 | 3 (1, 10) | 2 (0.9, 7) | 212 (48, 1011) | <0.001 |
| BUN, mmol/L (2.9–8.2) | 1414 | 5 (4, 6) | 5 (4, 6) | 14 (8, 23) | <0.001 |
| Scr, μmol/L (44–106) | 1414 | 70 (59, 83) | 69 (59, 81) | 100 (71, 211) | <0.001 |
| IL-2, pg/ml (0.1–4.1) | 344 | 3 (3, 4) | 3 (3, 4) | 3 (3, 5) | 0.849 |
| IL-4, pg/ml (0.1–3.2) | 380 | 3 (2, 4) | 3 (2, 4) | 2 (2, 4) | 0.804 |
| IL-6, pg/ml (0.1–2.9) | 659 | 10 (4, 37) | 9 (4, 30) | 71 (29, 442) | <0.001 |
| IL-10, pg/ml (0.1–5) | 380 | 4 (3, 6) | 4 (3, 5) | 11 (6, 30) | <0.001 |
| TNF α, pg/ml (0.1–23) | 380 | 3 (2, 6) | 4 (2, 6) | 3 (2, 4) | 0.183 |
| IFN γ, pg/ml (0.1–18) | 380 | 3 (2, 4) | 3 (2, 4) | 3 (2, 4) | 0.88 |
CRP C-reactive protein, LDH lactate dehydrogenase, ALT alanine aminotransferase, AST aspartate aminotransferase, BNP brain natriuretic peptide, BUN blood urea nitrogen, Scr serum creatinine, IL interleukin, TNF tumor necrosis factor, IFN interferon.
Dynamic changes in hematological co-variate
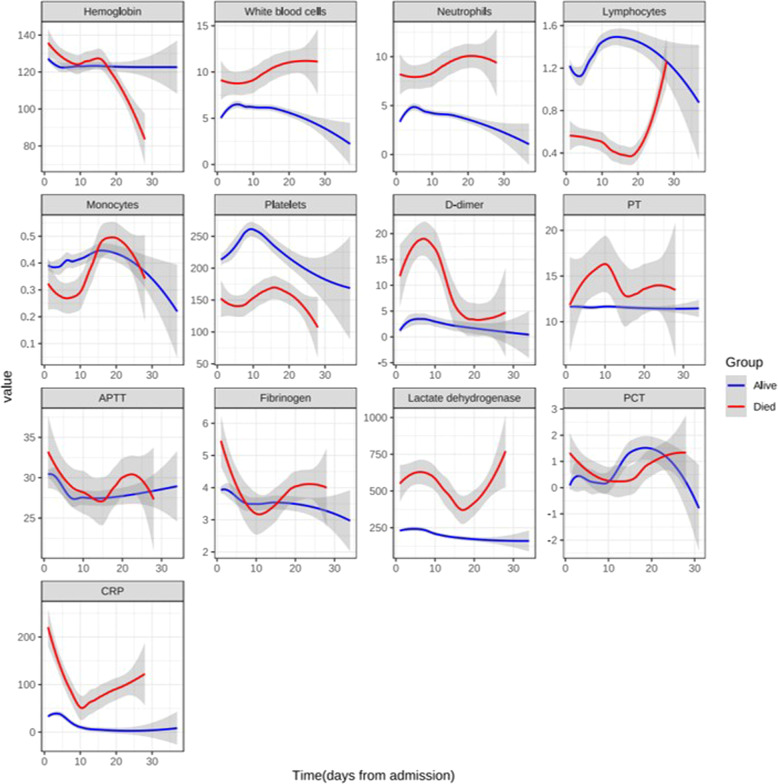
Next, we studied dynamic changes in hematological co-variates between survivors and non-survivors in 390 subjects from Wuhan Third Hospital with daily determinations (Supplementary Table 1). Subjects who died had higher concentrations of WBCs, neutrophils, D-dimmer, PT, LDH, and CRP but lower concentrations of lymphocytes and platelets throughout their hospitalization. These dynamic changes are displayed in Fig. 1.
Fig. 1. Dynamic changes of hematological variables in patients with COVID-19 during hospitalization.
The Y-axis “value” include units of all above data: ×10E + 9/L for white bloods cells, neutrophils, lymphocytes, monocytes, and platelets; g/L for hemoglobin, fibrinogen (FIB); mg/L for D-dimer; s for prothrombin time (PT), activated partial thromboplastin time (APTT); U/L for lactate dehydrogenase (LDH); ng/ml for procalcitonin (PCT); mg/L for C-reactive protein (CRP). Data are presented as medians (interquartile ranges, IQR).
Uni- and multivariable analyses
We analyzed admission hematological co-variates and their multiple measurements considering changes (Δ = Max − Min, Max − baseline, or baseline − Min) correlated with risk of death in all subjects (Supplementary Table 2). In a multivariable logistic regression model, age (Odds Ratio [OR] = 1.18 [1.02, 1.36]; P = 0.026), baseline D-dimer (OR = 3.18 [1.48, 6.82]; P = 0.003), Δ fibrinogen (OR = 6.45 [1.31, 31.69]; P = 0.022), Δ platelets (OR = 0.95 [0.90–0.99]; P = 0.029), Δ CRP (OR = 1.09 [1.01, 1.18]; P = 0.037) and Δ LDH (OR = 1.03 [1.01, 1.06]; P = 0.007) correlated with an increased risk of death (Table 6).
Table 6.
Multivariate analysis of hematological co-variates associated with in-hospital death of patients with COVID-19.
| Odds Ratio | 95% Confidence Interval | P value | |
|---|---|---|---|
| Age (years) | 1.18 | (1.02–1.36) | 0.026 |
| Baseline D-dimer (mg/L) | 3.18 | (1.48–6.82) | 0.003 |
| Δ Platelet (×10E + 9/L)a | 0.95 | (0.90–0.99) | 0.029 |
| Δ Neutrophil (×10E + 9/L)a | 1.31 | (0.99–1.72) | 0.058 |
| Δ Fibrinogen (g/L)b | 6.45 | (1.31–31.69) | 0.022 |
| Δ C-reactive protein (mg/L)c | 1.09 | (1.01–1.18) | 0.037 |
| Δ Lactate dehydrogenase (U/L)c | 1.03 | (1.01–1.06) | 0.007 |
aΔ = Max − Min.
bΔ = baseline − Min.
cΔ = Max − baseline.
Discussion
Our data indicate two baseline co-variates (age and D-dimer) on admission and four dynamic co-variates (Δs of concentrations of CRP, LDH, fibrinogen, and platelets) correlate with an increased risk of death in almost 1500 hospitalized persons with COVID-19. In our dataset, we could not confirm other co-variates such as male sex [19], and comorbidities of atherosclerotic cardiovascular disease [20] and hypertension [21] that had been excluded from the present study.
Two admission co-variates correlated with risk of death: age and D-dimer concentration. Similar correlates are reported by others in COVID-19 [8, 19, 20, 22–24] and in two other coronavirus infections, severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome [25, 26]. Age related immune deficiency may be the explanation of this association but is unproved [27]. High D-dimer concentration may result from the inflammation associated with COVID-19 and subsequent activation of coagulation [28]. Several potential risk factors during hospitalization, including disseminated intra-vascular coagulation, infection, dehydration, prolonged immobilization, mechanical ventilation, and central venous catheter use may further increase D-dimer concentrations [29, 30].
Four dynamic co-variates correlated with an increased risk of death including Δs of concentrations of CRP, LDH, fibrinogen, and platelets. Similar data are rarely reported in COVID-19-infection [7, 8, 31]. Higher admission LDH concentration was reported to be a risk factor for death by different studies [19, 23, 32]. However, we found the dynamics were more predictive. Han et al. report a dynamic decrease of CRP concentration in 17 subjects with COVID-19 who recovered [31]. There are few data on dynamics of fibrinogen concentration in persons with COVID-19. Tang et al. reported differences in dynamic fibrinogen between survivor and non-survivors, but this dynamic co-variate was not identified to be a risk factor for death [7]. Admission and dynamic platelets are not reported to correlate with risk of death in persons with COVID-19 [22, 33, 34].
Our study has important limitations. It was retrospective and researchers were not blinded to the outcome when they analyzed the data. Also, we have no external validation cohort. Finally, we did not adjust for multiple comparisons. As such, our conclusions should be interpreted as exploratory and descriptive. Because subjects more likely to die have profound changes in several of these co-variates the correlations we report should not be assumed to be cause-and-effect.
In conclusion, we show admission hematological co-variates except D-dimer concentration are not associated with an increased risk of death in a large cohort of subjects with COVID-19. However, dynamic measurements of platelets, fibrinogen, CRP, and LDH correlate with risk of death. We await validation of our conclusions.
Supplementary information
Acknowledgements
We thank patients, families, and health care providers participating in our study. Funded by the Natural Science Foundation of China (NSFC; 81974009 to QL, 81974221 and 81470330 to ZC) and the Fundamental Research Funds for the Central Universities (2020kfyXGYJ086 to QL). RPG acknowledges support from the National Institute of Health Research Biomedical Research Center funding scheme.
Author contributions
QL, ZC, and YH designed the study. QL, YC, LeC, DW, JY, HW, WH, LC, FD, WeiC, WenC, LL, QR, QL, WR, and FG collected the data. All authors had full access to the data, were involved in data interpretation, and vouch for the accuracy of the analyses. QL, YC, LeC, DW, and RPG prepared the typescript which all authors approved final approval and supported the decision to submit for publication.
Data availability
All data generated or analyzed during this study are included in this published article and its Supplementary Information files.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contribution equally: Qiubai Li, Yulin Cao, Lei Chen, Di Wu, Jianming Yu, Hongxiang Wang, Wenjuan He, Li Chen, Fang Dong, Weiqun Chen
Contributor Information
Qiubai Li, Email: qiubaili@hust.edu.cn.
Zhichao Chen, Email: chenzhichao@hust.edu.cn.
Yu Hu, Email: dr_huyu@126.com.
Supplementary information
The online version of this article (10.1038/s41375-020-0910-1) contains supplementary material, which is available to authorized users.
References
- 1.Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020. 10.1001/jama.2020.2648. [DOI] [PubMed]
- 2.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verity R, Okell LC, Dorigatti I, Winskill P, Whittaker C, Imai N, et al. Estimates of the severity of coronavirus disease 2019: a model-based analysis. Lancet Infect Dis. 2020;20:669–77. doi: 10.1016/S1473-3099(20)30243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–9. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–20. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–13. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang N, Li D, Wang X, Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J Thromb Haemost. 2020;18:844–7. doi: 10.1111/jth.14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–62. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang G, Zhang J, Wang B, Zhu X, Wang Q, Qiu S. Analysis of clinical characteristics and laboratory findings of 95 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a retrospective analysis. Respir Res. 2020;21:74. doi: 10.1186/s12931-020-01338-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qin C, Zhou L, Hu Z, Zhang S, Yang S, Tao Y, et al. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin Infect Dis. 2020. 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed]
- 11.Liu J, Li S, Liu J, Liang B, Wang X, Wang H, et al. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine. 2020;55:102763. doi: 10.1016/j.ebiom.2020.102763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He W, Chen L, Chen L, Yuan G, Fang Y, Chen W, et al. COVID-19 in persons with haematological cancers. Leukemia. 2020;34:1637–45. doi: 10.1038/s41375-020-0836-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li W, Wang D, Guo J, Yuan G, Yang Z, Gale RP. COVID-19 in persons with chronic myeloid leukaemia. Leukemia. 2020. 10.1038/s41375-020-0853-6. [DOI] [PMC free article] [PubMed]
- 14.WHO. Clinical management of severe acute respiratory infection when novel coronavirus (nCoV) infection is suspected: interim guidance. WHO. 2020. https://www.who.int/publications-detail/clinical-management-of-severe-acute-respiratory-infection-when-novel-coronavirus-(ncov)-infection-is-suspected. Accessed 25 Feb.
- 15.National Health Commission of China. The novel coronavirus pneumonia diagnosis and treatment program, 7th version. China. 2020. http://www.nhc.gov.cn/yzygj/s7653p/202003/46c9294a7dfe4cef80dc7f5912eb1989.shtml. Accessed 04 Apr.
- 16.Yang X, Yu Y, Xu J, Shu H, Xia JA, Liu H, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020;S2213-2600:30079–5. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Zhou L, Yang Y, Peng W, Wang W, Chen X. Therapeutic and triage strategies for 2019 novel coronavirus disease in fever clinics. Lancet Respir Med. 2020;8:e11–2. doi: 10.1016/S2213-2600(20)30071-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gale RP. Perspective: SARS-CoV-2, COVID-19 and haematologists. Acta Haematol. 2020. 10.1159/000508021. [DOI] [PMC free article] [PubMed]
- 19.Li X, Xu S, Yu M, Wang K, Tao Y, Zhou Y, et al. Risk factors for severity and mortality in adult COVID-19 inpatients in Wuhan. J Allergy Clin Immunol. 2020. 10.1016/j.jaci.2020.04.006. [DOI] [PMC free article] [PubMed]
- 20.Du RH, Liang LR, Yang CQ, Wang W, Cao TZ, Li M, et al. Predictors of mortality for patients with COVID-19 pneumonia caused by SARS-CoV-2: a prospective cohort study. Eur Respir J. 2020;55:2000524. 10.1183/13993003.00524-2020. [DOI] [PMC free article] [PubMed]
- 21.Guan WJ, Liang WH, Zhao Y, Liang HR, Chen ZS, Li YM, et al. Comorbidity and its impact on 1590 patients with Covid-19 in China: a Nationwide analysis. Eur Respir J. 2020. 10.1183/13993003.00547-2020 [DOI] [PMC free article] [PubMed]
- 22.Tang N, Bai H, Chen X, Gong J, Li D, Sun Z, et al. Anticoagulant treatment is associated with decreased mortality in severe coronavirus disease 2019 patients with coagulopathy. J Thromb Haemost. 2020;18:1094–9. doi: 10.1111/jth.14817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu C, Chen X, Cai Y, Xia J, Zhou X, Xu S, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med. 2020. 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed]
- 24.Chen R, Liang W, Jiang M, Guan W, Zhan C, Wang T, et al. Risk factors of fatal outcome in hospitalized subjects with coronavirus disease 2019 From a nationwide analysis in China. Chest. 2020. 10.1016/j.chest.2020.04.010. [DOI] [PMC free article] [PubMed]
- 25.Choi KW, Chau TN, Tsang O, Tso E, Chiu MC, Tong WL, et al. Outcomes and prognostic factors in 267 patients with severe acute respiratory syndrome in Hong Kong. Ann Intern Med. 2003;139:715–23. doi: 10.7326/0003-4819-139-9-200311040-00005. [DOI] [PubMed] [Google Scholar]
- 26.Hong KH, Choi JP, Hong SH, Lee J, Kwon JS, Kim SM, et al. Predictors of mortality in Middle East respiratory syndrome (MERS) Thorax. 2018;73:286–9. doi: 10.1136/thoraxjnl-2016-209313. [DOI] [PubMed] [Google Scholar]
- 27.Opal SM, Girard TD, Ely EW. The immunopathogenesis of sepsis in elderly patients. Clin Infect Dis. 2005;41:S504–12. doi: 10.1086/432007. [DOI] [PubMed] [Google Scholar]
- 28.Connors JM, Levy JH. COVID-19 and its implications for thrombosis and anticoagulation. Blood. 2020;135:1033–45. doi: 10.1182/blood.2020006000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang T, Chen R, Liu C, Liang W, Guan W, Tang R, et al. Attention should be paid to venous thromboembolism prophylaxis in the management of COVID-19. Lancet Haematol. 2020;7:E362–3. doi: 10.1016/S2352-3026(20)30109-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Terpos E, Ntanasis-Stathopoulos I, Elalamy I, Kastritis E, Sergentanis TN, Politou M, et al. Hematological findings and complications of COVID-19. American J Hematol. 2020. 10.1002/ajh.25829. [DOI] [PMC free article] [PubMed]
- 31.Han X, Cao Y, Jiang N, Chen Y, Alwalid O, Zhang X, et al. Novel coronavirus pneumonia (COVID-19) progression course in 17 discharged patients: comparison of clinical and thin-section CT features during recovery. Clin Infect Dis. 2020. 10.1093/cid/ciaa271 [DOI] [PMC free article] [PubMed]
- 32.Xie J, Hungerford D, Chen H, Abrams S, Li S, Wang G, et al. Development and external validation of a prognostic multivariable model on admission for hospitalized patients with COVID-19. SSRN. 2020. 10.2139/ssrn.3562456.
- 33.Xie J, Covassin N, Fan Z, Singh P, Gao W, Li G, et al. Association between hypoxemia and mortality in patients with COVID-19. Mayo Clin Proc. 2020;95:1138–47. doi: 10.1016/j.mayocp.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y, Du X, Chen J, Jin Y, Peng L, Wang HHX, et al. Neutrophil-to-lymphocyte ratio as an independent risk factor for mortality in hospitalized patients with COVID-19. J Infect. 2020. 10.1016/j.jinf.2020.04.002. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its Supplementary Information files.