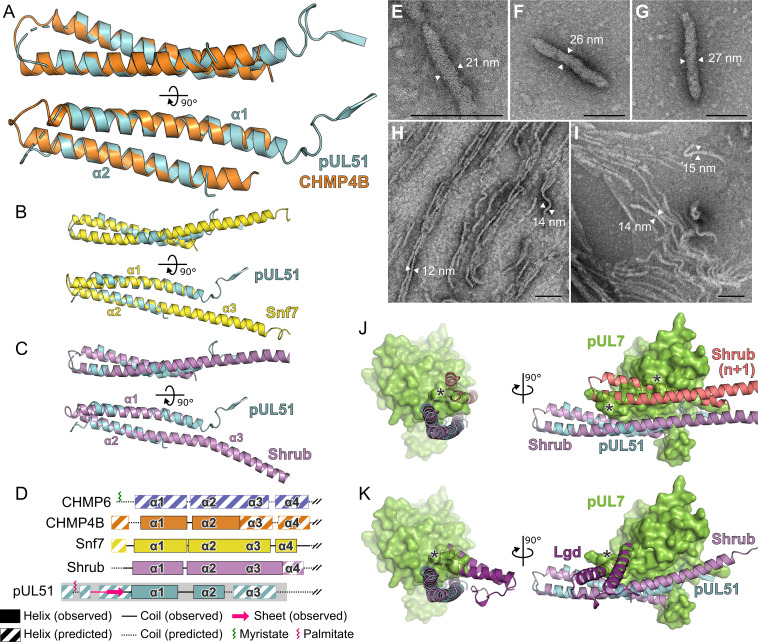
Figure 5. Structural similarity of HSV-1 pUL51 to cellular ESCRT-III proteins.
(A) pUL51 (cyan) is shown superposed on the helical hairpin (conserved helices α1 and α2) of human CHMP4B (orange; PDB 4ABM) (Martinelli et al., 2012). (B and C) pUL51 (cyan) superposed on conserved helices α1 and α2 of CHMP4B homologues (B) yeast Snf7 (yellow; PDB 5FD7) (Tang et al., 2015) and (C) fly Shrub (violet; PDB 5J45) (McMillan et al., 2016). Note helices α2 and α3 of the ESCRT-III core domains of that Snf7 and Shrub are elongated and continuous in polymeric forms of these proteins (Tang et al., 2015; McMillan et al., 2016). (D) Schematic representation of selected cellular ESCRT-III protein core domains and pUL51. Residues 1–150 of cellular ESCRT-III proteins and 1–190 of pUL51 are depicted. Secondary structure of crystal structures shown in panels A–C are in solid lines (coil) and solid boxes (helices). Predicted secondary structure (Petersen et al., 2009) outside these regions is shown as dotted lines (coil) and striped boxes (helices). The N-terminal region of pUL51 that forms a β-sheet with the pUL7 cloning tag, presumably an artefact of crystallization, and preceding residues are shown in pink. The region of pUL51 used for electron microscopy analysis is shaded in grey. Myristoylation (CHMP6) or palmitoylation (pUL51) sites are indicated by green and purple sticks, respectively. (E–I) Negative stain transmission electron microscopy images of pUL51 filaments. Scale bars, 100 nm. (E–G) Representative images of pUL51 proto-filaments formed when 100 µM pUL51(1–170) in 20 mM tris pH 8.5 was incubated on grids for 30 s before staining. (H, I) Representative images of pUL51 filaments formed when 10 µM pUL51(1–170) in 20 mM HEPES pH 7.5 was incubated on grids for 1–2 min before staining. (J and K) The pUL7:pUL51 core heterodimer is shown superposed onto (J) two subunits of the putative Shrub homopolymer (violet and pink; PDB 5J45) (McMillan et al., 2016), or (K) the complex of Shrub with the regulatory DM14-3 domain of Lgd (purple; PDB 5VO5) (McMillan et al., 2017). pUL7 is shown as a green molecular surface. Spatial overlap between pUL7 and (J) the second subunit of Shrub, or (K) the Lgd DM14-3 domain, is denoted by asterisks.