Abstract
PRAME (PReferentially expressed Antigen in MElanoma) is a melanoma-associated antigen. While diffuse immunoreactivity for PRAME is found in most primary cutaneous melanomas, melanocytic nevi express PRAME usually only in a subpopulation of tumor cells or not at all. Hence, testing for PRAME expression has the potential to provide useful information for the assessment for diagnostically ambiguous melanocytic neoplasms. Many of the latter tumors are currently studied by cytogenetic methods for ancillary evidence in support of or against a diagnosis of melanoma. In this study we analyzed 110 diagnostically problematic melanocytic tumors comparing results for PRAME IHC with those from fluorescence in situ hybridization (FISH) and/or single nucleotide polymorphism (SNP)-array, and each with the final diagnostic interpretation. In 90% of cases there was concordance between PRAME IHC and cytogenetic tests results, and in 92.7% concordance between PRAME IHC and the final diagnosis. The high concordance between PRAME IHC and cytogenetic test results as well as the final diagnosis supports the use of PRAME IHC as an ancillary test in the evaluation of ambiguous primary cutaneous melanocytic neoplasms, especially given its practical advantage of lower cost and faster turnaround over cytogenetic or gene expression studies. However, our results indicate that PRAME IHC and cytogenetic tests for melanocytic tumors are not entirely interchangeable and on occasion each type of test may yield false-negative or false-positive results.
Keywords: PRAME, immunohistochemistry, melanoma, nevus, FISH, SNP-array
INTRODUCTION
PRAME (PReferentially expressed Antigen in MElanoma) is a tumor-associated antigen that was identified through autologous T-cell epitope cloning in a patient with metastatic cutaneous melanoma[1]. It was subsequently found that PRAME is not only expressed in cutaneous melanoma, but also ocular melanoma and various non-melanocytic malignant neoplasms, including non-small cell lung cancer, breast carcinoma, renal cell carcinoma, ovarian carcinoma, leukemia, synovial sarcoma and myxoid liposarcoma[2–11]. PRAME expression in normal healthy tissues is essentially restricted to testis, ovary, placenta, adrenals, and endometrium[1, 12]. Because of its expression profile, PRAME is a member of the family of cancer testis antigens (CTA), and considered a potential target for immunotherapy [7, 13].
PRAME is also a component of a 23-gene array diagnostic assay for cutaneous melanoma[14, 15], and one of the two genes used in a non-invasive molecular assay for guiding clinicians on the need for biopsy of a melanocytic lesion[16].
We have previously documented the detection of PRAME expression by immunohistochemistry in a cohort of 400 melanocytic tumors with histomorphology that allowed unequivocal classification as malignant melanoma or benign nevi. We found that 88 to 94% of non-spindle cell primary cutaneous melanomas show diffuse immunoreactivity for PRAME while most nevi (86%) are negative for PRAME with a few benign lesions (~14%) showing staining for PRAME in a subset (generally <50%) of lesional cells. Only one case (<1%) corresponding to a pigmented junctional Spitz nevus in a child showed diffuse PRAME labeling. [17]
While most melanocytic tumors can be classified as benign or malignant based on histomorphology and clinical findings, occasionally ambiguous microscopic features and clinical context decrease the certainty of classification and complicate the diagnostic assessment. Cytogenetic tests, including FISH for melanoma and genome-wide array-CGH, have been proven valuable in the assessment of challenging melanocytic tumors.[18–23] However, cost, turnaround time, relatively limited availability, as well as limitations in the sensitivity and specificity of these tests can be problematic and limit their use in certain cases. In current practice, optimal interpretation of ambiguous melanocytic tumors rests in the integration of expert histomorphological assessment, results of ancillary tests, and clinical context.
Herein we evaluate the use of PRAME IHC in 110 melanocytic tumors with ambiguous features by assessing percentage agreement with results of FISH for melanoma and/or single nucleotide polymorphism (SNP)-array and final diagnostic interpretation.
MATERIALS AND METHODS
Case Selection
110 melanocytic tumors with ambiguous features upon H&E examination by at least 2 dermatopathologists were retrieved from our cancer center’s pathology archive under an IRB-approved protocol. All cases had FISH for melanoma and/or SNP-array and PRAME IHC performed as part of the evaluation of the tumors for clinical care.
Immunohistochemical Analysis
Five-micron thick tissue sections were cut from formalin fixed and paraffin embedded (FFPE) tissue blocks of all cases (n=110). A commercially available anti-PRAME monoclonal antibody (mAb EPR20330; Abcam, #219650) was used on a Leica-Bond-3 autostainer platform. The staining of PRAME IHC was recorded as the percentage of immunoreactive tumor cells with nuclear labeling per total number of tumor cells. 0 indicated no staining at all. Staining of 1 – 25% of tumor cells was scored as 1+. Labeling of 26 – 50% of tumor cells was scored as 2+. If 51–75% of tumor cells were positive, it was designated as 3+. If 76% or more of the tumor cells were positive, it was recorded as 4+ or “diffuse”.
FISH for melanoma
FISH analysis was performed on FFPE tissue sections using locus-specific probes and centromere probes (CEP) (Abbott Molecular, Inc., Des Plaines, IL) in 77 cases. The four-color FISH panel consists of locus-specific probes targeting the RREB1 gene (6p25), MYB gene (6q23), and CCND1 gene (11q13) and the centromere of chromosome 6 (CEP6) probe, with additional probe sets for CDKN2A(9p21)/CEP9. FFPE sections (4 μm) generated from FFPE blocks of tumor specimens were pretreated by deparaffinizing in xylene and dehydrating in ethanol. Dual-color FISH was performed according to the Vysis protocol for FFPE sections (Abbott Molecular) with a few minor modifications. FISH analysis and signal capture were performed using fluorescence microscopes (Axio; Carl Zeiss AG, Jena, Germany) coupled with an ISIS FISH Imaging System (MetaSystems GmbH, Altlussheim, Germany). Thirty tumor cell interphase nuclei were evaluated for each specimen. Cutoff values for positive results are as follows: gains of RREB1 > 29%, or relative gains of RREB1/CEP 6 > 55%, or relative loss of MYB/CEP 6 > 40%, or gains of CCND1 in > 38%, or homozygous loss of CDKN2A (p16) > 33% of the cells examined.
SNP array
SNP array using the Affymetrix OncoScan assay (Affymetrix, Santa Clara, CA) as previously described[24] was performed on 40 cases.
Criteria for interpretation of ancillary tests results
Interpretation of PRAME IHC as “positive or in favor of a diagnosis of melanoma” was defined as immunoreactivity in >75% of lesional cells (4+). PRAME IHC scores of 0 to 3+ were interpreted as “negative or not supportive of melanoma”. Results of FISH for melanoma were interpreted as “positive or negative” according to previously published cut-off values.[25] Results of SNParray were considered “negative or not supportive of melanoma” when no unbalanced genomic aberrations were detected or when only an isolated aberration known to occur in indolent lesions (e.g. isolated gain of 11p in Spitz nevi) were found. Results of SNP-array were recorded as “positive or in favor of a diagnosis of melanoma” when ≥ 3 segmental chromosomal aberrations or a combination typical of melanoma (e.g. gain in 6p, loss of 6q) were detected.[18, 19, 21, 22, 26] Of the total of 40 cases where SNParray results were available, 30 cases could be classified as “positive” or “negative” according the aforementioned criteria. The remainder 10 cases showed abnormal SNP-array results of uncertain significance that could not readily be classified as “positive” or “negative” as per the criteria above, and thus were excluded from inter-test agreement calculations between PRAME IHC and cytogenetics studies, and from agreement calculations between cytogenetic test results and final diagnosis.
Final diagnostic interpretation with integration of all available morphologic, clinical, and ancillary tests data was performed by consensus of at least 2 dermatopathologists.
RESULTS
Clinical and morphologic characteristics of ambiguous melanocytic neoplasms
110 cutaneous melanocytic neoplasms with ambiguous histomorphology were evaluated. Patient ages ranged from 2 to 90 years (mean = 41.1, median =41.5). 57 were female (51.8%), 53 were male (48.2%). Tumors arose in the trunk (31.8%), head and neck (24.5%), lower extremity (23.6%), upper extremity (17.3%), perineum (1.8%), and site was unknown in one case (0.9%). All lesions were either intradermal or compound (with epidermal and dermal components). They were grouped by the diagnostic challenge they posed as atypical Spitz tumor/nevus versus spitzoid melanoma (n=42), dysplastic nevus versus melanoma (n=26), and traumatized or mitotically active nevus versus nevoid melanoma (n=33). Additional 9 cases involved the following diagnostic problems: combined nevus versus melanoma (n=3), deep penetrating nevus versus melanoma (n=2), pigmented epithelioid melanocytoma versus melanoma (n=2), sclerosing blue nevus versus desmoplastic melanoma (n=1), and acral nevus versus melanoma (n=1) (Table 1).
Table 1.
Summary of characteristics of 110 ambiguous melanocytic tumors.
| Gender (%) | Age range (mean; median) | PRAME IHC | FISH | SNP-array | Dx | PRIHC & FISH/SNPa agreement (%) | PRIHC & Dx agreement (%) | |
|---|---|---|---|---|---|---|---|---|
| Spitzoid neoplasm (n=42) | F: 23 (54.8) M: 19 (45.2) |
2–78 (27.3; 19) |
4+: 6 0–3+: 36 |
Pos: 4 Neg: 12 |
Pos: 4 Neg: 17 Ab: 8 |
MM: 7 Ind: 35 |
31/34 (91.2) |
39/42 (92.9) |
| DysN vs MM (n=26) |
F: 9 (34.6) M: 17 (65.4) |
19–81 (50.6; 47.5) |
4+: 5 0–3+: 21 |
Pos: 5 Neg: 20 |
Pos: 0 Neg: 0 Ab: 1 |
MM: 6 Ind: 20 |
23/25 (92) |
25/26 (96.2) |
| Nevoid (n=33) | F: 19 (57.6) M: 14 (42.4) |
13–90 (50.5; 51) |
4+: 9 0–3+: 24 |
Pos: 11 Neg: 18 |
Pos: 4 Neg: 4 Ab: 0 |
MM: 13 Ind: 20 |
28/33 (84.8) |
29/33 (87.9) |
| Combined nevus vs MM (n=3) | F: 3 (100) | 5–31 (21.3; 28) |
4+: 0 0–3+: 3 |
Pos: 0 Neg: 2 |
Pos: 0 Neg: 0 Ab: 1 |
MM: 0 Ind: 3 |
2/2 | 3/3 |
| DPN vs MM (n=2) |
F: 1 M: 1 |
35, 73 | 4+: 1 0–3+: 1 |
Pos: 1 Neg: 1 |
Pos: 0 Neg: 0 Ab: 0 |
MM: 1 Ind: 1 |
2/2 | 2/2 |
| PEM vs MM (n=2) |
F: 1 M: 1 |
25, 81 | 4+: 1 0–3+: 1 |
Pos: 0 Neg: 1 |
Pos: 1 Neg: 0 Ab: 0 |
MM: 1 Ind: 1 |
2/2 | 2/2 |
| Acral nevus vs MM (n=1) | F: 1 | 46 | 4+: 0 0–3+: 1 |
Pos: 0 Neg: 1 |
Pos: 0 Neg: 0 Ab: 0 |
MM: 0 Ind: 1 |
1/1 | 1/1 |
| Blue nevus vs MM (n=1) | M: 1 | 67 | 4+: 0 0–3+: 1 |
Pos: 0 Neg: 1 |
Pos: 0 Neg: 0 Ab: 0 |
MM: 0 Ind: 1 |
1/1 | 1/1 |
| Total (n=110) | F: 57 (51.8) M: 53 (48.2) |
2–90 (41.1; 41.5) |
4+: 22 0–3+: 88 |
Pos: 21 Neg: 56 |
Pos: 9 Neg: 21 Ab: 10* |
MM: 28 Ind: 82 |
90/100*
(90) |
102/110 (92.7) |
DysN= dysplastic nevus, MM= malignant melanoma, DPN= deep penetrating nevus, PEM= pigmented epithelioid melanocytoma, F= female, M= male, IHC= immunohistochemistry, FISH= fluorescence in situ hybridization for melanoma, Pos= positive, Neg= negative, SNPa= SNP-array, Ab= abnormal SNP-array result of uncertain significance, Dx= diagnosis, Ind= indolent (including nevi and low risk AST), PRIHC= immunohistochemistry for PRAME
10 cases with abnormal SNP-array results of uncertain significance are excluded from agreement calculations between PRAMEIHC and cytogenetic test results
PRAME IHC, FISH for melanoma and/or SNP-array inter-test concordance
Immunohistochemistry for PRAME was performed in all lesions (110 cases). As defined in the Methods section, only diffuse (4+) immunoreactivity for PRAME (i.e. >75% of lesional cells staining) was regarded as positive, i.e., in favor of melanoma. FISH for melanoma was performed in 77/110 cases. SNP-array was performed in 40/110 cases. In 7 cases results for both FISH for melanoma and SNP-array were available with all 7 cases showing FISH and SNP array inter-test agreement in support or against melanoma. In 10 cases SNP-array results were abnormal, but in a way that could not be confidently classified as “in favor of melanoma or not” (FISH for melanoma was not performed in these cases). Because the observed genomic aberrations were of uncertain significance, these 10 cases were excluded from percentage agreement calculations of cytogenetic results with PRAME IHC as well as from percentage agreement calculations of cytogenetic results with final diagnosis.
There was inter-test agreement between PRAME IHC and FISH for melanoma and/or SNP-array results in 90/100 (90%) of cases. Of the 10 cases with discordant results, 8 cases had cytogenetic findings (6 by FISH only; 2 by FISH and SNP array) in favor of melanoma and non-diffuse PRAME immunoreactivity (score of 0–3+). Two cases with negative FISH results showed diffuse 4+ PRAME immunoreactivity (Table 2).
Table 2.
Correlation of PRAME IHC with FISH and/or SNP-array (SNPa) results
| FISH/ SNPa positive |
FISH/ SNPa negative |
Total cases | ||
|---|---|---|---|---|
| PRAME IHC | 100 | Inter-test agreement 90% | ||
| 4+ | 18 | 2 | ||
| 0–3+ | 8 | 72 |
IHC= immunohistochemistry, FISH= fluorescence in situ hybridization for melanoma, SNPa= SNP-array
Comparison of final diagnostic interpretation with PRAME IHC and cytogenetic tests results
There was agreement in PRAME IHC and final diagnostic interpretation in 102/110 (92.7%) of cases. Of the 28 cases with final diagnosis of melanoma, 21 showed 4+ PRAME IHC (75% sensitivity). Of the 82 tumors interpreted as benign or indolent, 81 cases showed non-diffuse PRAME IHC (98.8% specificity) (Table 3; Figures 1, 2, and 3).
Table 3.
Correlation of PRAME IHC with final diagnostic interpretation
| Favor malignant | Favor indolent | Total cases | ||
|---|---|---|---|---|
| PRAME IHC | 110 | Agreement 92.7% | ||
| 4+ | 21 | 1 | Sensitivity 75% | |
| 0–3+ | 7 | 81 | Specificity 98.8% |
IHC= immunohistochemistry
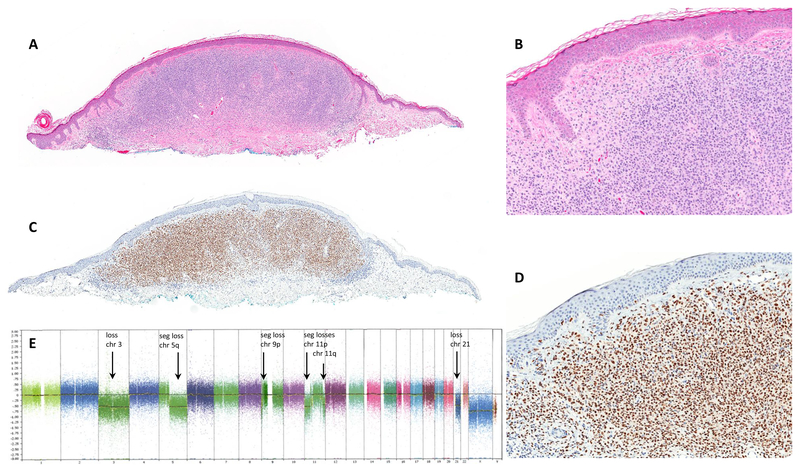
Figure 1.
(A, B) Nevoid melanoma, H&E stain. (C, D) Diffuse (4+) PRAME immunoreactivity. (E) SNParray showing multiple aberrations including loss of chromosome 3, segmental loss in 5q, segmental loss in 9p, segmental losses in 11p, segmental loss in 11q, loss of chromosome 21.
chr= chromosome, seg= segmental
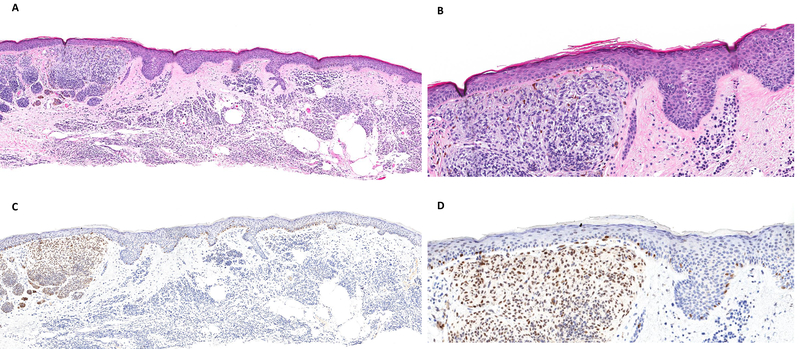
Figure 2.
(A, B) Nevoid melanoma, H&E stain. (C) Tumor cells are completely negative for PRAME IHC. (E) FISH showed gains in RREB1 (red probe) and CCND1 (green probe) meeting cut offs supportive of melanoma.
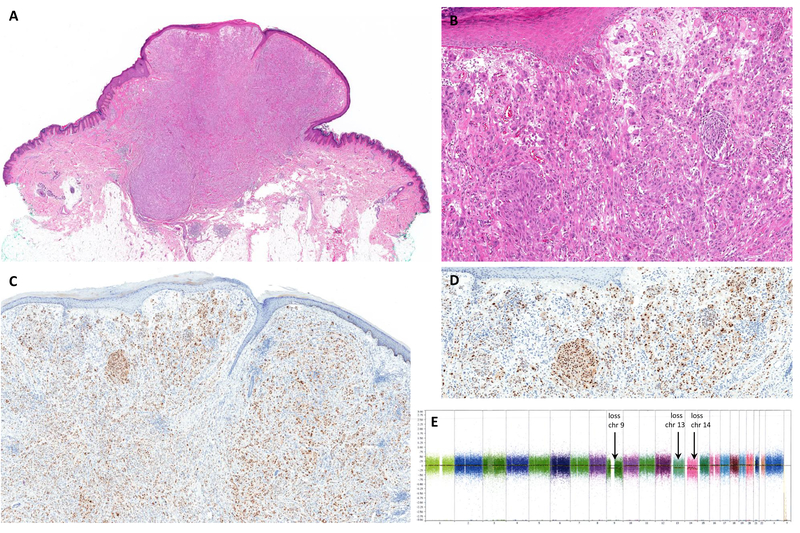
Figure 3.
(A, B) Atypical Spitz tumor arising in the upper extremity of a 6-year-old child, H&E stain. (C, D) Tumor cells are diffusely positive for PRAME IHC. (E) SNParray showed aberrations of uncertain significance including loss of whole chromosomes 9, 13, and 14. No homozygous deletion of CDKN2A (9p21) was detected in tumor cells by FISH (not shown). chr= chromosome
There was agreement between cytogenetic tests results (FISH for melanoma and/or SNP array) and final diagnostic interpretation in 96/100 cases (96%). 24 out 26 cases with final diagnosis of melanoma showed cytogenetic abnormalities classified as “positive or in favor of melanoma” (92.3% sensitivity). In the remainder 74 cases judged as indolent 72 cases lacked cytogenetic aberrations of melanoma (97.3% specificity).
Of note, two cases with negative FISH test results were interpreted as melanoma. One of them corresponded to a melanoma with nevoid features and 4+ PRAME immunoreactivity and the other corresponded to a melanoma arising in a nevus. The latter lesion was composed of a biphenotypic population of melanocytes with diffuse immunoreactivity for PRAME detected in the atypical population of cells with distinct morphology from the PRAME-negative associated nevus (Figure 4). In 2 lesions, both from 3-year-old children, cytogenetic abnormalities were found (1 case by FISH only; 1 case by FISH and SNP array), but based on the clinical context and histopathologic features, the findings were felt to be insufficient for a malignant Spitz tumor, and a diagnosis of atypical Spitz tumor was favored. Both of these tumors were completely negative for PRAME.
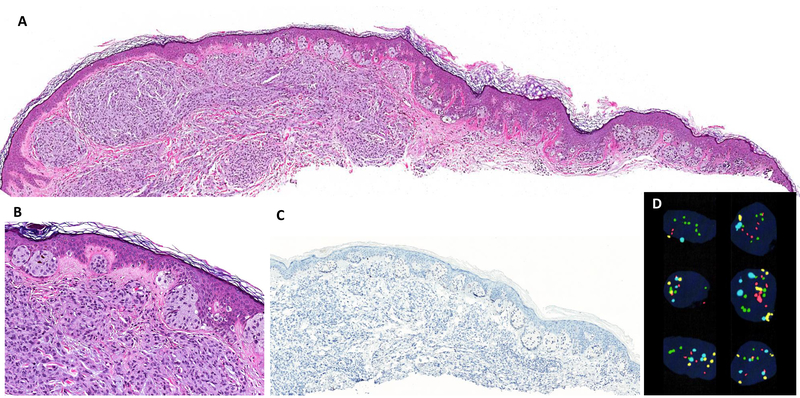
Figure 4.
(A, B) Posterior shoulder of 80-year-old male showing a biphenotypic melanocytic neoplasm that corresponds to melanoma arising in a nevus, H&E stain. (C, D) PRAME IHC is diffusely positive in the markedly atypical epithelioid cells of the lesion that show distinct expansile growth in the superficial dermis, while smaller melanocytes showing maturation with depth are negative for PRAME. Results of the FISH assay did not meet criteria for melanoma (not shown).
DISCUSSION
PRAME, as a cancer testis antigen (CTA), shows an expression profile predominantly restricted to various cancer cells, especially melanoma [7, 13, 27]. However, PRAME does not map to the chromosome X and its mRNA is also found in non-neoplastic cells, such as testis, ovary, placenta, adrenals, and endometrium qualifying PRAME as a non-classical CTA [1, 28–30]. Having an antibody to visualize PRAME expression in tissue sections by immunohistochemistry, we previously demonstrated markedly different results in benign versus malignant melanocytic neoplasms. We documented that 88–94% of non-spindle cell cutaneous melanomas show nuclear immunoreactivity for PRAME in >75% of tumor cells. Conversely, we found that benign nevi express PRAME by immunohistochemistry in a minority of cases (13.1%) and when immunoreactivity is present, this is generally focal (present in <50% of cells).[17]
Given prior evidence of PRAME as biomarker and its expression profile we reasoned that IHC for PRAME could be useful in the assessment of challenging melanocytic neoplasms. While routine H&E histomorphology remains the gold standard for the evaluation of melanocytic tumors, there is recognized value in incorporating data of cytogenetic and other ancillary tests as an additional layer of evidence for diagnostic interpretation of difficult cases. We then sought to compare results of PRAME IHC to those of the FISH for melanoma assay and SNP-array, and correlate them with final diagnostic interpretation in a cohort of 110 melanocytic tumors with challenging histomorphology.
With a threshold of 4+ staining (>75% of tumor cells showing immunoreactivity) to regard PRAME IHC as in support of a diagnosis of melanoma, we demonstrated that there is high agreement (90% concordance) between results of PRAME IHC, FISH for melanoma, and SNP-array. Of 10 cases with discordant PRAME IHC and cytogenetic results, 6 corresponded to lesions with non-diffuse PRAME immunoreactivity (score 0 to 3+) in which cytogenetic results and final interpretation favored melanoma. In 2 cases with diffuse 4+ PRAME IHC and negative cytogenetic results, the final diagnostic interpretation favored malignant melanoma. Conversely, in 2 cases with final diagnostic interpretation favoring an indolent Spitz tumor in pediatric patients, PRAME IHC was negative while cytogenetic studies were suggestive of melanoma.
Sensitivity and specificity of the FISH for melanoma assay has been reported at around 85% and 95%, respectively, and varies in different cohorts and subsets of melanocytic tumors. Array CGH has an estimated sensitivity of >90% and specificity of ~95% for melanoma.[23, 24] However, in clinical practice besides test performance metrics additional relevant considerations on the use of ancillary studies are tumor volume requirement, cost, turnaround time, and availability, which can be problematic for molecular cytogenetic testing.
Immunohistochemistry offers advantages of more rapid turn around time, lower cost, and availability in most pathology laboratories. However, PRAME IHC, can yield false negative and false positive results. As previously documented, bona fide primary cutaneous and metastatic melanomas can be completely negative for PRAME by IHC or display only focal immunoreactivity. Although very rarely in our experience, diffuse PRAME staining can occur in benign nevi. Our previous results showed that diffuse (4+) immunoreactivity for PRAME has a sensitivity of ~90% and specificity of ~99% for unequivocal non-spindle cell primary cutaneous melanomas.[17] Bearing in mind the unusual set of cases in this series, the sensitivity of diffuse (4+) PRAME expression for detecting melanoma in this cohort was only 75%, with specificity of 98.8%. Overall agreement between PRAME IHC results and final diagnostic interpretation was 92.7%.
A gene-profiling assay (Myriad Mypath) that includes detection of PRAME mRNA as one of the targets of its panel[14, 15, 31] has been previously compared with results of FISH for melanoma in a cohort of challenging melanocytic lesions. Sensitivity and specificity were found to be similar in the group of spitzoid melanocytic lesions with overall agreement with final interpretation of ~87% for both tests, whereas FISH was reported to show better performance -sensitivity of 67% against 50%, and agreement with final interpretation of 75% versus 54%- in nevoid lesions.[32] Our results show high overall agreement between results of PRAME protein detection by IHC and cytogenetic test results (concordance of 90%) and with final diagnostic interpretation (concordance 92.7%).
A special challenge is encountered in lesions where a diagnosis of melanoma arising from or immediately adjacent to a nevus is suspected. In these cases, additional considerations apply to the interpretation of PRAME IHC as it does to other ancillary studies including cytogenetic tests. In this regard, diffuse immunoreactivity for PRAME in the most cytologically and/or architecturally atypical component of a biphenotypic tumor would lend support to the suspicion of melanoma (Figure 4 illustrates an example). Such scenarios emphasize the importance of interpreting PRAME IHC in conjunction with careful routine H&E examination: recognition of a morphologically distinct population of atypical cells allows interpretation of PRAME IHC as 4+ in the population of concern, in support of melanoma associated to nevus.
In this cohort, 3 cases showed 3+ immunoreactivity for PRAME i.e. lesions with over 50% to 75% of tumor cells positive for PRAME IHC (as defined in the methods section), a result that can be problematic in terms of reproducibility of test interpretation. Although not common in this cohort, we acknowledge that most dermatopathologists would likely find such 3+ PRAME IHC results to be either non-informative or too ambiguous to inform their interpretation of challenging lesions for clinical care. Indeed, we agree that in the context of current knowledge on this marker it would be prudent to consider obtaining further evidence to support a diagnosis in such cases.
Limitations of this study include the relatively low number of cases with a final diagnosis of melanoma (28/110), but this reflects the composition of most cohorts of ambiguous tumors – they tend to be enriched with atypical but indolent tumors. For the majority of cases, PRAME IHC and cytogenetic studies were performed concurrently as part of the evaluation of each lesion prior to rendering a final diagnostic interpretation with dermatopathologists having access to all available test results (non-blinded study). Furthermore, there is an inherent element of subjectivity in the histomorphologic examination of challenging melanocytic neoplasms, which in turn can limit reproducibility of diagnostic interpretation even amongst expert dermatopathologists.[33]
In conclusion, we have documented that PRAME IHC shows good concordance with results of FISH for melanoma and/or SNP-array and with final diagnostic interpretation in a cohort of challenging melanocytic lesions. Our results support the value of PRAME IHC as an ancillary tool in the evaluation of challenging melanocytic lesions to be used in combination with expert morphologic examination, other available ancillary test results, and clinical context. Additional studies by other groups and evaluation of cohorts with long-term follow up are needed to further assess sensitivity and specificity of PRAME IHC in ambiguous melanocytic tumors and to provide insight on its limitations.
ACKNOWLEDGEMENTS
The authors would like to thank the team of laboratory technicians in Immunohistochemistry and Cytogenetics for their excellent work. They also thank Fernando Garcia, Yesenia Gonzalez, and Maria Sanchez for their assistance with digital images.
Disclosures: Research reported in this publication was supported in part by the Cancer Center Support Grant of the National Institutes of Health/National Cancer Institute under award number P30CA008748. The authors have disclosed that they have no significant relationships with, or financial interest in, any commercial companies pertaining to this article.
REFERENCES
- [1].Ikeda H, Lethe B, Lehmann F, et al. Characterization of an antigen that is recognized on a melanoma showing partial HLA loss by CTL expressing an NK inhibitory receptor. Immunity. 1997; 6:199–208. [DOI] [PubMed] [Google Scholar]
- [2].Hemminger JA, Toland AE, Scharschmidt TJ, et al. Expression of cancer-testis antigens MAGEA1, MAGEA3, ACRBP, PRAME, SSX2, and CTAG2 in myxoid and round cell liposarcoma. Mod Pathol. 2014; 27:1238–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Iura K, Kohashi K, Hotokebuchi Y, et al. Cancer-testis antigens PRAME and NY-ESO-1 correlate with tumour grade and poor prognosis in myxoid liposarcoma. J Pathol Clin Res. 2015; 1:144–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Iura K, Maekawa A, Kohashi K, et al. Cancer-testis antigen expression in synovial sarcoma: NY-ESO-1, PRAME, MAGEA4, and MAGEA1. Hum Pathol. 2017; 61:130–9. [DOI] [PubMed] [Google Scholar]
- [5].Neumann E, Engelsberg A, Decker J, et al. Heterogeneous expression of the tumor-associated antigens RAGE-1, PRAME, and glycoprotein 75 in human renal cell carcinoma: candidates for T-cell-based immunotherapies? Cancer Res. 1998; 58:4090–5. [PubMed] [Google Scholar]
- [6].Oberthuer A, Hero B, Spitz R, et al. The tumor-associated antigen PRAME is universally expressed in high-stage neuroblastoma and associated with poor outcome. Clin Cancer Res. 2004; 10:4307–13. [DOI] [PubMed] [Google Scholar]
- [7].Pujol JL, De Pas T, Rittmeyer A, et al. al. Safety and Immunogenicity of the PRAME Cancer Immunotherapeutic in Patients with Resected Non-Small Cell Lung Cancer: A Phase I Dose Escalation Study. J Thorac Oncol. 2016; 11:2208–17. [DOI] [PubMed] [Google Scholar]
- [8].Roszik J, Wang W-L, Ravi V, et al. Expression and clinical correlations of PRAME in sarcoma subtypes. Journal of Clinical Oncology. 2016; 34:11067-. [Google Scholar]
- [9].Sanchez MI, Field MG, Kuznetsov JN, et al. The role of PRAME in promoting uveal melanoma metastasis [abstract]. In: proceedings of the American Association for Cancer Research Annual Meeting 2017 Washington, DC, Philadelphia, PA: AACR; 2017;77(suppl 13) 4861. [Google Scholar]
- [10].Zhang W, Barger CJ, Eng KH, et al. PRAME expression and promoter hypomethylation in epithelial ovarian cancer. Oncotarget. 2016; 7:45352–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Epping MT, Wang L, Edel MJ, et al. The human tumor antigen PRAME is a dominant repressor of retinoic acid receptor signaling. Cell. 2005; 122:835–47. [DOI] [PubMed] [Google Scholar]
- [12].Goodison S, Urquidi V. The cancer testis antigen PRAME as a biomarker for solid tumor cancer management. Biomark Med. 2012; 6:629–32. [DOI] [PubMed] [Google Scholar]
- [13].Gutzmer R, Rivoltini L, Levchenko E, et al. Safety and immunogenicity of the PRAME cancer immunotherapeutic in metastatic melanoma: results of a phase I dose escalation study. ESMO Open. 2016; 1:e000068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Clarke LE, Flake DD, 2nd, Busam K, et al. An independent validation of a gene expression signature to differentiate malignant melanoma from benign melanocytic nevi. Cancer. 2017; 123:617–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ko JS, Matharoo-Ball B, Billings SD, et al. Diagnostic Distinction of Malignant Melanoma and Benign Nevi by a Gene Expression Signature and Correlation to Clinical Outcomes. Cancer Epidemiol Biomarkers Prev. 2017; 26:1107–13. [DOI] [PubMed] [Google Scholar]
- [16].Ferris LK, Jansen B, Ho J, et al. Utility of a Noninvasive 2-Gene Molecular Assay for Cutaneous Melanoma and Effect on the Decision to Biopsy. JAMA Dermatol. 2017; 153:675–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lezcano C, Jungbluth AA, Nehal KS, et al. PRAME Expression in Melanocytic Tumors. Am J Surg Pathol. 2018; 42:1456–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bastian BC, LeBoit PE, Hamm H, et al. Chromosomal gains and losses in primary cutaneous melanomas detected by comparative genomic hybridization. Cancer Res. 1998; 58:2170–5. [PubMed] [Google Scholar]
- [19].Bastian BC, Olshen AB, LeBoit PE, et al. Classifying melanocytic tumors based on DNA copy number changes. Am J Pathol. 2003; 163:1765–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bauer J, Bastian BC. Distinguishing melanocytic nevi from melanoma by DNA copy number changes: comparative genomic hybridization as a research and diagnostic tool. Dermatol Ther. 2006; 19:40–9. [DOI] [PubMed] [Google Scholar]
- [21].Gerami P, Jewell SS, Morrison LE, et al. Fluorescence in situ hybridization (FISH) as an ancillary diagnostic tool in the diagnosis of melanoma. Am J Surg Pathol. 2009; 33:1146–56. [DOI] [PubMed] [Google Scholar]
- [22].Gerami P, Li G, Pouryazdanparast P, et al. A highly specific and discriminatory FISH assay for distinguishing between benign and malignant melanocytic neoplasms. Am J Surg Pathol. 2012; 36:808–17. [DOI] [PubMed] [Google Scholar]
- [23].Gerami P, Zembowicz A. Update on fluorescence in situ hybridization in melanoma: state of the art. Arch Pathol Lab Med. 2011; 135:830–7. [DOI] [PubMed] [Google Scholar]
- [24].Wang L, Rao M, Fang Y, et al. A genome-wide high-resolution array-CGH analysis of cutaneous melanoma and comparison of array-CGH to FISH in diagnostic evaluation. J Mol Diagn. 2013; 15:581–91. [DOI] [PubMed] [Google Scholar]
- [25].Fang Y, Dusza S, Jhanwar S, et al. Fluorescence in situ hybridization (FISH) analysis of melanocytic nevi and melanomas: sensitivity, specificity, and lack of association with sentinel node status. Int J Surg Pathol. 2012; 20:434–40. [DOI] [PubMed] [Google Scholar]
- [26].Stark M, Hayward N. Genome-wide loss of heterozygosity and copy number analysis in melanoma using high-density single-nucleotide polymorphism arrays. Cancer Res. 2007; 67:2632–42. [DOI] [PubMed] [Google Scholar]
- [27].Gezgin G, Luk SJ, Cao J, et al. PRAME as a Potential Target for Immunotherapy in Metastatic Uveal Melanoma. JAMA Ophthalmol. 2017; 135:541–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chang AY, Dao T, Gejman RS, et al. A therapeutic T cell receptor mimic antibody targets tumor-associated PRAME peptide/HLA-I antigens. J Clin Invest. 2017; 127:2705–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wadelin F, Fulton J, McEwan PA, et al. Leucine-rich repeat protein PRAME: expression, potential functions and clinical implications for leukaemia. Mol Cancer. 2010; 9:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Simpson AJ, Caballero OL, Jungbluth A, et al. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005; 5:615–25. [DOI] [PubMed] [Google Scholar]
- [31].Clarke LE, Pimentel JD, Zalaznick H, et al. Gene expression signature as an ancillary method in the diagnosis of desmoplastic melanoma. Hum Pathol. 2017; 70:113–20. [DOI] [PubMed] [Google Scholar]
- [32].Reimann JDR, Salim S, Velazquez EF, et al. Comparison of melanoma gene expression score with histopathology, fluorescence in situ hybridization, and SNP array for the classification of melanocytic neoplasms. Mod Pathol. 2018; 31:1733–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Gerami P, Busam K, Cochran A, et al. Histomorphologic assessment and interobserver diagnostic reproducibility of atypical spitzoid melanocytic neoplasms with long-term follow-up. Am J Surg Pathol. 2014; 38:934–40. [DOI] [PubMed] [Google Scholar]