To the Editor,
The Lombardy Region (10 million inhabitants) in Northern Italy has recently been involved in the largest CoVID-19 (coronavirus infectious disease-2019) outbreak outside China. Indeed, since Thursday, 20 February, when the first case of SARS-CoV-2 infection was recorded in our region, up to 15 March, 13 272 CoVID-19 cases have been registered. Among those, 24% (3220/13 272) have been detected at our Regional Reference Laboratory (Molecular Virology Unit, Fondazione IRCCS Policlinico San Matteo Pavia, Italy). The Lombardy Region policy to combat the spread of SARS-CoV-2 includes the screening of all symptomatic suspected cases and their contacts, according to the World Health Organization case definitions. The nasal swabs from all suspected cases were collected during the medical examination and the strategy for sampling in adults and children has been the same.
According to World Health Organization suggestions, nasal swabs (UTM viral transport®, Copan Italia S.p.a) from all suspected cases and their contacts were tested with at least two real-time reverse transcription PCR (RT-PCR) assays targeting different genes (E and RdRp) of SARS-CoV-2 [1]. In addition, nasal swabs were also evaluated with a novel quantitative RT-PCR targeting an additional SARS-CoV-2 gene (M) developed in our laboratory (details provided upon request).
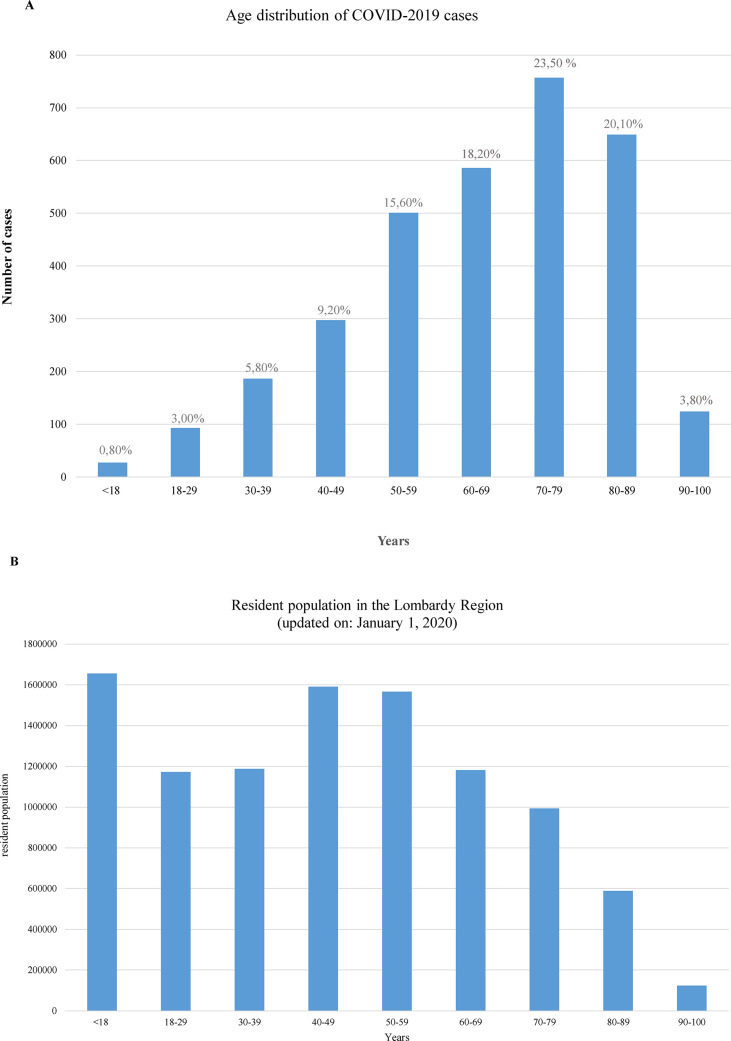
Strikingly, among a total of 3220 SARS-CoV-2 positive patients, only 0.80% (27/3220) were paediatric (<18 years), while 99.2% were adult (>18 years). A complete picture of the age distribution of all positive cases is reported in Fig. 1 A,B. The age of paediatric patients ranged from 4 months to 17 years (median, 11 years). Forty-eight per cent (13/27) were aged 12–17 years, 26% (7/27) were aged 6–11 years, and 26% (7/27) were <6 years.
Fig. 1.
(A) Age distribution of COVID-2019 cases, Lombardy Region, Italy 2020. (B) Resident population in the Lombardy Region (updated 1 January 2020), data collected from the Italian National Institute of Statistic (www.istat.it).
In addition to the low frequency of paediatric CoVID-19 cases, it must also be noted that the majority of them were clinically mild (70%, 19/27), and SARS-CoV-2 detection was possible because of the proactive search in contacts of more evident CoVID-19 presentations. At the time of writing, none of the paediatric patients with confirmed SARS-CoV-2 infection diagnosed in our laboratory required Intensive Care Unit (ICU) medical admission.
Our report agrees with the data already reported in China [[2], [3], [4]], supporting the hypothesis that clinically evident CoVID-19 is less frequent and milder in the paediatric population [5]. The data presented here, collected within the context of an organized surveillance programme, corroborate previous observations and underline the need to understand better the clinical spectrum of CoVID-19 in the paediatric population.
Indeed, all other viral respiratory infections are mostly concentrated in the paediatric population. CoVID-19 affects children and young adults less severely than the middle-aged or aged population, which contrasts with experience from previous viral pandemics, including influenza A. Furthermore, other respiratory viral infections, including influenza, respiratory syncytial virus, human metapneumovirus, adenovirus, parainfluenza virus, measles, picornaviruses and other coronaviruses, tend to cause more severe disease in the paediatric population. A common explanation for this phenomenon is the lack of pre-existing immunity to most of these viral families, with subsequent maturation of the immunological repertoire after each subsequent exposure to viral pathogens. In the context of SARS-CoV-2, lack of antecedent immunity to viral pathogens does not appear to be detrimental for severe disease. Recently, an age-dependent expression of the SARS-CoV-2 receptor, angiotensin-converting enzyme 2 (ACE 2), has been demonstrated in the nasal epithelium [6]. In particular, children had a lower ACE 2 expression than adults. These results may help explain the lower rates of SARS-CoV-2 infection in children. Much has still to be explained regarding this new and mysterious beta-coronavirus.
Transparency declaration
There were no conflict of interest. This study was supported by funds from Lombardy Region, Milan, Italy. Data collection and analysis of cases and contacts was part of a continuing public health outbreak investigation and were thus considered exempt from institutional review board approval.
Acknowledgements
We thank Daniela Sartori for manuscript editing.
The San Matteo Pavia COVID-19 Task Force: R. Bruno, M. Mondelli, E. Brunetti, A. Di Matteo, E. Seminari, L. Maiocchi, V. Zuccaro, L. Pagnucco, B. Mariani, S. Ludovisi, R. Lissandrin, A. Parisi, P. Sacchi, SFA Patruno, G. Michelone, R. Gulminetti, D. Zanaboni, S. Novati, R. Maserati, P. Orsolini, M. Vecchia(ID Staff); M Sciarra, E. Asperges, M. Colaneri, A. Di Filippo, M. Sambo, S. Biscarini, M. Lupi, S. Roda, TC Pieri, I. Gallazzi, M. Sachs, P. Valsecchi(ID Resident); S. Perlini, C. Alfano, M. Bonzano, F. Briganti, G. Crescenzi, AG Falchi, R. Guarnone, B. Guglielmana, E. Maggi, I. Martino, P. Pettenazza, S. Pioli di Marco, F. Quaglia, A. Sabena, F. Salinaro, F. Speciale, I. Zunino (ECU Staff Emergency Care Unit); M. De Lorenzo, G. Secco, L. Dimitry, G. Cappa, I. Maisak, B. Chiodi, M. Sciarrini, B. Barcella, F. Resta, L. Moroni, G. Vezzoni, L. Scattaglia, E. Boscolo, C. Zattera, MF Tassi, V. Capozza, D. Vignaroli, M. Bazzini (ECU Resident Emergency Care Unit); G. Iotti, F. Mojoli, M. Belliato, L. Perotti, S. Mongodi, G. Tavazzi (Intensive Care Unit); G. Marseglia, A. Licari, I. Brambilla (Pediatric Unit); DBarbarini, A. Bruno, P. Cambieri, G. Campanini, G. Comolli, M. Corbella, R. Daturi, M. Furione, B. Mariani, R. Maserati, E. Monzillo, S. Paolucci, M. Parea, E. Percivalle, A. Piralla,F Rovida, A. Sarasini, M. Zavattoni (Virology Staff); G. Adzasehoun, L. Bellotti, E. Cabano, G. Casali, L. Dossena, G. Frisco, G. Garbagnoli, A. Girello, V. Landini, C. Lucchelli, V. Maliardi, S. Pezzaia, M. Premoli (Virology Technical staff); A. Bonetti, G. Caneva, I. Cassaniti, A. Corcione, R. Di Martino, A. Di Napoli, A. Ferrari, G. Ferrari, L. Fiorina, F. Giardina, A. Mercato, F. Novazzi, G. Ratano, B. Rossi, IM Sciabica, M. Tallarita, E. Vecchio Nepita (Virology Resident); M. Calvi, M. Tizzoni (Pharmacy Unit); C. Nicora, A. Triarico, V. Petronella, C. Marena, A. Muzzi, P. Lago (Hospital Management).
Editor: L. Leibovici
Contributor Information
San Matteo Pavia COVID-19 Task Force:
R. Bruno, M. Mondelli, E. Brunetti, A. Di Matteo, E. Seminari, L. Maiocchi, V. Zuccaro, L. Pagnucco, B. Mariani, S. Ludovisi, R. Lissandrin, A. Parisi, P. Sacchi, S.F.A. Patruno, G. Michelone, R. Gulminetti, D. Zanaboni, S. Novati, R. Maserati, P. Orsolini, M. Vecchia, M. Sciarra, E. Asperges, M. Colaneri, A. Di Filippo, M. Sambo, S. Biscarini, M. Lupi, S. Roda, T.C. Pieri, I. Gallazzi, M. Sachs, P. Valsecchi, S. Perlini, C. Alfano, M. Bonzano, F. Briganti, G. Crescenzi, A.G. Falchi, R. Guarnone, B. Guglielmana, E. Maggi, I. Martino, P. Pettenazza, S. Pioli di Marco, F. Quaglia, A. Sabena, F. Salinaro, F. Speciale, I. Zunino, M. De Lorenzo, G. Secco, L. Dimitry, G. Cappa, I. Maisak, B. Chiodi, M. Sciarrini, B. Barcella, F. Resta, L. Moroni, G. Vezzoni, L. Scattaglia, E. Boscolo, C. Zattera, M.F. Tassi, V. Capozza, D. Vignaroli, M. Bazzini, G. Iotti, F. Mojoli, M. Belliato, L. Perotti, S. Mongodi, G. Tavazzi, G. Marseglia, A. Licari, I. Brambilla, D. Barbarini, A. Bruno, P. Cambieri, G. Campanini, G. Comolli, M. Corbella, R. Daturi, M. Furione, B. Mariani, R. Maserati, E. Monzillo, S. Paolucci, M. Parea, E. Percivalle, A. Piralla, F. Rovida, A. Sarasini, M. Zavattoni, G. Adzasehoun, L. Bellotti, E. Cabano, G. Casali, L. Dossena, G. Frisco, G. Garbagnoli, A. Girello, V. Landini, C. Lucchelli, V. Maliardi, S. Pezzaia, M. Premoli, A. Bonetti, G. Caneva, I. Cassaniti, A. Corcione, R. Di Martino, A. Di Napoli, A. Ferrari, G. Ferrari, L. Fiorina, F. Giardina, A. Mercato, F. Novazzi, G. Ratano, B. Rossi, I.M. Sciabica, M. Tallarita, E. Vecchio Nepita, M. Calvi, M. Tizzoni, C. Nicora, A. Triarico, V. Petronella, C. Marena, A. Muzzi, and P. Lago
References
- 1.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu Y., Li X., Zhu B., Liang H., Fang C., Gang Y., et al. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat Med. 2020:1–4. doi: 10.1038/s41591-020-0817-4. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guan W., Ni Z., Hu Y., Liang W., Ou C., He J., et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020 doi: 10.1056/NEJMoa2002032. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., et al. Early transmission dynamics in Wuhan, China, of novel Coronavirus-infected pneumonia. N Engl J Med. 2020 doi: 10.1056/NEJMoa2001316. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castagnoli R., Votto M., Licari A., Brambilla I., Bruno R., Perlini S., et al. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection in children and adolescents: a systematic review. JAMA Pediatr. 2020 doi: 10.1001/jamapediatrics.2020.1467. in press. [DOI] [PubMed] [Google Scholar]
- 6.Bunyavanich S., Do A., Vicencio A. Nasal gene expression of angiotensin-converting enzyme 2 in children and adults. JAMA. 2020 doi: 10.1001/jama.2020.8707. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]