Abstract
Antiviral resistance frequently complicates the treatment of herpes simplex virus (HSV) infections in immunocompromised patients. Here we present the case of an adolescent boy with dedicator of cytokinesis 8 (DOCK8) deficiency, who experienced recurrent infections with resistant HSV-1. We used both phenotypic and genotypic methodologies to characterize the resistance profile of HSV-1 in the patient and conclude that genotypic testing outperformed phenotypic testing. We also present the first analysis of intrahost HSV-1 evolution in an immunocompromised patient. While HSV-1 can remain static in an immunocompetent individual for decades, the virus from this patient rapidly acquired genetic changes throughout its genome. Finally, we document a likely case of transmitted resistance in HSV-1 between the patient and his brother, who also has DOCK8 deficiency. This event demonstrates that resistant HSV-1 is transmissible among immunocompromised persons.
Keywords: HSV-1, DOCK8 deficiency, immunodeficiency, phenotypic antiviral resistance, genotypic antiviral resistance, intrahost evolution, transmitted resistance
We show that genotypic antiviral resistance testing for HSV-1 can outperform phenotypic testing, which remains the clinical standard. We also demonstrate that HSV-1 can evolve rapidly in an immunocompromised host and that resistant HSV-1 can be transmitted between immunocompromised persons.
Up to 10% of herpes simplex virus (HSV) infections in immunocompromised patients are due to acyclovir-resistant viruses [1]. Resistant HSV infections are particularly common among allogenic hematopoietic stem cell transplant patients, in whom they cause nearly half of all HSV infections [1]. In immunocompetent patients, resistant HSV infections are less commonly encountered but are responsible for 6% of all HSV keratitis cases [2].
The clinical management of antiviral-resistant HSV infections is complicated by a number of factors. Treatment options are limited by the small number of effective antivirals available and the toxicities associated with therapies like foscarnet, cidofovir, and interferon. Additionally, the development of resistance may be rapid, emerging with as little as 2 days of drug exposure [3]. Finally, HSV is the only virus for which resistance testing is routinely performed phenotypically in clinical settings. These tests have long turn-around times and high interlaboratory variability [4] and are available only for acyclovir, ganciclovir, and foscarnet.
Here we describe the case of an adolescent boy with hyper-IgE syndrome due to dedicator of cytokinesis 8 (DOCK8) deficiency. DOCK8 functions in the regulation of actin cytoskeletons and mutations in this gene result in numerous defects in the innate and adaptive immune systems, including impaired function, trafficking, and survival of B, T, NK, innate lymphoid, and dendritic cells, decreased cytokine production and responsiveness, and abnormal antibody production [5]. Cutaneous viral infections are a hallmark of DOCK8 deficiency, due to the failure of T and NK cells to migrate properly in skin [5]. The described patient experienced recurrent skin lesions due to resistant HSV-1, and our efforts to treat him demonstrate the clinical challenges associated with the management of antiviral-resistant HSV infections. We compare the results of phenotypic and genotypic antiviral-resistance testing for HSV-1 from this patient and conclude that results of genotypic testing are more consistent with his previous antiviral exposures and his clinical response to certain antiviral agents. We also conducted the first study of HSV-1 evolution in an immunocompromised host by examining the genomes of longitudinal samples from the patient. We find that the patient’s HSV-1 rapidly acquired changes throughout its genome, challenging the paradigm that HSV-1 is a slowly evolving virus in all contexts. Finally, we document a probable case of transmission of resistant HSV-1 between the patient and his brother, who also has DOCK8 deficiency.
METHODS
Sample Collection
The patient’s parents provided written consent for sample collection from the patient and his brother and for the publication of certain medical information, including photographs. This study was approved by the institutional review board of the University of Washington. Twelve HSV-1 samples were collected from the patient and 1 sample was collected from his younger brother during a 4-year period. All samples were collected from clinically apparent, cutaneous lesions. An aliquot from each sample was grown in culture to confirm the presence of HSV-1.
Phenotypic Analysis
Seven of the patient’s HSV-1 samples and 1 sample from the patient’s brother were sent to a commercial laboratory (ARUP Laboratories) for phenotypic resistance testing. This laboratory uses a chemiluminescence-based assay to determine half maximal effective concentration (IC50) values for acyclovir and foscarnet for HSV-1 samples. This method has been described previously [6]. Acyclovir sensitivity was defined as IC50 < 2µg/mL and resistance as IC50 ≥ 2µg/mL. Foscarnet sensitivity was defined as IC50 < 100µg/mL and resistance as IC50 ≥ 100µg/mL. An indeterminant result indicates failure of the assay results for a particular sample and a particular drug to meet quality control criteria (Supplementary Note 1).
Genome Sequencing and Analysis
We sequenced the genomes of 8 HSV-1 samples from the patient and 1 sample from his brother. DNA was extracted for sequencing directly from lesion swabs without amplification in culture. Instead a probe-capture next-generation sequencing technique described previously [7] was used to isolate and selectively amplify HSV genomic material. Consensus genomes and alignment files were generated from raw sequencing reads using a publicly available computational pipeline [7]. The average read depth across all 9 genomes was 351.8×. When the terminal and internal repeat regions and intragenic regions were removed from consideration, average read depth increased to 380.4× across all sequenced samples (Supplementary Table 1).
Consensus genomes were aligned to an HSV-1 reference sequence (strain 17, NC_001806.2) [8] with the multiple alignment using fast Fourier transform (MAFFT) algorithm [9]. Terminal repeats and intragenic regions were removed from the alignment. Single nucleotide variants (SNVs) were called relative to the reference using Geneious [10] with manual review. Variants were not called at sites with read depth of less than 10. Minor variants present in at least 10% of reads were also called using Geneious [10] with manual review. Minor variants were not called at sites with read depth less than 10, and we required that both alleles for minor variants be present on at least 2 reads. Antiviral resistance mutations were ascertained by comparing the SNVs and minor variants identified above to an online database of known HSV resistance mutations [11].
Data Sharing
Consensus genomes and short read archives have been submitted to GenBank under Bioproject PRJNA562737.
RESULTS
Phenotypic Resistance Testing Frequently Indeterminant or Inconsistent With Previous Antiviral Exposures
The patient was diagnosed with DOCK8 deficiency by genetic testing as a toddler. Oral acyclovir was started at the time of this diagnosis. Higher doses were administered for treatment of suspected or confirmed HSV disease and the patient was otherwise maintained on a lower, prophylactic dose. The patient was not exposed to any antivirals other than acyclovir prior to the study period (Supplementary Table 2).
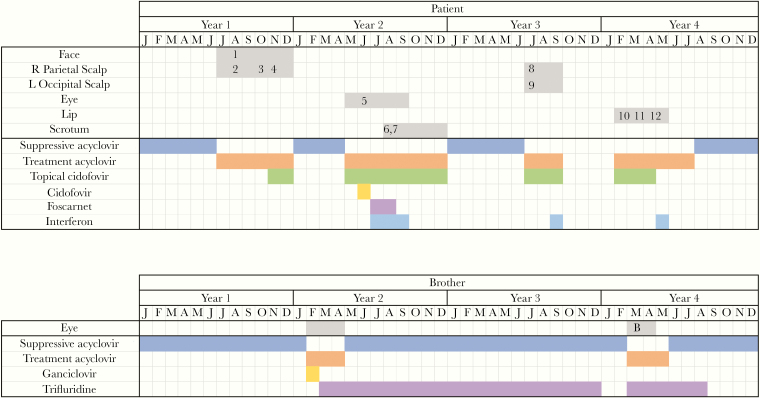
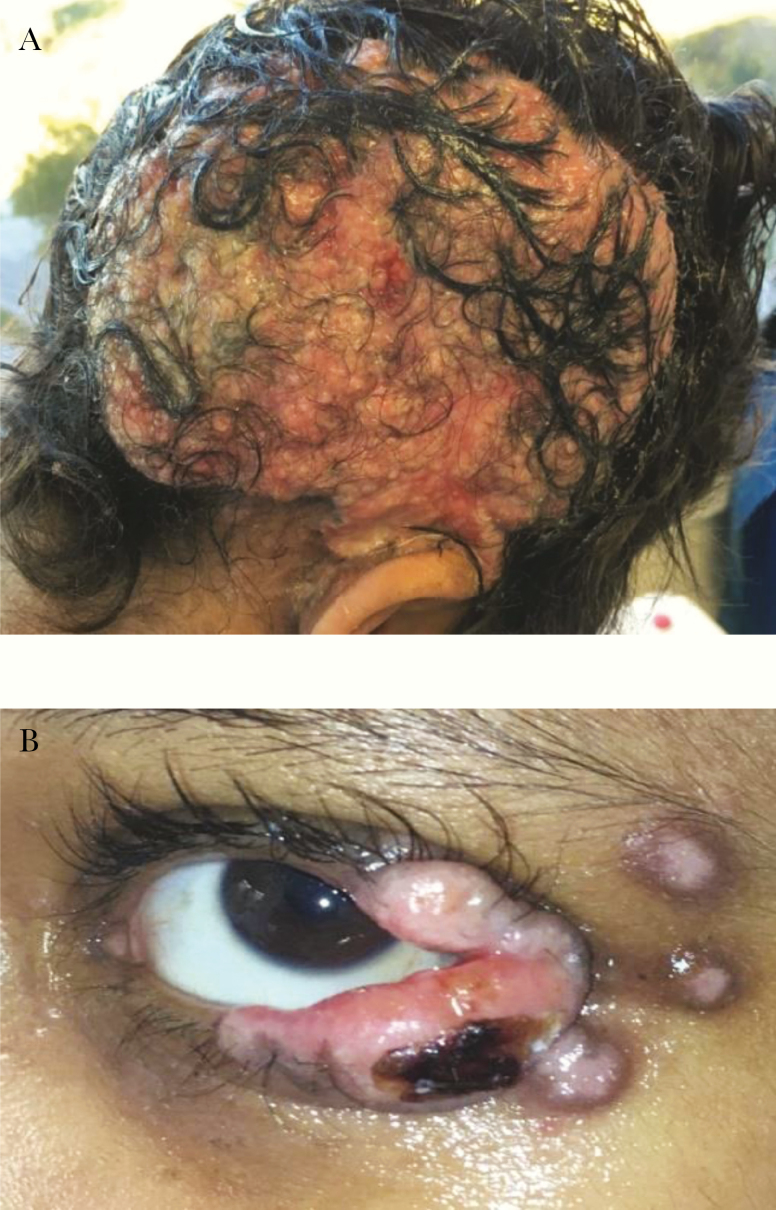
During the study period (patient aged 11 to 15 years), the patient experienced multiple culture-confirmed HSV-1 infections at different cutaneous sites (Table 1 and Figure 1). The first of these infections involved the right parietal scalp, ear, and face. A viral sample from the scalp was sent for phenotypic resistance testing. Results were indeterminant for acyclovir but showed foscarnet resistance, although the patient had no previous exposure to foscarnet. The patient initially improved on acyclovir (Supplementary Table 3) but the scalp lesion later worsened, developing into a large pink exudative plaque histologically consistent with herpes vegetans (Figure 2 and Figure 3). A second sample was sent for phenotypic testing and this result indicated acyclovir resistance and foscarnet sensitivity. Topical 2% cidofovir cream was added to the patient’s antiviral regimen and the lesions subsequently began to improve.
Table 1.
Samples Collected From the Patient and his Brother and Results of Phenotypic and Genotypic Testing for Resistance
| Result of Phenotypic Resistance Testing, IC50 | ||||||
|---|---|---|---|---|---|---|
| Sample Number | Subject | Site | Collection Date | Acyclovir | Foscarnet | Resistance Indicated by Genomic Analysis |
| 1 | Patient | Face | August, year 1 | Acyclovir | ||
| 2 | Patient | Right parietal scalp | August, year 1 | Acyclovir | ||
| 3 | Patient | Right parietal scalp | October, year 1 | I | R 101.6 | Acyclovir |
| 4 | Patient | Right parietal scalp | November, year 1 | R 10.6 | S 75.0 | Acyclovir |
| 5 | Patient | Eye | June, year 2 | I | I | |
| 6 | Patient | Scrotum | August, year 2 | Acyclovir, cidofovir | ||
| 7 | Patient | Scrotum | August, year 2 | R 11.1 | I | |
| 8 | Patient | Right parietal scalp | July, year 3 | R >64.0 | I | Acyclovir |
| 9 | Patient | Left occipital scalp | July, year 3 | Acyclovir | ||
| 10 | Patient | Lip | February, year 4 | S 1.7 | S 33.0 | |
| 11 | Patient | Lip | March, year 4 | Acyclovir | ||
| 12 | Patient | Lip | April, year 4 | R 3.1 | S 31.3 | |
| Brother | Eye | March, year 4 | I | S 12.9 | Acyclovir | |
Abbreviations: I, indeterminant; IC50, half maximal effective concentration; S, sensitive; R, resistant.
Figure 1.
Timeline of HSV-1 infections and antiviral regimens. Numbers correspond to samples numbers in Table 1. “B” denotes the brother’s sample in the timeline.
Figure 2.
A, Herpes vegetans of right parietal scalp. B, Exophytic lesions adjacent to patient’s left eye.
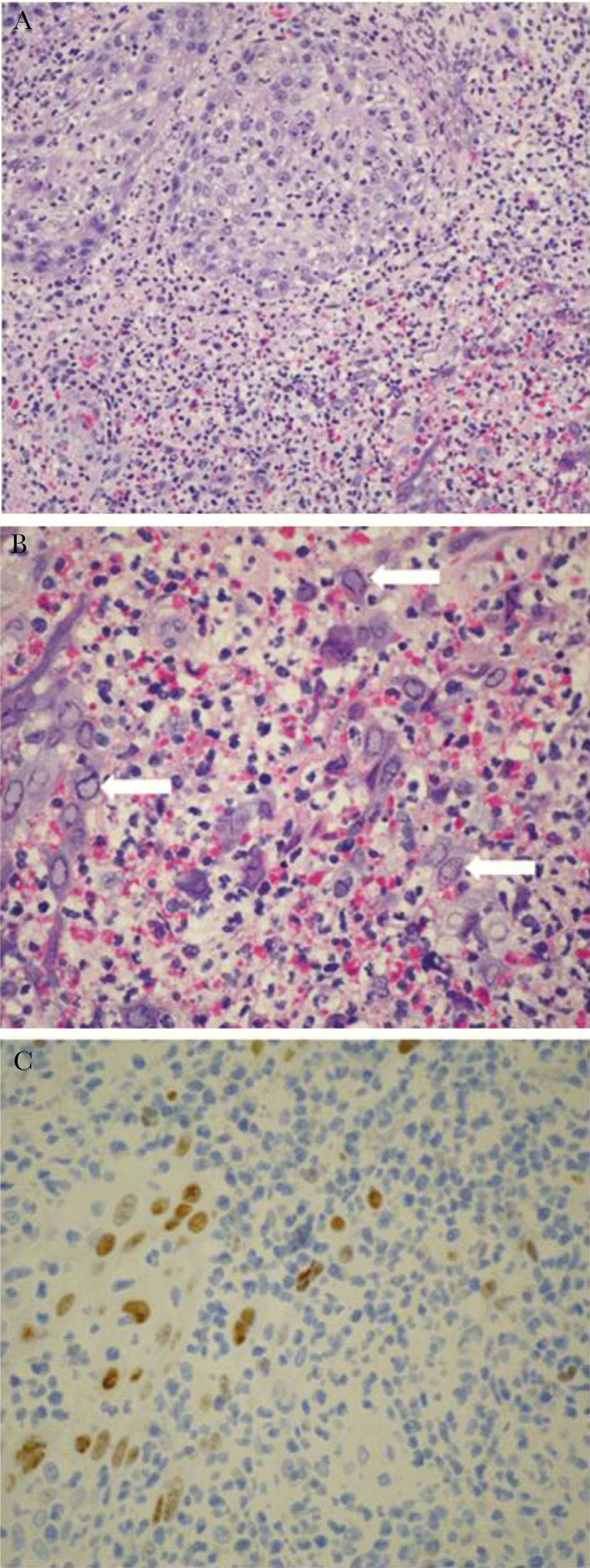
Figure 3.
A, Biopsy of herpes vegetans lesion revealing skin ulceration with pseudoepitheliomatous hyperplasia and intense dermal inflammation (H&E staining, ×200 magnification). B, Same scalp biopsy sample as in (A) under higher-power magnification with numerous nuclear viral inclusions (white arrows) characteristic of herpes viruses seen among the inflammatory cells (H&E staining, ×400 magnification). C, Immunohistochemistry for herpes simplex virus-1 performed on scalp biopsy sample showing presence of virus in tissue (×400 magnification).
Several months later, the patient developed purulent conjunctivitis of the left eye with adjacent exophytic papules and was treated with acyclovir. However, the periorbital lesions continued to worsen and systemic cidofovir was added. This regimen, too, failed to control the infection, so intravenous (IV) cidofovir was replaced with foscarnet and a dose of pegylated interferon α-2b was administered. After these treatment changes, the eye lesions began to improve. Phenotypic testing of virus from the periorbital lesions was performed but results were indeterminant for both acyclovir and foscarnet.
As the eye lesions healed, the patient developed ulcerated nodules of the scrotum. Phenotypic testing of a sample from these ulcers showed acyclovir resistance but was indeterminant for foscarnet.
The nodules were treated with a combination of acyclovir, interferon, topical cidofovir, and foscarnet. The latter 2 agents subsequently had to be stopped due to toxicity, but resolution of the nodules was eventually achieved with acyclovir and interferon.
For about 6 months after the scrotal lesions resolved, the patient had no evidence of active HSV-1 infection. The patient then developed a new lesion on the left occipital scalp and a smaller lesion on the right parietal scalp at the site of his previous infection. A sample from the right scalp was sent for phenotypic testing, which indicated resistance to acyclovir but was indeterminant for foscarnet. The scalp lesions were treated with acyclovir and topical cidofovir with some improvement. However, the addition of interferon was again required to achieve resolution.
Between each of the 4 HSV-1 infections described above, the patient was maintained on low-dose, suppressive acyclovir. However, acyclovir was discontinued after resolution of the scalp lesions in the hope that the patient’s HSV-1 would become more susceptible. About 5 months later, the patient developed ulcerations and a pustule on his upper lip. Two samples were sent from these lesions for phenotypic testing. The first showed acyclovir sensitivity while the second demonstrated acyclovir resistance. Both resulted as sensitive to foscarnet. The lip lesions were initially treated with acyclovir and topical cidofovir. The latter subsequently had to be stopped due to cutaneous irritation at the application site with associated bleeding. It was replaced with interferon. After this substitution, the lip lesions began to improve.
Patient’s Younger Brother Also Experienced Infections Due to Resistant HSV-1
The patient’s younger brother presented as an infant with a papulopustular eruption that tested positive for Candida. Given his brother’s history, he underwent genetic testing, which confirmed DOCK8 deficiency, and he was started on prophylactic oral acyclovir to prevent HSV disease. Despite prophylaxis, he developed HSV gingivostomatitis with lesions that spread to the face at age 5 years. These lesions resolved after weeks of treatment with IV acyclovir. Several years later at age 10 years, he developed HSV keratitis and was treated with acyclovir and ganciclovir eye drops. When he failed to improve on this regimen, the ganciclovir was discontinued and he was treated with a prolonged course of trifluridine drops. Two years later, he experienced a recurrence of HSV keratitis, which was again treated with acyclovir and trifluridine drops. A sample was sent for phenotypic testing. The test for acyclovir resistance was indeterminant but the sample was found to be sensitive to foscarnet.
Genotypic Testing for Antiviral Resistance Consistent With History of Antiviral Exposures
To better understand the resistance profile of the patient’s HSV-1, we performed a retrospective genomic analysis on 8 of his viral samples. Consistent with the patient’s chronic exposure to acyclovir, all sequenced samples were found to carry a single nucleotide consensus change in the thymidine kinase gene known to confer acyclovir resistance (c.527G > A, p.Arg176Gln) [12–14]. HSV-1 from the patient’s brother also carried this same acyclovir resistance mutation. Additionally, the sample collected from the patient’s scrotal ulcers was found to carry a single nucleotide consensus change in the DNA polymerase gene known to confer both acyclovir and cidofovir resistance (c.2462C > T, p.Thr821Met) [15, 16]. This mutation may also confer foscarnet resistance [15, 16]. This sample was collected 1 month after the patient was exposed to systemic cidofovir and after initiation of foscarnet therapy. No other genetic variants known to confer antiviral resistance were observed in any of the 9 sequenced samples as consensus changes or as minor variants (Supplementary Table 4 and 5) [11].
Samples From the Patient Rapidly Accumulated Genetic Changes Across the Genome
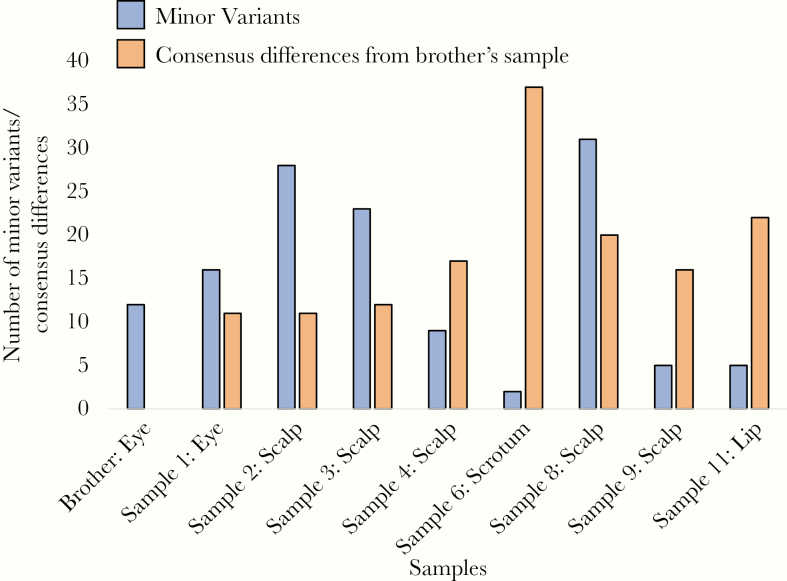
To examine how HSV-1 evolves in an immunocompromised host over time, we next evaluated the genic regions of the 8 sequenced HSV-1 genomes from the patient in their entirety. While 1 pair of samples had no consensus differences (Supplementary Table 6), the average number of SNV pairwise differences between the patient’s samples was 18 and the highest number observed was 43. The genomic loci that varied among the patient’s samples (both as consensus changes and minor variants) were distributed throughout the genome. At least 1 consensus SNV or minor variant was present in 76% of all genes and 59% of all genes contained at least 1 nonsynonymous SNV or minor variant. Anatomic site appeared to play a role in the evolution of HSV-1 in the patient. The most divergent of the patient’s samples came from the scrotal ulcers, which were physically distant from all other lesions. The scrotal sample was also more homogeneous than the other samples with only 2 minor variants observed (Figure 4) and was the only sample from the patient to carry a second resistance mutation. HSV-1 from the patient closely resembled HSV-1 from his younger brother. Only 11 SNV differences were observed between the brother’s sample and the first sequenced sample from the patient. The brother’s HSV-1 also carried the same acyclovir-resistance mutation in thymidine kinase that was observed in the patient, suggesting that the patient and his brother were infected with closely related viruses. Finally, we noted that later samples from the patient tended to be less similar to the brother’s HSV-1 sample than earlier ones, indicating that the patient’s HSV-1 gradually accumulated new consensus changes over time.
Figure 4.
The number of minor variants observed in each sequenced sample. The number of consensus differences between each of the patient’s samples and the brother’s sample is also shown.
Discussion
Phenotypic testing is currently the standard method for assessing antiviral resistance in HSV in clinical settings. Its utility is limited by its long turn-around times (Supplementary Table 7), its unavailability for some antivirals, and its frequent failure to provide definitive results. For the 8 samples that we sent for phenotypic testing, the average turn-around time from sample collection to receipt of resistance results was 29 days for acyclovir and 35 days for foscarnet. Five out of these 8 samples had indeterminant results for at least 1 antiviral and no resistance testing was available for cidofovir, which we frequently used to treat the patient. Additionally, foscarnet resistance was reported for the first sample sent for testing. In retrospect, we suspect that this result was inaccurate, given that the patient had no previous exposure to this drug and that he subsequently responded well to it. We also noted that the foscarnet IC50 for sample 3 was just above the cutoff for resistance. Because of the phenotypic testing result suggesting resistance, we avoided using foscarnet to treat the herpes vegetans of the right parietal scalp and delayed its use for the periorbital lesions until other agents had been tried. Thus, inaccurate results from phenotypic testing can lead to avoidance or delay in administration of an effective therapy.
The main limitation of genetic antiviral resistance testing for HSV is that some variants in the thymidine kinase and DNA polymerase genes have not yet been phenotypically characterized. Nonetheless, in this case, genotypic testing predicted a resistance profile for the sequenced samples that was consistent with the patient’s previous exposures and his clinical responses to antiviral agents. In particular, the acyclovir resistance mutation found in all samples was consistent with the patient’s long history of exposure to the drug and explained why the patient’s infections did not respond well to acyclovir as monotherapy. Genotypic testing also indicated the presence of a second resistance mutation (c.2462C > T, p.Thr821Met) in the scrotal sample. This mutation may have been selected for because of the patient’s frequent exposure to acyclovir. However, as this mutation confers cidofovir and possibly foscarnet resistance, its appearance may also be the result of the patient having been recently exposed to systemic cidofovir and foscarnet for the first time. Given these observations, we think that genotypic testing outperformed phenotypic testing for this patient, although we acknowledge that our assessment of clinical response to antivirals may have been confounded by the patient’s medical complexity and the effects of agents he received to treat other issues.
In addition to the information they provided about antiviral resistance, the HSV-1 genomes from the patient offered an unprecedented look at HSV-1 evolution in an immunocompromised host. HSV-1 can remain genomically static in immunocompetent hosts over decades [17]. This stasis stands in stark contrast to the multiple SNV changes and minor variants that we observed in our patient’s samples (Supplementary Notes 2 and 3). This difference in the rate of viral genomic change between immunocompetent and immunocompromised hosts is the likely genesis of the difference in the frequency of antiviral resistance between the 2 groups. The high mutation rate of HSV-1 in immunocompromised persons has important implications for the future management of antiviral resistance in HSV. First, all identified antiviral resistance mutations for antivirals currently in use for HSV are located in either the thymidine kinase or DNA polymerase genes. However, consensus sequence changes were observed across the genome in our immunocompromised patient, suggesting that if antiviral resistance mutations are ever identified in other genes, these mutations may also be observed at higher frequencies in immunocompromised hosts. Secondly, the high mutation rate explains why immunocompromised hosts do not require prolonged exposure to an antiviral to develop resistance. Finally, this high mutation rate can lead to the presence of genetically distinct populations of HSV-1 with different antiviral resistance profiles coexisting within a single patient, further complicating the management of HSV-1 infections in immunocompromised hosts.
The HSV-1 sample collected from the patient’s scrotal ulcers was unique among the patient’s samples for carrying a second antiviral resistance mutation. It was also the most divergent of the patient’s samples and the most homogeneous. All of the patient’s other cutaneous lesions were located on the head, suggesting that they were seeded by viral populations emerging from the trigeminal ganglia. The scrotal HSV-1 population may have instead been seeded by inoculation from another cutaneous lesion. The low population diversity seen in the scrotal sample relative to the patient’s other samples may indicate that the scrotal HSV-1 population was founded by a small number of viruses. This observation suggests that the patient was either chronically shedding HSV-1 from the face and scalp, thereby allowing these populations to accumulate significant diversity, or that each face and scalp lesion was founded by relatively large and diverse populations of virus emerging from the ganglionic reservoir.
Finally, we observed that the patient’s HSV-1 was genetically similar to an HSV-1 sample from his brother and that the patient’s and brother’s viruses shared the same acyclovir resistance mutation (c.527G > A, p.ARG176Gln). As there are numerous mutations that confer acyclovir resistance, the presence of this shared resistance mutation suggests its transmission either from the patient to his younger brother or from a third individual to both boys. To our knowledge, this is the first description of probable transmitted resistance in HSV-1. Despite this observation, many questions still remain about the transmissibility of resistant HSV-1. All of the sampled viruses described here were viable in their DOCK8-deficient hosts and when grown in vitro on Vero cells (Supplementary Note 4). However, the fitness of these viruses relative to wildtype viruses and the extent of their ability to infect immunocompetent and other immunocompromised hosts remains unclear. Answering these questions will be key to formulating clinical and public health approaches to antiviral resistance in HSV going forward.
One limitation of the study is that most of the samples had phenotypic resistance information or genotypic resistance information but not both. Of the 13 samples that were used in this study, only 4 underwent both genotypic and phenotypic antiviral resistance testing. This introduces some uncertainty into our comparison of the results of phenotypic and genotypic testing. This uncertainty is perhaps most relevant for sample 10. Phenotypic testing indicated that this sample was sensitive to both acyclovir and foscarnet. We suspect that the result of the phenotypic assay for acyclovir sensitivity in this case was inaccurate, because an acyclovir resistance mutation was fixed in all sequenced samples from the patient. However, we acknowledge that we cannot be certain about this, as we have no genotypic data from sample 10. We would emphasize that genotypic and phenotypic resistance testing results were concordant for 3 of the 4 samples that underwent both types of testing. For the fourth, foscarnet sensitivity was suggested by genotypic testing while the phenotypic assay indicated foscarnet resistance. In this case, we think that the absence of previous exposure of the patient to foscarnet leaves the phenotypic result in question, though it remains possible that the lack of concordance in this case was due to a shortcoming of the genotypic testing. However, the latter explanation seems unlikely to us as all known foscarnet resistance mutations are in the DNA polymerase and no variants were observed in this gene in Sample 3 that were not present in other samples.
In conclusion, we have demonstrated how the challenges of treating resistant HSV in immunocompromised hosts can be compounded by the limitations of phenotypic antiviral resistance testing. We have also demonstrated that genetic testing for resistance using next-generation sequencing is a promising alternative, although one that has not yet been approved for clinical use. We also showed that in the context of immunocompromise, HSV-1 rapidly accumulates changes throughout its genome. This finding explains the observed ability of HSV to quickly develop resistance in immunocompromised patients. Finally, our analysis of HSV-1 from the 2 immunocompromised subjects of this study revealed the first likely case of transmitted resistance for this virus, an observation that raises interesting questions about the fitness and transmissibility of resistant HSV-1 and impact of antiviral resistance on the broader HSV population.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgements. We thank the patient and his family for their courage, strength, and dignity in all that they endured.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases (NIAID) (grant number 5T32AI118690-04 T32 Training Grant to A. M. C. at the Vaccine and Infectious Diseases Division, Fred Hutchinson Cancer Research Institute, Seattle, Washington, USA); and the NIAID Intramural Research Program.
Potential conflicts of interest. A. L. G. has consulted for Abbott Molecular. D. E. Y. has been an investigator on antiviral studies initiated and sponsored by Chimerix, Inc., antifungal studies initiated and sponsored by Astellas, an antifungal study initiated and sponsored by Merck, an antifungal study initiated and sponsored by Pfizer, and an investigator-initiated fungal diagnostics study supported by Viracor-Eurofins. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Presented in part: International Herpes Workshop, Knoxville, TN, 20–24 July 2019.
References
- 1. Frobert E, Burrel S, Ducastelle-Lepretre S, et al. Resistance of herpes simplex viruses to acyclovir: an update from a ten-year survey in France. Antiviral Res 2014; 111:36–41. [DOI] [PubMed] [Google Scholar]
- 2. Piret J, Boivin G. Antiviral resistance in herpes simplex virus and varicella-zoster virus infections: diagnosis and management. Curr Opin Infect Dis 2016; 29:654–62. [DOI] [PubMed] [Google Scholar]
- 3. Danve-Szatanek C, Aymard M, Thouvenot D, et al. Surveillance network for herpes simplex virus resistance to antiviral drugs: 3-year follow-up. J Clin Microbiol 2004; 42:242–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bacon TH, Levin MJ, Leary JJ, Sarisky RT, Sutton D. Herpes simplex virus resistance to acyclovir and penciclovir after two decades of antiviral therapy. Clin Microbiol Rev 2003; 16:114–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Biggs CM, Keles S, Chatila TA. DOCK8 deficiency: insights into pathophysiology, clinical features and management. Clin Immunol 2017; 181:75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tardif KD, Jorgensen S, Langer J, Prichard M, Schlaberg R. Simultaneous titration and phenotypic antiviral drug susceptibility testing for herpes simplex virus 1 and 2. J Clin Virol 2014; 61:382–6. [DOI] [PubMed] [Google Scholar]
- 7. Greninger AL, Roychoudhury P, Xie H, et al. Ultrasensitive capture of human herpes simplex virus genomes directly from clinical samples reveals extraordinarily limited evolution in cell culture. mSphere 2018; 3:pii: e00283-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Davison AJ. Evolution of sexually transmitted and sexually transmissible human herpesviruses. Ann N Y Acad Sci 2011; 1230:E37–49. [DOI] [PubMed] [Google Scholar]
- 9. Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 2013; 30:772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kearse M, Moir R, Wilson A, et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012; 28:1647–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sauerbrei A, Bohn-Wippert K, Kaspar M, Krumbholz A, Karrasch M, Zell R. Database on natural polymorphisms and resistance-related non-synonymous mutations in thymidine kinase and DNA polymerase genes of herpes simplex virus types 1 and 2. J Antimicrob Chemother 2016; 71:6–16. [DOI] [PubMed] [Google Scholar]
- 12. Chibo D, Druce J, Sasadeusz J, Birch C. Molecular analysis of clinical isolates of acyclovir resistant herpes simplex virus. Antiviral Res 2004; 61:83–91. [DOI] [PubMed] [Google Scholar]
- 13. Bestman-Smith J, Schmit I, Papadopoulou B, Boivin G. Highly reliable heterologous system for evaluating resistance of clinical herpes simplex virus isolates to nucleoside analogues. J Virol 2001; 75:3105–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kussmann-Gerber S, Kuonen O, Folkers G, Pilger BD, Scapozza L. Drug resistance of herpes simplex virus type 1—structural considerations at the molecular level of the thymidine kinase. Eur J Biochem 1998; 255:472–81. [DOI] [PubMed] [Google Scholar]
- 15. Andrei G, Fiten P, Froeyen M, De Clercq E, Opdenakker G, Snoeck R. DNA polymerase mutations in drug-resistant herpes simplex virus mutants determine in vivo neurovirulence and drug-enzyme interactions. Antivir Ther 2007; 12:719–32. [PubMed] [Google Scholar]
- 16. Bestman-Smith J, Boivin G. Drug resistance patterns of recombinant herpes simplex virus DNA polymerase mutants generated with a set of overlapping cosmids and plasmids. J Virol 2003; 77:7820–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pandey U, Renner DW, Thompson RL, Szpara ML, Sawtell NM. Inferred father-to-son transmission of herpes simplex virus results in near-perfect preservation of viral genome identity and in vivo phenotypes. Sci Rep 2017; 7:13666. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.