ABSTRACT
China has a large area of saline-alkaline land that can be utilized for the cultivation of transgenic rice. Therefore, the growth and reproductive behavior of transgenic rice are not only a problem for production that needs to be resolved, but also an important aspect of environmental risk assessment for saline alkali soil. In the present study, an insect-resistant transgenic cry1C* rice, T1C-19, was grown in farmland and saline-alkaline soils. The transcription and translation of the exogenous cry1C*, and vegetative and reproductive fitness, such as plant height, tiller number, biomass, filled grain number and weight per plant, were assessed. Our findings indicated that the transcription and translation of exogenous cry1C* gene in T1C-19 rice grown in saline-alkaline soil were lower than that grown in farmland; however, the correlation was not significant. The vegetative and reproductive growth abilities of T1C-19 were lower than that of the parental rice, Minghui63 (MH63), in farmland. In alkaline-saline soil, except for tiller number and biomass, there were no significant differences between T1C-19 and MH63 in other vegetative indices. In contrast, the reproductive indices of T1C-19 were significantly higher than those of MH63. The results suggested that T1C-19 had a strong reproductive capacity, and significantly reduced the loss of yield caused by insects, thereby leading to a higher yield than that of MH63 grown in saline-alkaline soils. This may promote the cultivation of saline-alkaline soil to permit farming of T1C-19 in China in the future, despite the possible increased ecological risks.
KEYWORDS: Saline-alkaline soil, T1C-19 rice, Fitness, Transcription and translation of cry1C*, Vegetative and reproductive growth abilities
Introduction
Genetically modified (GM) crops have produced significant economic, environmental and social benefits in terms of guaranteeing agricultural production, improving crop yield and quality, saving labor, and reducing the environmental pollution. These benefits have greatly promoted its commercialization worldwide, with transgenic crops, including cotton, corn, soybean, and oilseed rape, commercially cultivated for almost 20 years on a large scale.1 Rice (Oryza sativa) is one of the most important staple food crops; the combination of different insect-resistant genes and promoters has significantly increased the stable and efficient expression of various insecticidal proteins in rice, with numerous insect-resistant transgenic rice lines successfully developed in the past 20 years.2–9 On October 22, 2009, the Ministry of Agriculture of China issued biosafety certificates for the cultivation of the insect-resistant transgenic rice lines Huahui 1 (HH1) and Bt Shanyou 63 (Bt-SY63) in the Hubei Province, and were renewed in December 2014, indicating that GM rice is also approaching commercialization. Therefore, there is an urgent need for a scientific evaluation of the potential ecological risks of insect-resistant transgenic rice after environmental release, especially with regard to the potential ecological risk of the spread of transgenic rice to wasteland, saline-alkali land, and weeds and other natural ecosystems outside farmland, through seed spraying or gene flow into wild rice. Zhang et al.10 reported that potential transgene flow may occur between adjacent fields where the smallholder farming systems exist.
The natural ecosystem is different from the farmland ecosystem, with strong artificial selection, and the characteristics of natural selection, such as poor soil nutrition, low target insect pressure, and high weed competition. Therefore, different selection pressures of the two ecosystems may lead to a different fitness. In the farmland ecosystem, the exogenous Bt gene effectively reduced pest damage and conferred significant yield advantages to insect-resistant transgenic rice and wild rice and weedy rice expressing insect-resistant genes under high insect pressure. Jiang et al.11 reported that under conditions of high target insect pressure in the field, the yield of cry1C* and cry2A* transgenic rice lines was 8.4% and 25.4% higher than that of the parental rice line MH63, respectively. In addition, the Bt/CpTI transgene was still effective in later generations derived from crosses between transgenic rice and weedy rice or wild rice, resulting in lower insect damage and increased fecundity under high insect pressure.8,12,13 However, previous studies have indicated that the transgenic rice and hybrid progeny with the exogenous Bt gene usually had an obvious yield disadvantage compared with the parental rice line in the field or greenhouse with relatively low insect pressure conditions.14–18 Liang et al.19 found that the empty grain number per spike of transgenic cry1Ab/c rice HH1 was significantly higher than that of parent rice MH63 under the field conditions with low insect pressure sprayed with insecticide, and the grain filling rate of HH1 was lower than that of MH63. Therefore, at present, the study of the fitness of different transgenic rice in the farmland ecosystem is relatively well established, but corresponding studies on the change of the fitness and the potential environmental risk after escaping into the natural ecosystem are lacking.
Soil salinization has become an important factor restricting the sustainable development of world agriculture, which is unbeneficial to the vegetative and reproductive growth of crops. China has a large area of saline-alkali land (~3.6 × 107 hm2), major rice-producing areas exist in saline-alkali land in China, including the northeast, northwest, and north China, and coastal areas.15,20 Owing to improper irrigation and drainage, the area of secondary salinization has continued to expand, and the damage has continued to spread. In addition, China is the origin of wild rice, that wild rice exists primarily in natural ecosystems containing saline-alkali soil, and that the planting area of wild rice overlaps with cultivated rice. Chinese rice scientists have studied the cultivation of rice in coastal shoal areas in recent years.21 It is predicted that saline-alkali land may be available as a potential arable land resource for rice, large-scale commercial planting of GM rice may occur in the future in fields with high salinity or left in saline-alkali land around farmland by seed or gene flow, the evaluation of rice fitness and speculation of the long-term ecological risk of continued cultivation in this saline-alkali land will provide a valuable resource, particular in terms of biosafety, for the commercialized planting of transgenic rice.
Considering there is increased concern that the widespread adoption of Bt crops may lead to the development of resistance to the insecticidal genes in pest populations.22 In 2006, an insect-resistant cry1C* transgenic rice line T1C-19 was successfully developed with high target insect resistance. However, the expression of exogenous Bt protein was significantly lower than that of the cry1Ab/c transgenic rice lines HH1 and Bt-SY63, the cry2A* transgenic rice line T2A-1, and other transgenic rice lines with insect resistance,2,23,24 thereby reducing the effect of overexpression of exogenous proteins in transgenic crops.25 Moreover, there are no common shared binding sites between the cry1C* and cry1A Bt genes commonly used in transgenic rice lines for target pests, and the polymerization of these type of genes can potentially prolong the resistance to target insects.26 Therefore, T1C-19 is considered to represent another promising commercially transgenic rice line for the future. Under both farmland and saline-alkaline soil conditions in a greenhouse, the present study comprised the following: (1) the transcription and translation of exogenous cry1C* in transgenic rice T1C-19 were detected during different growth stages, and the correlation between them was analyzed; (2) the vegetative and reproductive growth abilities of transgenic rice T1C-19 and parental MH63 rice were measured; (3) the fitness of transgenic rice T1C-19 and parental MH63 rice was investigated, and the possible causes of fitness difference were explored. The aim was to provide a theoretical basis to evaluate the environmental safety of insect-resistant cry1C* transgenic rice, T1C-19, after commercial planting in the future.
Materials and Methods
Plant Material
The transgenic cry1C* rice T1C-19, an indica rice strain, and its non-transgenic counterpart Minghui-63 (MH63), an elite cytoplasmic male sterile (CMS) restorer line with the highest cumulative planting area in China, were used in the present study. These lines were provided by the National Key Laboratory of Crop Genetic Improvement, Wuhan, China. The cry1C* gene was synthesized by using the wild-type cry1Ca5 gene of Bacillus thuringiensis. T1C-19 rice exhibited high δ-endotoxin expression and had insecticidal activity against borers such as Chilo suppressalis and Tryporyza incertulas.6
Soil
Saline-alkaline soil (topsoil, 0–30 cm layer) for the pot experiments was obtained from a typical saline-alkaline region in the surrounding coastal wetland of the National Nature Reserve for Rare Birds in Yancheng, Jiangsu, China (32°48′47″–34°29′28″N, 119°53′45″–121°18′12″E), where there is a large area of rice paddy fields within 10 km. Farmland soil (topsoil, 0–30 cm layer) for the pot experiments was obtained from a conventional paddy field located in Luhe District, Nanjing, Jiangsu, China (32°11′–32°27′N, 118°34′–119°03′E) and mixed with nutrient soil in a 1:1 ratio. The physical and chemical properties of the two soils are shown in Fig. S1. The total nitrogen, total phosphorus, available phosphorus, and available potassium content of saline-alkaline soil were significantly lower than in the farmland soil, and the salt content,27 pH, and organic matter content were significantly higher than in the farmland soil. In the present study, the saline-alkaline soil was considered to have a medium-high saline-alkaline content, according to the salinity grading of coastal saline-alkaline soils.28 These methods had been described similarly in a previously published paper.23
Experimental Design
A pot experimental was arranged in the glasshouse of the Nanjing Environmental Science Institute of the Ministry of Ecology and Environment, and the isolation conditions were good. The experimental site was located on a rooftop surrounded by offices and residential buildings, and no farmland with planted rice, vegetables, and other crops was within 5 km, ensuring that pot plants were exposed to low insect pressure without target insects. We did not spray insecticides to exclude insects completely. The present study mainly focused on the difference in fitness of cry1C* transgenic rice T1C-19 under simulated saline-alkali and farmland soils described above. The pot experiment mainly comprised of two plots: no stress (farmland soil) and stress (saline-alkali soil). The rice lines with consistent growth from a small breeding plate were transplanted into large pots (840 mm length × 560 mm width × 360 mm height) filled with saline-alkali soil or farmland soil at the five-leaf stage, respectively. Ten replicate pots of each soil were set up for the two rice lines: 12 individuals were arranged in four rows and three columns with 19–21 cm intervals uniformly distributed in each replicate pot; in total, 480 plants were randomly placed in the glasshouse. For the farmland soil, the rice was watered frequently, and weeds occurring in the pots were removed by hand during any growth stage to avoid yield loss. For the saline-alkali soil, the water layer was maintained at approximately 1 cm and a constant salt content was ensured. In addition, no agricultural operations were performed. All the remaining test materials were burned or inactivated after the completion of the trial. These methods had been described similarly in a previously published paper.23
Quantification of Relative mRNA Expression of cry1C* by Real-time Fluorescence Quantitative PCR
Rice leaves and stems were collected at the tilling (July 20, 2017), jointing (August 5, 2017), heading (September 5, 2017), filling (September 25, 2017), and maturing (October 25, 2017) stages. To minimize the differences in Cry1C* protein expression due to environmental factors, five samples per plot were pooled and five replicate pots were randomized. After collection, the samples were immersed in liquid nitrogen, and stored at -80°C in an ultralow temperature freezer until RNA extraction. The RNeasy plant mini kit (69104, Qiagen, Germany) was used to extract and purify RNA in accordance with the manufacturer’s instructions. One microgram of RNA was used as a template, and cDNA was synthesized from all samples by reverse transcription using the PrimeScript RT Reagent Kit with gDNA Eraser (RR047A, Takara, Japan). The size of the designed quantitative primer was 100–300 bp, and the rice actin gene, with stable expression, was selected as the housekeeping gene (Gene-specific primers were shown in Table S2). After all, data were normalized by using the Ct value of the housekeeping gene, the target gene expression was evaluated among samples by using the 2−∆∆ct method. Quantitative PCR was performed by using the Bio-Rad CFX-96 Real-Time PCR system with TB Green Premix Ex Taq II (Tli RNaseH Plus; RR820A, Takara, Japan) and the gene-specific primers cry1C*-F and cry1C*-R. The qRT-PCR reaction volume was 20.00 µL, comprising 10.00 µL TB Green Premix Ex Taq II (Tli RNaseH Plus), 0.40 µL forward primer, 0.40 µL reverse primer, 2.00 µL cDNA, and 7.20 µL ddH2O. The two-step quantitative PCR procedure was 40 cycles of 95°C, 30 s; 95°C, 5 s; 60°C, 35 s. Finally, the dissolution curve of the quantitative PCR was determined.
Quantification of Cry1C* Protein by Enzyme-linked Immunosorbent Assay
The same batch samples for RNA extraction were used to detect the expression of exogenous proteins by ELISA. The expression of Cry1C* protein in the transgenic rice T1C-19 was quantified using the QualiPlate Kit for Cry1C* (EnviroLogix Inc., Portland, ME, USA). Approximately 20 mg of tissue sample was weighed and its mass was recorded. They were pulverized in liquid nitrogen with a tissue crusher (TissueLyser II, Qiagen, Hilden, Germany). One milliliter Cry1C* 1× extraction buffer and the milled samples were added to 2-mL microreaction tubes. The samples were incubated at 4°C in a shaker for 30 min at 150 rpm, and then centrifuged at 10,000 × g for 5 min at 4°C. The supernatant was collected and diluted 12.5–100-folds with 1× extraction buffer. The level of Cry1C* protein was quantified according to the instructions of the manufacturer. The absorbance was measured at 450 nm using a Microplate Reader (Infinite M2000, Tecan Group Inc., Männedorf, Switzerland). The reader was calibrated by plotting a standard curve of OD against protein concentration using the following concentration of Cry1C* protein standards: 0.5, 0.6, 1, 1.5, 2, and 2.5 ng mL−1. The Cry1C* protein level in the fresh rice samples was determined using a standard curve and the dilution ratios of the extraction solution. The Cry1C* standards were prepared using the QualiPlate kit for Cry1C*.
Measurement of Fitness Component Based on Vegetative and Reproductive Growth
To estimate vegetative growth abilities at four vegetative growth stages, samples were collected, excluding the edge rows to avoid edge effects; 20 individuals were randomly chosen to measure the number of tillers and plant height at the tilling, heading, filling, and maturing stages. Simultaneously, a chlorophyll meter (SPAD-502, Minolta Camera Co., Osaka, Japan) was used to obtain the SPAD values of the 20 most fully expanded flag leaves that were randomly measured at the heading, filling, and maturing stages, and calculating the leaf areas by leaf length × leaf width × 0.75. Samples were collected by cutting them from the surface of soil, and were oven-dried at 80°C until a constant weight was reached; subsequently, the determination of biomass at the tilling, heading, filling, and maturing stages was measured.
To estimate the reproductive growth abilities at the maturing stage of T1C-19 rice and MH63 rice, the following traits were determined: (1) effective number of panicles per plant determined as the number of filled grains more than 5 grains; (2) spike length; (3) spike weight measured by the weight of effective number of panicles per plant; (4) number of filled grains per plant; (5) total number of grains per plant calculated by the number of filled grains per plant plus number of empty grains per plant; (6) filled grain weight per plant measured by the weight of filled grains per plant; (7) 1000-grain weight measured by the weight of 1000 grains randomly; and (8) seed setting rate calculated as the ratio of the number of filled grains per plant to the total number of grains per plant. In all treatments, we calculated the average value of plants of the same genotype in each pot, and six pots were randomly selected.
Data Collection and Analyses
Independent t-tests were performed to identify any significant differences between the farmland and saline-alkaline soil treatments in terms of the transcription and translation of exogenous cry1C* for each growth stage. Under each soil condition, Duncan’s multiple comparison test was used to analyze the temporal and spatial changes in the transcription and translation of exogenous cry1C * during different growth stages. Two-way ANOVA was applied to estimate the effect of two soil conditions and five growth stages on the transcription and translation of exogenous cry1C*.
Relative differences between T1C-19 and MH63 rice in terms of vegetative and reproductive growth were compared with an independent t-test. Three-way ANOVA was applied to estimate the effect of two soil conditions, two rice lines, and three or four growth stages on vegetative performance. Two-way ANOVA was applied to estimate the effect of two soil conditions and two rice lines on reproductive performance.
According to the method of Song et al.29, for estimation of fitness of vegetative and reproductive growth ability in T1C-19, the fitness per trait of the MH63 rice was defined as “1.00”, and T1C-19 was then assigned a fitness value based on its ratio of per trait in T1C-19 compared with those of the MH63 control. The fitness composites were calculated according to the average fitness value of all traits associated with a stage or all stages. Independent t-tests were used to analyze the differences in fitness. All the statistical analyses were computed by using SPSS v.16.0 for Windows (IBM Corp., Armonk, NY, USA).
Results
Present study did not observe dead heart and leaf rolling caused by rice stem borers (Scirpophaga incertulas, C. suppressalis, and Sesamia inferens) and rice leaf borers (Cnaphalocrocis medinalis) of Bt transgenic rice under glasshouse. However, there are very few non-target insects due to the absence of pesticides, including spiders (Araneida), ladybugs (Coccinellidae), and locusts (Locusta migratoria manilensis). The present study indicated that the pot experiments lacked the target insect pressure.
The mRNA Transcription and Expression of cry1C* Gene
The mRNA transcription and protein expression of cry1C* in the T1C-19 rice leaves in farmland soil were higher than that those in saline-alkaline soil at the same growth stage (P < .05, Fig. 1).
Figure 1.
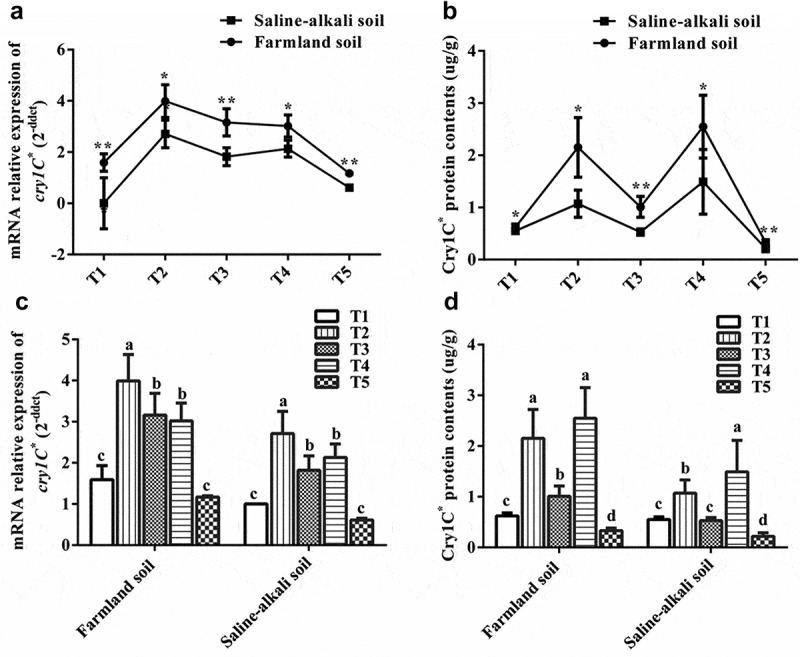
The transcription and translation of exogenous cry1C* gene in T1C-19 rice leaf grown in farmland and saline-alkaline soils at five stages (T1: Tillering, T2: Jointing, T3: Heading, T4: Filling, T5: Maturing); Farmland soil values with * and ** were significantly different from those of the saline-alkaline soil according to the t-test (P < .05 and P < .01, respectively) in the leaf; a, b, c, and d indicated significant differences between the five growth stages of T1C-19 rice leaf grown on the same soil according to Duncan’s multiple range test (P < .05).
The mRNA transcription of cry1C* gene in T1C-19 varied temporally and spatially among different growth stages under the same soil conditions. Initially, transcription increased and then decreased with growth in both soils. It reached the maximum level at the jointing stage, with a significant decrease to a minimum level at maturity. Two-way ANOVA indicated that the soil and growth stages significantly affected the mRNA transcription of cry1C* (Table 1).
Table 1.
Effects of soil condition (two levels), growth stage (five levels), and interactions among them, the transcription and translation of the exogenous cry1C* in transgenic T1C-19 rice.
| Cry1C* protein (μg g−1) |
cry1C* mRNA relative expression (2−ddct) |
|||||
|---|---|---|---|---|---|---|
| Df | F | p | Df | F | p | |
| Soil | 1 | 78.57 | 0.00 | 1 | 57.51 | 0.00 |
| Growth stage | 4 | 106.47 | 0.00 | 4 | 53.77 | 0.00 |
| Soil×Growth stage | 4 | 9.95 | 0.00 | 4 | 1.814 | 0.14 |
p < .05 indicated significant differences.
The expression of the Cry1C* protein in T1C-19 varied temporally and spatially among different growth stages under the same soil conditions. It initially increased and then decreased with growth in both soils. It reached the maximum level of 2.55 (farmland soil) and 1.49 μg g−1 FW (saline-alkaline soil) at the filling stage, but significantly decreased to 0.33 (farmland soil) and 0.22 μg g−1 FW (saline-alkaline soil) at maturity (Fig. 1c). Two-way ANOVA indicated that the soil, growth stage, and their interaction significantly affected the expression of Cry1C* (Table 1).
However, there was no significant correlation between mRNA transcription and protein expression of exogenous cry1C* in the same soil conditions (Table 2).
Table 2.
Correlation coefficients between insecticidal protein content and mRNA relative expression of cry1C* in rice leaf in farmland and saline-alkali soils.
| Correlation coefficients | p | |
|---|---|---|
| Farmland soil | 0.79 | 0.10 |
| Saline-alkali soil | 0.78 | 0.12 |
p < .05 indicated significant difference.
Vegetative Growth Indices
Plant Height
There were significant differences in the plant height of T1C-19 rice and MH63 rice in each soil (P < .01; Fig. 2). The height of the same rice line was significantly higher in farmlands than in saline-alkaline soils, by approximately 30–48% in the four key growth stages.
Figure 2.
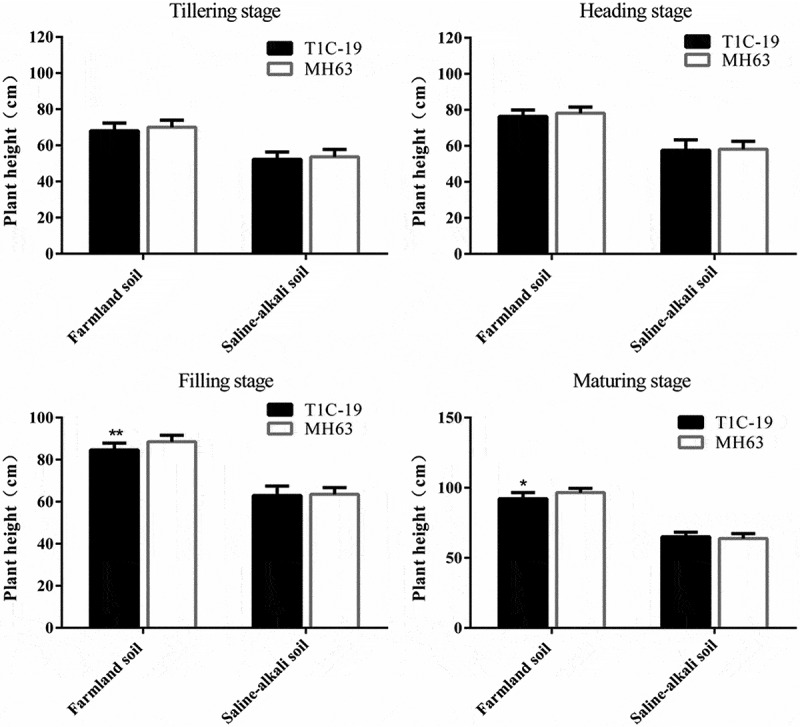
Plant heights (mean ± SEM) of T1C-19 rice and MH63 rice at four stages grown in farmland and saline-alkaline soils. The values of T1C-19 rice with * and ** were significantly different from those of MH63 according to the t-test (P < .05 and P < .01, respectively).
Under both the soils, the height of T1C-19 and MH63 was similar throughout the growth stage, increasing gradually with growth. In farmlands, the height of T1C-19 was significantly lower than that of MH63, by approximately 4% at the filling and maturing stages (P < .01). In the saline-alkaline soils, the height of T1C-19 was not significantly different from that of MH63 at the four key growth stages (P > .05), and the magnitude of this fitness cost was significantly lower than that measured for these two rice lines grown in farmland soil (Table 3). Three-way ANOVA indicated that the soil, exogenous gene, growth stage, interaction between exogenous gene and soil, and interaction between soil and growth stage significantly affected plant height (Table S1).
Table 3.
Fitness for vegetative and reproductive indices of T1C-19 vs. MH63 rice grown on farmland and saline-alkali soils.
| Farmland soil | Saline-alkali soil | |||
|---|---|---|---|---|
| Vegetative indices | Fitness | Plant height (cm) | 0.97 | 0.98 |
| Tiller number | 0.84* | 0.65* | ||
| Biomass(g) | 0.87* | 0.79* | ||
| Composite fitness | 0.89* | 0.81* | ||
| Reproductive indices | Fitness | Effective panicle number per plant | 0.79* | 1.00 |
| Panicle length (cm) | 0.96* | 1.08 | ||
| Panicle weight (g) | 0.94* | 1.69* | ||
| Total grain number per plant | 0.89* | 0.71* | ||
| Filled grain number per plant | 0.87* | 3.36* | ||
| Filled grain weight per plant (g) | 0.87* | 3.36* | ||
| Thousand grain weight (g) | 1.00 | 1.00 | ||
| Seed-setting rate (%) | 0.97 | 4.56* | ||
| Composite fitness | 0.91* | 2.10* |
Fitness ratio was defined as agronomic traits of T1C-19 vs. MH63 rice; Composite fitness was defined by the average value of all fitness for vegetative traits or reproductive traits. *indicates fitness significantly more than or less than 1.00 according to t-test (p < 0.05).
Tiller Number
There were significant differences in tiller number of T1C-19 rice and MH63 rice in each soil (P < .01; Fig. 3). For the same rice line, the tiller number of plants grown in farmland soil was significantly higher (approximately 1.83–3.59-fold) than those plants grown in saline-alkaline soil at the four key growth stages.
Figure 3.
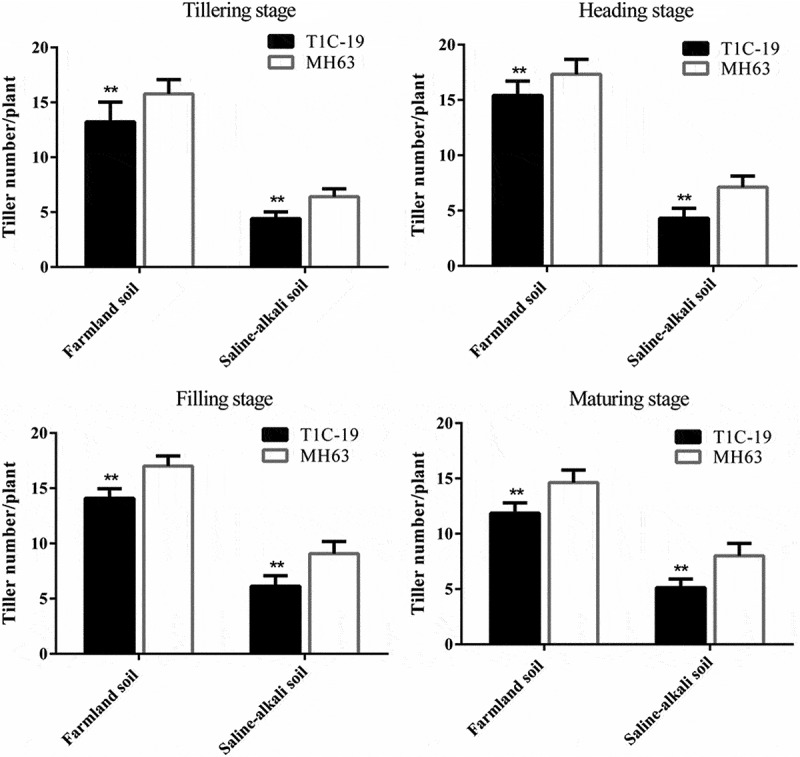
Tiller number per plant (mean ± SEM) of T1C-19 and MH63 rice at four stages grown in farmland and saline-alkaline soils. The values of T1C-19 rice with ** were significantly different from those of MH63 rice according to the t-test (P < .01).
In both the soil types, the variation of tiller number between T1C-19 and MH63 rice lines throughout the growth period was similar; it initially increased and then decreased with growth. In farmland soil, the tiller number of T1C-19 was significantly lower than that of MH63, by approximately 11.02–18.89% at the tillering, heading, filling, and maturing stages (P < .01). In saline-alkaline soil, the tiller number of T1C-19 was significantly lower than that of MH63 grown in farmlands, by approximately 31.25–39.66%, at the four growth stages (P < .01). There was a significant fitness cost in the number of tillers between T1C-19 and MH63 grown in saline-alkaline soil, and the magnitude of this fitness cost was higher than that measured for the two rice lines grown in farmland soil (Table 3). Three-way ANOVA indicated that the soil, exogenous gene, growth stage, and interaction between soil and growth stage significantly affected the number of tillers (Table S1).
Leaf Length, Width, and Area
There were significant differences in the leaf length, width, and area of T1C-19 rice and MH63 rice in each soil (P < .01; Fig. 4). For the same rice line, these parameters were significantly higher in plants grown in farmland than those plants grown in saline-alkaline soil, by approximately 1.86–2.06-, 1.46–1.58-, and 2.56–3.16-fold, respectively.
Figure 4.
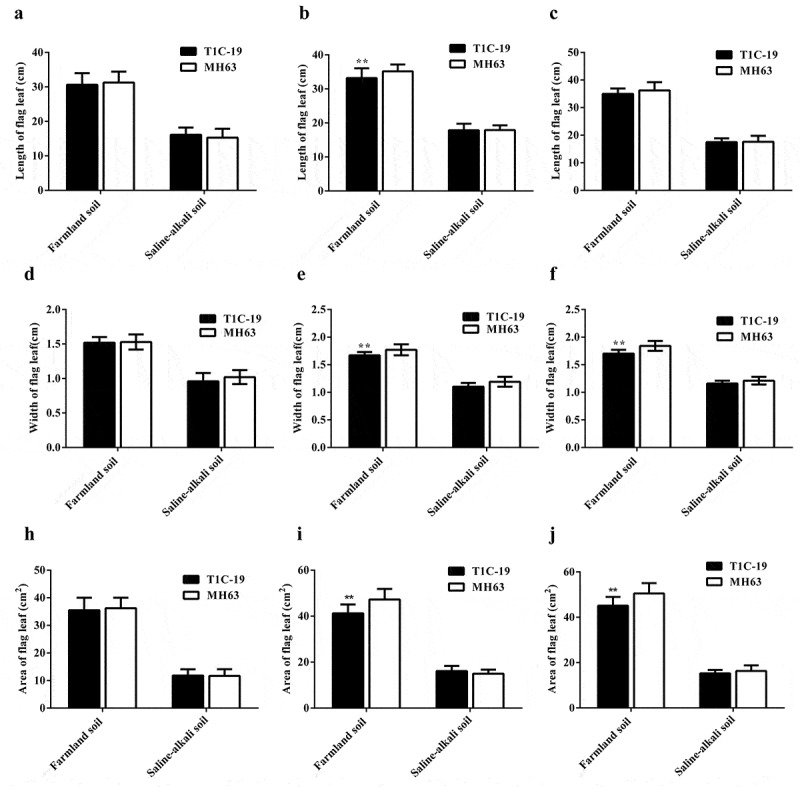
Flag leaf length (mean ± SEM), width (mean ± SEM), and area index (mean ± SEM) of T1C-19 and MH63 rice grown in farmland and saline-alkaline soils. (a,d,h). Heading stage; (b,e,i). Filling stage; (c,f,j). Maturing stage. The values of T1C-19 rice with ** were significantly different from those of MH63 rice according to the t-test (P < .01).
The variation in leaf length between T1C-19 and MH63 at all growth stages was similar under the two soil conditions: it gradually increased with the growth of plant. Under farmland soil conditions, the leaf length of T1C-19 was significantly lower than that of MH63, by approximately 5.55% at the filling stage. However, there was no significant difference in leaf length between T1C-19 and MH63 at the three key growth stages when grown in saline-alkaline soils.
With respect to leaf width, the variation between T1C-19 and MH63 was consistent with that of leaf length under the two soil conditions. Under farmland soil conditions, the leaf width of T1C-19 was significantly lower than that of MH63 by approximately 5.65% and 7.61% at the filling and maturing stages, respectively (P < .01). However, the leaf width did not differ significantly between T1C-19 and MH63 at the three key growth stages in saline-alkaline soil.
The variation in leaf area between T1C-19 and MH63 was consistent with that of leaf length and width under the two soil conditions. Under farmland soil condition, the leaf area of T1C-19 was significantly lower than that of MH63, by approximately 12.8% and 10.69% at the filling and maturing stages, respectively (P < .01). However, the leaf area did not differ significantly between T1C-19 and MH63 at the three key growth stages in saline-alkaline soil. Three-way ANOVA indicated that the soil, exogenous gene, growth stage, interaction between soil and exogenous gene, and interaction between soil and growth stage significantly affected the leaf area (Table S1).
SPAD Value
There were significant differences in the SPAD value of T1C-19 rice and MH63 rice in each soil (P < .01; Fig. 5). For the same rice line, the SPAD value of plants grown in farmland soil was higher than that of plants grown in saline-alkaline soil by approximately 21–58%.
Figure 5.
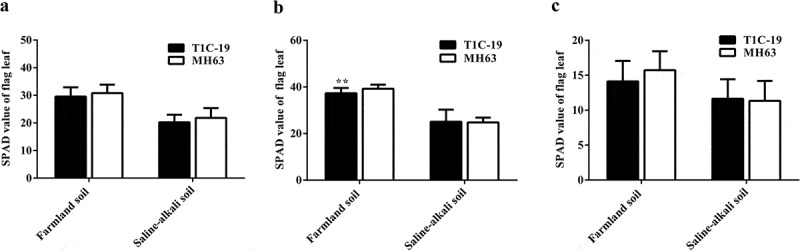
SPAD indices (mean ± SEM) of T1C-19 and MH63 rice grown in farmland and saline-alkaline soils. (a). Heading stage, (b). Filling stage, and (c). Maturing stage. The values of T1C-19 rice with ** were significantly different from those of MH63 rice according to the t-test (P < .01).
In both soil types, the variation in the SPAD value between T1C-19 and MH63 was similar: it initially increased and then decreased with growth. Under farmland soil conditions, the SPAD value of T1C-19 and MH63 was significantly lower than that of MH63, by approximately 4.84%, only at the filling stage (P < .01). However, there was no significant difference in the SPAD value between T1C-19 and MH63 at the three key growth stages in saline-alkaline soil. Three-way ANOVA indicated that the soil, exogenous gene, growth stage, and interaction between soil and growth stage significantly affected the SPAD value (Table S1).
Biomass
There were significant differences in biomass of T1C-19 rice and MH63 rice in each soil (P < .01; Fig. 6). For the same rice line, the biomass of plants grown in farmland soil was significantly higher than that of plants grown in saline-alkaline soil, by approximately 2.57–3.67-fold at the four key growth stages.
Figure 6.
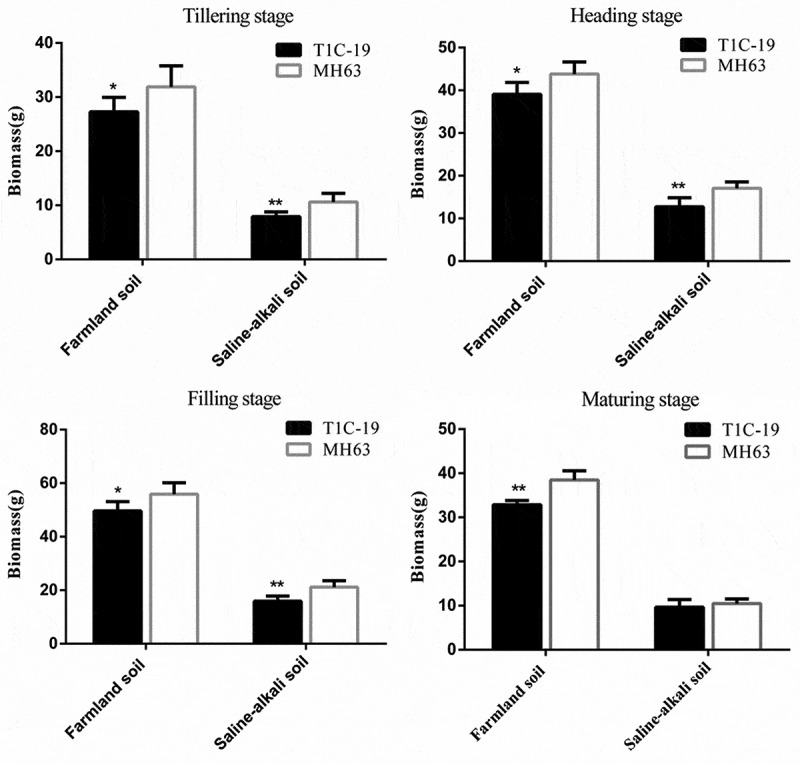
Biomass (mean ± SEM) of T1C-19 and MH63 rice grown in farmland and saline-alkaline soils at four stages. The values of T1C-19 rice with * and ** were significantly different from those of MH63 rice according to the t-test (P < .05 and P < .01, respectively).
For plants grown in farmland soil, the variation in biomass between T1C-19 and MH63 was similar throughout the growth stage: it initially increased and then decreased with growth. The biomass of T1C-19 was significantly lower than that of MH63, by approximately 10.84–14.62% at the tillering, heading, filling, and maturing stages (P < .05). In saline-alkali soil, the biomass of T1C-19 was significantly lower than that of MH63, by approximately 24.21–25.31% at the tillering, heading, and filling stages (P < .05). Furthermore, the magnitude of this fitness cost was higher than that in farmland soil (Table 2). Notably, these results significantly correlated with the lower tiller number of T1C-19 than those of MH63 at the main growth stage. Three-way ANOVA indicated that the soil, exogenous gene, growth stage, and interaction between soil and growth stage significantly affected the biomass of plants (Table S1).
Days to 50% Flowering
The time taken to complete 50% of flowering was affected by the soil. MH63 and T1C-19 rice heading both occurred at 95 days under farmland soil, and their heading occurred at 103 days under saline-alkaline soil, which was a delay of 8 days in the growth stage. The time taken to complete 50% flowering of MH63 rice and T1C-19 rice was the same in both soils.
Reproductive Growth Indices
There were significant differences between the reproductive growth indices of T1C-19 and MH63 in each soil <0.01; Fig. 7). For the same rice line, compared with the farmland soil, the reproductive growth indices were decreased significantly in saline-alkaline soil.
Figure 7.
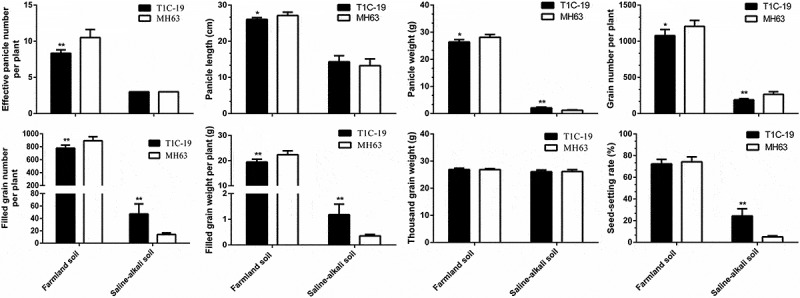
Reproductive traits (mean ± SEM) of T1C-19 rice and MH63 rice grown on farmland and saline-alkaline soils. The values of T1C-19 rice with * and ** were significantly different from those of MH63 rice according to the t-test (P < .05 and P < .01, respectively).
In farmland soil, the effective panicle number, panicle length, panicle weight, total grains number per plant, filled grains number per plant, and filled grain weight per plant of T1C-19 were significantly lower than those of MH63 by approximately 20.63%, 4.00%, 6.25%, 10.56%, 12.93%, and 12.93%, respectively (P < .05). There was a significant cost with respect to most of the yield-related traits between T1C-19 and MH63 grown in farmland soil (Table 2). In saline-alkaline soil, the panicle weight, filled number grains per plant, filled grain weight per plant, and seed-setting rate of T1C-19 were significantly higher than those of MH63 by 1.69-, 3.36-, 3.36- and 4.56-fold, respectively (P < .01). However, both T1C-19 and MH63 were similar in terms of effective panicle number, panicle length, and 1000-grain weight. There were significant fitness benefits for most of the yield-related traits between T1C-19 and MH63 grown in saline-alkaline soil (Table 3). Two-way ANOVA indicated that soil conditions, exogenous gene, and their interaction significantly affected most of the yield-related traits (Table 4).
Table 4.
Two-way ANOVA showing the effects of soil conditions (two levels) and rice line (two levels) on reproductive growth indices.
| Reproductive traits | Soil |
Rice line |
Soil × Rice line |
||||||
|---|---|---|---|---|---|---|---|---|---|
| df | F | p | df | F | p | df | F | p | |
| Effective panicle number/plant | 1 | 745.79 | 0.00 | 1 | 21.26 | 0.00 | 1 | 21.26 | 0.00 |
| Panicle length | 1 | 743.37 | 0.00 | 1 | 1.94 | NS | 1 | 0.64 | NS |
| Panicle weight | 1 | 7,772.09 | 0.00 | 1 | 2.46 | NS | 1 | 20.07 | 0.00 |
| Total grain number/plant | 1 | 2,370.56 | 0.00 | 1 | 5.36 | 0.03 | 1 | 7.76 | 0.01 |
| Filled grain number/plant | 1 | 2,964.83 | 0.00 | 1 | 13.72 | 0.00 | 1 | 35.66 | 0.00 |
| Filled grain weight/plant | 1 | 4,847.75 | 0.00 | 1 | 27.41 | 0.00 | 1 | 5.37 | 0.03 |
| 1,000 grain weight | 1 | 8.67 | 0.01 | 1 | 0.01 | NS | 1 | 0.07 | NS |
| Seed-setting rate | 1 | 817.28 | 0.00 | 1 | 12.26 | 0.00 | 1 | 35.10 | 0.00 |
p< .05 indicated significant difference; NS indicated no significant difference.
Discussion
Expression of Exogenous Cry1C* Protein
Present study indicated that the transcription and translation of cry1C* gene in T1C-19 rice varied temporally and spatially during whole growth stages in farmland and saline-alkali soils, although there was no significant correlation between transcription and translation of cry1C* in T1C-19 rice during the whole growth stage in either soil, and the correlation was not influenced by the stress of saline-alkali soil. Similar studies have shown that the correlation between transcription and translation of the exogenous gene in Bt cotton and Bt maize was weak under stress growth conditions.30,31 Posttranslational modification of genes might help to understand the mechanism for the lack of significant correlation transcription and translation of cry1C* in T1C-19 rice.32 Previous studies have shown that the exogenous protein expression could provide more direct and accurate messages than those the gene expression level for assessing and monitoring the safety in GM crops.30,31 The expression of exogenous proteins in natural ecosystems might also determine the fitness performance and ecological consequences of transgenic crops.33 In saline-alkali soil, we showed the expression of the Cry1C* protein in T1C-19 was significantly lower than that in farmland soil. Luo et al.27 and Zhang et al.31 found that exogenous protein expression in Bt cotton was negatively correlated with saline stress conditions. Similar studies indicated that the expression of exogenous protein in insect-resistant transgenic Bt cotton lines, Bt corn lines negatively correlated with salt, flooding, and low-temperature stressful growth conditions.30,34–37 To some extent, the expression of exogenous Bt proteins were affected by the growth process, the physical and chemical properties of the soil, and the external stress environment in transgenic crops. Considering that the expression of Cry1C* protein in T1C-19 at most growth stages was 0.22–2.55 μg·g−1 FW, which was significantly lower than that of the exogenous Cry1Ab/c protein in HH1 rice (4.28–10.15 μg·g−1 FW) under the same farmland and saline-alkali soils.23 This might reduce the fitness cost of transgenic rice T1C-19 owing to the overexpression of exogenous proteins.25In conclusion, Present study indicated the expression of Cry1C* in T1C-19 was significantly inhibited by saline-alkaline soil, although it still effectively resisted the target insects, such as S. incertulas and C. suppressalis in saline-alkaline soil (data not shown). Therefore, the expression of Cry1C* protein might bring fitness benefits to T1C-19 in saline-alkaline soil with high target insect pressure. In contrast, it might confer the fitness cost toward vegetative and reproductive growth abilities of T1C-19 in saline-alkaline soil with low target insect pressure, mainly because the expression of Cry1C* protein might have consumed considerable amount of material and energy from the host cell. Previous studies have indicated that the exogenous protein expression of transgenic crops may be related to an underlying fitness cost in the absence of, or under low, relevant selective pressures.33,38
Effect of Transgene Expression on Fitness Components in Farmland Soil
Present study indicated that, under farmland soil conditions without target insect pressure, the important vegetative growth indices, such as plant height, number of tillers, biomass, and important reproductive growth indices, such as filled grain number per plant, filled grain weight per plant, of T1C-19 all were significantly lower than those of MH63 in the same soil. Therefore, there was a significant fitness cost of vegetative and reproductive growth abilities in T1C-19. Chen et al.14, Xia et al.18 and Jiang et al.39 showed that the tiller number of transgenic Bt/CpT1 rice or the biomass of transgenic cry1C* rice were significantly lower than that of the parental rices under greenhouse or field conditions with very low insect pressure. Similar studies also have shown the total yield and seed-setting rate of the transgenic cry1C* and cry2A* rice lines were significantly lower than those of the parental rice line MH63 under farmland with low insect pressure.11,24,40 These findings indicated that the exogenous Bt genes usually had an obvious vegetative and reproductive growth disadvantage compared with the parental rice line under field or greenhouse with low insect pressure or the absence of insect pressure.16–18 An alternative explanation is that exogenous protein overexpression might have adversely affected plant vegetative and reproductive growth in the absence of a target or at low target insect pressure.23,33
Effect of Transgene Expression on Fitness Components in Saline-alkali Soil
Present study indicated that the vegetative and reproductive growth abilities of T1C-19 and MH63 rices were significantly lower in saline-alkaline soil than those in farmland soil. Jiang et al.41 have showed that the SPAD value of flag leaves, biomass, and yield of Bt rice and parental rice were significantly lower under drought stress than that of normal, flood condition. Similar studies also have shown in Bt transgenic maize and Bt transgenic cotton under drought conditions.35,37 We showed that, under saline-alkaline soil stress condition without target insect pressure, except for the number of tillers and biomass of T1C-19, which were significantly lower than those of MH63, there were no significant differences in other vegetative growth indices between T1C-19 and MH63. Jiang et al.41 showed that, under the drought stress condition with strictly controlled pests and diseases, the SPAD value and biomass of insect-resistant transgenic rice Bt-MH63 harboring the cry1C*, cry2A*, cry1Ab/c genes all were significantly lower than those of MH63. However, the important reproductive indices, such as filled grains number per plant and filled grain weight per plant, of T1C-19 were significantly higher than those of MH63 under saline-alkaline soil condition without target insect pressure. Therefore, there was a significant fitness benefit. Fang et al.42 showed that, under the conditions of high temperature and drought without glyphosate, the full-grain number per plant of Arabidopsis thaliana with transfected with the epsps gene was significantly higher than that of parent Arabidopsis thaliana. Present study indicated that the reproductive abilities and ecological risk of T1C-19 in saline-alkali soil were different from those in farmland soil. A possible explanation is that the low expression of Cry1C* protein and less energy consumption leads to a negligible fitness effect on the reproductive ability of T1C-19 under saline-alkaline soil. Moreover, saline-alkaline soil might lead to the trade-off between vegetative growth and reproductive growth in T1C-19, resulting in significant differences from the parental line MH.6314,33 These results suggest that, from the perspective of agricultural production, the insect-resistant cry1C* transgenic rice T1C-19 has a stronger reproductive ability than the parental rice line MH63, and that this can significantly reduce the yield loss caused by target insects, leading to higher yield advantages compared with that of MH63 under saline-alkaline soil conditions. This will help promote the large-scale application of saline-alkaline land for the cultivation of T1C-19 in the future in China. If T1C-19 escapes from the tillage system into the natural saline-alkaline ecosystem, it may be conducive to the establishment of its own population, and therefore brings potential ecological risks.
Conclusions
Present study indicated that T1C-19 had significantly weaker vegetative and reproductive growth abilities, with higher fitness costs than those of its parental line MH63 in farmland soil. In contrast, T1C-19 showed weaker vegetative growth and stronger reproductive abilities than those of MH63 under saline-alkaline soil (Fig. 8). This would promote the use of saline-alkaline soil for T1C-19 cultivation in China, although this might have higher ecological risks. However, these conclusions were drawn from data obtained over 1 year from saline-alkaline areas in the absence of target insect pressure. In this model system, it was easier to detect the real fitness effects of exogenous genes on rice vegetative and reproductive growth abilities in the absence of target insect pressure, but ignore a single factor stress could not exist under the natural ecosystem. Furthermore, considering the environmental biosafety assessments should be conducted for longer than 2 years. In the future, we plan to conduct the tests of three stress factors, including saline-alkali, target insect, or saline-drought with weed competition for more than 2 years. Rice volunteers may also frequently appear in the wastelands, marsh, orchards or other habitats near farmland due to seed movement, gene flow or seedlings dispersal during the farming. Thus, it is necessary to permanently and systematically assess the fitness of T1C-19 rice under simulated conditions of these natural habitats. This will provide a valuable reference on environmental safety for the long-term natural ecological risks that may be occurred in response to the commercialized planting of transgenic rice.
Figure 8.
Higher fitness benefit in saline-alkaline soil and lower fitness cost in farmland soil of transgenic T1C-19 rice compared to parental MH63 rice.
In addition, the present study speculated that the reproductive abilities and ecological risk of T1C-19 in saline-alkali soil were different from those in farmland soil that may relate to the expression of exogenous proteins. To date, no relevant studies have shown that the tissue culture process may cause the unintentional mutation of insect-resistant transgenic rice T1C-19, while a recent study found that the location effect of exogenous gene insertion may not cause significant unintended effects.43 However, we can not ignore that the tissue culture process and insertion sites may cause some unintended effects. Therefore, we are currently using molecular biological techniques, such as genome sequencing, to detect whether there are other unintended gene mutations, with the exception of exogenous genes. We are doing this with insect-resistant transgenic rice (T1C-19) and non-transgenic parental rice (MH63).
Funding Statement
This work was supported by the “Basic Scientific Research Program in National Nonprofit Scientific Research Institutes (GYZX190103)”; “National Major Special Projects for Genetically Modified Organisms (2016ZX08012-005)”.
Acknowledgments
This work was funded by grants from the Basic Scientific Research Program in National Nonprofit Scientific Research Institutes(GYZX190103) and the National Major Special Projects for Genetically Modified Organisms (2016ZX08012-005). The authors thank Editage for revising the language and style.
Authors’ Contributions
J.M.F designed and carried out the experiments, and wrote the manuscript. B.L. designed and helped to calibrate the manuscript.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
Availability of Data and Materials
The data sets supporting the conclusions of this article are included within the article and its additional files.
References
- 1.ISAAA . Global status of commercialized biotech/GM Crops: 2017. ISAAA. 2017; Brief 52. [Google Scholar]
- 2.Chen H, Tang W, Xu CG, Li XH, Lin YJ, Zhang QF.. Transgenic indica rice plants harboring a synthetic cry2A* gene of Bacillus thuringiensis exhibit enhanced resistance against lepidopteran rice pests. Theor Appl Genet. 2005;111:1330–37. [DOI] [PubMed] [Google Scholar]
- 3.Jiang GH, Xu CG, Tu JM, Li XH, He YQ, Zhang QF.. Pyramiding of insect- and disease-resistant genes into an elite indica, cytoplasm male sterile restorer line of rice, ‘Minghui 63ʹ. Plant Breed. 2010;123:112–16. [Google Scholar]
- 4.Lin XF, Liu ZM, Liu DP, Hao WY, Tang KX. Introduction double insect resistance genes cry1a(a)-pta to japonica rice and assessment of resistance to rice stem borer. Mol Plant Breed. 2006;4:345–50. [Google Scholar]
- 5.Shu QY, Ye GY, Cui HR, Cheng XY, Xiang YB, Wu DX, Gao MW, Xia YW, Cui H, Sardana R. Transgenic rice plants with a synthetic cry1Ab gene from Bacillus thuringiensis were highly resistant to eight lepidopteran rice pest species. Mol Breed. 2000;6:433–39. [Google Scholar]
- 6.Tang W, Chen H, Xu CG, Li XH, Lin YJ, Zhang QF. Development of insect-resistant transgenic indica rice with a synthetic cry1C* gene. Mol Breed. 2006;18:1–10. [Google Scholar]
- 7.Tu JM, Datta K, Firoz AM, Fan Y, Singh KG, Kumar DS. Expression and function of a hybrid Bt toxin gene in transgenic rice conferring resistance to insect pest. Plant Biotechnol. 1998;15:195–203. [Google Scholar]
- 8.Yang X, Li L, Cai XX, Wang F, Su J, Lu BR. Efficacy of insect-resistance Bt/CpTI transgenes in F5-F7 generations of rice crop-weed hybrid progeny: implications for assessing ecological impact of transgene flow. Sci Bull. 2015;60:1563–71. [Google Scholar]
- 9.Yao FY, Zhu CX, Li GX, Wen FJ. Identification of Bt rice for resistance to stripe stem borer and genetic analysis of transgenes. Scientia Agricultura Sinica. 2002;35:142–45. [Google Scholar]
- 10.Zhang CJ, Yook MJ, Park HR, Lim SH, Kim JW, Nah G, Song HR, Jo BH, Roh KH, Park S, et al. Assessment of potential environmental risks of transgene flow in smallholder farming systems in Asia: brassica napus, as a case study in Korea. Sci Total Environ. 2018;641:688–95. [DOI] [PubMed] [Google Scholar]
- 11.Jiang Y, Pan SG, Cai ML, Li CF, Zhan M, Wang JP, Mohamed I, Cao CG. Assessment of yield advantages of Bt-MH63 with cry1C* or cry2A* genes over MH63 (Oryza sativa L.) under different pest control modes. Field Crop Res. 2014;155:153–58. [Google Scholar]
- 12.Dong SS, Xiao MQ, Ouyang DX, Rong J, Lu BR, Su J, Wang F, Chen JK, Song ZP. Persistence of transgenes in wild rice populations depends on the interaction between genetic background of recipients and environmental conditions. Ann Appl Biol. 2017;171:202–13. [Google Scholar]
- 13.Li L, Yang X, Wang L, Yan HX, Su J, Wang F, Lu BR. Limited ecological risk of insect-resistance transgene flow from cultivated rice to its wild ancestor based on life-cycle fitness assessment. Sci Bull. 2016;61:1440–50. [Google Scholar]
- 14.Chen LY, Snow AA, Wang F, Lu BR. Effects of insect-resistance transgenes on fecundity in rice (Oryza sativa, Poaceae): a test for underlying costs. Am J Bot. 2006;93:94–101. [Google Scholar]
- 15.Jiang XJ, Tang L, Liu XJ, Cao WX, Zhu Y. Spatial and temporal characteristics of rice potential productivity and potential yield increment in main production regions of China. J Integr Agr. 2013;12:45–56. [Google Scholar]
- 16.Kim S, Kim C, Li W, Kim T, Li Y, Zaidi MA, Altosaar I. Inheritance and field performance of transgenic Korean Bt rice lines resistant to rice yellow stem borer. Euphytica. 2008;164:829–39. [Google Scholar]
- 17.Shu QY, Cui HR, Ye GY, Wu DX, Xia YM, Gao M, Altosaar I. Agronomic and morphological characterization of Agrobacterium-transformed Bt rice plants. Euphytica. 2002;127:345–52. [Google Scholar]
- 18.Xia H, Chen LY, Wang F, Lu BR. Yield benefit and underlying cost of insect-resistance transgenic rice: implication in breeding and deploying transgenic crops. Field Crop Res. 2010;118:215–20. [Google Scholar]
- 19.Liang YY, Liu F, Li JS, Cheng ZX, Chen HF, Wang XM, Xiao NW, Liu YB. Coexistence of Bt-transgenic and conventional rice affects insect abundance and plant fitness in fields. Pest Manag Sci. 2018;74:1646–53. [DOI] [PubMed] [Google Scholar]
- 20.Yang JS. Development and prospect of the research on sail-affected soils in china. Acta Pedologica Sinica. 2008;45:837–45. [Google Scholar]
- 21.Chen Y, Wang P, Wang KX, Chen N, Zhao L. The strategic choice of sea rice industry development in China. J Ocean U China. 2018;1:50–54. [Google Scholar]
- 22.Wang XJ, Liu QS, Meissle M, Peng YF, Li YH. Bt rice could provide ecological resistance against non-target planthoppers. Plant Biotechnol J. 2018;16:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu JM, Song XL, Liu B, Shen WJ, Fang ZX, Zhang L. Fitness cost of transgenic cry1Ab/c rice under saline-alkaline soil condition. Front Plant Sci. 2018;9:1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ling L, Jiang Y, Meng JJ, Cai LM, Cao GC. Phloem transport capacity of transgenic rice T1c-19 (Cry1C*) under several potassium fertilizer levels. PLoS One. 2018;13:e0195058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rawat P, Singh AK, Ray K, Chaudhary B, Kumar S, Gautam T, Kanoria S, Kaur G, Kumar P, Pental D. Detrimental effect of expression of Bt endotoxin Cry1Ac on in vitro regeneration, in vivo growth and development of tobacco and cotton transgenics. J Biosci. 2011;36:363. [DOI] [PubMed] [Google Scholar]
- 26.Tang H, Chen G, Chen FJ, Han LZ, Peng YF. Development and relative fitness of Cry1C resistance in Chilo suppressalis. Pest Manag Sci. 2018;74:590–97. [DOI] [PubMed] [Google Scholar]
- 27.Luo JY, Zhang S, Peng J, Zhu XZ, Lv LM, Wang CY, Li CH, Zhou ZG, Cui JJ. Effects of soil salinity on the expression of Bt toxin (Cry1Ac) and the control efficiency of Helicoverpa armigera in field-grown transgenic Bt cotton. PLoS One. 2017;12:e0170379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dong HZ, Xin CS, Li WJ. Soil salinity grading of cotton field in coastal saline area. Shandong Agric Sci. 2012;3:36–39. [Google Scholar]
- 29.Song ZP, Lu BR, Wang B, Chen JK. Fitness estimation through performance comparison of F1 hybrids with their parental species Oryza rufipogon and O.sativa. Ann Bot. 2014;93:311–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trtikova M, Wikmark OG, Zemp N, Widmer A, Hilbeck A. Transgene expression and bt protein content in transgenic bt maize (MON810) under optimal and stressful environmental conditions. PLoS One. 2015;10:e0123011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X, Wang J, Peng S, Li Y, Tian XF, Wang GC, Zhang ZN, Dong ZD, Chen Y, Chen DH. Effects of soil water deficit on insecticidal protein expression in boll shells of transgenic bt cotton and the mechanism. Front Plant Sci. 2017;8:2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J, Islam F, Li L, Long MJ, Yang C, Jin XL, Ali B, Mao BZ, Zhou WJ. Complementary RNA-sequencing based transcriptomics and iTRAQ proteomics reveal the mechanism of the alleviation of quinclorac stress by salicylic acid in Oryza sativa ssp. Japonica. Int J Mol Sci. 2017;18:1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu YB, Ge F, Liang YY, Wu G, Li JS. Characterization of competitive interactions in the coexistence of Bt-transgenic and conventional rice. BMC Biotechnol. 2015;15:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Addison SJ, Rogers DJ. Potential impact of differential production of the Cry2ab and Cry1ac proteins in transgenic cotton in response to cold stress. J Econ Entomol. 2010;103:1206–15. [DOI] [PubMed] [Google Scholar]
- 35.Jiang LJ, Duan LS, Tian XL, Wang BM, Zhang HF, Zhang MC, Li ZH. NaCl salinity stress decreased Bacillus thuringiensis (Bt) protein content of transgenic Bt cotton (Gossypium hirsutum L.) seedlings. Environ Exp Bot. 2006;55:315–20. [Google Scholar]
- 36.Luo Z, Dong HZ, Li WJ, Ming Z, Zhu YQ. Individual and combined effects of salinity and waterlogging on Cry1Ac expression and insecticidal efficacy of Bt cotton. Crop Prot. 2008;27:1485–90. [Google Scholar]
- 37.Traore SB, Carlson RE, Pilcher CD, Rice ME. Bt and Non-Bt maize growth and development as affected by temperature and drought stress. Agron J. 2000;92:1027. [Google Scholar]
- 38.Zeller SL, Kalinina O, Brunner S, Keller B, Schmid B. Transgene × environment interactions in genetically modified wheat. PLoS One. 2010;5:e11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang Y, Huang SQ, Cai ML, Li CF, Kong X, Zhang F, Mohamed I, Cao CG. Yield changes of Bt-MH63 with cry1C* or cry2A* genes compared with MH63 (Oryza sativa) under different nitrogen levels. Field Crop Res. 2013;151:101–06. [Google Scholar]
- 40.Wang F, Jian ZP, Nie LX, Cui KR, Peng SB, Lin YJ, Huang JL. Effects of N treatments on the yield advantage of Bt-SY63 over SY63 (Oryza sativa) and the concentration of Bt protein. Field Crop Res. 2012;129:39–45. [Google Scholar]
- 41.Jiang Y, Ling L, Zhang LL, Wang KX, Li XX, Cai ML, Zhan M, Li CF, Wang JP, Cao CG. Comparison of transgenic Bt rice and their non- Bt counterpart in yield and physiological response to drought stress. Field Crop Res. 2018;217:45–52. [Google Scholar]
- 42.Fang J, Nan P, Gu ZY, Ge XC, Feng YQ, Lu BR. Overexpressing exogenous 5-enolpyruvylshikimate-3-phosphate synthase (epsps) genes increases fecundity and auxin content of transgenic Arabidopsis plants. Front Plant Sci. 2018;9:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Betts SD, Basu S, Bolar J, Booth R, Chang S, Cigan AM, Farrell J, Gao H, Harkins K, Kinney A, et al. Uniform expression and relatively small position effects characterize sister transformants in maize and soybean. Front Plant Sci. 2019;10:1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets supporting the conclusions of this article are included within the article and its additional files.


