Abstract
Recent advances in computer technology have generated a new area of research known as radiomics. Radiomics is defined as the high throughput extraction and analysis of quantitative features from imaging data. Radiomic features provide information on the gray-scale patterns, inter-pixel relationships, as well as shape and spectral properties of radiological images. Moreover, these features can be used to develop computational models that may serve as a tool for personalized diagnosis and treatment guidance. Although radiomics is becoming popular and widely used in oncology, many problems such as overfitting and reproducibility issues remain unresolved. In this review, we will outline the steps of radiomics used for oncology, specifically addressing applications for breast cancer patients and focusing on technical issues.
Keywords: Radiomics, Informatics, Texture, Breast cancer, Quantitative imaging
INTRODUCTION
Radiological imaging is mandatory for diagnosis, staging, treatment planning, post-operative surveillance, and response evaluation during the routine management of cancer. These imaging methods include simple X-ray radiography, ultrasound (US), computed tomography (CT), positron emission tomography, and magnetic resonance imaging (MRI). It has been postulated that these radiological images contain more information that is not visible to the human eye. This information is regarded as radiological texture and can provide additional details on the targeted tissue. Researchers have suggested that this invisible information can be extracted from images using advanced texture and shape analysis. Recent advancements in computer technology used to investigate these texture and shape analyses have led to the development of a new area of research termed radiomics.
Radiomics is based on the assumption that extracted imaging data are the product of mechanisms occurring at a genetic and molecular level linked to the genotypic and phenotypic characteristics of the tissue (1,2). A high-throughput extraction of quantitative features from radiological images creating a high-dimensional data set followed by data mining for potentially improved decision support is typically performed (3,4,5). Traditionally, radiomics features provide information regarding gray-scale patterns, inter-pixel relationships, shape, and spectral properties within regions of interest (ROIs) on radiological images (6,7,8). Incorporated with clinicopathological data, radiomics is well suited to the concept of personalized medicine and has drawn considerable attention in radiology (3,9).
In this review, we will outline the steps of radiomics used in oncology focusing on MRI, in addition to mammography, digital breast tomosynthesis (DBT), and US, specifically addressing potential applications in breast cancer imaging and focusing on technical issues.
Steps in Radiomics Analysis
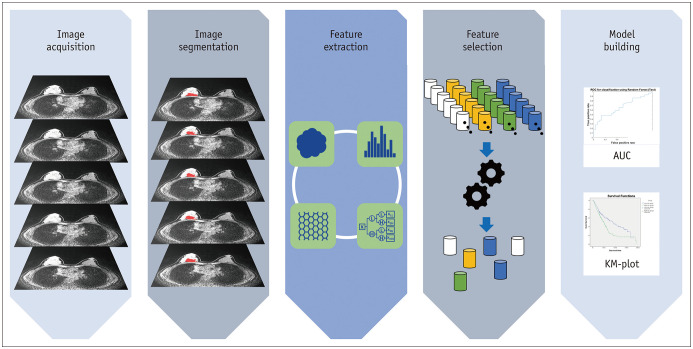
Radiomics analysis is an analytical framework applicable to various target sites and imaging modalities. This section describes the typical steps involved in radiomics analysis. An overview of the steps undertaken in the studies on radiomics is presented in Figure 1.
Fig. 1. Overview of steps in radiomics studies.
AUC = area under curve, KM = Kaplan-Meier
Image Acquisition
For any study, the first step is to acquire the appropriate images. MRI, US, DBT, and mammography are widely used for breast imaging, each providing distinct information. All of these modalities produce potentially different raw data depending on the scanner model and imaging parameters (10). These differences reduce the reproducibility of radiomics studies and make direct comparisons among radiomics features difficult. Recently, the Quantitative Imaging Biomarker Alliance and Quantitative Imaging Network have defined standardized imaging protocols and recommendations in the field of quantitative imaging (11). This standardization effort is expected to improve the reproducibility of future radiomics research. It is good practice to acquire all imaging data using the same scanner and imaging parameters in order to ensure the stability of the features of radiomics (10,12,13,14,15). In summary, accurate reporting of the imaging parameters is necessary to improve the reproducibility of future studies.
Specification of ROIs
The next step is to specify (or segment) the ROI, from which radiomics features are computed. ROIs are specified to limit the spatial extents of the analysis and can be specified manually, semi-automatically, or automatically (16,17). Among the methods of tumor segmentation, automated or semi-automated methods have been reported to be superior to manual methods for segmenting the tumor (18,19). If the target region has a well-defined boundary, all three methods of ROI specification are possible. For target regions that require expertise to specify, manual segmentation is necessary.
Feature Extraction
The third step is feature extraction, which computes hundreds or thousands of features from the ROIs. The features are defined using mathematical formulas and are thus objective imaging features. The features are broadly classified into four categories: morphological, histogram-based, textural, and transform-based features (9). Morphological features conventionally reflect the shape and physical characteristics of the ROI. Notable features in this category include the compactness, sphericity, surface area, convexity, and surface-to-volume ratio (9,20). Histogram-based features extract information from the intensity histogram of the ROI. Notable features in this category are the median, inter-quartile range, entropy, uniformity, skewness, and kurtosis. These features involve specifying the binning parameters (e.g., number of bins). Many studies have commonly adopted 256 bins. To rationalize the number of bins, a statistical formula such as the Freedman-Diaconis rule could be applied. Existing breast imaging studies reported the entropy, mean, minimum, and maximum as important features (17). These features do not consider the spatial neighborhood information of the voxels. In comparison, textural features consider voxels and their neighbors and are the main features that have propelled many radiomics studies. The gray-level co-occurrence matrix (GLCM)-based features are widely used to specify neighborhood information (6). The GLCM models intensity pairs (not the intensity of a single voxel) and the relative frequency of the pair in the neighborhood. Many radiomics studies have noted the entropy (different from the histogram-based feature), contrast, and homogeneity of the GLCM as important features related to tumor heterogeneity (21,22). Gray-level size zone matrix (GLSZM)-based features are also widely used. The GLSZM assumes that the ROI is made of many blobs of varying intensity and size (23). Transform-based features involve transforming the original image with a user-selected transform. A new image is created after the transform and the features mentioned in the previous categories are computed (24). Multi-scale transforms such as wavelet and Laplacian of Gaussian transforms are widely used. Figure 2 contains a summary of the typical radiomics feature categories.
Fig. 2. Summary of typical radiomics features in four categories.
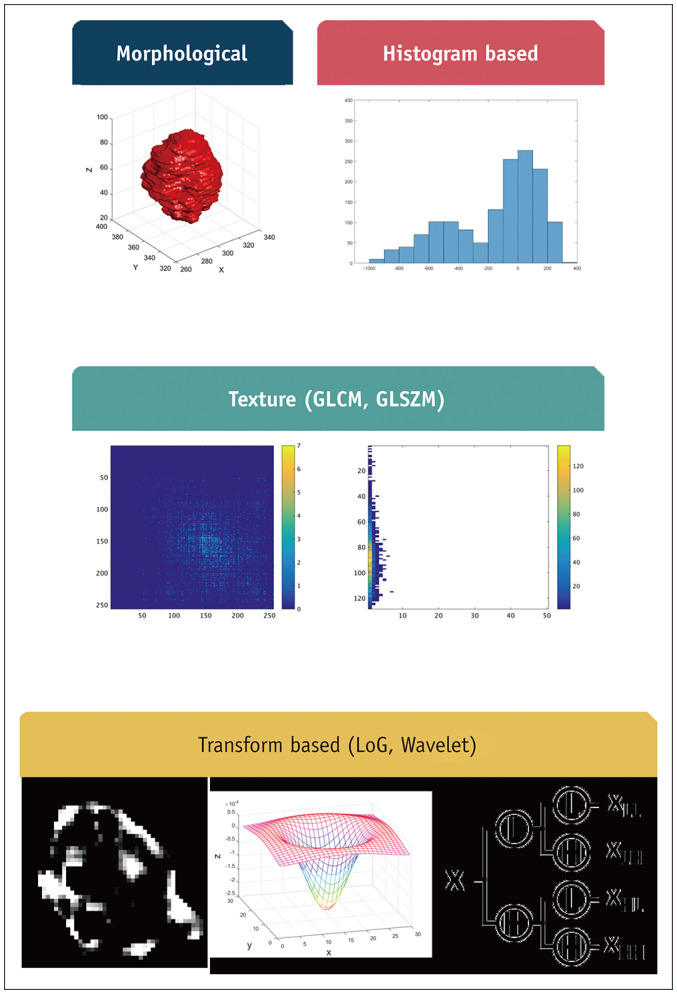
GLCM = gray-level co-occurrence matrix, GLSZM = gray-level size zone matrix
Feature Selection
As hundreds or thousands of features are computed in the previous step, we need to select a few features that reflect our desired response variable. The desired response variable differs based on the study, and various types of information are used including disease diagnosis, survival, and recurrence. Numerous studies have employed machine learning for the selection of relevant features. In radiomics, the least absolute shrinkage and selection operator (LASSO), its variant elastic net, principal component analysis (PCA), and random forest approaches are widely used (25,26).
Model Building
Next, models are built using the selected features to suit the goal of the study. For classification goals, various classifiers are used including support vector machine (SVM), random forest, and XGBoost classifiers (25,27). To predict continuous variables, various regression methods including linear regression, regularized linear regression, and random forest are used. Some of the feature selection approaches iteratively combine the fourth and fifth steps to determine the features that lead to good model performance.
Technical Issues in Radiomics Analysis
Issue of Overfitting and Validation
Most radiomics studies involve applying machine learning algorithms in the model building step (28). Modern machine learning approaches are highly flexible with very high degrees of freedom and thus could lead to overfitting to the given training data. Therefore, performance measures derived from the training data cannot be trusted and validation using independent test data is necessary. However, many breast radiomics studies are limited to single-center data lacking external validation. In such cases, researchers can adopt cross-validation or bootstrapping using parts of the data as training data and the remaining parts as validation data (29,30,31). In cross-validation, leave-one-out validation and k-fold cross-validation are widely used. Leave-one-out validation is more susceptible to overfitting than k-fold cross-validation as we use most data for training and reserve only one sample for validation in each iteration. In bootstrapping, we randomly sample data into training and validation datasets many times (typically thousands of times), and thus can derive a distribution of performance results.
The optimal method of validation uses independent validation, which is typically found in multi-center studies. A radiomics model is trained using the data from one center and then further validated using the data of another center; this is always the preferred method of validation. However, acquiring multi-center data could be difficult for many researchers in practice. One solution is to leverage an open database such as The Cancer Imaging Archive (TCIA) to acquire the external validation data (32). As of January 2020, there are 14 collections of breast imaging hosted on TCIA spanning MRI, mammogram, and CT images.
Feature Reproducibility
The reproducibility of radiomics analysis is the biggest issue, and can occur in every step of radiomic research. The retrospective nature of the studies, the broad heterogeneity of software used, and the variability of the radiomics features used in the different studies raise legitimate concerns regarding the potential lack of reproducibility in radiomics research. Radiomics studies consider hundreds or thousands of features (33). They are derived from imaging data and any factors affecting the imaging data such as scanner types or scanning parameters might lead to unwanted variability in features. It is good practice to acquire imaging data using standardized settings (3,34). Data obtained under such settings could be shared across studies for validation. Slice thickness and voxel shape (i.e., rectangle vs. square) are well-known imaging factors affecting feature reproducibility (35). Several early investigators have reported that many features were often unstable (36,37,38). In a study of 219 radiomics features, only 66 reported intraclass correlation coefficient values of more than 0.90 (36,37). More recently, Berenguer et al. (39) conducted a phantom study to identify the most reproducible and nonredundant radiomics features for CT. Their results indicated that many radiomics features were redundant and exhibited poor reproducibility. For retrospective studies, we could face different imaging settings that can be partially mitigated by using different imaging settings as co-variates in the regression framework.
ROI segmentation is another layer that could hinder reproducibility. Automatic methods are preferred as they offer excellent reproducibility, but they can only be applied for well-defined tumor regions. The manual approach is time-consuming and is potentially limited by degraded reproducibility. Semi-automatic methods are the middle ground between the manual and automatic methods, and they allow human experts to correct mistakes in the automatic results (40). Manual methods would be better used only in cases in which the target boundaries are difficult to discern, and thus expert interpretation is required.
Many of the radiomics features are computed from histograms. Histograms are affected by the binning parameters (i.e., the number of bins and bin width) (41). Researchers should select these parameters to ensure that they can estimate the underlying intensity distribution effectively. For a large ROI, using many bins is appropriate; however, for a small ROI, using many bins is problematic as there could be insufficient samples in a given bin to estimate the underlying distribution. The issue of histogram parameters is more important for textural features, as the histogram estimates a two-dimensional distribution (i.e., intensity pair), in which there is a higher probability of insufficient samples.
There are many software tools for the ROI specification and feature extraction stages. Many software tools implement the most well known formulas, but there are many hidden nuance parameters. It is good practice to use widely used tools, thereby allowing other researchers to reproduce these studies elsewhere. For ROI specification, the Medical Image Processing, Analysis, and Visualization (MIPAV, version 9.0.0, https://mipav.cit.nih.gov/) and 3D slicer (version 4.10.2, https://www.slicer.org/) tools are widely used (42,43). For feature extraction, PyRadiomics and Imaging Biomarker Explorer (IBEX) are widely used (44,45). Different software could result in different values on the same lesion.
Radiomic Application in Breast Cancer Imaging
A radiomics methodology was first applied to head and neck, and lung cancer imaging (9,46,47) and has been more recently applied to breast imaging (48).
Radiomics appears capable of offering imaging biomarkers, which are useful not only for diagnosing breast cancer, but also for predicting the treatment response and risk of recurrence. With regard to breast cancer, radiomics approaches have been investigated mainly with MRI. However, some studies have explored the potential of radiomics with different imaging modalities, including standard mammography, DBT, and US. We will briefly review the current and potential role of radiomics for predicting malignancy, the response to neoadjuvant chemotherapy (NAC), axillary lymph node metastasis, pathologic or prognostic factors, and cancer recurrence. To enhance the readers' understanding, we have roughly divided our results into characterization and prediction, and then described more specific indications. Representative radiomics studies are summarized in Table 1.
Table 1. Summary of Representative Radiomics Research in Breast Imaging.
| Reference | Indication | Modality | Patients (Number) | Radiomics Features (Number) | Findings |
|---|---|---|---|---|---|
| Bickelhaupt et al. (49) | Malignancy prediction | MRI: DWI, T2WI | 222 | 359 | Radiomics features are better than only using ADC alone |
| Nie et al. (50) | Malignancy prediction | MRI: DCE | 71 | 18 | Quantitative morphologic and texture features analysis showed reasonably high accuracy |
| Wang et al. (51) | Malignancy prediction | MRI: DCE | 99 | 30 | Radiomics features and pharmacokinetic factors differentiated benign and malignant masses |
| Cai et al. (52) | Malignancy prediction | MRI: DCE, DWI | 234 | 28 | Developed GLCM-based features from DCE-MRI with ADC as well as kinetic and morphological features |
| Parekh & Jacobs (53) | Malignancy prediction | MRI: DCE, T2WI, DWI | 124 | 30 | Entropy RFMs were found to be most reliable |
| Garra et al. (54) | Malignancy prediction | US | 80 | 14 | Sensitivity of 100% and specificity of 80% were found |
| Luo et al. (55) | Malignancy prediction | US | 315 | 1044 | Radiomics nomograms showed better discrimination than radiomics scores or BI-RADS category |
| Zhang et al. (56) | Malignancy prediction | US: conventional, | 117 | 364 | Results of sonoelastomic features showed AUC of 0.917 and accuracy of 88% in validation set |
| Drukker et al. (57) | Malignancy prediction | Mammogram: conventional, three-compartment (water, lipid, protein) image from dual energy mammogram | 109 | 5 | Combined mammography radiomics plus quantitative three-compartment image analysis prospectively showed better PPV3 |
| Li et al. (58) | Malignancy prediction | Mammogram | 182 | 32 | Combining contralateral normal breast radiomic features with those of lesion showed better performance |
| Tagliafico et al. (59) | Malignancy prediction | DBT | 40 | 104 | Radiomics analysis of DBT could be used to facilitate cancer detection and characterization in multicenter prospective study |
| Holli et al. (60) | Differentiation between ILC and IDC | MRI: DCE, | 20 | 300 | Entropy-based GLCM 2020-06-09features and first subtraction were most effective |
| Waugh et al. (61) | Differentiation between ILC and IDC | MRI: DCE | 200 | 220 | Entropy was significantly different between IDC |
| Li et al. (62) | Correlation with pathology | MRI: DCE | 91 | 38 | MRI-based phenotypes were significantly associated with receptor status and heterogeneity was important feature to discriminate different subtypes |
| Liang et al. (63) | Ki-67 correlation | MRI: DCE, T2WI | 318 | 10207 | Rad-score from T2WI was significantly associated with Ki-67 status |
| Marino et al. (64) | Correlation with pathology | Mammogram: contrast-enhanced | 100 | 300 | Radiomics analysis with CEM has potential for differentiating tumors with different pathologic findings |
| Ahmed et al. (65) | NAC response | MRI: DCE | 100 | 16 | Texture features showed significant differences between non-responders and partial responders |
| Braman et al. (66) | NAC response | MRI: DCE | 117 | 99 | Peritumoral radiomics contributed to accurate response prediction |
| Braman et al. (67) | NAC response | MRI: DCE | 209 | 495 | Peritumoral radiomics were useful in characterizing HER2+ tumors and estimating response to HER2-targeted therapy |
| Liu et al. (68) | NAC response | MRI: T2WI, DWI, DCE | 586 | 13950 | Radiomics of multiparametric MRI yielded better performance to predict pCR than clinical model |
| Dong et al. (69) | LN metastasis prediction | MRI: T2WI, DWI | 146 | 10962 | Radiomics features from DWI showed higher correlation with SLN metastases than those from ADC mapping |
| Yang et al. (70) | LN metastasis predcition | Mammogram | 147 | 45 | Radiomics nomogram can predict LN metastasis |
| Yu et al. (71) | LN metastasis prediction | US | 426 | 96 | Radiomics nomogram can predict LN metastasis |
| Chan et al. (72) | Cancer recurrence prediction | MRI: DCE | 563 | 322 | Radiomics model discriminate between patients at low risk and those at high risk of recurrence |
| Park et al. (23) | Cancer recurrence prediction | MRI: DCE | 294 | 156 | Higher rad-score was correlated with worse disease-free survival |
ADC = apparent diffusion coefficient, AUC = area under curve, BI-RADS = breast imaging reporting and data system, CEM = contrast-enhanced mammography, DBT = digital breast tomosynthesis, DCE = dynamic contrast-enhanced, DWI = diffusion-weighted imaging, GLCM = gray-level co-occurrence matrix, HER2 = human epidermal growth factor receptor 2, IDC = invasive ductal carcinoma, ILC = invasive lobular carcinoma, LN = lymph node, MRI = magnetic resonance imaging, NAC = neoadjuvant chemotherapy, pCR = pathologic complete response, PPV3 = positive predictive value 3, RFM = radiomics feature maps, SLN = sentinel lymph node, T1WI = T1-weighted image, T2WI = T2 weighted image, US = ultrasound
Characterization
Discrimination of Benign and Malignant Lesions
As a typical and representative indication, radiomic features have been demonstrated to be useful for discriminating benign and malignant tissues in many disease types.
MRI
Several early trials performed to differentiate benign and malignant breast tumors (73,74,75) in a relatively small number of patients with limited numbers of radiomic features reported excellent results. A recent trend is to use a special sequence or multiparametric MRI.
In 2017, a retrospective study aimed to establish the potential ability of a diffusion MRI radiomic signature to determine the malignant nature of suspicious breast lesions detected on screening mammography (49). They employed radiomics methodology on two unenhanced MRI sequences: diffusion-weighted imaging (DWI) and T2-weighted sequences. Two radiomics classifiers allowed benign and malignant lesions to be distinguished more accurately than the mean apparent diffusion coefficient (ADC) parameter alone.
Nie et al. (50) reported that combining shape-based, volume-based, and GLCM textural features of post-contrast MR images of 71 patients using an artificial neural network (ANN) may be used to differentiate malignant from benign tumors in breast cancer (50). To focus on more technical aspects, the same group compared the two-feature selection and classification methods of logistic regression and the ANN to distinguish malignant breast tumors from benign tumors (76)-there was no significant difference in the results obtained from the two methods.
In a recent study, Wang et al. (51) used morphological and GLCM textural features in combination with pharmacokinetic parameters obtained from dynamic contrast-enhanced (DCE)-MRI to distinguish benign and malignant breast tumors. The authors found that the textural features of GLCM entropy, GLCM energy, and compactness, along with the pharmacokinetic parameters of rate constant (kep) and volume of plasma (vp), are the most discriminative with a sensitivity and specificity of 91% and 92%, respectively. Cai et al. (52) used GLCM-based textural features from DCE-MRI in combination with the ADC, kinetic curve features, and morphological features to distinguish benign and malignant breast tumors in a cohort of 234 patients.
Recently, Parekh & Jacobs (53) evaluated a correlation between radiomics features and different breast tissue of interest, and generated radiomics feature maps (RFMs) for the visualization and evaluation of radiological images. The radiomics features were then correlated with different breast tissues and were compared with quantitative values of radiological parameters. Malignant lesions exhibited higher values of entropy; the entropy RFM was most reliable when distinguishing malignant and benign lesions, thereby reflecting the tumor heterogeneity and its vascular status.
US
Garra et al. (54) performed texture analysis (histogram, GLCM, and fractal dimension) on breast US in a cohort of 80 patients. The authors were able to identify malignant lesions with a sensitivity of 100% and specificity of 80%. Multiple studies have differentiated benign and malignant breast lesions using texture analysis on US (77,78,79).
A recent study by Luo et al. (55) developed nomograms incorporating radiomics and a breast imaging reporting and data system (BI-RADS) for predicting breast cancer in BI-RADS US category 4 or 5 lesions. In their study, comprising 315 pathologically confirmed breast lesions, nomograms combining the radiomics score and BI-RADS category exhibited better discrimination of benign and malignant lesions than either the radiomics score or the BI-RADS category.
In contrast to prior studies, Zhang et al. (56) focused on sonoelastograms and radiomics. Their results indicated that some sonoelastic features might help to discriminate benign and malignant breast tumors.
Mammography and DBT
Multiple studies have investigated the use of texture analysis on mammograms for the detection of masses (80,81,82), which was achieved using variable textural features according to the study design.
Drukker et al. (57) prospectively investigated the combination of mammography radiomics and three-compartment (water, lipid, protein) breast (3CB) image analysis of dual-energy mammography. In their study, the positive predictive value 3 of combined mammography radiomics plus quantitative 3CB image analysis was higher than that of conventional digital mammography (48.5% vs. 32.1%, respectively), while the sensitivity was slightly decreased.
In a retrospective study of 182 patients (106 malignant and 76 benign) conducted by Li et al. (58), the radiomic features of contralateral normal breasts were used alongside those of diseased breasts to improve the accuracy of digital mammography. The performance of the combined lesion and parenchyma classifier in the differentiation of malignant and benign mammographic lesions was better than that which only used the lesion features.
A multi-center and prospective study applied a radiomics approach to DBT for the first time to differentiate normal breast tissue from malignant breast tissue in patients with dense breasts (59). Twenty patients with negative standard mammography findings who had had DBT-detected and histologically confirmed breast cancer were enrolled. Further, 20 patients of similar age and breast density with negative DBT and US were matched as a control group. From 104 radiomics features extracted, three (skewness, entropy, and 90th percentile) were found to differ significantly between the two groups. The results also revealed that the energy, entropy, and dissimilarity correlated significantly with the tumor size; the entropy also correlated with the receptor status. Despite the small patient number and the biased selection of features, their study indicated that a radiomics analysis of DBT images could be used to facilitate cancer detection and to acquire a better characterization of the detected lesion.
Correlation with Pathologic or Prognostic Factors
Recent studies have extended the concept of radiomics to correlate pathologic or prognostic factors with radiomic features, hypothesizing that underlying tumor biological characteristics may be presented as different radiomic values. In an attempt to distinguish invasive lobular carcinoma from invasive ductal carcinoma, Holli et al. (60) applied radiomic analysis (histogram, GLCM, gray-level run-length) on T1-weighted pre-contrast, post-contrast, and subtraction breast MRI datasets from 20 patients. The authors identified the entropy-based GLCM features to be the most effective features and yielded a maximum accuracy of 100% using linear-discriminant analysis (LDA) and nonlinear-discriminant analysis (NDA) on the first subtraction and contrast images. Waugh et al. (61) made similar observations that entropy, a texture measure of pixel distribution randomness, significantly differed between lobular and ductal lesions.
Numerous studies have applied a radiomics approach to predict the molecular subtype of breast cancers, which is essential in establishing a strategy for patient treatment (83). Furthermore, the integration between radiomics and genomic features, known as radiogenomics, has revealed promising results in oncology, providing opportunities to better understand tumors and thus improve diagnosis and prognosis (9); however, this could be a different topic for review. A retrospective study published in 2016 explored the correlation between quantitative features and cancer receptor status (estrogen receptor, progesterone receptor, human epidermal growth factor receptor 2 [HER2]) (62). It was demonstrated that MR image-based tumor phenotypes were significantly associated with the receptor status and that heterogeneity is an important feature for discriminating different subtypes, which, in the near future, might be possible using a radiomics predictive signature that would serve as a virtual biopsy.
The Ki-67 labeling index is routinely used as a prognostic marker in breast cancer patients to estimate both cell proliferation and the therapeutic response. Liang et al. (63) proposed a new, noninvasive Ki-67 predictor status based on breast MRI. They retrospectively analyzed 318 MR images of breast cancer patients (200 for the training dataset and 118 for the validation dataset), whose Ki-67 status was known. The authors selected 30 features and composed a rad-score for each patient following the analysis of the T2-weighted images and enhanced T1-weighted images. The rad-score calculated on the T2-weighted images was significantly associated with the Ki-67 status, in both the training and validation sets. These results suggest that a new radiomics marker, obtained with routinely performed unenhanced MRI sequences, might pre-operatively predict Ki-67 expression in breast cancer.
Interestingly, Marino et al. (64) applied radiomics analysis on contrast-enhanced mammography (CEM) to determine the breast cancer invasiveness, hormone receptor status, and tumor grade. They retrospectively reviewed the CEM of 100 patients with 103 breast cancers and tried various combinations of radiomics features and selection methods.
Prediction
Prediction of the Response to NAC
Most radiomic studies have focused on predicting the response to NAC, particularly the pathologic complete response (pCR), using MRI.
In early research, using histogram-based features, Parikh et al. (84) evaluated whether changes in MRI textural features could predict the pCR to NAC. In their study, comprising 36 consecutive primary breast cancer patients, an increase in T2-weighted MRI uniformity and a decrease in T2-weighted MRI entropy after NAC was found to be possibly helpful for the earlier prediction of the pCR as compared to tumor size change.
Ahmed et al. (65) produced GLCM-based features for breast MRI (pre-contrast and 1–5 minutes post-contrast) to predict the response in 100 breast cancer patients. The authors found that the texture features significantly differed between non-responders (decrease in tumor diameter of less than 50%) and partial responders (decrease in tumor diameter over 50%) to chemotherapy when implemented on post-contrast images. There were no differences in pre-contrast images.
Interestingly, Braman et al. (66) evaluated the ability of radiomic textural analysis of peri-tumoral regions as well as intra-tumoral regions on pre-treatment DCE-MRI to predict the pCR to NAC in 117 breast cancer patients. Their results demonstrated that peri-tumoral radiomics contributed to the successful prediction of the pCR from pre-treatment imaging. In their subsequent study, Braman et al. (67) reconfirmed the value of peri-tumoral radiomic features for characterizing HER2-positive tumors and estimating the response to HER2-targeted NAC.
Recently, researchers utilized multiple MRI sequences and radiomics signatures (rad-scores) calculated from numerous radiomic features. For example, Liu et al. (68) conducted a multi-center retrospective study to evaluate the performance of radiomics of multiparametric MRI (RMM) for predicting the pCR. A total of 586 patients were enrolled and a rad-score was calculated using 13950 radiomics features from T2-weighted, T1-weighted, diffusion-weighted, and contrast-enhanced T1-weighted imaging before NAC. Their results indicated that RMM yielded a better performance for predicting the pCR than the clinical model.
Prediction of Lymph Node Metastases
Determining the axillary lymph node status remains a mandatory requirement of the diagnostic process. In 2017, Dong et al. (69) reported that radiomics features extracted from DWI sequences were more highly correlated with sentinel lymph node (SLN) metastases than those extracted from ADC. To predict axillary lymph node metastases pre-operatively, some researchers have applied radiomics nomograms based on mammography (70) or US (71). They built the rad-score from a large number of radiomic features and then incorporated additional radiological and clinicopathological findings. They reported that the radiomics nomogram was a reliable method for noninvasively predicting SLN metastases.
Regarding the technical aspect, Cui et al. (85) compared the performance of three classifiers (SVM, k-nearest neighbor [KNN], LDA) and reported that the effect of the SVM classifier for predicting breast axillary lymph node metastases was significantly higher than that of the KNN or LDA classifiers.
Although these results need further validation, they may be useful for clinical decision-making with respect to axillary surgery, potentially avoiding invasive procedures in patients at low risk of SLN metastases.
Prediction of Cancer Recurrence
Several early studies have evaluated the value of texture analysis in predicting the long-term outcome of cancer patients in pre-operative (86) or NAC settings (87). In pre-operative settings, Kim et al. (86) evaluated the relationship between MRI textural features and survival outcomes in 203 patients with primary breast cancer. They only used histogram-based uniformity and entropy in T2-weighted images and contrast-enhanced T1 subtraction images. They concluded that patients with breast cancers that appeared more heterogeneous on T2-weighted images (higher entropy) and those that appeared less heterogeneous on contrast-enhanced T1-weighted subtraction images (lower entropy) had poorer outcomes. Similarly, in NAC settings, Pickles et al. (87) demonstrated that higher entropy in DCE-MRI was associated with poorer outcomes. Recently, in a study by Chan et al. (72), the potential of a radiomics model to discriminate 563 early breast cancer patients at low risk from those at high risk of recurrence during long-term follow-up was identified, based on radiomic features of wash-in and washout images from pre-treatment MRI.
A recent retrospective study proposed a radiomics approach based on pre-operative MRI to develop a radiomic signature (rad-score) associated with breast cancer recurrence (23). They enrolled 294 patients affected by invasive breast cancer presenting as a mass on contrast-enhanced MRI. A total of 156 features were extracted, and a rad-score was calculated. Patients were divided into high-risk or low-risk categories based on the rad-score itself. Subsequently, a nomogram including the rad-score and MRI and clinicopathological findings was designed to predict cancer recurrence. The results revealed that a higher rad-score correlated with poorer disease-free survival (DFS) and that the DFS estimation was more accurate when clinicopathological data were included in the evaluation.
Future Directions
Role of Deep Learning
Radiomics studies are heavily dependent on machine learning and this field has developed into deep learning. Thus, recent radiomics studies are increasingly adopting deep learning methods (16,88). The most notable difference between conventional machine learning and deep learning is the use of handcrafted features (89). In conventional machine learning, researchers compute handcrafted features that are pre-defined using domain expertise. This implies that we need to devise different features for different tasks. For example, imaging features optimized for breast imaging might not work well for other organs such as the lung. In deep learning, the deep learning network can learn the specific features from the data themselves and thus there is no need to specify pre-defined features. This implies that one could apply the same deep learning methodology to solve many different tasks. However, the deep learning networks can properly learn features from the data only if you have sufficient data to train the network. This could be difficult to apply in practice as the sample size requirements for deep learning could be very large (90).
In the image acquisition step, deep learning could be used to mitigate the effects of different imaging settings. One could adopt deep learning-based image synthesis to match different imaging settings (91). For example, given data with mixed magnetic field strengths (i.e., 1.5T vs. 3T), we could generate 3T images from 1.5T images, thereby matching the imaging settings (92).
In the ROI specification step, segmentation of the ROI could be enhanced with deep learning compared to conventional machine learning approaches. There has been significant growth in deep learning-based segmentation. A few of the recent deep learning segmentation algorithms were able to perform difficult segmentation tasks and possibly eliminate the need for manual segmentation (93,94).
In the feature extraction step, deep learning might provide insights for new features (95). Current radiomics features are “hand-crafted” features, which are formed by expert opinions (9). Deep learning approaches are data-driven and capable of learning relevant features from the data themselves. Thus, we could develop new features by careful analysis of the deep learning framework.
The widespread adoption of deep learning is primarily driven by superior performance in various tasks in radiology. However, deep learning methods suffer from the “interpretability” issue (96). It is unclear why this method works well. It essentially functions as a difficult to interpret “black box” that performs well. Many efforts have been made to address this interpretability issue, including class activation mapping and attention mechanisms. Another deep learning issue in medical imaging is the scarcity of pre-trained deep learning models. Deep learning models inherently have high degrees of freedom and we need to solve for thousands of millions of parameters. This requires massive amounts of training data. Thus, a common approach is to transfer a model already trained from a similar domain to the target domain (16,97). This allows the optimization of the parameters to become feasible. There are many well-established deep learning models for natural images (dogs, cats, flowers, etc.), but there is a scarcity of established pre-trained models in medical imaging, particularly breast imaging. Active research is being conducted to provide pre-trained models specific to medical imaging.
Data Sharing and Open Science
Radiomics studies, particularly those adopting deep learning, have very high degrees of freedom. Thus, these studies require considerably more samples than conventional imaging studies. A team of researchers is limited by the samples they can collect. The only solution to this problem is large-scale data sharing (3,33,34). We, as a research community, should establish a plan to share our data on proper power radiomics studies. One source of larger-scale data is the research databases hosted by government agencies, such as TCIA (35,98). More government resources should be allocated to create new databases. Some government funding agencies are even enforcing the sharing of data produced from research grants.
Radiomics research involves the development and application of computer codes throughout the process. To improve the reproducibility of these studies, researchers should share all the computer codes used in their studies via code-sharing sites. This is a common practice in the computer science field, and research in radiology should follow this as well.
CONCLUSION
The application of radiomics in breast cancer imaging is an expanding research topic and has the potential to be used as a surrogate marker in precision medicine. The studies presented in our review have reported that radiomics is promising for various purposes with different modalities. However, the application of the proposed radiomics approaches in real clinical practice is still hampered by the several concerns we have described herein. In one rapid review to assess the overall quality of the studies (99), the authors used the radiomics quality score (RQS), which evaluates sixteen studies relevant to a radiomics-specific context. The results of the RQS score showed that the overall quality of the studies tended to be modest and limited. However, further advances in technology and efforts to standardize the methodology among researchers would make radiomics a more robust field and boost confidence in its results.
Footnotes
Conflicts of Interest: The authors have no potential conflicts of interest to disclose.
References
- 1.Limkin EJ, Sun R, Dercle L, Zacharaki EI, Robert C, Reuzé S, et al. Promises and challenges for the implementation of computational medical imaging (radiomics) in oncology. Ann Oncol. 2017;28:1191–1206. doi: 10.1093/annonc/mdx034. [DOI] [PubMed] [Google Scholar]
- 2.Khangura S, Konnyu K, Cushman R, Grimshaw J, Moher D. Evidence summaries: the evolution of a rapid review approach. Syst Rev. 2012;1:10. doi: 10.1186/2046-4053-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar V, Gu Y, Basu S, Berglund A, Eschrich SA, Schabath MB, et al. Radiomics: the process and the challenges. Magn Reson Imaging. 2012;30:1234–1248. doi: 10.1016/j.mri.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haralick RM, Shanmugam K, Dinstein I. Textural features for image classification. IEEE Trans Syst Man Cybern. 1973;SMC-3:610–621. [Google Scholar]
- 7.Mandelbrot BB. The fractal geometry of nature. San Francisco: Freeman and co.; 1982. [Google Scholar]
- 8.Amadasun M, King R. Textural features corresponding to textural properties. IEEE Trans Syst Man Cybern Syst. 1989;19:1264–1274. [Google Scholar]
- 9.Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larue RT, Defraene G, De Ruysscher D, Lambin P, van Elmpt W. Quantitative radiomics studies for tissue characterization: a review of technology and methodological procedures. Br J Radiol. 2017;90:20160665. doi: 10.1259/bjr.20160665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aerts HJ. The potential of radiomic-based phenotyping in precision medicine: a review. JAMA Oncol. 2016;2:1636–1642. doi: 10.1001/jamaoncol.2016.2631. [DOI] [PubMed] [Google Scholar]
- 12.Peerlings J, Woodruff HC, Winfield JM, Ibrahim A, Van Beers BE, Heerschap A, et al. Stability of radiomics features in apparent diffusion coefficient maps from a multi-centre test-retest trial. Sci Rep. 2019;9:4800. doi: 10.1038/s41598-019-41344-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Timmeren JE, Leijenaar RTH, van Elmpt W, Wang J, Zhang Z, Dekker A, et al. Test-retest data for radiomics feature stability analysis: generalizable or study-specific? Tomography. 2016;2:361–365. doi: 10.18383/j.tom.2016.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larue RTHM, van Timmeren JE, de Jong EEC, Feliciani G, Leijenaar RTH, Schreurs WMJ, et al. Influence of gray level discretization on radiomic feature stability for different CT scanners, tube currents and slice thicknesses: a comprehensive phantom study. Acta Oncol. 2017;56:1544–1553. doi: 10.1080/0284186X.2017.1351624. [DOI] [PubMed] [Google Scholar]
- 15.Mackin D, Fave X, Zhang L, Fried D, Yang J, Taylor B, et al. Measuring computed tomography scanner variability of radiomics features. Invest Radiol. 2015;50:757–765. doi: 10.1097/RLI.0000000000000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu Y, Hosny A, Zeleznik R, Parmar C, Coroller T, Franco I, et al. Deep learning predicts lung cancer treatment response from serial medical imaging. Clin Cancer Res. 2019;25:3266–3275. doi: 10.1158/1078-0432.CCR-18-2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang SY, Franc BL, Harnish RJ, Liu G, Mitra D, Copeland TP, et al. Exploration of PET and MRI radiomic features for decoding breast cancer phenotypes and prognosis. NPJ Breast Cancer. 2018;4:24. doi: 10.1038/s41523-018-0078-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rios Velazquez E, Aerts HJ, Gu Y, Goldgof DB, De Ruysscher D, Dekker A, et al. A semiautomatic CT-based ensemble segmentation of lung tumors: comparison with oncologists' delineations and with the surgical specimen. Radiother Oncol. 2012;105:167–173. doi: 10.1016/j.radonc.2012.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heye T, Merkle EM, Reiner CS, Davenport MS, Horvath JJ, Feuerlein S, et al. Reproducibility of dynamic contrast-enhanced MR imaging. Part II. Comparison of intra- and interobserver variability with manual region of interest placement versus semiautomatic lesion segmentation and histogram analysis. Radiology. 2013;266:812–821. doi: 10.1148/radiol.12120255. [DOI] [PubMed] [Google Scholar]
- 20.Grove O, Berglund AE, Schabath MB, Aerts HJ, Dekker A, Wang H, et al. Quantitative computed tomographic descriptors associate tumor shape complexity and intratumor heterogeneity with prognosis in lung adenocarcinoma. PLoS One. 2015;10:e0118261. doi: 10.1371/journal.pone.0118261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nam KJ, Park H, Ko ES, Lim Y, Cho HH, Lee JE. Radiomics signature on 3T dynamic contrast-enhanced magnetic resonance imaging for estrogen receptor-positive invasive breast cancers: preliminary results for correlation with Oncotype DX recurrence scores. Medicine (Baltimore) 2019;98:e15871. doi: 10.1097/MD.0000000000015871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee SH, Cho HH, Lee HY, Park H. Clinical impact of variability on CT radiomics and suggestions for suitable feature selection: a focus on lung cancer. Cancer Imaging. 2019;19:54. doi: 10.1186/s40644-019-0239-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park H, Lim Y, Ko ES, Cho HH, Lee JE, Han BK, et al. Radiomics signature on magnetic resonance imaging: association with disease-free survival in patients with invasive breast cancer. Clin Cancer Res. 2018;24:4705–4714. doi: 10.1158/1078-0432.CCR-17-3783. [DOI] [PubMed] [Google Scholar]
- 24.Aerts HJ, Grossmann P, Tan Y, Oxnard GR, Rizvi N, Schwartz LH, et al. Defining a radiomic response phenotype: a pilot study using targeted therapy in NSCLC. Sci Rep. 2016;6:33860. doi: 10.1038/srep33860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parmar C, Grossmann P, Bussink J, Lambin P, Aerts HJWL. Machine learning methods for quantitative radiomic biomarkers. Sci Rep. 2015;5:13087. doi: 10.1038/srep13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang C, Huang Y, He L, Chen X, Ma Z, Dong D, et al. The development and validation of a CT-based radiomics signature for the preoperative discrimination of stage I-II and stage III-IV colorectal cancer. Oncotarget. 2016;7:31401–31412. doi: 10.18632/oncotarget.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen W, Liu B, Peng S, Sun J, Qiao X. Computer-aided grading of gliomas combining automatic segmentation and radiomics. Int J Biomed Imaging. 2018;2018:2512037. doi: 10.1155/2018/2512037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Napel S, Mu W, Jardim-Perassi BV, Aerts HJWL, Gillies RJ. Quantitative imaging of cancer in the postgenomic era: radio(geno)mics, deep learning, and habitats. Cancer. 2018;124:4633–4649. doi: 10.1002/cncr.31630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho H, Lee S, Kim J, Park H. Classification of the glioma grading using radiomics analysis. PeerJ. 2018;6:e5982. doi: 10.7717/peerj.5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y, Zhang Y, Cheng R, Liu S, Qu F, Yin X, et al. Radiomics analysis of apparent diffusion coefficient in cervical cancer: a preliminary study on histological grade evaluation. J Magn Reson Imaging. 2019;49:280–290. doi: 10.1002/jmri.26192. [DOI] [PubMed] [Google Scholar]
- 31.Coroller TP, Agrawal V, Narayan V, Hou Y, Grossmann P, Lee SW, et al. Radiomic phenotype features predict pathological response in non-small cell lung cancer. Radiother Oncol. 2016;119:480–486. doi: 10.1016/j.radonc.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging. 2013;26:1045–1057. doi: 10.1007/s10278-013-9622-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Traverso A, Wee L, Dekker A, Gillies R. Repeatability and reproducibility of radiomic features: a systematic review. Int J Radiat Oncol Biol Phys. 2018;102:1143–1158. doi: 10.1016/j.ijrobp.2018.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14:749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 35.Zhao B, Tan Y, Tsai WY, Qi J, Xie C, Lu L, et al. Reproducibility of radiomics for deciphering tumor phenotype with imaging. Sci Rep. 2016;6:23428. doi: 10.1038/srep23428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Balagurunathan Y, Gu Y, Wang H, Kumar V, Grove O, Hawkins S, et al. Reproducibility and prognosis of quantitative features extracted from CT images. Transl Oncol. 2014;7:72–87. doi: 10.1593/tlo.13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balagurunathan Y, Kumar V, Gu Y, Kim J, Wang H, Liu Y, et al. Test-retest reproducibility analysis of lung CT image features. J Digit Imaging. 2014;27:805–823. doi: 10.1007/s10278-014-9716-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tixier F, Hatt M, Le Rest CC, Le Pogam A, Corcos L, Visvikis D. Reproducibility of tumor uptake heterogeneity characterization through textural feature analysis in 18F-FDG PET. J Nucl Med. 2012;53:693–700. doi: 10.2967/jnumed.111.099127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berenguer R, Pastor-Juan MDR, Canales-Vázquez J, Castro-García M, Villas MV, Mansilla Legorburo F, et al. Radiomics of CT features may be nonreproducible and redundant: influence of CT acquisition parameters. Radiology. 2018;288:407–415. doi: 10.1148/radiol.2018172361. [DOI] [PubMed] [Google Scholar]
- 40.Park JE, Park SY, Kim HJ, Kim HS. Reproducibility and generalizability in radiomics modeling: possible strategies in radiologic and statistical perspectives. Korean J Radiol. 2019;20:1124–1137. doi: 10.3348/kjr.2018.0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leijenaar RT, Nalbantov G, Carvalho S, van Elmpt WJ, Troost EG, Boellaard R, et al. The effect of SUV discretization in quantitative FDG-PET radiomics: the need for standardized methodology in tumor texture analysis. Sci Rep. 2015;5:11075. doi: 10.1038/srep11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin JC, Pujol S, et al. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn Reson Imaging. 2012;30:1323–1341. doi: 10.1016/j.mri.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bazin PL, Cuzzocreo JL, Yassa MA, Gandler W, McAuliffe MJ, Bassett SS, et al. Volumetric neuroimage analysis extensions for the MIPAV software package. J Neurosci Methods. 2007;165:111–121. doi: 10.1016/j.jneumeth.2007.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ger RB, Cardenas CE, Anderson BM, Yang J, Mackin DS, Zhang L, et al. Guidelines and experience using Imaging Biomarker Explorer (IBEX) for radiomics. J Vis Exp. 2018;(131):57132. doi: 10.3791/57132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77:e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson R, Devaraj A. Radiomics of pulmonary nodules and lung cancer. Transl Lung Cancer Res. 2017;6:86–91. doi: 10.21037/tlcr.2017.01.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang Y, Liu Z, He L, Chen X, Pan D, Ma Z, et al. Radiomics signature: a potential biomarker for the prediction of disease-free survival in early-stage (I or II) non-small cell lung cancer. Radiology. 2016;281:947–957. doi: 10.1148/radiol.2016152234. [DOI] [PubMed] [Google Scholar]
- 48.Rahbar H, McDonald ES, Lee JM, Partridge SC, Lee CI. How can advanced imaging be used to mitigate potential breast cancer overdiagnosis? Acad Radiol. 2016;23:768–773. doi: 10.1016/j.acra.2016.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bickelhaupt S, Jaeger PF, Laun FB, Lederer W, Daniel H, Kuder TA, et al. Radiomics based on adapted diffusion kurtosis imaging helps to clarify most mammographic findings suspicious for cancer. Radiology. 2018;287:761–770. doi: 10.1148/radiol.2017170273. [DOI] [PubMed] [Google Scholar]
- 50.Nie K, Chen JH, Yu HJ, Chu Y, Nalcioglu O, Su MY. Quantitative analysis of lesion morphology and texture features for diagnostic prediction in breast MRI. Acad Radiol. 2008;15:1513–1525. doi: 10.1016/j.acra.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang TC, Huang YH, Huang CS, Chen JH, Huang GY, Chang YC, et al. Computer-aided diagnosis of breast DCE-MRI using pharmacokinetic model and 3-D morphology analysis. Magn Reson Imaging. 2014;32:197–205. doi: 10.1016/j.mri.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 52.Cai H, Peng Y, Ou C, Chen M, Li L. Diagnosis of breast masses from dynamic contrast-enhanced and diffusion-weighted MR: a machine learning approach. PLoS One. 2014;9:e87387. doi: 10.1371/journal.pone.0087387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parekh VS, Jacobs MA. Integrated radiomic framework for breast cancer and tumor biology using advanced machine learning and multiparametric MRI. NPJ Breast Cancer. 2017;3:43. doi: 10.1038/s41523-017-0045-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garra BS, Krasner BH, Horii SC, Ascher S, Mun SK, Zeman RK. Improving the distinction between benign and malignant breast lesions: the value of sonographic texture analysis. Ultrason Imaging. 1993;15:267–285. doi: 10.1177/016173469301500401. [DOI] [PubMed] [Google Scholar]
- 55.Luo WQ, Huang QX, Huang XW, Hu HT, Zeng FQ, Wang W. Predicting breast cancer in breast imaging reporting and data system (BI-RADS) ultrasound category 4 or 5 lesions: a nomogram combining radiomics and BI-RADS. Sci Rep. 2019;9:11921. doi: 10.1038/s41598-019-48488-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Q, Xiao Y, Suo J, Shi J, Yu J, Guo Y, et al. Sonoelastomics for breast tumor classification: a radiomics approach with clustering-based feature selection on sonoelastography. Ultrasound Med Biol. 2017;43:1058–1069. doi: 10.1016/j.ultrasmedbio.2016.12.016. [DOI] [PubMed] [Google Scholar]
- 57.Drukker K, Giger ML, Joe BN, Kerlikowske K, Greenwood H, Drukteinis JS, et al. Combined benefit of quantitative three-compartment breast image analysis and mammography radiomics in the classification of breast masses in a clinical data set. Radiology. 2019;290:621–628. doi: 10.1148/radiol.2018180608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H, Mendel KR, Lan L, Sheth D, Giger ML. Digital mammography in breast cancer: additive value of radiomics of breast parenchyma. Radiology. 2019;291:15–20. doi: 10.1148/radiol.2019181113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tagliafico AS, Valdora F, Mariscotti G, Durando M, Nori J, La Forgia D, et al. An exploratory radiomics analysis on digital breast tomosynthesis in women with mammographically negative dense breasts. Breast. 2018;40:92–96. doi: 10.1016/j.breast.2018.04.016. [DOI] [PubMed] [Google Scholar]
- 60.Holli K, Lääperi AL, Harrison L, Luukkaala T, Toivonen T, Ryymin P, et al. Characterization of breast cancer types by texture analysis of magnetic resonance images. Acad Radiol. 2010;17:135–141. doi: 10.1016/j.acra.2009.08.012. [DOI] [PubMed] [Google Scholar]
- 61.Waugh SA, Purdie CA, Jordan LB, Vinnicombe S, Lerski RA, Martin P, et al. Magnetic resonance imaging texture analysis classification of primary breast cancer. Eur Radiol. 2016;26:322–330. doi: 10.1007/s00330-015-3845-6. [DOI] [PubMed] [Google Scholar]
- 62.Li H, Zhu Y, Burnside ES, Huang E, Drukker K, Hoadley KA, et al. Quantitative MRI radiomics in the prediction of molecular classifications of breast cancer subtypes in the TCGA/TCIA data set. NPJ Breast Cancer. 2016;2:16012. doi: 10.1038/npjbcancer.2016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liang C, Cheng Z, Huang Y, He L, Chen X, Ma Z, et al. An MRI-based radiomics classifier for preoperative prediction of Ki-67 status in breast cancer. Acad Radiol. 2018;25:1111–1117. doi: 10.1016/j.acra.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 64.Marino MA, Pinker K, Leithner D, Sung J, Avendano D, Morris EA, et al. Contrast-enhanced mammography and radiomics analysis for noninvasive breast cancer characterization: initial results. Mol Imaging Biol. 2019 Aug 28; doi: 10.1007/s11307-019-01423-5. [Epub] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ahmed A, Gibbs P, Pickles M, Turnbull L. Texture analysis in assessment and prediction of chemotherapy response in breast cancer. J Magn Reson Imaging. 2013;38:89–101. doi: 10.1002/jmri.23971. [DOI] [PubMed] [Google Scholar]
- 66.Braman NM, Etesami M, Prasanna P, Dubchuk C, Gilmore H, Tiwari P, et al. Intratumoral and peritumoral radiomics for the pretreatment prediction of pathological complete response to neoadjuvant chemotherapy based on breast DCE-MRI. Breast Cancer Res. 2017;19:57. doi: 10.1186/s13058-017-0846-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Braman N, Prasanna P, Whitney J, Singh S, Beig N, Etesami M, et al. Association of peritumoral radiomics with tumor biology and pathologic response to preoperative targeted therapy for HER2 (ERBB2)-positive breast cancer. JAMA Netw Open. 2019;2:e192561. doi: 10.1001/jamanetworkopen.2019.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu Z, Li Z, Qu J, Zhang R, Zhou X, Li L, et al. Radiomics of multiparametric MRI for pretreatment prediction of pathologic complete response to neoadjuvant chemotherapy in breast cancer: a multicenter study. Clin Cancer Res. 2019;25:3538–3547. doi: 10.1158/1078-0432.CCR-18-3190. [DOI] [PubMed] [Google Scholar]
- 69.Dong Y, Feng Q, Yang W, Lu Z, Deng C, Zhang L, et al. Preoperative prediction of sentinel lymph node metastasis in breast cancer based on radiomics of T2-weighted fat-suppression and diffusion-weighted MRI. Eur Radiol. 2018;28:582–591. doi: 10.1007/s00330-017-5005-7. [DOI] [PubMed] [Google Scholar]
- 70.Yang J, Wang T, Yang L, Wang Y, Li H, Zhou X, et al. Preoperative prediction of axillary lymph node metastasis in breast cancer using mammography-based radiomics method. Sci Rep. 2019;9:4429. doi: 10.1038/s41598-019-40831-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu FH, Wang JX, Ye XH, Deng J, Hang J, Yang B. Ultrasound-based radiomics nomogram: a potential biomarker to predict axillary lymph node metastasis in early-stage invasive breast cancer. Eur J Radiol. 2019;119:108658. doi: 10.1016/j.ejrad.2019.108658. [DOI] [PubMed] [Google Scholar]
- 72.Chan HM, van der Velden BHM, Loo CE, Gilhuijs KGA. Eigentumors for prediction of treatment failure in patients with early-stage breast cancer using dynamic contrast-enhanced MRI: a feasibility study. Phys Med Biol. 2017;62:6467–6485. doi: 10.1088/1361-6560/aa7dc5. [DOI] [PubMed] [Google Scholar]
- 73.Sinha S, Lucas-Quesada FA, DeBruhl ND, Sayre J, Farria D, Gorczyca DP, et al. Multifeature analysis of Gd-enhanced MR images of breast lesions. J Magn Reson Imaging. 1997;7:1016–1026. doi: 10.1002/jmri.1880070613. [DOI] [PubMed] [Google Scholar]
- 74.Gibbs P, Turnbull LW. Textural analysis of contrast-enhanced MR images of the breast. Magn Reson Med. 2003;50:92–98. doi: 10.1002/mrm.10496. [DOI] [PubMed] [Google Scholar]
- 75.Ertaş G, Gülçür HO, Tunaci M. Improved lesion detection in MR mammography: three-dimensional segmentation, moving voxel sampling, and normalized maximum intensity-time ratio entropy. Acad Radiol. 2007;14:151–161. doi: 10.1016/j.acra.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 76.McLaren CE, Chen WP, Nie K, Su MY. Prediction of malignant breast lesions from MRI features: a comparison of artificial neural network and logistic regression techniques. Acad Radiol. 2009;16:842–851. doi: 10.1016/j.acra.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sivaramakrishna R, Powell KA, Lieber ML, Chilcote WA, Shekhar R. Texture analysis of lesions in breast ultrasound images. Comput Med Imaging Graph. 2002;26:303–307. doi: 10.1016/s0895-6111(02)00027-7. [DOI] [PubMed] [Google Scholar]
- 78.Chen DR, Chang RF, Kuo WJ, Chen MC, Huang YL. Diagnosis of breast tumors with sonographic texture analysis using wavelet transform and neural networks. Ultrasound Med Biol. 2002;28:1301–1310. doi: 10.1016/s0301-5629(02)00620-8. [DOI] [PubMed] [Google Scholar]
- 79.Chen DR, Chang RF, Huang YL, Chou YH, Tiu CM, Tsai PP. Texture analysis of breast tumors on sonograms. Semin Ultrasound CT MR. 2000;21:308–316. doi: 10.1016/s0887-2171(00)90025-8. [DOI] [PubMed] [Google Scholar]
- 80.Karahaliou A, Skiadopoulos S, Boniatis I, Sakellaropoulos P, Likaki E, Panayiotakis G, et al. Texture analysis of tissue surrounding microcalcifications on mammograms for breast cancer diagnosis. Br J Radiol. 2007;80:648–656. doi: 10.1259/bjr/30415751. [DOI] [PubMed] [Google Scholar]
- 81.Wei D, Chan HP, Petrick N, Sahiner B, Helvie MA, Adler DD, et al. False-positive reduction technique for detection of masses on digital mammograms: global and local multiresolution texture analysis. Med Phys. 1997;24:903–914. doi: 10.1118/1.598011. [DOI] [PubMed] [Google Scholar]
- 82.Chan HP, Sahiner B, Lam KL, Petrick N, Helvie MA, Goodsitt MM, et al. Computerized analysis of mammographic microcalcifications in morphological and texture feature spaces. Med Phys. 1998;25:2007–2019. doi: 10.1118/1.598389. [DOI] [PubMed] [Google Scholar]
- 83.Goldhirsch A, Winer EP, Coates AS, Gelber RD, Piccart-Gebhart M, Thürlimann B, et al. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the primary therapy of early breast cancer 2013. Ann Oncol. 2013;24:2206–2223. doi: 10.1093/annonc/mdt303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Parikh J, Selmi M, Charles-Edwards G, Glendenning J, Ganeshan B, Verma H, et al. Changes in primary breast cancer heterogeneity may augment midtreatment MR imaging assessment of response to neoadjuvant chemotherapy. Radiology. 2014;272:100–112. doi: 10.1148/radiol.14130569. [DOI] [PubMed] [Google Scholar]
- 85.Cui X, Wang N, Zhao Y, Chen S, Li S, Xu M, et al. Preoperative prediction of axillary lymph node metastasis in breast cancer using radiomics features of DCE-MRI. Sci Rep. 2019;9:2240. doi: 10.1038/s41598-019-38502-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kim JH, Ko ES, Lim Y, Lee KS, Han BK, Ko EY, et al. Breast cancer heterogeneity: MR imaging texture analysis and survival outcomes. Radiology. 2017;282:665–675. doi: 10.1148/radiol.2016160261. [DOI] [PubMed] [Google Scholar]
- 87.Pickles MD, Lowry M, Gibbs P. Pretreatment prognostic value of dynamic contrast-enhanced magnetic resonance imaging vascular, texture, shape, and size parameters compared with traditional survival indicators obtained from locally advanced breast cancer patients. Invest Radiol. 2016;51:177–185. doi: 10.1097/RLI.0000000000000222. [DOI] [PubMed] [Google Scholar]
- 88.Choe J, Lee SM, Do KH, Lee G, Lee JG, Lee SM, et al. Deep learning-based image conversion of CT reconstruction kernels improves radiomics reproducibility for pulmonary nodules or masses. Radiology. 2019;292:365–373. doi: 10.1148/radiol.2019181960. [DOI] [PubMed] [Google Scholar]
- 89.Goodfellow I, Bengio Y, Courville A. Deep learning. Cambridge: MIT Press; 2016. [Google Scholar]
- 90.Shen G, Dwivedi K, Majima K, Horikawa T, Kamitani Y. End-to-end deep image reconstruction from human brain activity. Front Comput Neurosci. 2019;13:21. doi: 10.3389/fncom.2019.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shin HC, Tenenholtz NA, Rogers JK, Schwarz CG, Senjem ML, Gunter JL, et al. Medical image synthesis for data augmentation and anonymization using generative adversarial networks; International workshop on simulation and synthesis in medical imaging; 2018 September 16; Granada, Spain. [Google Scholar]
- 92.Nie D, Trullo R, Lian J, Wang L, Petitjean C, Ruan S, et al. Medical image synthesis with deep convolutional adversarial networks. IEEE Trans Biomed Eng. 2018;65:2720–2730. doi: 10.1109/TBME.2018.2814538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Işın A, Direkogğlu C, Şah M. Review of MRI-based brain tumor image segmentation using deep learning methods. Procedia Comput Sci. 2016;102:317–324. [Google Scholar]
- 94.Havaei M, Davy A, Warde-Farley D, Biard A, Courville A, Bengio Y, et al. Brain tumor segmentation with deep neural networks. Med Image Anal. 2017;35:18–31. doi: 10.1016/j.media.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 95.Afshar P, Mohammadi A, Plataniotis KN, Oikonomou A, Benali H. From handcrafted to deep-learning-based cancer radiomics: challenges and opportunities. IEEE Signal Process Mag. 2019;36:132–160. [Google Scholar]
- 96.Gilpin LH, Bau D, Yuan BZ, Bajwa A, Specter M, Kagal L. Explaining explanations: an overview of interpretability of machine learning; 2018 IEEE 5th international conference on data science and advanced analytics (DSAA); 2018 October 1–3; Turin, Italy. [Google Scholar]
- 97.Hosny A, Parmar C, Coroller TP, Grossmann P, Zeleznik R, Kumar A, et al. Deep learning for lung cancer prognostication: a retrospective multi-cohort radiomics study. PLoS Med. 2018;15:e1002711. doi: 10.1371/journal.pmed.1002711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Parmar C, Barry JD, Hosny A, Quackenbush J, Aerts HJWL. Data analysis strategies in medical imaging. Clin Cancer Res. 2018;24:3492–3499. doi: 10.1158/1078-0432.CCR-18-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Valdora F, Houssami N, Rossi F, Calabrese M, Tagliafico AS. Rapid review: radiomics and breast cancer. Breast Cancer Res Treat. 2018;169:217–229. doi: 10.1007/s10549-018-4675-4. [DOI] [PubMed] [Google Scholar]